Abstract
Rationale
Recent studies have revealed that the lung microbiota of critically ill patients is altered and predicts clinical outcomes. The incidence of invasive fungal infections, namely, invasive pulmonary aspergillosis (IPA), in immunocompromised patients is increasing, but the clinical significance of variations in lung bacterial communities is unknown.
Objectives
To define the contribution of the lung microbiota to the development and course of IPA.
Methods and measurements
We performed an observational cohort study to characterise the lung microbiota in 104 immunocompromised patients using bacterial 16S ribosomal RNA gene sequencing on bronchoalveolar lavage samples sampled on clinical suspicion of infection. Associations between lung dysbiosis in IPA and pulmonary immunity were evaluated by quantifying alveolar cytokines and chemokines and immune cells. The contribution of microbial signatures to patient outcome was assessed by estimating overall survival.
Main results
Patients diagnosed with IPA displayed a decreased alpha diversity, driven by a markedly increased abundance of the Staphylococcus, Escherichia, Paraclostridium and Finegoldia genera and a decreased proportion of the Prevotella and Veillonella genera. The overall composition of the lung microbiome was influenced by the neutrophil counts and associated with differential levels of alveolar cytokines. Importantly, the degree of bacterial diversity at the onset of IPA predicted the survival of infected patients.
Conclusions
Our results reveal the lung microbiota as an understudied source of clinical variation in patients at risk of IPA and highlight its potential as a diagnostic and therapeutic target in the context of respiratory fungal diseases.
Keywords: aspergillus lung disease, critical care, opportunist lung infections, respiratory infection
Key messages.
What is the key question?
Does the lung microbiota modulate susceptibility to invasive pulmonary aspergillosis (IPA) and predict its clinical outcome?
What is the bottom line?
We show that variation in the lung microbiota is associated with the development of IPA in critically ill patients. Patients with IPA display a microbiota signature in the lower airways characterised by a decreased bacterial diversity that predicts clinical outcomes after infection. Moreover, we describe associations between lung dysbiosis in IPA and pulmonary antifungal immunity, including alveolar cytokines and chemokines and neutrophil counts.
Why read on?
Recent studies have unveiled alterations in the lung microbiota of critically ill patients, which were associated with alveolar and systemic inflammation to predict disease outcomes. IPA is frequently observed in critically ill patients. However, no study to date has determined whether the lung microbiota is altered in patients with IPA or predict clinical outcomes after infection.
Introduction
In the past decade, advances in molecular methods for the quantification and sequencing of bacterial DNA have revealed that the lungs, previously considered sterile, instead harbour complex and dynamic communities of bacteria.1 The lung microbiome is known to diverge significantly across healthy2 and diseased states3 4 and is associated with variations in both alveolar and systemic immunity.5 The structure and composition of the lung microbiome are predictive of disease outcomes across numerous chronic respiratory diseases. For example, the diversity of sputum microbiota predicts exacerbations in bronchiectasis,6 disease progression and lung function decline in cystic fibrosis (CF)7 and mortality after acute exacerbations of chronic obstructive pulmonary disease (COPD).8
Although lung dysbiosis is established as a major trigger of exacerbations in inflammatory airway diseases, only a few studies have addressed the mechanisms whereby specific bacterial communities contribute to disease severity. Nonetheless, mounting evidence supports a key contribution of the lung microbiota to the activation and regulation of lung immunity. For instance, the dysbiosis in the lower respiratory tract of patients with COPD revealed an expansion of streptococcal genera that, by suppressing the antimicrobial peptide cathelicidin, impaired the pulmonary clearance of bacteria.4 Moreover, the disruption of the lung microbiota in patients with idiopathic pulmonary fibrosis was described to promote increased levels of alveolar profibrotic cytokines underlying local inflammation and aberrant repair to drive disease progression.9
The lung microbiome of critically ill patients also differs profoundly from that of healthy individuals10–12 and is associated with alveolar inflammation and injury.10 11 Such features of lung dysbiosis were found to predict the clinical outcome of patients with sepsis and acute respiratory distress syndrome (ARDS).13 14 A history of smoking was related to the composition of the lung microbiome in intensive care patients, and development of ARDS was associated with a respiratory microbial community structure characterised by a relative enrichment of taxa more abundant in smokers at baseline.11 Collectively, these observations highlight the composition of the lung microbiome as a key factor involved in the development of complications in the intensive care setting.
Owing to advances in critical care medicine and the introduction of broad-spectrum antibiotics, the incidence of invasive fungal infections in critical care is on the rise, namely, among haematological patients under chemotherapy or patients undergoing solid organ or allogeneic haematopoietic stem-cell transplantation (HSCT).15 In particular, invasive pulmonary aspergillosis (IPA) remains a major cause of mortality in these clinical settings with rates estimated between 20% and 30%16 17 and more than 80% when involving cases of infection with azole-resistant strains.18 The relevance of these numbers is further highlighted by the lack of any licensed vaccine and limitations in currently available tests for the diagnosis of IPA. An improved understanding of the pathogenesis of IPA is therefore required for the development of more effective diagnostic or control measures for this infection.
Although the importance of the lung microbiome has already been put forward in the context of IPA,19 20 no study to date has determined whether the lung microbiota is altered in patients with IPA and whether it predicts clinical outcomes after infection. In light of these observations, we hypothesised that variation in the lung microbiota is a relevant factor contributing to the development of IPA and its outcome in immunocompromised patients. We report that the dysbiosis of the lung microbiota in immunocompromised patients is associated with the development of IPA and that a decreased bacterial diversity in IPA predicts poorer outcomes. Moreover, we describe associations between lung dysbiosis and pulmonary immunity, as revealed by the differential effects on the levels of alveolar cytokines and the number of neutrophils. This study highlights the pivotal contribution of the lung microbiota to the regulation of pulmonary immunity and susceptibility to IPA in immunocompromised patients.
