Abstract
The DNA colony hybridization test with the polynucleotide probe for Vibrio parahaemolyticus toxR gene was performed. All 373 strains of V. parahaemolyticus gave positive results, and the strains belonging to four other Vibrio species including Vibrio alginolyticus gave weakly positive results, suggesting that toxR sequence variation may reflect the phylogenetic relationships of Vibrio species. We then established a toxR-targeted PCR protocol for the specific detection of V. parahaemolyticus.
Vibrio parahaemolyticus is a marine bacterium, and some strains can cause gastroenteritis in humans through the consumption of contaminated seafood. Molecular epidemiological studies revealed a strong correlation between the possession of particular hemolysin genes (tdh, trh, or both) and the ability to cause disease, supporting the fact that these genes are important virulence genes (4, 16). However, a small portion of clinical strains carried neither of the virulence genes (4, 16). Therefore, the isolation of the organism in the clinical setting and from food samples suspected of being the source of infection, followed by identification of the isolated strains, is still the standard procedure in investigations of gastroenteritis due to V. parahaemolyticus. If a PCR method allows detection of the nucleotide sequence specific to V. parahaemolyticus, it can facilitate identification of the organism.
The genes encoding two hemolysins designated TLH and δ-VPH of V. parahaemolyticus were examined by Taniguchi et al. (18, 19). These genes appear to be fairly specific to V. parahaemolyticus. Lee et al. (6, 7) cloned a 0.76-kb nucleotide sequence of unknown function and claimed that the nucleotide sequence is specific to V. parahaemolyticus by hybridization and PCR methods (6, 7). However, only limited numbers of strains were examined in those studies, and the relationships of the hemolysin genes and the 0.76-kb sequence with the phylogeny of V. parahaemolyticus are not known. For identification purposes, it is ideal to use a nucleotide sequence that is well conserved and that reflects the phylogenetic relationship. rRNA sequences are often used for this purpose. However, the rRNA sequence homologies between V. parahaemolyticus and related species are so high that the rRNA sequence does not appear to be suitable for the purpose described above. For example, the 16S rRNA sequences of V. parahaemolyticus and Vibrio alginolyticus are >99% identical (5, 15). The gyrB gene encodes the B subunit of DNA gyrase, which is essential for DNA replication. The homology of the gyrB sequences between V. parahaemolyticus and V. alginolyticus is 86.8% (20). For this reason, a PCR procedure targeting the gyrB gene was recently developed for the specific detection of V. parahaemolyticus in shrimp (20).
The toxR gene was first discovered as the regulatory gene of the cholera toxin operon, but it was later shown to be involved in the regulation of many other genes in Vibrio cholerae (1, 9). We subsequently found the toxR gene in V. parahaemolyticus and demonstrated its regulatory function (8). The toxR gene sequences have also been cloned from Vibrio fisheri and at least two other species of Vibrio, and their sequences have been analyzed (13, 14). Therefore, the toxR gene appears to be well conserved among Vibrio species. The degree of homology of the toxR gene between V. parahaemolyticus and V. cholerae (52% identity) is much lower than that of the rRNA gene (91 to 92% identity) (5, 8). We therefore investigated in this study whether the toxR gene sequence can be used to develop a PCR method for the specific identification V. parahaemolyticus.
Phenotypic characteristics of bacterial strains.
The bacterial strains used in this study are listed in Table 1. Clinical strains listed as V. parahaemolyticus, listed as having an unknown identification (non-V. parahaemolyticus), and identified as known Vibrio species (11 strains; see below) were isolated from patients at hospitals in India, Bangladesh, and Korea and at quarantine stations in Japan during the period between 1977 and 1997 (11, 12). The nonclinical strains were isolated from seawater in Korea in the summer of 1996. The seawater samples were plated, after enrichment in alkaline peptone water, onto thiosulfate-citrate-bile salts-sucrose agar (Eiken Chemical, Co., Ltd., Tokyo, Japan), and blue-green colonies were selected. All clinical and nonclinical strains examined in this study were tested for the characteristics listed in Table 2 by standard procedures (3) except that the NaCl concentration of the test medium was adjusted to 1.5%.
