Abstract
The σ2 receptor has attracted intense interest in cancer imaging1, psychiatric disease2, neuropathic pain3–5, and other areas of biology6,7. We determined the crystal structure of this receptor in complex with the clinical candidate roluperidone2 and the tool compound PB288. These structures templated a large-scale docking screen of 490 million virtual molecules, of which 484 compounds were synthesized and tested. 127 new chemotypes with affinities superior to 1 μM were identified, 31 of which had affinities superior to 50 nM. Hit rate fell smoothly and monotonically with docking score. We optimized three hits for potency and selectivity, achieving affinities ranging from 3 to 48 nM with up to 250-fold selectivity versus the σ1 receptor. Crystal structures of two new ligands bound to the σ2 receptor confirmed the docked poses. To investigate the contribution of the σ2 receptor in pain, two potent σ2-selective ligands and one potent σ1/σ2 non-selective ligand were tested for efficacy in a mouse neuropathic pain model. All three ligands demonstrated time-dependent decreases in mechanical hypersensitivity in the spared nerve injury model9, supporting a role for the σ2 receptor in nociception. This study illustrates the opportunities for rapid discovery of in vivo probes to study under-explored areas of biology using structure-based screens of diverse, ultra-large libraries.
Introduction
The σ receptors are integral membrane proteins widely expressed in both the central nervous system (CNS) and in peripheral tissues10. They are divided into σ1 and σ2 subtypes based on differences in tissue distribution and in pharmacological profile11, but despite their names, the two proteins are sequence-unrelated. Cloned in 1996, the σ1 receptor has no paralog within the human genome; its closest homolog of known function is the yeast Δ8,7 sterol isomerase ERG212. Studies conducted on σ1 knockout mouse tissue13 showed that the σ2 is not a splice variant or modified form of σ1, but rather derives from an unrelated gene. The molecular identity of the σ2 receptor remained unknown until we purified it from calf liver tissue14 and showed that it is TMEM97, an ER-resident membrane protein that regulates the sterol transporter NPC115,16. TMEM97 is predicted to be a four-helix bundle protein with both amino and carboxy termini facing the cytoplasm. An EXPERA family17 member, the σ2 receptor is distantly related to emopamil-binding protein (EBP), the mammalian Δ8,7 sterol isomerase required for cholesterol synthesis, and to TM6SF2, which regulates liver lipid homeostasis18.
The σ2 receptor is overexpressed in proliferating cells and in many tumors19, and labeled σ2 ligands have been proposed as tools for cancer diagnosis and therapy1. A ternary complex between the σ2 receptor, PGRMC1, and the LDL receptor was reported to increase the rate of LDL internalization7. Consistent with its high expression in the CNS, the σ2 receptor has also been proposed as a target for the treatment of CNS disorders. The σ2 receptor ligand Elayta (CT1812) is in clinical trials for mild to moderate Alzheimer’s disease6, and roluperidone (MIN-101) is in clinical development for schizophrenia2. When tested in animal models, σ2 receptor ligands reduce alcohol-withdrawal symptoms5,20 and have a neuroprotective effect in brain injury21. Finally, recent studies have found σ2 ligands to be anti-allodynic in models of neuropathic pain3–5. As this is also true of σ1 ligands, and because most σ2 ligands cross-react with the σ1 receptor, probe ligands selective for σ2 over σ1 would help illuminate σ2 biology and could be leads for novel therapeutics. However, little is known of the receptor’s molecular architecture or the structural bases for ligand recognition, stunting the discovery of selective ligands22,23. Here, we employed a biochemical and structural approach combined with computational docking to address these issues.
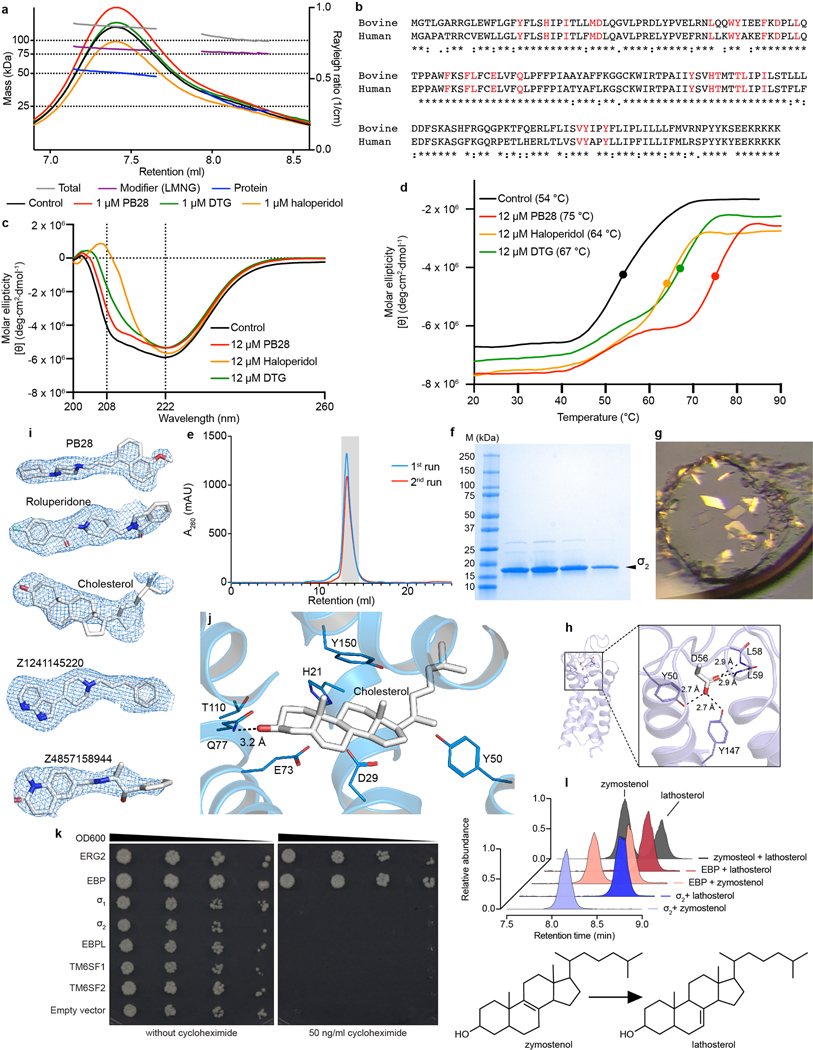
Structure determination
The human σ2 receptor was expressed in Sf9 insect cells, extracted with detergent, and purified14. Size exclusion chromatography multi-angle light scattering (SEC-MALS) showed that the receptor is a dimer in solution. Intriguingly, all members of the EXPERA family are either dimers or pseudo-dimers, although the functional role of dimerization remains unknown. Unlike the σ1 receptor, which can change oligomeric state in response to ligand binding24, the presence of ligands did not perturb the oligomeric state of σ2 (Extended Data Fig. 1a). As the human σ2 receptor was not tractable in structural studies, further experiments were performed with the homologous bovine σ2 receptor (Extended Data Fig. 1b). Circular dichroism (CD) experiments showed that the bovine σ2 receptor is 74% helical (Extended Data Fig. 1c). Thermal unfolding demonstrated that the receptor is remarkably stable, with a midpoint of the unfolding transition (Tm) of 54 °C (Extended Data Fig. 1d). Crystals of the σ2 receptor were grown by the lipidic cubic phase method25 (Extended Data Fig. 1e–g). Three datasets were collected for the receptor in complex with PB288, roluperidone2, and a ligand tentatively modeled as cholesterol (Extended Data Table 1). Molecular replacement was performed using a model derived from the structure of EBP26 (see Methods).