Methods
Study design
The study population comprised adult patients (≥18 years of age) hospitalised between 2013 and 2018 at the Leuven University Hospitals, Leuven, Belgium. Cases of ‘probable’ or ‘proven’ IPA were identified according to the 2008 criteria from the European Organisation for Research and Treatment of Cancer/Mycoses Study Group.21 The control group included patients with no evidence for the presence of Aspergillus spp in the bronchoalveolar lavage (BAL) sample (negative culture, negative microscopy and a galactomannan optical density index <0.5). Control patients were randomly selected from the patients indicated for bronchoscopy at around the same time as the cases of IPA to obtain a 1:1 case–control ratio. Patients with ‘possible’ disease were excluded from the study, and no mold-active antifungals were administered by the treating physician(s) before sample collection. This study was approved by the Ethics Committee for Research in Life and Health Sciences of the University of Minho, Portugal, and the Ethics Committee of the University Hospitals of Leuven, Belgium (IRB number 126/014). Written informed consent was obtained from the patient or a representative prior to collection of airway samples. Additional details, including sequencing and quality control checks (online supplemental file 1), are provided in the online supplemental material.
thoraxjnl-2020-216179supp001.pdf (999.7KB, pdf)
Statistical analyses
The global effects of metadata variables on the microbiome composition of samples were tested using the adonis function from the vegan R package (V.2.5–5),22 which runs a permutational multivariate ANOVA by fitting linear models to distance matrices. Each variable was tested using each of the distance matrices that were calculated in order to test the association of the variable with the differing effects of each beta diversity calculation. Principal coordinates analysis was performed on the indicated distance matrix using the pcoa function from the ape R package (V.5.3).23 The linear model for each variable also included three covariates, namely, case/control, as this was the most relevant and influential variable, and age and gender to account for systematic variation from these factors. The sequencing date, of which there were two for these data, was also considered as the ‘strata’ argument to account for a potential batch effect. The associations between variables and particular taxa were tested using the lm function from the stats R package (V.3.6.3).24 The function fits a linear model, using the same covariates as fixed effects in the PERMANOVA model, also including the sequencing date. Additional details regarding statistical methods, including model assumptions and correction for multiple testing, are provided in the supplementary material. Complete model outputs for the significant associations are provided in the online supplemental file 2).
Results
The composition and diversity of the lung microbiome are altered in IPA
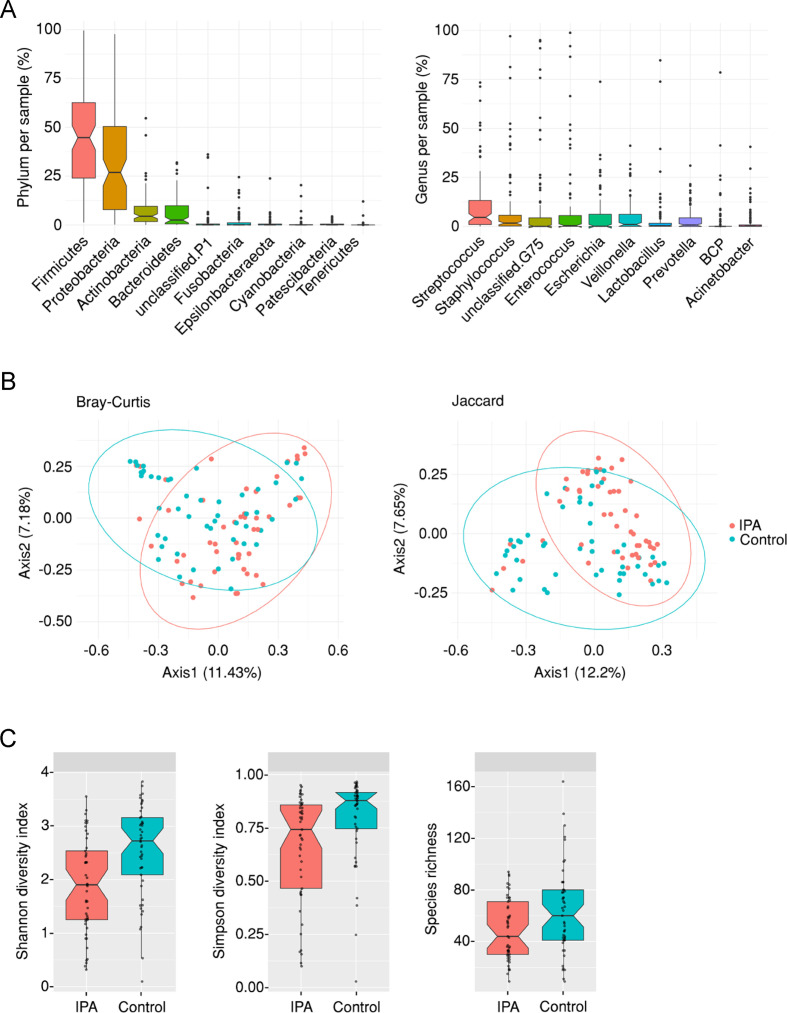
Previous studies have demonstrated a shift in the composition of the bacterial communities in the lower airways from critically ill patients undergoing pneumonia or sepsis.13 14 The first aim of our study was therefore to compare the overall composition of the lung microbiome in patients with IPA and uninfected, disease-matched controls. We analysed BAL specimens collected on clinical suspicion of infection from 104 immunocompromised patients, including 54 cases diagnosed with probable/proven IPA and 50 controls without IPA. The demographics and underlying clinical conditions of both groups are shown in table 1. In the overall cohort, Firmicutes, Proteobacteria, Actinobacteria and Bacteroidetes were the most abundant phyla, whereas Streptococcus, Staphylococcus, Enterococcus, Escherichia, Veillonella, Lactobacillus and Prevotella were the most common genera (figure 1A).
Table 1.