TABLE 1.
Results of tests for toxR gene
| Species | Source or strain no.a | Detection of toxR geneb by the following assay:
|
|
|---|---|---|---|
| DNA probe | PCR | ||
| V. parahaemolyticus | C | + (363/363) | + (363/363) |
| V. parahaemolyticus | N | + (10/10) | + (10/10) |
| V. cholerae O1 | C | − (12/12) | − (12/12) |
| V. cholerae O139 | C | − | − |
| V. cholerae non-O1, non-O139 | C | − (11/11) | − (11/11) |
| V. mimicus | C | − (18/18) | − (18/18) |
| V. fluvialis | C | − (12/12) | − (12/12) |
| V. hollisae | C | − (7/7) | − (7/7) |
| V. vulnificus | C | − (7/7) | − (7/7)c |
| V. vulnificus | N | − (4/4) | − (4/4)d |
| V. furnisii | C | − (2/2) | − (2/2) |
| V. furnisii | N | − | − |
| V. alginolyticus | N | +W (10/10) | − (10/10) |
| V. aestiarianus | ATCC 35048 | − | − |
| V. anguillarum | PT87050 | − | − |
| V. campbellii | ATCC 25920 | +W | − |
| V. carchariae | ATCC 35084 | +W | − |
| V. cincinnatiensis | ATCC 35912 | − | − |
| V. damsela | ATCC 33539 | − | − |
| V. diazotrophicus | ATCC 33466 | − | − |
| V. gazogenes | ATCC 29988 | − | − |
| V. harveyi | ATCC 14126 | +W | − |
| V. ichthyoenteri | IFO15847 | − | − |
| V. iliopiscarius | ATCC 51760 | − | − |
| V. logei | ATCC 15382 | − | − |
| V. mediterranei | ATCC 43341 | − | − |
| V. metschnikovii | RIMD2208006 | − | − |
| V. mytili | ATCC 51288 | − | − |
| V. nereis | ATCC 25917 | − | − |
| V. navarrensis | ATCC 51183 | − | − |
| V. nigripulchritudo | ATCC 27043 | − | − |
| V. ordalii | ATCC 33509 | − | − |
| V. orientalis | ATCC 33934 | − | − |
| V. mediterranei | ATCC 43341 | − | − |
| V. pelagius | ATCC 25916 | − | − |
| V. penaeicida | IFO15640 | − | − |
| V. proteolyticus | NCMB1326 | − | − |
| V. splendisdus | ATCC 33125 | − | − |
| V. tubiashii | ATCC 19109 | − | − |
| Aeromonas hydrophila | C | − (10/10) | − (10/10) |
| Aeromonas sobria | C | − (10/10) | − (10/10) |
| Plesiomonas shigelloides | C | − (10/10) | − (10/10) |
| Escherichia coli | Ce | − (20/20) | − (20/20) |
| Shigella dysenteriae | C | − (2/2) | − (2/2) |
| Shigella boydii | C | − (2/2) | − (2/2) |
| Shigella sonnei | C | − (3/3) | − (3/3) |
| Salmonella choleraesuis | Cf | − (10/10) | − (10/10) |
| Staphylococcus aureus | Cg | − (4/4) | − (4/4) |
| Unknownh | C | − (22/22) | − (22/22) |
| N | − (86/86) | − (86/86) | |
C, clinical (human); N, nonclinical (environmental).
+, detected; +W, weak reaction detected; −, not detected. The values in parentheses indicate the number of positive strains/number of strains tested for multiple strains.
Three strains produced amplicons larger than the specific amplicons.
Two strains produced amplicons larger than the specific amplicons.