Overall structure of the σ2 receptor
The three σ2 receptor crystal structures are similar, with a backbone root mean square deviation (RMSD) of 0.75 Å. As anticipated from SEC-MALS, the structures showed that σ2 is an intimately associated homodimer, burying 890 Å2 of surface area in a dimer interface mainly formed by transmembrane helix 3 (TM3; Fig. 1a). The two protomers adopt the same conformation (backbone RMSD of 0.34 Å, 160 residues), with each adopting the expected four-helix bundle fold.
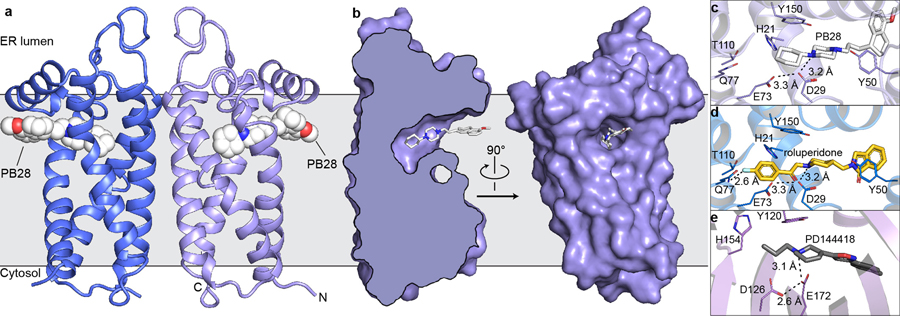
Figure 1 |. Structure of the σ2 receptor and binding site ligand recognition.
a, Structure of the σ2 receptor bound to PB28. Amino- and carboxy-termini are indicated. Membrane boundaries were calculated using the PPM server31. b, Cross-section of the σ2 receptor binding pocket (left) and view of the entrance to the binding pocket from the membrane (right). c, View of PB28 binding pose, showing charge–charge interaction with Asp29 (black dotted line) and contacts with other binding pocket residues. d, Analogous structure of the roluperidone binding pose. e, Structure of the σ1 receptor bound to PD144418 (PDB ID: 5HK1). Amino acids that serve similar roles and positioned in a similar orientation to amino acids in the σ2 receptor are indicated.
The four transmembrane helices of the protein are all kinked due to the presence of proline residues in each, creating a ligand-binding cavity near the center of the protein. This cavity is entirely occluded from solvent by extracellular loops 1 and 2, which form a well-ordered cap over the luminal surface of the protein. Asp56, which is crucial for ligand binding14, bridges extracellular loop 1 to TM helix 4 using a hydrogen bond network (Extended data Fig. 1h). Hence, Asp56 is likely important for receptor folding and not directly for ligand recognition14. Rather than opening to the ER lumen, the pocket opens laterally into the lipid bilayer (Fig. 1b), reminiscent of lipid-binding G protein-coupled receptors28, and its opening is lined with hydrophobic and aromatic residues. Ligands may enter through this opening in their neutral, deprotonated form and then become protonated in the binding site, forming a salt bridge with the conserved Asp29 (Fig. 1c–d). A second conserved acidic residue, Glu73, is located 3 Å away from Asp29, suggesting that these residues are hydrogen-bonded to each another, with Glu73 likely protonated.
The two σ receptors are not homologs and do not share the same fold; the σ2 receptor is a four-helix bundle, while the σ1 receptor has a β-barrel cupin fold29. Nevertheless, the binding pockets of the two receptors are remarkably similar (Fig. 1c–e), placing functionally similar amino acids in cognate spatial positions, which is perhaps the result of convergent evolution and explains how two very different folds can share closely overlapping ligand recognition profiles.
Both σ receptors are homologs of proteins that catalyze the same step in sterol biosynthesis. The σ1 receptor is a homolog of ERG2, the fungal Δ8,7 sterol isomerase; the σ2 receptor is a homolog of EBP, the mammalian Δ8,7 sterol isomerase. Both EBP and ERG2 rely on two interacting acidic residues in their active site for catalysis, which occurs by protonation of the substrate at carbon 9 (C9) followed by proton abstraction from C7, shifting the double bond into the C8-C7 position. All necessary components for catalysis appear to be present in σ2 receptor, yet it doesn’t catalyze sterol isomerization. It can neither function in vivo to rescue a strain of yeast that lacks ERG2 (Extended Data Fig. 1k) nor can it function in vitro to convert zymostenol to lathosterol (Extended Data Fig. 1l). The same is true for the σ1 receptor, which also has all the conserved residues required for catalysis but cannot rescue yeast that lack a sterol isomerase12 (Extended Data Fig. 1k). It was recently reported that Δ8–9 sterols can serve as signaling molecules30, which may hint at a possible physiological function of the σ receptors as sensors of these molecules evolved from enzymes that would modify them.