Demographics and clinical characteristics of the study cohort
| Variables |
IPA
(n=54) |
Controls
(n=50) |
P value |
| Age, mean±SD | 63.6±12.6 | 60.9±13.1 | 0.26 |
| Man, no (%) | 34 (63) | 35 (70) | 0.53 |
| Underlying disease, no (%)* | |||
| Acute leukaemia | 16 (29.6) | 14 (28) | 0.08 |
| Allogeneic HSCT | 10 (18.5) | 16 (32) | |
| Chronic lymphoproliferative diseases | 8 (14.8) | 10 (20) | |
| SOT | 5 (9.3) | 4 (8) | |
| Influenza A (H1N1) | 8 (14.8) | 0 (0) | |
| Solid tumours | 2 (3.7) | 4 (8) | |
| Liver cirrhosis | 3 (5.6) | 2 (4) | |
| COPD | 2 (3.7) | 0 (0) | |
| ICU admission | 28 (51.9) | 12 (24) | 0.009 |
| Mechanical ventilation | 18 (33.3) | 5 (10) | 0.005 |
| Severe neutropenia, no (%)† | 20 (37) | 11 (22) | 0.13 |
| BAL cell counts, mean (range)‡ | |||
| Neutrophils | 4.9 (0–30.3) | 4.0 (0–24.6) | 0.28 |
| Lymphocytes | 4.0 (0–175.7) | 1.3 (0.1–17.4) | 0.004 |
| CMV IgG serostatus | |||
| Positive | 18 (33.3) | 20 (40) | 0.80 |
| Negative | 14 (25.9) | 12 (24) | |
| Unknown | 22 (40.7) | 18 (36) | |
| Immunosuppression, no (%) | |||
| Steroids | 25 (46.3) | 14 (28) | 0.06 |
| Other immunosuppressive regimens | 4 (7.4) | 10 (20) | |
| None | 25 (46.3) | 26 (52) | |
| Antibiotherapy, no (%) | |||
| β-lactams§ | 50 (92.6) | 33 (66) | 0.003 |
| Other antibiotics | 2 (3.7) | 8 (16) | |
| None | 2 (3.7) | 9 (18) |
P values were calculated by Fisher’s exact probability t-test for categorical variables or by Student’s t-test or Mann-Whitney U test for continuous variables.
*SOT included lung, heart, kidney and liver transplants. Solid tumours included patients with lung, breast and liver cancer.
†Severe neutropenia was defined as ≤0.5×109 cells/L.
‡Cell counts in BAL were expressed as the number ×103 cells/µL.
§Carbapenems and piperacillin–tazobactam were included in the β-lactams category.
BAL, bronchoalveolar lavage; CMV, cytomegalovirus; COPD, chronic pulmonary obstructive disease; HSCT, haematopoietic stem-cell transplantation; ICU, intensive care unit; IPA, invasive pulmonary aspergillosis; SOT, solid organ transplantation.
Figure 1.
The lung microbiome is altered in patients with invasive pulmonary aspergillosis (IPA). (A) Relative abundance (%) of operational taxonomical units assigned at the phylum and genus levels in each bronchoalveolar lavage sample from the overall cohort. (B) Principal coordinate analysis of cases of IPA and controls based on the Bray-Curtis dissimilarity metric and the Jaccard distance. Axes represent the first two principal coordinates, with the variation explained by the indicated variable shown as percentage. (C) Boxplots illustrating the alpha diversity (measured by the Shannon and Simpson Diversity Indexes) and the species richness of the lung microbiome in cases of IPA and controls. Median values and IQRs are indicated in the plots.
We next compared the composition of bacterial communities in IPA and control patients using complementary approaches. We first visualised communities using principal component analysis and the Bray-Curtis dissimilarity metric and the Jaccard coefficient (figure 1B and online supplemental files 3 and 4). While considerable taxonomical overlap was found across IPA and control patients, these analyses revealed a strong effect of the infection status on the overall composition of the lung microbiome. This collective difference in community composition was also reflected by a markedly lower alpha diversity (measured by both the Shannon and Simpson Diversity Indexes) and species richness of the bacterial taxa in patients with IPA than uninfected controls (figure 1C and online supplemental file 5). Despite the heterogeneity in the clinical conditions of the patients, our analyses discarded a major influence of the underlying disease on the overall composition of the lung microbiome, although individual taxa, namely, Delftia, were more abundant in specific disease groups as the result of a few outlier samples (online supplemental figure S1).
Patients with IPA exhibit a differential abundance of specific bacterial taxa
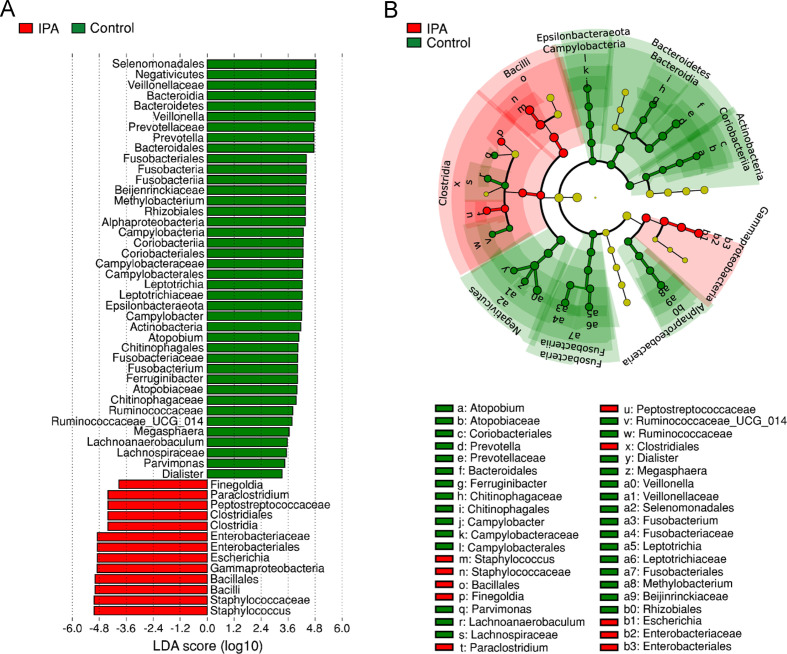
To identify the specific taxa responsible for the collective differences in community composition, we next compared the relative abundance of prominent taxa across patients with IPA and controls using the linear discriminant analysis effect size method. We detected a total of 53 taxonomical clades whose relative abundance had the most significant effect size on the lung microbiome (figure 2A). Among these clades, the Staphylococcus, Escherichia, Paraclostridium and Finegoldia genera were more abundant in cases of IPA, whereas the Prevotella and Veillonella genera were the most abundant in control patients. Besides Veillonella, other members of the Veillonellaceae family, including Dialister and Megasphaera, were also enriched in control patients. Accordingly, the phylogenetic mapping of the differentially abundant bacterial taxa revealed an enrichment of the Bacilli, Clostridia and Gammaproteobacteria classes in patients with IPA, whereas the taxa that differed the most among controls belonged to the Bacteroidetes, Actinobacteria, Fusobacteria and Epsilonbacteraeota phyla and the Alphaproteobacteria and Negativicutes classes (figure 2B).
Figure 2.