Included enteropathogenic (7 strains), enteroinvasive (3 strains), enterotoxigenic (3 strains), enterohemorrhagic (6 strains), and enteroaggregative (1 strain) types.
Included six different O serovars.
The S. aureus strains produced enterotoxin type A (2 strains) and type B (2 strains).
Explained in the text.
TABLE 2.
Characteristics of the 373 strains of V. parahaemolyticus identified
| Test | Resulta | Test | Result | |
|---|---|---|---|---|
| TSI | K/A | Voges-Proskauer | − | |
| H2S (on TSI) | − | Simmons citrate | + (98.7) | |
| Lysine decarboxylase | + (99.5) | Oxidation-fermentation | F | |
| Indole | + (99.4) | Acid from: | ||
| Mannitol | + | |||
| Motility | + | Inositol | − | |
| Glucose | + | |||
| Growth in: | Arabinose | + (72.2) | ||
| 0% NaCl | − | Lactose | − | |
| 3% NaCl | + | Maltose | + | |
| 7% NaCl | + | Rhamnose | − (95.4) | |
| 10% NaCl | − | Sucrose | − | |
| Mannose | + | |||
| Oxidase | + | |||
| Arginine dihydrolase | − | |||
| Ornithine decarboxylase | + (98.1) | |||
| Methyl red | + (96.8) |
K/A, alkaline top and acidic bottom; +, positive; −, negative; F, fermentative. If one or more strains gave a variable result, the percentage of the strains that gave the result indicated by the sign is given in the parentheses.
The phenotypic characteristics of these 494 strains were examined in two steps. The following characteristics were examined in the initial screening step. The strains that showed an alkaline top, an acidic bottom, and no H2S production in the TSI reaction and that gave positive results in the lysine decarboxylase, indole, and motility tests were selected. These strains were further examined for the other characteristics listed in Table 2. Three hundred sixty-five strains that gave the results listed in Table 2 were identified as V. parahaemolyticus. Seven, two, two, and one strain were identified as Vibrio mimicus, Vibrio furnisii, V. alginolyticus, and V. cholerae non-O1 and non-O139, respectively, and are included in Table 1 as such. The other strains could not be assigned to the species listed above from the characteristics examined. These strains were not characterized further and are thus included in Table 1 as strains with unknown identifications.
The test strains that were not selected in the initial screening step were not characterized further and, except for eight strains, are included in Table 1 with the strains with unknown identifications. The eight exceptional strains were negative by the lysine decarboxylase test (two strains) or the indole test (six strains). However, these strains carried the toxR gene (described below), and therefore, the other characteristics of these strains were examined. These strains had the characteristics listed in Table 2 and carried the V. parahaemolyticus gyrB gene (described below). Therefore, these strains were identified as V. parahaemolyticus and are included in Table 1 as such.
Most of the strains belonging to other Vibrio species and other genera were our laboratory stock strains or strains supplied by other workers for this study, and their phenotypic characteristics were not examined.
DNA colony hybridization.
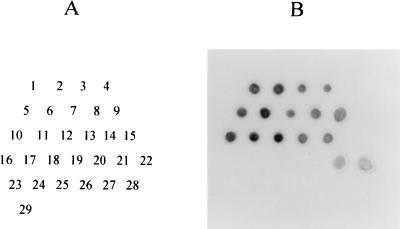
We examined 373 strains of V. parahaemolyticus and 290 strains belonging to non-V. parahaemolyticus species by the DNA colony hybridization test with the 678-bp V. parahaemolyticus toxR gene probe as described previously (8). Marine Agar 2216 (Difco Laboratories, Detroit, Mich.) and an incubation temperature of 25°C were used in place of Luria-Bertani (LB) agar and an incubation temperature of 37°C to grow the standard strains belonging to 26 species of the genus Vibrio (those with specific strain numbers in Table 1). The results are summarized in Table 1. All clinical and nonclinical strains of V. parahaemolyticus gave clearly positive results. Of the non-V. parahaemolyticus strains, those belonging to four Vibrio species including V. alginolyticus gave positive results, although the hybridization signals were weaker than those for the V. parahaemolyticus strains. The other non-V. parahaemolyticus strains yielded negative results (Table 1). The hybridization signals of representative strains of Vibrio species (reference strains; Table 3) are presented in Fig. 1. The results indicate that the V. parahaemolyticus toxR sequence is perfectly conserved among V. parahaemolyticus strains and that some other Vibrio species carry nucleotide sequences that are fairly homologous to that of the V. parahaemolyticus toxR gene. These non-V. parahaemolyticus species are phylogenetically closely related to V. parahaemolyticus (5, 15).