Docking against the σ2 receptor
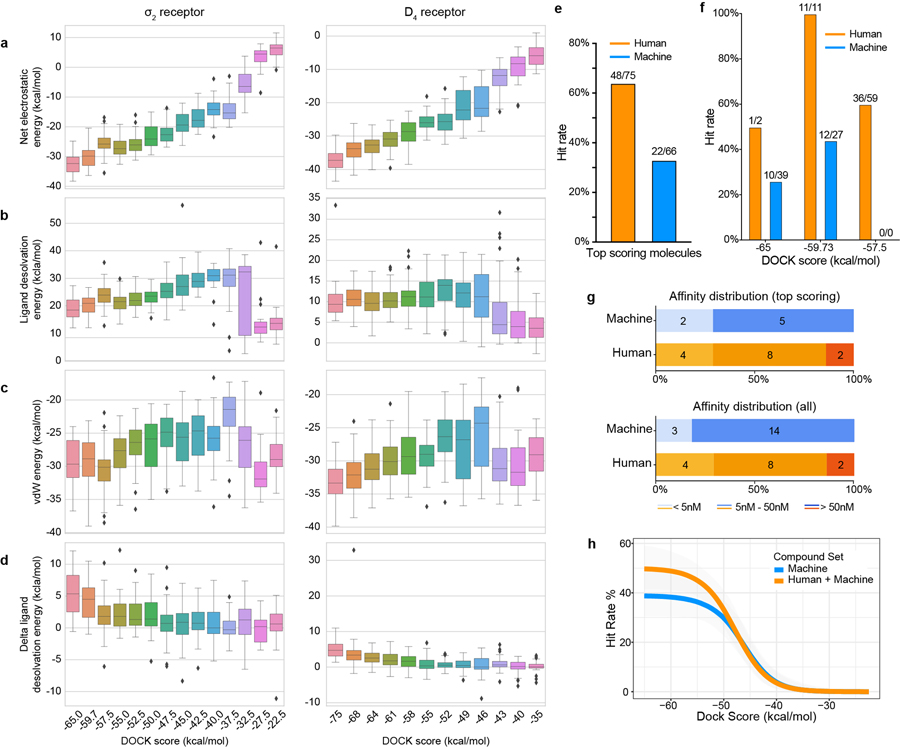
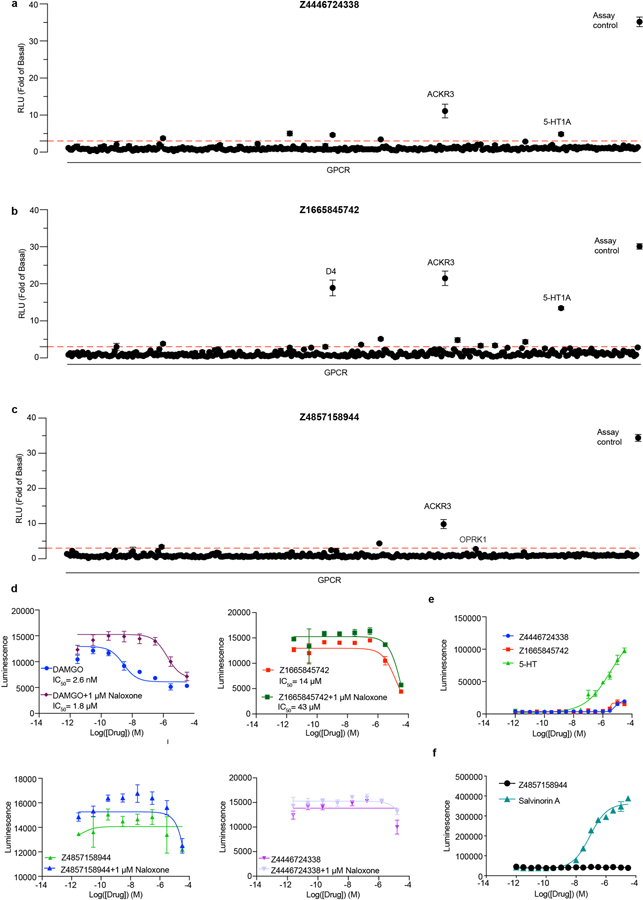
Docking against the σ2 receptor had two goals: discovering novel and σ2-selective chemotypes, and investigating how docking scores predict binding likelihood. This has been explored only once before at scale, against the dopamine receptor, revealing a sigmoidal relationship between hit-rate (active ligands/number-tested) and score32. The promiscuous σ2 site promised a higher hit-rate, increasing the dynamic range of any relationship observed. Guided by score alone for most molecules picked, supplemented by manual selection among the highest-ranking docked molecules, we prioritized 577 molecules for synthesis, spread among 14 scoring bins, of which 484 compounds were successfully produced. We tested compounds at 1 μM and defined as “hits” those that displaced greater than 50% [3H]-DTG σ2 binding. 127 of 484 molecules qualified, accounting for 26% of compounds over the full scoring range and a 60% hit-rate among the top-ranked molecules (Fig. 2a). Hit-rates fell monotonically with score, as with the dopamine receptor32, with a slope of −4.2%/(kcal/mol) in the inflection region, with one exception (below). The curve dropped from a hit rate of 61% at a docking score of about −60 kcal/mol to 0% at the four lowest scoring bins (−40 to −22.5 kcal/mol) (Fig. 2b, Supplementary Fig. 1).
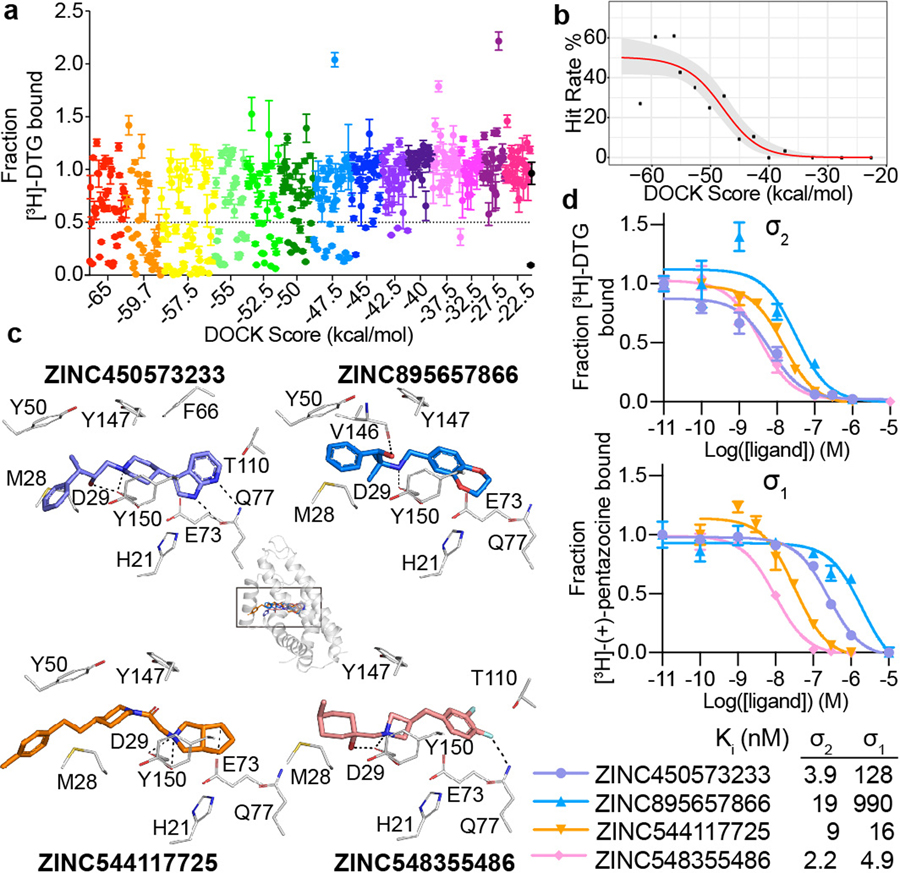
Figure 2 |. Docking 490 million molecules against the σ2 receptor.