Invasive pulmonary aspergillosis (IPA) affects the abundance of selected bacterial taxa. (A) Histogram of taxonomical clades with differential abundance between cases of IPA and controls based on linear discriminant analysis (LDA) scores (log10). Taxa enriched in IPA are indicated with a negative LDA score (red), and taxa enriched in controls have a positive score (green). Only taxa meeting an LDA score >3 or <−3 are shown. (B) Cladogram illustrating the distribution of taxa with differential abundance between cases of IPA and controls. Each circle represents a taxon, with those more abundant in cases of IPA coloured in red and those more abundant in controls in green. Yellow circles represent non-significant differences. The diameter of each circle is proportional to the taxon abundance.
Because the altered microbiome profiles observed in IPA could be attributed to a variable use of antibiotics, we next assessed their impact, most frequently involving β-lactams in our cohort, on the lung microbiome of patients with IPA and controls. The administration of β-lactams (from 2 weeks before until the date of bronchoscopy) induced a decrease in the diversity of the bacterial communities in both cases of infection and controls (online supplemental figure S2). Among the bacterial taxa that showed a differential abundance in the IPA and control groups, only Prevotella was significantly affected by β-lactams, an effect that was however independent of the infection status. In these conditions, IPA promoted a far greater inhibitory effect on the abundance of Prevotella than β-lactams. In addition, the impact of IPA on the abundance of Prevotella was further emphasised by the removal of the natural tendency towards an increasing abundance of Prevotella with age. The use of steroids did not have a significant effect on the overall composition of the microbiome by any of the distance measures tested.
The lung microbiome in IPA is associated with altered pulmonary immunity
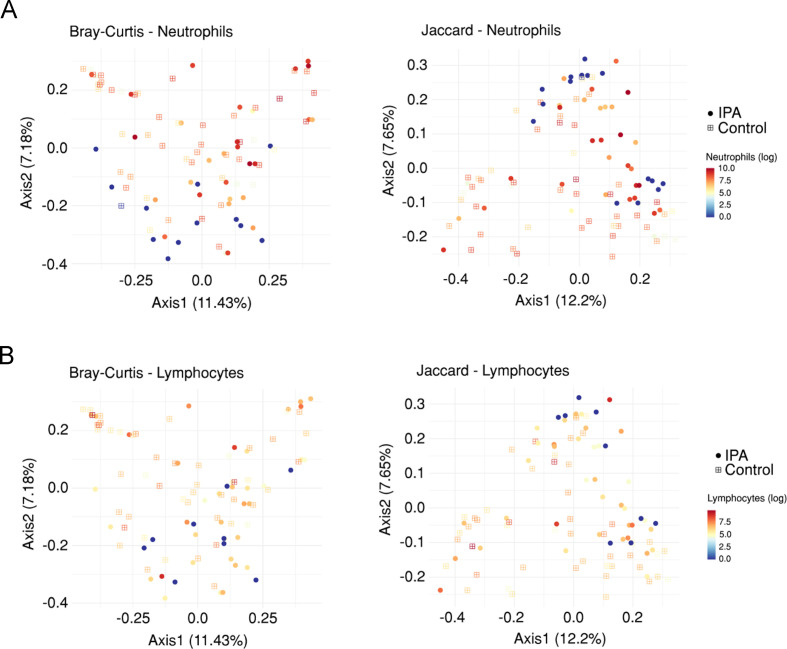
The microbiome has the capacity to regulate the antimicrobial immunity in the lung.25 Because the immune status of the host represents one major factor predisposing to IPA,26 we evaluated the association between lung bacterial communities and alveolar immunity, namely, the number of neutrophils and lymphocytes, and the levels of cytokines and chemokines in the BAL. We started by analysing bacterial communities using a PERMANOVA analysis based on both the Bray-Curtis and the Jaccard distance matrices. We observed a noticeable gradient in the number of neutrophils (figure 3A), but not lymphocytes (figure 3B), indicating a shift in the overall composition of the lung microbiome according to neutrophil counts. The alpha diversity was, however, not affected by the number of either immune cell type (online supplemental figure S3). The finding that neutrophil counts were also positively associated with bacterial reads unclassified at the phylum level suggests that the abundance of rare and understudied bacterial taxa increases with higher neutrophil counts.
Figure 3.
Neutrophil counts influence the lung microbiome. Principal coordinate analysis based on the Bray-Curtis dissimilarity metric and the Jaccard distance, coloured according to the number of (A) neutrophils and (B) lymphocytes in each bronchoalveolar lavage sample from the overall cohort. Axes represent the first two principal coordinates, with the variation explained by the indicated variable shown as percentage. Circles indicate cases of invasive pulmonary aspergillosis (IPA), and squares indicate controls.
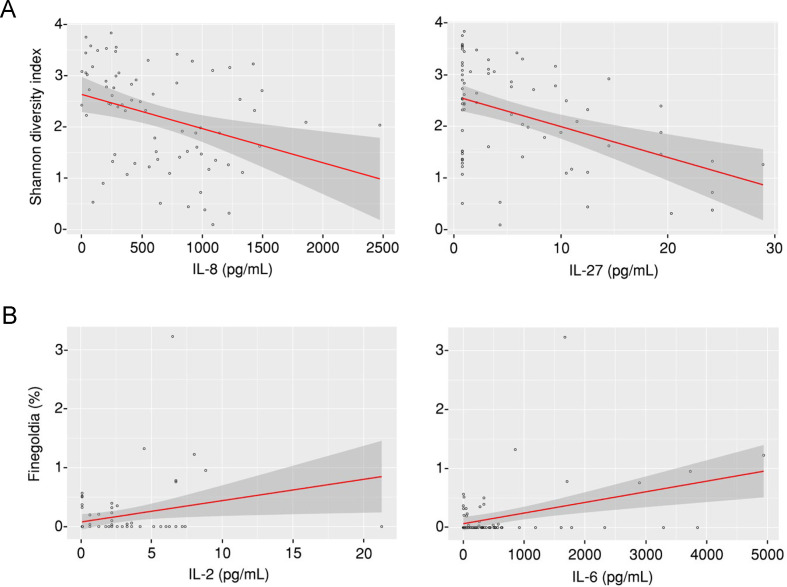
To explore the relationship between the lung microbiome and local immunity, we measured the levels of 34 cytokines and chemokines in the BAL in a subgroup of 79 patients within the overall cohort, including 38 cases of IPA (online supplemental table S1), corresponding to all the patients for which enough sample was available also for these measurements. Among the different analytes tested, interleukin (IL) 8 and IL-27 were negatively associated with the bacterial alpha diversity (measured by the Shannon Diversity Index) in the overall cohort (figure 4A), although the effects for IL-8 were observed in control patients only (online supplemental figure S4). Moreover, associations were commonly found between the abundance of specific bacterial taxa and the levels of alveolar cytokines (online supplemental table S2). Among the taxa found to be enriched in patients with IPA, the abundance of Finegoldia was positively associated with the levels of both IL-2 and IL-6 (figure 4B), with the association with IL-2 being specifically observed in cases of IPA, but not controls (online supplemental figure S4). No associations were observed for Prevotella and Veillonella, the two genera most abundantly detected in controls.