TABLE 3.
Reference strains of Vibrio species used in this study
| Refer-ence no. | Species | Straina |
|---|---|---|
| 1 | V. parahaemolyticus | AQ3855 (O6:K18, tdh+, trh1+, trh2−) |
| 2 | V. parahaemolyticus | VP-170 (O3:K6, tdh+, trh1−, trh2−) |
| 3 | V. parahaemolyticus | AQ4537 (O3:K72, tdh+, trh1+, trh2−) |
| 4 | V. parahaemolyticus | AQ4781 (O1:KUT, tdh+, trh1−, trh2+) |
| 5 | V. parahaemolyticus | AQ4889 (O4:K12, tdh+, trh1+, trh2−) |
| 6 | V. parahaemolyticus | KX-V231 (O3:K6, tdh+, trh1−, trh2−) |
| 7 | V. parahaemolyticus | K-16 (O10:K71, tdh+, trh1+, trh2−) |
| 8 | V. parahaemolyticus | K-102 (O2:KUT, tdh−, trh1−, trh2−) |
| 9 | V. parahaemolyticus | VP3 (O2:K3, tdh+, trh1−, trh2−) |
| 10 | V. parahaemolyticus | VP87 (O1:K1, tdh+, trh1−, trh2+) |
| 11 | V. parahaemolyticus | VP121 (O3:K6, tdh+, trh1−, trh2−) |
| 12 | V. parahaemolyticus | W-9802 (O4:K8, tdh+, trh1−, trh2−) |
| 13 | V. parahaemolyticus | Y-27669 (O8:K22, tdh+, trh1−, trh2−) |
| 14 | V. parahaemolyticus | AM-8626 (O3:K6, tdh+, trh1−, trh2−) |
| 15 | V. cholerae O1 | NIH41 |
| 16 | V. cholerae O1 | NIH35A3 |
| 17 | V. cholerae O139 | MO45 |
| 18 | V. cholerae non-O1, non-O139 | AM2 |
| 19 | V. hollisae | 525-82 |
| 20 | V. anguillarum | PT-87050 |
| 21 | V. alginolyticus | 219 |
| 22 | V. alginolyticus | 220 |
| 23 | V. mimicus | RIMD2218002 |
| 24 | V. vulnificus | RIMD2219022 |
| 25 | V. fluvialis | RIMD2220002 |
| 26 | V. furnisii | RIMD2223001 |
| 27 | V. damsela | RIMD2222001 |
| 28 | V. metchnikovii | RIMD2208006 |
The O:K serotype (KUT:K untypeable) and presence (+) or absence (−) of the tdh, trh1, and trh2 genes are indicated in parentheses for the V. parahaemolyticus strains.
FIG. 1.
DNA colony hybridization test with the V. parahaemolyticus toxR gene probe for reference strains of Vibrio species. (A) Location of the inoculated strains. The numbers correspond to those of the reference strains listed in Table 3. Strain 24 was Escherichia coli HB101 (a negative control). (B) Hybridization signals of the test strains detected on X-ray film.