a, Displacement of the radioligand [3H]-DTG by each of the 484 molecules tested at 1 μM (mean ± SEM of three technical replicates). The molecules are colored and grouped by docking score. Dashed line indicates 50% radioligand displacement. Dots below the dashed line represent confirmed binders, whose numbers diminish with worsening docking score. b, The hit-rate of 484 experimentally tested compounds was plotted against docking energy. The docking score (dock50) and slope at the maximum (slope50) are −48 kcal mol−1 and −4.2% per kcal mol−1, respectively. The gray band represents the 95% credible interval. c, Docked poses of four representative ligands from different scaffolds. d, Competition binding curves of the four molecules in c. against the σ2 receptor (upper panel) and the σ1 receptor (lower panel). The data are the mean ± SEM from three technical replicates.
The highest scoring bin had a hit rate of 27%, much lower than the 61% hit-rate observed in the 2nd-best scoring bin. This dip in the hit-rate curve illuminates defects in the scoring function. Many of the molecules in the top scoring bin had unexpectedly low desolvation penalties (Extended Data Fig. 2a,b, left column). DOCK3.7 pre-calculates these energies from one conformation among hundreds docked, not necessarily the highest scoring conformation against a target. Indeed, recalculating ligand desolvation using the docked conformation for molecules tested against σ2 and dopamine receptors increased desolvation penalties for molecules in the top-scoring bin, reducing their ranking and so suggesting a method to improve the scoring function (Extended Data Fig. 2d).
To supplement molecules prioritized by score alone, we picked a comparable number of high-ranking molecules by human inspection32,33. In the top three scoring bins (139 molecules) the human-prioritized hit rate (67%) was higher than that by docking score alone (33%) (Extended Data Fig. 2e and 2f), and the human-prioritized molecules reached higher affinities (Extended Data Fig. 2g,h). Broadly, these patterns reflect what was observed against the dopamine receptor.
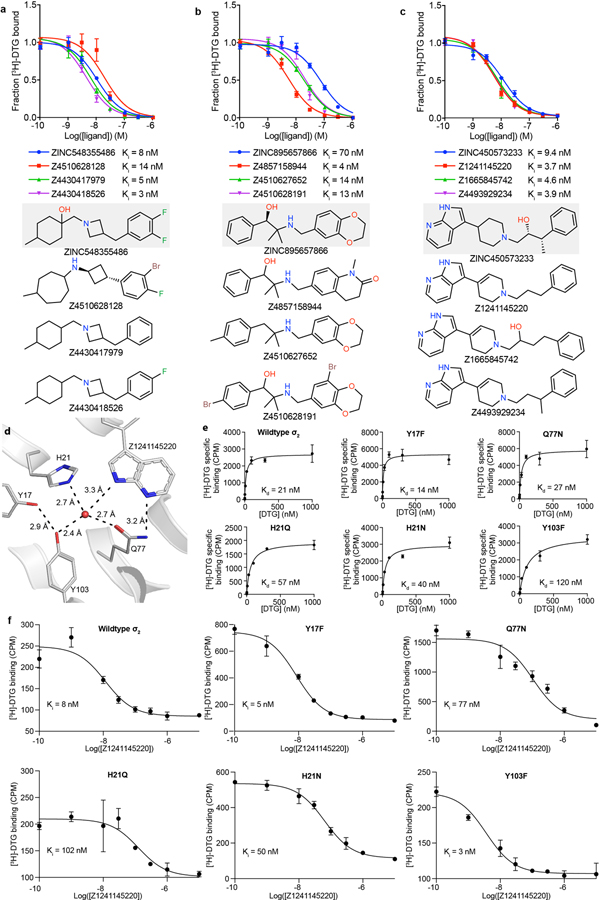
Seeking selective probes for the σ2 receptor, we measured competition binding curves for 14 docking hits with high radioligand displacement at 1 μM. Ki values ranged from 2.4 to 68 nM. In competition binding versus σ1 receptor (Fig. 2d, Extended Data Table 2, and Supplementary Table 1), several of these had substantial selectivity for σ2 over σ1, including ZINC450573233 and ZINC895657866, which were 30- and 46-fold selective, respectively.
We sought to improve the affinities of three potent ligands, each representing a different scaffold (Extended Data Fig. 3a–c). 20,000 analogs identified in SmallWorld (https://sw.docking.org/) from a 28 billion virtual library were docked into the σ2 site (Methods, Supplementary Table 1). Of these, 105 were synthesized and tested, improving the affinity of each scaffold by 2- to 18-fold (Extended Data Fig. 3a–c, Supplementary Table 1); for two chemotypes, σ2 selectivity improved 47- and >250-fold (Z1665845742 and Z4857158944), respectively.