Figure 4.
Taxa enriched in invasive pulmonary aspergillosis are associated with increased inflammation. (A) Association analysis between the alpha diversity (measured by the Shannon Diversity Index) and the levels of interleukin (IL) 8 and IL-27 in each bronchoalveolar lavage (BAL) sample from the overall cohort. (B) Association analysis between the abundance of Finegoldia (%) and the levels of IL-2 and IL-6 in each BAL sample from the overall cohort.
Diversity of the lung microbiome predicts the outcome of IPA
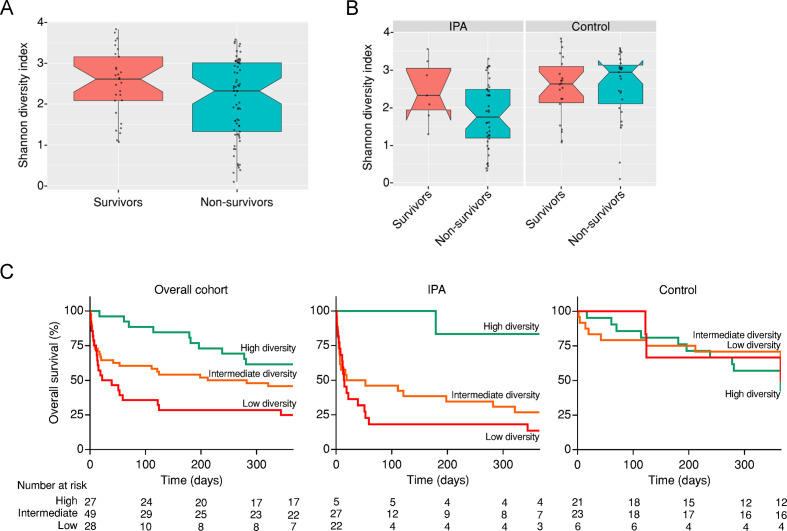
In our cohort of immunocompromised patients with suspected infection, a total of 46 (85.2%) patients with IPA died, whereas only 27 (54.0%) patients without IPA failed to survive. Likewise, the time to death or last follow-up visit was markedly shorter for cases of IPA than control patients (online supplemental figure S5). We first analysed whether the bacterial diversity (measured by the Shannon Diversity Index) in the lung microbiome could discriminate survivors from non-survivors. We found that the alpha diversity was not different between both groups in the overall cohort (figure 5A). This effect was however masked by the infection status of the patients, since patients diagnosed with IPA that did not survive presented instead a markedly decreased alpha diversity compared with IPA survivors (figure 5B).
Figure 5.
The diversity of the lung microbiome predicts survival in invasive pulmonary aspergillosis (IPA). Boxplot illustrating the alpha diversity (measured by the Shannon Diversity Index) of the lung microbiome among (A) survivors and non-survivors in the overall cohort and (B) survivors and non-survivors grouped according to the infection status (IPA and controls). Median values and IQRs are indicated in the plots. (C) Kaplan-Meier estimates of 1-year overall survival in the overall cohort (log rank p=0.004), cases of IPA (log rank p=0.02) and controls (log rank p=0.46), according to low, intermediate and high levels of alpha diversity. The number of patients at risk at baseline and selected time points is shown.
To evaluate whether the diversity in bacterial taxa at baseline could predict the outcome of the patients, we classified the subjects from the overall cohort into groups based on the percentiles of the individual Shannon Diversity Indexes. At the time of BAL collection, high microbial diversity (index >3.02) was observed in 27 (26.0%) patients, intermediate diversity (index >1.46 and<3.02) was observed in 49 (47.1%) patients and low diversity (index <1.46) was observed in 28 (26.9%) patients. We next analysed the 1-year overall survival of the patients using Kaplan-Meier estimates for each of the diversity groups. The estimated survival was 62% for the high-diversity group, 46% for the intermediate-diversity group and 25% for the low-diversity group (figure 5C). The contribution of the bacterial diversity to survival was even more evident among patients with IPA, in which the estimated survival was 83% for the high-diversity group, 27% for the intermediate-diversity group and 14% for the low-diversity group. Accordingly, cases of IPA within the low-diversity group (compared with the high-diversity group) had a fourfold increased risk of death after infection (HR, 4.2; 95% CI, 1.34–13.1; p=0.014). No differences were found in the survival of control patients according to the defined alpha diversity groups (HR, 1.3; 95% CI, 0.41–3.89; p=0.68).
Discussion
Understanding how the lung microbiome interacts with the immune system is essential to uncover the molecular and cellular mechanisms underlying the pathogenesis of IPA and may offer crucial insights into a vast network of signals that may be amenable to therapeutic targeting.19 Here, we analysed the contribution of the lung microbiota to the establishment and outcome of IPA in immunocompromised patients. We provide compelling evidence for marked alterations in the community composition and diversity of the lung microbiome in patients with IPA. Our data further demonstrate that defined airway microbial states underlie functionally relevant immunological shifts that relate to distinct profiles of progression and outcome of IPA.
Our study is the first to demonstrate the existence of a microbiome signature characterised by a significant loss of bacterial diversity in the airways of patients with IPA. These dynamic changes are likely to occur over time in patients following hospital admission, which, in turn, elicit inflammatory signals contributing to the onset of IPA and further progression of infection. An identical process occurs in HIV infection, where the concerted action of immune defects and chronic inflammation alters the microbiome composition in the lower airways towards an enrichment in pneumonia-associated bacteria.27 28 While the selection pressure by antibiotics could explain, at least in part, the airway dysbiosis and support the establishment of a microenvironment permissive for specific pathogens, our study discarded a major effect of antibiotic therapy on the lung microbiome signature of patients with IPA.