The presence or absence of the tdh, trh1, and trh2 genes in the test strain was also examined by the DNA colony hybridization test with polynucleotide probes as described previously (10). Of the 363 clinical strains of V. parahaemolyticus, 14 strains (4%) carried neither the tdh, trh1, nor trh2 gene. These results encouraged us to develop a PCR protocol that allows the specific detection of the V. parahaemolyticus toxR sequence.
PCR.
To develop a PCR method specific for the V. parahaemolyticus toxR gene, various oligonucleotide primer sets were tested with the reference strains. These strains included 14 strains of V. parahaemolyticus and 14 strains of non-V. parahaemolyticus species (Table 3). The primer sequences were selected from the regions not conserved between the V. parahaemolyticus toxR and the V. cholerae toxR sequences (8). Five forward primers and two reverse primers (data not shown) were selected, and all 10 combinations of these primers were tested. The dilution of the boiled culture supernatant, the annealing temperature, and the numbers of amplification cycles of the PCR were varied and the results were compared. Amplicons of the expected sizes were also detected in strains of V. alginolyticus and V. vulnificus when various PCR primer sets were tested with the reference strains under low annealing temperatures (data not shown). These results supported our hypothesis that the toxR gene is well conserved in Vibrio species and that variation in the toxR sequences may reflect the phylogenetic relationship of Vibrio species.
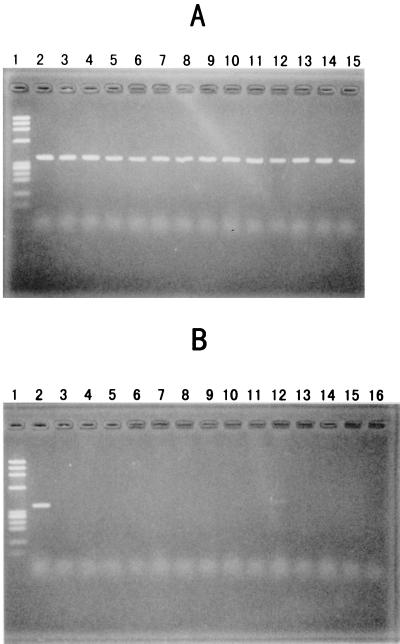
After extensive efforts to optimize the PCR conditions, the following method was shown to allow the specific detection of the V. parahaemolyticus toxR gene. The test strain was grown in LB broth containing 1% NaCl at 37°C with shaking (160 rpm) overnight. One milliliter of the culture was boiled for 5 min, and the supernatant was obtained by centrifugation (13,000 rpm) on a tabletop centrifuge (Centrifuge 5415C; Eppendorf, Hamburg, Germany) at room temperature. The supernatant was diluted 10-fold in distilled water. The PCR conditions were as follows. The reaction mixture consisted of 3 μl of the template (supernatant of the boiled culture diluted 1:10), 5 μl of 10× buffer containing 20 mM MgCl2 (Ex Taq buffer; Takara, Tokyo, Japan), 0.25 μl of Taq polymerase (Ex Taq; Takara), 4 μl of 2.5 mM deoxynucleoside triphosphate, 2 μl of each primer (10 pmol/μl), and 33.75 μl of distilled water. The amplification conditions were 20 cycles of amplification consisting of denaturation at 94°C for 1 min, annealing at 63°C for 1.5 min, and extension at 72°C for 1.5 min. The primer sequences were 5′-GTCTTCTGACGCAATCGTTG-3′ (forward) and 5′-ATACGAGTGGTTGCTGTCATG-3′ (reverse). The sequences of the forward and reverse primers correspond to base positions 609 to 629 and 956 to 958, respectively, of the previously described V. parahaemolyticus toxR sequence (8). Ten microliters of the reaction mixture was mixed with 2 μl of the dye solution (0.07% bromophenol blue, 20% Ficoll), and the mixture was resolved by electrophoresis in 2% agarose. The 368-bp amplicons of the reference strains obtained by this method are shown in Fig. 2A and B. The culture condition described above allowed enough bacterial growth so that only 20 cycles of amplification was enough to achieve clearly visible amplicon bands.