Structures of σ2 in complex with analogs
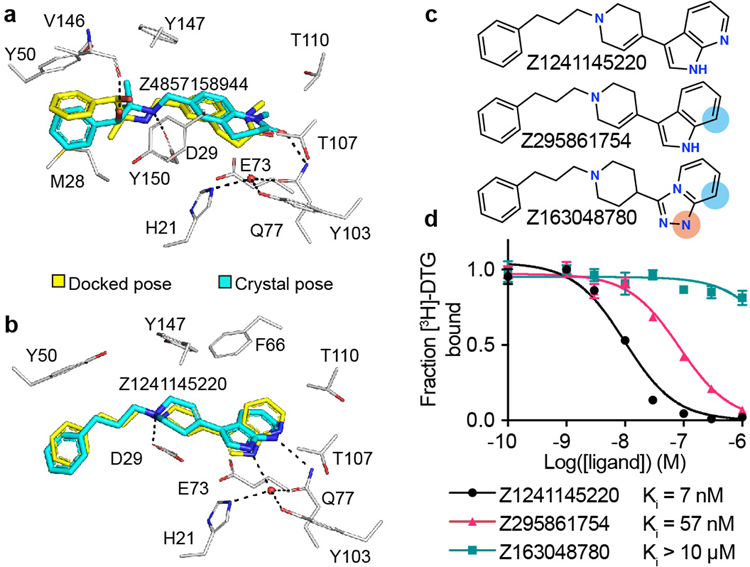
To test our docking poses, we determined the crystal structures of σ2 bound to two high-affinity ligands Z1241145220 (σ2 Ki = 3.7 nM; PDB ID: 7M95) and Z4857158944 (σ2 Ki = 4 nM; PDB ID: 7M96). Electron density maps confirmed the docking predictions, with RMSD values between the crystallized and docked poses of 0.88 and 1.4 Å, respectively (Fig. 3a–b, Extended Data Table 1, and Extended Data Fig. 1i). Newly predicted hydrogen-bond interactions with the backbone carbonyl of Val146, which was not seen in the roluperidone or PB28 complexes, corresponded well between docked and crystallographic poses. A hydrogen bond interaction with Gln77 is also found in the roluperidone and cholesterol complexes (Fig. 1d, Extended Data Fig. 1j). The higher resolution of this structure, 2.4 Å, also revealed an ordered water molecule in one of the binding sub-sites, coordinated by residues His21, Tyr103, and Gln77, and by an azaindole nitrogen in Z1241145220 (Fig. 3b).
Figure 3 |. High structural fidelity between docked and crystallographic poses of novel σ2 receptor ligands.
Ligand crystal poses (carbons in cyan) overlaid with respective docked poses (yellow). σ2 receptor carbons are in grey, oxygens in red, nitrogens in blue, sulfurs in yellow, hydrogen bonds are shown as black dashed lines. a, Z4857158944-bound complex (PDB ID: 7M96; RMSD = 1.4 Å). b, Z1241145220-bound complex (PDB ID: 7M95; RMSD = 0.88 Å). c, Two Z1241145220 analogues that disrupt the hydrogen bonds with Gln77 and the structural water. Blue and apricot circles depict differences between the analogues and the parent compound. d, Competition binding curve of compounds from c. The data are the mean ± SEM from three technical replicates.
This water was not modeled in the docked structure, so to investigate its role in ligand recognition we tested two analogues that were designed to disrupt the hydrogen bonds between Gln77 and the water (Fig. 3c). Z295861754, which should only hydrogen-bond with the water but not with Gln77, suffered an 8-fold decrease in affinity, whereas Z163048780, which should not hydrogen bond with either Gln77 or the water, had a Ki value > 10 μM (Fig. 3d), indicating a crucial role of the water for Z1241145220. We further generated a series of σ2 mutants in which the coordination of this water molecule was disrupted. Competition binding assays with Z1241145220 showed that mutating either His21 or Gln77 reduces the affinity by about 10-fold (Extended data Fig. 3d–f). Taken together, these results demonstrate that the ordered water is an integral part of the binding pocket and is required for high-affinity binding of Z1241145220, and likely other ligands.
σ2 ligands active in mouse pain model
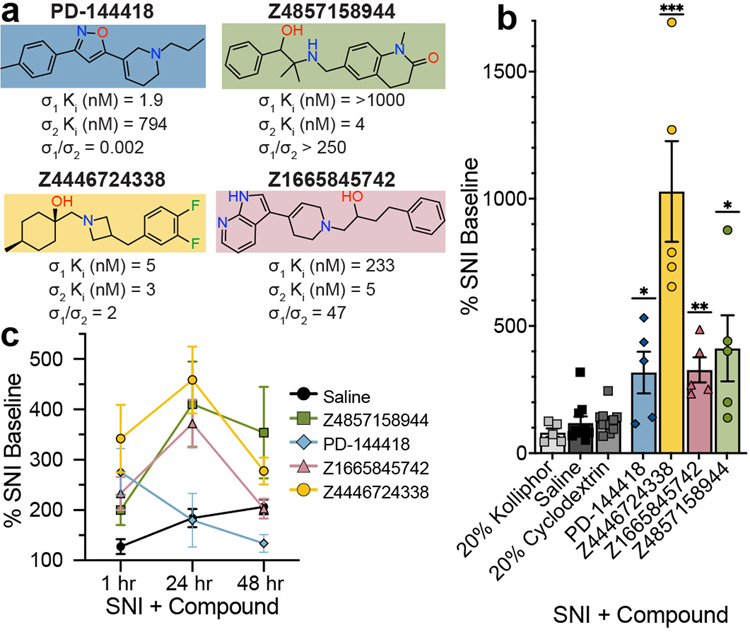