The loss of bacterial diversity observed in patients with IPA was concurrent to an overgrowth of bacteria described to cause pneumonia under favourable conditions. For example, Staphylococcus spp are common inhabitants of the upper airways that can become ‘accidental’ pathogens as the result of complex interactions with other microbes and the immune status of the host.29 In addition, certain bacterial taxa, especially those within the Gammaproteobacteria class, for example, Escherichia, encode the metabolic capacity to use inflammatory by-products to survive and prosper.30 During chronic inflammation, these bacteria can thrive and outcompete others that lack the metabolic capacity to benefit from such inflammation. This has been suggested in HSCT recipients, in which a Gammaproteobacteria domination in the gut was associated with pulmonary complications.31 Similar mechanisms appear to be at play in the respiratory tract since an increased abundance of these taxa in HSCT recipients also contributed to impaired pulmonary function and alveolar concentrations of inflammatory cytokines.32 Our finding that lung dysbiosis worsens the prognosis of patients with IPA is compatible with a recent study reporting that the enrichment of the lung microbiome with gut-associated bacteria, for example, Enterobacteriaceae, allows for the prediction of clinical outcomes in critically ill patients.13 Other studies have also found that the lung microbiome predicts disease outcomes in idiopathic pulmonary fibrosis,9 33 COPD8 and bronchiectasis.6
Besides the dominance by specific bacterial taxa, IPA was characterised by a decreased abundance of anaerobic commensals from the Prevotella and Veillonella genera. This observation is in line with the increased risk of pneumonia in elderly and young adults in the absence of these taxa.34 In contrast, an increased abundance of these bacteria has been associated with higher levels of lymphocyte and neutrophil inflammation in the lower airways,35 and an airway microbiome dominated by Prevotellaceae has been proposed to elicit lung inflammation with a T helper 17 phenotype.36 Although we confirmed a relationship between the lung microbiome and local inflammation in IPA, our data suggest instead that the fungus prospers in an environment with a poor abundance of these bacteria in a context of exuberant inflammation. In fact, the dysbiosis in IPA appears to impact preferentially innate rather than T-cell-derived cytokines, a finding in line with the impact of neutrophils, but not lymphocytes, on alpha diversity. These findings are supported not only by previous reports showing increased alveolar levels of proinflammatory cytokines in patients with IPA37 but also by the ability of these cytokines to predict survival in haematological patients with suspected mould infection.38
Patients with HIV undergoing antiretroviral therapy display an increased and persistent abundance of the Prevotella and Veillonella genera in the lower-airway microbiome,27 suggesting that the control of the infection allows the replenishment of the microbiome with anaerobes usually found in healthy states. How the functional activity of these bacteria could elicit protection in the context of IPA remains to be addressed. One possible mechanism involves complex immune and metabolic interactions between the different components of the microbiome and the immune system of the host.39 The production of short-chain fatty acids (SCFAs), end products of bacterial anaerobic metabolism, has been associated with increased susceptibility to tuberculosis in HIV patients as the result of direct inhibitory effects on T-cell function.40 Although the regulation of immunological mechanisms by bacterial metabolites in the context of IPA remains to be elucidated, bacterial producers of SCFAs have been found to dampen allergic airway responses in mice by inducing the expansion of T regulatory cells,41 with the latter being critical in maintaining pulmonary immune homeostasis in mouse models of CF subjected to experimental allergic aspergillosis.42
Despite its unbiased nature and the considerable number of patients enrolled, our study has certain limitations. The most relevant regards the clinical heterogeneity of the patient population (eg, underlying disease, mechanical ventilation and degree of immunocompromise) and the inability to conclude whether the observed alterations in the lung microbiome are a cause or a consequence of the development of IPA or merely coincide with the disease status. Indeed, the identified associations represent hypotheses on possible direct or indirect causal relationships that should be confirmed in distinct patient cohorts and experimental models other than humans. In addition, our study focused specifically on the bacterial microbiome, and it remains to be addressed whether significant changes also occur in other microbial compartments, for example, the mycobiome and the virome. In fact, admission to the intensive care unit was associated with marked changes in the oral mycobiome,43 and thus, variation in the fungal communities may also help to explain the susceptibility to IPA. Finally, the analysis of additional control groups, for example, patients with ARDS without IPA or non-immunocompromised patients with community-acquired or hospital-acquired pneumonia could expand further the relevance of our conclusions.
Our data, however, also raise interesting questions that warrant further investigation. While our study focused on the lower-airway microbiome, there are reports demonstrating that the microbiome of distant mucosal sites may impact the risk of pneumonia.31 Future studies should interrogate whether the contribution of the microbiota to IPA and its outcome is more likely to result from distant or local host–microbiome interactions. In addition, non-invasive approaches, for example, oropharyngeal washes, would allow the longitudinal analysis of patients at risk of IPA and unveil whether specific alterations in the airway microbiome precede the onset of the infection and could be used in risk prediction tools.
Our results provide support to studies in larger cohorts evaluating the prognostic and diagnostic potential of the airway microbiome and its immune interactions, particularly in combination with fungal surrogate markers and integrating the genetic risk profile of the patient.44 An improved understanding of microbiome dynamics in patients at risk of IPA is expected to pave the way towards the implementation of individualised approaches using antimicrobials or immunotherapies to regulate bacterial community composition, microbial metabolism or the immune response.
Footnotes
AH and JRW contributed equally.
Contributors: Conception and design: AC, TG and CC. Collection of clinical specimens and acquisition of patient-level data: TM, KL and JM. Acquisition of data: AH, JRW, TM, SMG and RG. Analysis and interpretation of data: AH, JRW, TM, AC, TG and CC. Drafting or revising of manuscript: AH, JRW, TM, KL, SMG, RG, JM, AC, TG and CC. Final approval of manuscript: AH, JRW, TM, KL, SMG, RAG, JM, AC, TG and CC.