FIG. 2.
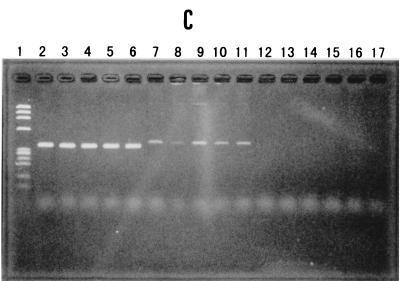
PCR assay detects the toxR gene of V. parahaemolyticus. The reference strains are described in Table 3. (A) Reference strains of V. parahaemolyticus; lane 1, φX174 phage DNA digested with HaeIII (molecular weight markers); lanes 2 to 15, reference strains 1 to 14 (Table 3), respectively. (B) Reference strains of non-V. parahaemolyticus species; lane 1, φX174 phage DNA digested with HaeIII; lane 2, reference strain V. parahaemolyticus 13 (a positive control); lanes 3 to 16, reference strains 15 to 28 (non-V. parahaemolyticus species), respectively. (C) Comparison of selected strains of V. parahaemolyticus and all test strains of V. vulnificus; lane 1, φX174 phage DNA digested with HaeIII; lanes 2 to 6, V. parahaemolyticus strains (positive controls); lanes 7 to 17, test strains of V. vulnificus.
The PCR protocol established with the reference strains was applied to the examination of all test strains (Table 1). All strains of V. parahaemolyticus gave only specific amplicons, whereas non-V. parahaemolyticus strains did not. Nonspecific amplicons were generated by only 5 of 11 strains of V. vulnificus. These nonspecific amplicons were larger and less evident than the specific amplicons and thus were distinguishable from the specific amplicons (Fig. 2C, lanes 7 to 11). The isolation of V. vulnificus from patients with diarrhea appears to be a very rare event, but this possibility must not be ruled out (2). The nonspecific amplicons of some strains of V. vulnificus can be distinguished from the specific amplicon if the positive control is included in the gel electrophoresis analysis. Therefore, we do not think that it is necessary to run additional tests to exclude the possibility that the amplicon resulted from amplification of the V. vulnificus nucleotide sequence. However, if desired, such additional tests are available. This can be a simple biochemical test, for example, growth in 8% NaCl (3). Alternatively, the modification of the PCR protocol described above, in which the PCR primer set is substituted by the primer set 5′-AGCCCGCTTTCTTCAGACTC-3′ and 5′-AACGAGTCTTCTGCATGGTG-3′ or the primer set 5′-CGCTTTCTTCAGACTCAAGC-3′ and 5′-AACGAGTCTTCTGCATGGTG-3′, can be used; all strains of V. parahaemolyticus produced amplicons of 399 or 394 bp, respectively; none of the V. vulnificus strains produced any amplicons; some strains of V. alginolyticus, however, produced the amplicons (data not shown).
Growth of the test strain in LB broth at 37°C was used in the test described above. This growth condition was chosen for the organisms of clinical significance and is not suitable for most nonpathogenic Vibrio species in the marine environment. To confirm the specificity of the primer set, the standard strains belonging to 26 species of the genus Vibrio (those with specific strain numbers in Table 1) were also grown to good turbidity in Marine Broth 2216 (Difco Laboratories) at 25°C and were subjected to the PCR assay. All these strains gave negative results, whereas the V. parahaemolyticus strains grown under the same conditions gave positive results.