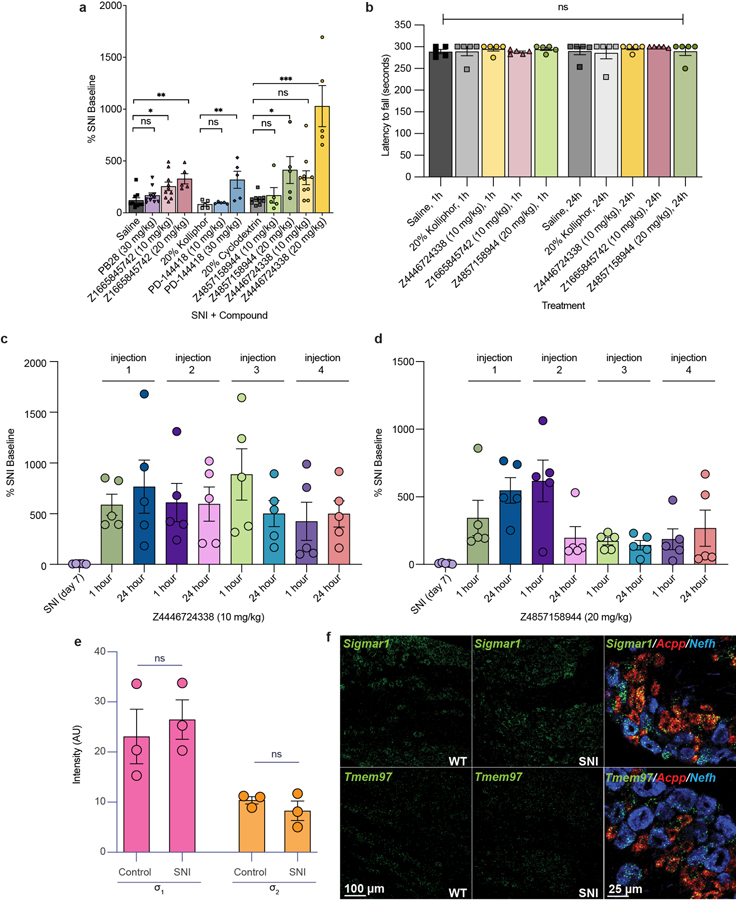
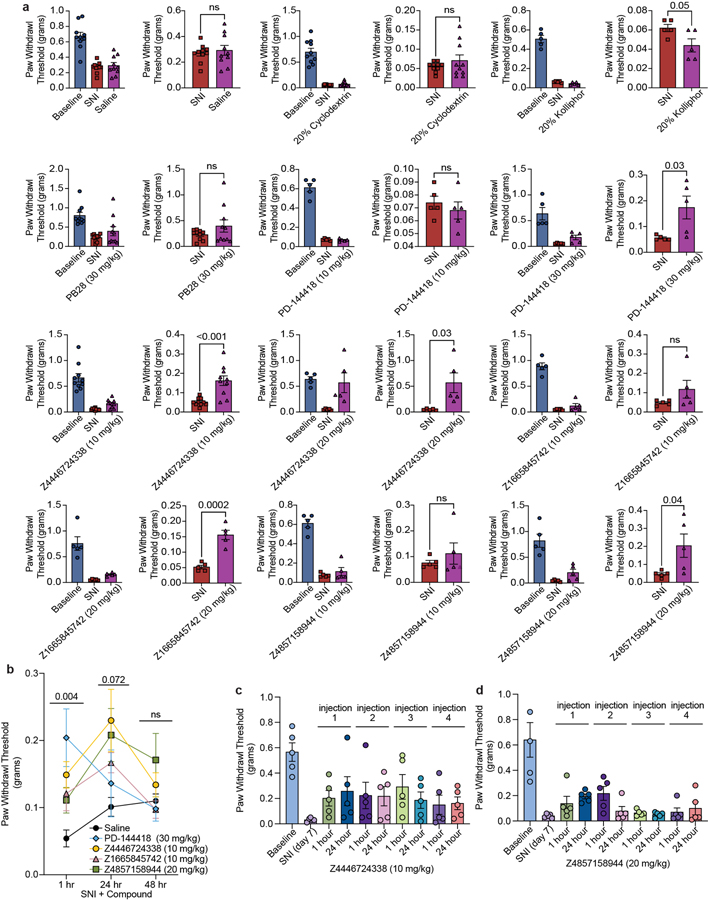
Genetic34,35 and pharmacological36–38 evidence supports a role of σ1 in chronic pain39. The discovery of the gene encoding for σ214, made understanding and distinguishing the roles of σ2 and σ1 in this indication3,4 fully possible. However, the limited availability of selective σ2 probes4 hinders the ability to distinguish the effect of the two receptors. Accordingly, we treated mice with three high-affinity σ2 ligands with differing degrees of σ2/σ1 selectivity: Z4857158944 (4 nM; >250-fold selective), Z1665845742 (5 nM; 47-fold selective), and Z4446724338 (3 nM non-selective) (Fig. 4a). We also treated with PB28, a well-established 5 nM non-selective ligand8. In pharmacokinetics experiments, the three docking-derived ligands had substantial brain permeability, with brain to plasma ratios ranging from 3 to 16, and brain half-lives ranging from 1.2 to 12 hours (Extended Data Table 3). PB28 also had high brain permeability and a relatively long half-life, though its brain Cmax was 3- to 8-fold lower than that of the new compounds. The high brain exposures of all four compounds encouraged us to examine them in a neuropathic pain model in mice.
Figure 4 |. σ1/2 ligands are anti-allodynic in a model of neuropathic pain.
a, Selectivity of four ligands at σ1 and σ2. PD-144418 values from the litrature47. b, Response of mice to a von Frey filament after spared nerve injury (SNI). Ligands are compared to their vehicles (PD-144418 30 mg/kg (n = 5) vs. kolliphor (n = 5), one-way ANOVA, F(2, 12) = 7.49, p = 0.008; Z4446724338 20 mg/kg (n = 5) vs cyclodextrin (n = 10), one-way ANOVA, F(2, 22) = 25.12, p = 0.0000021; Z4857158944 20 mg/kg (n = 5) vs cyclodextrin (n = 10), one-way ANOVA, F(2, 17) = 5.10, p = 0.02; Z1665845742 20 mg/kg (n = 5) vs saline (n = 10), one-way ANOVA, F(3, 31) = 6.18, p = 0.002; asterisks define individual group differences to respective vehicle control using Dunnett’s multiple comparisons Post-hoc test; kolliphor vs. PD-144418 30 mg/kg (p = 0.009); cyclodextrin vs. Z4446724338 20 mg/kg (p < 0.001); cyclodextrin vs. Z4857158944 20 mg/kg (p = 0.01); saline vs. Z1665845742 20 mg/kg (p = 0.002); * p < 0.05, ** p < 0.01, *** p < 0.001). Data shown are mean ± SEM. Also see Extended Data Fig. 4a. c, The anti-allodynic effects of σ2, but not σ1, ligands peak at 24 hours post-injection (two-way ANOVA; time × treatment interaction: F(8,80) = 2.25, p = 0.03; time: F(2,76) = 5.09, p = 0.009; treatment: F(4,40) = 5.40, p = 0.001; four treatment groups (n = 10) except PD-144418 (n = 5); asterisks define difference between Z4446724338 and saline at 1 hr (p = 0.03), 24 hr (p = 0.008), and 48 hr (p = 0.11) for simplicity; ns = not significant, * p < 0.05, ** p < 0.01). Data shown are mean ± SEM.