Funding: This work was supported by the Fundação para a Ciência e a Tecnologia (FCT) (PTDC/SAU-SER/29635/2017 to CC and PTDC/MED-GEN/28778/2017 to AC). Additional support was provided by FCT (UIDB/50026/2020 and UIDP/50026/2020); the Northern Portugal Regional Operational Programme (NORTE 2020), under the Portugal 2020 Partnership Agreement, through the European Regional Development Fund (ERDF) (NORTE-01-0145-FEDER-000013 and NORTE-01-0145-FEDER-000023); the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement number 847507 (to AC); and the “la Caixa” Foundation (ID 100010434) and FCT under the agreement LCF/PR/HR17/52190003 (to AC). Individual support was provided by FCT (SFRH/BD/136814/2018 to SMG, CEECIND/03628/2017 to AC and CEECIND/04058/2018 to CC). The TG group acknowledges support from the Spanish Ministry of Science and Innovation for grant PGC2018-099921-B-I00, cofounded by the ERDF, the CERCA Programme/Generalitat de Catalunya, the Catalan Research Agency (AGAUR) SGR423, the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement ERC-2016-724173 and an INB Grant (PT17/0009/0023—ISCIII-SGEFI/ERDF).
Competing interests: KL reports personal fees and non-financial support from Pfizer and MSD and personal fees from SMB Laboratoires, Gilead and FUJIFILM Wako, outside the submitted work. JM reports personal fees and non-financial support from MSD, Cidara, F2G and Scynexis; grants, personal fees and non-financial support from Pfizer and Gilead; and non-financial support from Bio-Rad, outside the submitted work.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information. The accession number for all sequencing data reported in this paper is NCBI Bioproject PRJNA647539.
Ethics statements
Patient consent for publication
Not required.
References
- 1. Dickson RP, Erb-Downward JR, Martinez FJ, et al. The microbiome and the respiratory tract. Annu Rev Physiol 2016;78:481–504. 10.1146/annurev-physiol-021115-105238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dickson RP, Erb-Downward JR, Freeman CM, et al. Bacterial topography of the healthy human lower respiratory tract. mBio 2017;8. 10.1128/mBio.02287-16. [Epub ahead of print: 14 Feb 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jorth P, Ehsan Z, Rezayat A, et al. Direct lung sampling indicates that established pathogens dominate early infections in children with cystic fibrosis. Cell Rep 2019;27:1190–204. 10.1016/j.celrep.2019.03.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Singanayagam A, Glanville N, Cuthbertson L, et al. Inhaled corticosteroid suppression of cathelicidin drives dysbiosis and bacterial infection in chronic obstructive pulmonary disease. Sci Transl Med 2019;11. 10.1126/scitranslmed.aav3879. [Epub ahead of print: 28 Aug 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Budden KF, Shukla SD, Rehman SF, et al. Functional effects of the microbiota in chronic respiratory disease. Lancet Respir Med 2019;7:907–20. 10.1016/S2213-2600(18)30510-1 [DOI] [PubMed] [Google Scholar]
- 6. Rogers GB, Zain NMM, Bruce KD, et al. A novel microbiota stratification system predicts future exacerbations in bronchiectasis. Ann Am Thorac Soc 2014;11:496–503. 10.1513/AnnalsATS.201310-335OC [DOI] [PubMed] [Google Scholar]
- 7. Acosta N, Heirali A, Somayaji R, et al. Sputum microbiota is predictive of long-term clinical outcomes in young adults with cystic fibrosis. Thorax 2018;73:1016–25. 10.1136/thoraxjnl-2018-211510 [DOI] [PubMed] [Google Scholar]
- 8. Leitao Filho FS, Alotaibi NM, Ngan D, et al. Sputum microbiome is associated with 1-year mortality after chronic obstructive pulmonary disease hospitalizations. Am J Respir Crit Care Med 2019;199:1205–13. 10.1164/rccm.201806-1135OC [DOI] [PubMed] [Google Scholar]
- 9. O'Dwyer DN, Ashley SL, Gurczynski SJ, et al. Lung microbiota contribute to pulmonary inflammation and disease progression in pulmonary fibrosis. Am J Respir Crit Care Med 2019;199:1127–38. 10.1164/rccm.201809-1650OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dickson RP, Singer BH, Newstead MW, et al. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol 2016;1:16113. 10.1038/nmicrobiol.2016.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Panzer AR, Lynch SV, Langelier C, et al. Lung microbiota is related to smoking status and to development of acute respiratory distress syndrome in critically ill trauma patients. Am J Respir Crit Care Med 2018;197:621–31. 10.1164/rccm.201702-0441OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zakharkina T, Martin-Loeches I, Matamoros S, et al. The dynamics of the pulmonary microbiome during mechanical ventilation in the intensive care unit and the association with occurrence of pneumonia. Thorax 2017;72:803–10. 10.1136/thoraxjnl-2016-209158 [DOI] [PubMed] [Google Scholar]
- 13. Dickson RP, Schultz MJ, van der Poll T, et al. Lung microbiota predict clinical outcomes in critically ill patients. Am J Respir Crit Care Med 2020;201:555–63. 10.1164/rccm.201907-1487OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kyo M, Nishioka K, Nakaya T, et al. Unique patterns of lower respiratory tract microbiota are associated with inflammation and hospital mortality in acute respiratory distress syndrome. Respir Res 2019;20:246. 10.1186/s12931-019-1203-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brown GD, Denning DW, Gow NAR, et al. Hidden killers: human fungal infections. Sci Transl Med 2012;4:165rv13. 10.1126/scitranslmed.3004404 [DOI] [PubMed] [Google Scholar]
- 16. Herbrecht R, Patterson TF, Slavin MA, et al. Application of the 2008 definitions for invasive fungal diseases to the trial comparing voriconazole versus amphotericin B for therapy of invasive aspergillosis: a collaborative study of the mycoses Study Group (MSG 05) and the European organization for research and treatment of cancer infectious diseases group. Clin Infect Dis 2015;60:713–20. 10.1093/cid/ciu911 [DOI] [PubMed] [Google Scholar]
- 17. Maertens JA, Raad II, Marr KA, et al. Isavuconazole versus voriconazole for primary treatment of invasive mould disease caused by Aspergillus and other filamentous fungi (secure): a phase 3, randomised-controlled, non-inferiority trial. Lancet 2016;387:760–9. 10.1016/S0140-6736(15)01159-9 [DOI] [PubMed] [Google Scholar]
- 18. van der Linden JWM, Snelders E, Kampinga GA, et al. Clinical implications of azole resistance in Aspergillus fumigatus, the Netherlands, 2007-2009. Emerg Infect Dis 2011;17:1846–54. 10.3201/eid1710.110226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gonçalves SM, Lagrou K, Duarte-Oliveira C, et al. The microbiome-metabolome crosstalk in the pathogenesis of respiratory fungal diseases. Virulence 2017;8:673–84. 10.1080/21505594.2016.1257458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kolwijck E, van de Veerdonk FL. The potential impact of the pulmonary microbiome on immunopathogenesis of Aspergillus-related lung disease. Eur J Immunol 2014;44:3156–65. 10.1002/eji.201344404 [DOI] [PubMed] [Google Scholar]
- 21. De Pauw B, Walsh TJ, Donnelly JP, et al. Revised definitions of invasive fungal disease from the European organization for research and treatment of Cancer/Invasive fungal infections cooperative group and the National Institute of allergy and infectious diseases Mycoses Study Group (EORTC/MSG) consensus group. Clin Infect Dis 2008;46:1813–21. 10.1086/588660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oksanen J, Blanchet F, Friendly M. Vegan: community ecology package (R package version 2.5–5) 2019.