All 373 strains identified as V. parahaemolyticus carried the toxR gene, as described above. Of these strains, 47 strains showed atypical biochemical characteristics such as negative results by tests for lysine or ornithine decarboxylation or indole production or the Simmons citrate test and positive results by the rhamnose fermentation test (Table 2). To support the identification, these rare strains were examined for the presence of the V. parahaemolyticus gyrB gene by the PCR method. The PCR for the specific detection of the V. parahaemolyticus gyrB gene was carried out essentially as described by Venkateswaran et al. (20). The boiled culture supernatant was prepared as described above, diluted to 1:10, and used as the template solution. We used an annealing temperature of 60°C rather than 58°C, which was specified by Venkateswaran et al. (20), because in our hands V. alginolyticus strains gave the amplicons of the expected size at the latter annealing temperature. All strains tested gave positive results.
In conclusion, the PCR protocol for the V. parahaemolyticus toxR gene established in this study is specific and rapid (amplification can be achieved in 2 h). Simultaneous use of this PCR method and the PCR method for the detection of virulence genes like tdh and trh (17) would be useful for the rapid investigation of suspected V. parahaemolyticus strains isolated from clinical specimens and food samples implicated as sources of infection for patients with cases of food poisoning.
Acknowledgments
This research was supported in part by funds from the Ohyama Health Foundation and Heiwa Nakajima Foundation, by a Grant-in Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan, by the U.S.-Japan Cooperative Medical Science Program, Cholera and Related Diarrheal Diseases, Japanese Panel, and by Pusan National University’s Overseas Research Program for Professors.
We are grateful to the following individuals in Japan who kindly supplied bacterial strains: Takeshi Honda of Osaka University, Masanori Ishibashi of Osaka Prefectural Public Health, Sumio Shinoda of Okayama University, Tatsunosuke Nakamura of Chiba University, and Toshihiro Nakai of Hiroshima University. We thank Yohko Takeda for technical assistance.
REFERENCES
- 1.DiRita V J. Co-ordinate expression of virulence genes by ToxR in Vibrio cholerae. Mol Microbiol. 1992;6:451–458. doi: 10.1111/j.1365-2958.1992.tb01489.x. [DOI] [PubMed] [Google Scholar]
- 2.Janda J M, Powers C, Bryant R G, Abbott S. Current perspectives on the epidemiology and pathogenesis of clinically significant Vibrio spp. Clin Microbiol Rev. 1988;1:245–267. doi: 10.1128/cmr.1.3.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelly M T, Hickman-Brenner F W, Farmer J J., III . Vibrio. In: Balows A, Hausler W J Jr, Herrmann K L, Isenberg H D, Shadomy H J, editors. Manual of clinical microbiology. 5th ed. Washington, D.C: American Society for Microbiology; 1991. pp. 384–395. [Google Scholar]
- 4.Kishishita M, Matsuoka N, Kumagai K, Yamasaki S, Takeda Y, Nishibuchi M. Sequence variation in the thermostable direct hemolysin-related hemolysin (trh) gene of Vibrio parahaemolyticus. Appl Environ Microbiol. 1992;58:2449–2457. doi: 10.1128/aem.58.8.2449-2457.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kita-Tsukamoto K, Oyaizu H, Nanba K, Shimidu U. Phylogenetic relationships of marine bacteria, mainly members of the family Vibrionaceae, determined on the basis of 16S rRNA sequences. Int J Syst Bacteriol. 1993;43:8–19. doi: 10.1099/00207713-43-1-8. [DOI] [PubMed] [Google Scholar]
- 6.Lee C Y, Chen C H, Chou Y W. Characterization of a cloned pR72H probe for Vibrio parahaemolyticus detection and development of a nonisotopic colony hybridization assay. Microbiol Immunol. 1995;39:177–183. doi: 10.1111/j.1348-0421.1995.tb02186.x. [DOI] [PubMed] [Google Scholar]
- 7.Lee C Y, Pan S F, Chen C H. Sequence of a cloned pR72H fragment and its use for detection of Vibrio parahaemolyticus in shellfish with the PCR. Appl Environ Microbiol. 1995;61:1311–1317. doi: 10.1128/aem.61.4.1311-1317.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin Z, Kumagai K, Baba K, Mekalanos J J, Nishibuchi M. Vibrio parahaemolyticus has a homolog of the Vibrio cholerae toxRS operon that mediates environmentally induced regulation of the thermostable direct hemolysin gene. J Bacteriol. 1993;175:3844–3855. doi: 10.1128/jb.175.12.3844-3855.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 10.Okuda J, Ishibashi M, Abbott S L, Janda J M, Nishibuchi M. Analysis of the thermostable direct hemolysin (tdh) gene and the tdh-related hemolysin (trh) genes in the urease-positive strains of Vibrio parahaemolyticus isolated on the West Coast of the United States. J Clin Microbiol. 1997;35:1965–1971. doi: 10.1128/jcm.35.8.1965-1971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okuda, J., M. Ishibashi, M. Hashimoto, M. J. Albert, and M. Nishibuchi. Unpublished data.