We tested the efficacy of these ligands in the spared nerve injury (SNI) mouse model of neuropathic pain, in which two out of three branches of the sciatic nerve are transected9, resulting in mechanical hypersensitivity (allodynia) transmitted by the uninjured peripheral (sural) nerve. In situ hybridization of dorsal root ganglia (DRG) sections, where the cell bodies of sensory neurons that transmit the “pain” message to the spinal cord reside, revealed expression of both σ1 and σ2 receptors in many DRG neurons, including myelinated and unmyelinated subsets (Extended Data Fig. 4). The expression of σ1 or σ2 did not change in DRG neurons seven days after SNI. When administered systemically to SNI mice, both σ2-selective ligands (Z1665845742 and Z4857158944) were anti-allodynic, increasing mechanical thresholds versus vehicle (Fig. 4b, Extended Data Fig. 4). This was comparable to the anti-allodynia conferred by a systemic injection of PD-144418, a σ1-selective ligand. Intriguingly, systemic injection of the non-selective Z4446724338 dose-dependently increased the mechanical thresholds of SNI mice (Fig 4b, Extended Data Fig. 4) with the highest dose completely reversing the SNI-induced mechanical allodynia (i.e., thresholds returned to pre-injury levels), a meaningfully higher level of anti-allodynia than observed with the selective σ2 ligands. Conversely, systemic injections of the non-selective PB288 produced mixed results, with anti-allodynic effects observed only in 60% of the mice (Extended Data Fig. 4). The much stronger anti-allodynia of Z4446724338 versus PB28 may reflect the former’s substantially higher brain permeability (Extended Data Table 3). Importantly, none of the new σ1 and σ2 ligands were sedative on the rotarod test (Extended Data Fig. 4), indicating that their anti-allodynic effect was not due to motor impairment.
The anti-allodynia of the σ2-selective ligands Z1665845742 and Z4857158944 suggest that this receptor is a potential target for managing neuropathic pain. However, because σ2 ligands are notoriously promiscuous, especially against GPCRs40,41, we counter-screened the three docking-derived ligands against potential off-targets. In a TANGO screen42 of 320 GPCRs, the molecules did not act as agonists or inverse agonists against most targets (Extended Data Fig. 5a–c), and the few cases where activity was observed did not replicate in concentration-response assays (Extended Data Fig. 5d–f, Supplementary Fig. 2–3). We also did not observe substantial activity at the μ-opioid receptor, a key pain target, in a G protein assay (Extended Data Fig. 5d). We further screened the compounds in binding assays against a panel of 19 targets including GPCRs, ion channels, and transporters; no binding was observed for any pain-related targets (Supplementary Table 2). These observations suggest that the primary mechanism of action of these ligands is via the σ2 receptor. The stronger activity of the σ1/2 ligand Z4446724338 suggests that σ1/2 polypharmacology may further increase anti-allodynia.
σ2 ligand effects peak after 24 hours
In earlier studies, σ1/2 ligands showed peak anti-allodynia up to 48 hours after dosing3. This unusual behavior was observed with ligands with mid-nanomolar potency and 9 to 14-fold selectivity vs. the σ1 receptor. We further explored this with the selective ligands, Z4857158944 and Z1665845742, and the non-selective ligand, Z4446724338. The molecules were tested post SNI, at 1, 24, and 48 hours after dosing. Supporting earlier reports, the anti-allodynia of the three new σ ligands increased over time, peaking 24 hours post-injection (Fig. 4c and Extended Data Fig. 6). In contrast, the anti-allodynia of the selective σ1 ligand PD-144418 was not sustained 24- or 48-hours post-injection. Furthermore, although the σ2-selective compounds exhibited reduced anti-allodynia efficacy at early time points versus the non-selective ligand Z4446724338, all three compounds conferred similar antinociception by 24 hours. This long-term activity cannot be easily explained by pharmacokinetics, as the brain half-life of all three compounds suggests minimal exposure past 12 hours (Extended Data Table 3). Rather, this time course may reflect longer term signaling or regulatory effects3.
To investigate tolerance, we also examined the effects of repeated injections of two of the lead compounds, Z4446724338 and Z4857158944. The antinociceptive effect of Z4446724338 persisted for the first three test days, and decreased slightly on the fourth day (Extended Data Fig. 4c–d, 6c–d). More tolerance was observed for compound Z4857158944; by the third injection, the antinociceptive effect was lost. Taken together, these results suggest that polypharmacology at the σ1 and σ2 receptor underlies an enhanced antinociceptive effect compared to selectivity for the σ2.
Discussion
The σ2 receptor has been enigmatic for 30 years. Its involvement in diverse biological processes and the lack of molecular data has clouded its biological role. Four key observations from this study begin to illuminate these issues. First, high-resolution crystal structures of the σ2 receptor complexed with roluperidone and with PB28 reveal a ligand binding site deeply embedded in the membrane (Fig. 1a, b), suggesting the possibility of a lipid as an endogenous ligand. The evolutionary connection of σ2 to EBP and the structure of the receptor bound to cholesterol support an ability to recognize sterols. The structures explain the simple pharmacophore of σ2 ligands—a cationic amine that ion-pairs with Asp29, while flanking hydrophobic and aromatic moieties are recognized by nearby aromatic residues. The structures also identify nearby polar residues, Gln77 and Thr110 that may aid in recognizing the hydroxyl moiety of sterols. These residues are rarely exploited by classic σ2 ligands but may provide new selectivity determinants for ligand discovery (Fig. 1c,d, and Extended Data Fig. 1j). Second, by testing 484 compounds across ranks from a library of 490 million docked, a quantitative relationship emerged between docking score and the likelihood of binding (Fig. 2). Crystal structures of docking-derived ligands confirmed the docking predictions (Fig. 3a,b). Third, among the top-ranking docking hits were 31 novel scaffolds with potent affinities (Ki < 100 nM) (Extended Data Table 2). Optimization of two of these led to potent ligands with 47 to >250-fold selectivity for the σ2 over the σ1 receptor (Supplementary Table 1). Fourth, three potent new σ2 chemotypes were tested for efficacy in a mouse model for neuropathic pain. All three were antiallodynic (Fig. 4). The expression pattern of the receptor and the activity of the σ2-selective ligands confirm a contribution of this receptor in pain processing and suggest its potential relevance in pain management.
The σ2 and the σ1 receptors are promiscuous, both binding to cationic amphiphiles, leading to receptor cross-reactivity. Although many selective σ1 ligands, like PD-144418 and (+)-pentazocine, have been described, there are far fewer selective ligands4,43 for the σ2 receptor. We sought to optimize for such selectivity22,44,45 using structure-based analoging, ultimately leading to two selective chemotypes. We combined one of these with a close analog that is σ2 inactive, affording a “probe pair” (Z1665845742 and Z1665798906 available via Sigma-Millipore’s probe collection, Cat. Nos. SML3141 and SML3142, respectively) (Supplementary Fig. 8). Such pairs can interrogate the role of the σ2 receptor in indications for which it has been widely mooted, including cancer1,19, schizophrenia2, and Niemann-Pick disease15,16, with the activity of the non-binding member controlling for inevitable off-targets.