- 23. Paradis E, Schliep K. Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019;35:526–8. 10.1093/bioinformatics/bty633 [DOI] [PubMed] [Google Scholar]
- 24. R Core Team . R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2013. http://www.R-project.org/ [Google Scholar]
- 25. Maschirow L, Suttorp N, Opitz B. Microbiota-Dependent regulation of antimicrobial immunity in the lung. Am J Respir Cell Mol Biol 2019;61:284–9. 10.1165/rcmb.2019-0101TR [DOI] [PubMed] [Google Scholar]
- 26. Latgé J-P, Chamilos G. Aspergillus fumigatus and aspergillosis in 2019. Clin Microbiol Rev 2019;33. 10.1128/CMR.00140-18. [Epub ahead of print: 18 Dec 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Twigg HL, Knox KS, Zhou J, et al. Effect of advanced HIV infection on the respiratory microbiome. Am J Respir Crit Care Med 2016;194:226–35. 10.1164/rccm.201509-1875OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cribbs SK, Uppal K, Li S, et al. Correlation of the lung microbiota with metabolic profiles in bronchoalveolar lavage fluid in HIV infection. Microbiome 2016;4:3. 10.1186/s40168-016-0147-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Otto M. Staphylococcus epidermidis--the 'accidental' pathogen. Nat Rev Microbiol 2009;7:555–67. 10.1038/nrmicro2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Winter SE, Winter MG, Xavier MN, et al. Host-Derived nitrate boosts growth of E. coli in the inflamed gut. Science 2013;339:708–11. 10.1126/science.1232467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harris B, Morjaria SM, Littmann ER, et al. Gut microbiota predict pulmonary infiltrates after allogeneic hematopoietic cell transplantation. Am J Respir Crit Care Med 2016;194:450–63. 10.1164/rccm.201507-1491OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. O'Dwyer DN, Zhou X, Wilke CA, et al. Lung dysbiosis, inflammation, and injury in hematopoietic cell transplantation. Am J Respir Crit Care Med 2018;198:1312–21. 10.1164/rccm.201712-2456OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Molyneaux PL, Cox MJ, Willis-Owen SAG, et al. The role of bacteria in the pathogenesis and progression of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2014;190:906–13. 10.1164/rccm.201403-0541OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de Steenhuijsen Piters WAA, Huijskens EGW, Wyllie AL, et al. Dysbiosis of upper respiratory tract microbiota in elderly pneumonia patients. Isme J 2016;10:97–108. 10.1038/ismej.2015.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Segal LN, Alekseyenko AV, Clemente JC, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome 2013;1:19. 10.1186/2049-2618-1-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Segal LN, Clemente JC, Tsay J-CJ, et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat Microbiol 2016;1:16031. 10.1038/nmicrobiol.2016.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gonçalves SM, Lagrou K, Rodrigues CS, et al. Evaluation of bronchoalveolar lavage fluid cytokines as biomarkers for invasive pulmonary aspergillosis in at-risk patients. Front Microbiol 2017;8:2362. 10.3389/fmicb.2017.02362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rawlings SA, Heldt S, Prattes J, et al. Using interleukin 6 and 8 in blood and bronchoalveolar lavage fluid to predict survival in hematological malignancy patients with suspected pulmonary mold infection. Front Immunol 2019;10:1798. 10.3389/fimmu.2019.01798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Egland PG, Palmer RJ, Kolenbrander PE. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc Natl Acad Sci U S A 2004;101:16917–22. 10.1073/pnas.0407457101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Segal LN, Clemente JC, Li Y, et al. Anaerobic bacterial fermentation products increase tuberculosis risk in Antiretroviral-Drug-Treated HIV patients. Cell Host Microbe 2017;21:530–7. 10.1016/j.chom.2017.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Karimi K, Inman MD, Bienenstock J, et al. Lactobacillus reuteri-induced regulatory T cells protect against an allergic airway response in mice. Am J Respir Crit Care Med 2009;179:186–93. 10.1164/rccm.200806-951OC [DOI] [PubMed] [Google Scholar]
- 42. Iannitti RG, Carvalho A, Cunha C, et al. Th17/Treg imbalance in murine cystic fibrosis is linked to indoleamine 2,3-dioxygenase deficiency but corrected by kynurenines. Am J Respir Crit Care Med 2013;187:609–20. 10.1164/rccm.201207-1346OC [DOI] [PubMed] [Google Scholar]
- 43. Watkins RR, Mukherjee PK, Chandra J, et al. Admission to the intensive care unit is associated with changes in the oral Mycobiome. J Intensive Care Med 2017;32:278–82. 10.1177/0885066615627757 [DOI] [PubMed] [Google Scholar]
- 44. Campos CF, van de Veerdonk FL, Gonçalves SM, et al. Host genetic signatures of susceptibility to fungal disease. Curr Top Microbiol Immunol 2019;422:237–63. 10.1007/82_2018_113 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
thoraxjnl-2020-216179supp001.pdf (999.7KB, pdf)
Data Availability Statement
Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information. The accession number for all sequencing data reported in this paper is NCBI Bioproject PRJNA647539.