- 12.Okuda J, Ishibashi M, Hayakawa E, Nishino T, Takeda Y, Mukhopadhyay A K, Garg S, Bhattacharya S K, Nair G B, Nishibuchi M. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J Clin Microbiol. 1997;35:3150–3155. doi: 10.1128/jcm.35.12.3150-3155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okuda, J., and M. Nishibuchi. Unpublished data.
- 14.Reich K A, Schoolnik G K. The light organ symbiont Vibrio fisheri possesses a homolog of the Vibrio cholerae transmembrane transcriptional activator ToxR. J Bacteriol. 1994;176:3085–3088. doi: 10.1128/jb.176.10.3085-3088.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruimy R, Breittmayer V, Elbaze P, Lafay B, Boussemart O, Gauthier M, Christine R. Phylogenetic analysis and assessment of the genera Vibrio, Photobacterium, Aeromonas, and Plesiomonas deduced from small-subunit rRNA sequences. Int J Syst Bacteriol. 1994;44:416–426. doi: 10.1099/00207713-44-3-416. [DOI] [PubMed] [Google Scholar]
- 16.Shirai H, Ito H, Hirayama T, Nakabayashi Y, Kumagai K, Takeda Y, Nishibuchi M. Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect Immun. 1990;58:3568–3573. doi: 10.1128/iai.58.11.3568-3573.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tada J, Ohashi T, Nishimura N, Shirasaki Y, Ozaki H, Fukushima S, Takano J, Nishibuchi M, Takeda Y. Detection of thermostable direct hemolysin gene (tdh) and the thermostable direct hemolysin-related hemolysin gene (trh) of Vibrio parahaemolyticus by polymerase chain reaction. Mol Cell Probes. 1992;6:477–487. doi: 10.1016/0890-8508(92)90044-x. [DOI] [PubMed] [Google Scholar]
- 18.Taniguchi H, Hirano H, Kubomura S, Higashi K, Mizuguchi Y. Comparison of the nucleotide sequences of the genes for the thermostable direct hemolysin and the thermolabile hemolysin from Vibrio parahaemolyticus. Microb Pathog. 1986;1:425–432. doi: 10.1016/0882-4010(86)90004-5. [DOI] [PubMed] [Google Scholar]
- 19.Taniguchi H, Kubomura S, Hirano H, Mizue K, Ogawa M, Mizuguchi Y. Cloning and characterization of a gene encoding a new thermostable hemolysin from Vibrio parahaemolyticus. FEMS Microbiol Lett. 1990;67:339–346. doi: 10.1016/0378-1097(90)90020-q. [DOI] [PubMed] [Google Scholar]
- 20.Venkateswaran K, Dohmoto N, Harayama S. Cloning and nucleotide sequence of the gyrB gene of Vibrio parahaemolyticus and its application in detection of this pathogen in shrimp. Appl Environ Microbiol. 1998;64:681–687. doi: 10.1128/aem.64.2.681-687.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]