The very promiscuity of the σ2 receptor makes it a good template to investigate how docking score predicts binding likelihood, something only investigated once before at scale, with dopamine receptor32. As in that earlier study, a sigmoidal relationship between score and hit-rate emerged, here with hit rates peaking at over 60% (Fig. 2b). Unlike the dopamine receptor, which suffered from a long hit-rate plateau among the top-ranking molecules, σ2 hit rates continued to rise with docking score through most of the curve. The exception was among a thin slice of the very top scoring molecules, where hit rates actually dropped owing to a subset of molecules that “cheat” the scoring function (Extended Data Fig. 2), affording us the ability to improve it.
After completion of this study, a model of the σ2 receptor was released as part of the AlphaFold protein structure prediction database46. This model closely resembles the crystal structures solved here, with an overall backbone RMSD of 0.5 Å (Supplementary Fig. 4a). Importantly for ligand discovery, binding site residues have an all-atom RMSD value less than 2 Å (Supplementary Fig. 4b). Despite the high fidelity of the model to the experimental structure, the 484 new compounds from docking against the crystal structure scored relatively poorly against the AlphaFold model (Supplementary Fig. 4c), reflecting a slightly contracted pocket in the model. It may yet be true that other ligands could be found that fit the AlphaFold model well and bind to the receptor. To investigate this, new prospective docking will be informative.
Certain caveats bear airing. While our ultimate goal was to find σ2-selective ligands, a spectrum of affinities and selectivities for both σ receptors emerged, reflecting the similarities of their pockets and their well-known overlapping pharmacology (Fig. 1c–e). The high hit rates and potencies found here reflect a site unusually well-suited to ligand binding, something unlikely to translate to other targets. While the docking-predicted pose for Z4857158944 and for Z1241145220 were confirmed crystallographically, the important water-bridging interaction for Z1241145220 was missed.
The key observations of this work should not be obscured by these caveats. The crystal structures of σ2 receptors reveal the basis of its molecular recognition, and template structure-based campaigns for novel ligand discovery. From such campaigns emerged a predictive correlation between docking rank and likelihood of binding, and potent and selective σ2 ligands that may be used to probe receptor biology.
Methods
Protein expression and purification for crystallography.
The bovine σ2 receptor was cloned into pVL1392 with an N-terminal human protein C epitope tag followed by a 3C protease cleavage site. The construct was truncated after residue 168 to exclude the ER localization signal for better expression and to facilitate crystallization. This receptor construct was expressed in Sf9 insect cells (Expression Systems) using the BestBac baculovirus system (Expression Systems) according to manufacturer’s instruction. Infection was performed when cell density reached 4×106 cells per milliliter. Cells were shaken at 27 °C for 60 hours before harvest by centrifugation. Cell pellets were stored at −80 °C until purification.
During all purification steps ligands (PB28, roluperidone, Z1241145220, and Z4857158944) were present in all buffers at 1 μM. For the cholesterol-bound structure the protein was purified in the presence of 1 μM DTG. Cell paste was thawed and cells were disrupted by osmotic shock in 20 mM HEPES pH 8, 2 mM magnesium chloride, 1:100,000 (v:v) benzonase nuclease (Sigma Aldrich), and cOmplete EDTA-free Protease Inhibitor Cocktail (Roche). Lysed cells were centrifuged at 50,000 × g for 15 minutes. Following centrifugation, supernatant was discarded, and the membrane pellets were solubilized with a glass Dounce tissue homogenizer in 20 mM HEPES pH 8, 250 mM NaCl, 10% (v/v) glycerol, 1% (w/v) lauryl maltose neopentyl glycol (LMNG; Anatrace), and 0.1% (w/v) cholesterol hemisuccinate (CHS; Steraloids). Samples were stirred at 4 °C for 2 hours and then non-solubilized material was removed by centrifugation at 50,000 × g for 30 min. Supernatant was supplemented with 2 mM calcium chloride and filtered by a glass microfiber filter (VWR). Samples were then loaded by gravity flow onto 5 ml anti-protein C antibody affinity resin. Resin was washed with 10 column volumes of 20 mM HEPES pH 8, 250 mM NaCl, 2 mM calcium chloride, 1% (v/v) glycerol, 0.1% (w/v) LMNG, and 0.01% (w/v) CHS, and then with 10 column volumes of 20 mM HEPES pH 8, 250 mM NaCl, 2 mM calcium chloride, 0.1% (v/v) glycerol, 0.01% (w/v) LMNG, and 0.001% (w/v) CHS. The receptor was eluted with buffer containing 20 mM HEPES pH 8, 250 mM NaCl, 5 mM EDTA, 0.1% (v/v) glycerol, 0.01% (w/v) LMNG, 0.001% (w/v) CHS, and 0.2 mg/ml protein C peptide, in 1 ml fractions. Peak fractions were pulled and 3C protease was added (1:100 w:w) and incubated with the receptor at 4 °C overnight. Next the receptor was purified by size exclusion chromatography on a Sephadex S200 column (Cytiva) in 20 mM HEPES pH 8, 250 mM NaCl, 0.1% glycerol, 0.01% LMNG, and 0.001% CHS. Peak fractions were pulled, calcium chloride was added to 2 mM and the sample was reapplied on the anti-protein C resin to remove uncleaved receptor. The column was washed with 5 column volumes and flow-through and wash fractions were pulled, concentrated, and reapplied on SEC. Peak fractions were pulled, concentrated to 50 mg/ml, and aliquoted. Protein aliquots were flash frozen in liquid nitrogen and stored in −80 °C until use. Purity was evaluated by SDS-PAGE.
Crystallography and data collection.
Purified σ2 receptor was reconstituted into lipidic cubic phase (LCP) by mixing with a 10:1 (w:w) mix of monoolein (Hampton Research) with cholesterol (Sigma Aldrich) at a ratio of 1.5:1.0 lipid:protein by mass, using the coupled syringe reconstitution method25. All samples were mixed at least 100 times. The resulting phase was dispensed in 30–40 nl drops onto a hanging drop cover and overlaid with 800 nl of precipitant solution using a Gryphon LCP robot (Art Robbins Instruments). The PB28-bound crystals grew in 20–30% PEG 300, 0.1 M MES pH 6, 600 mM NaCl. The Roluperidone-bound crystals grew in 20% PEG 300, 0.1 M MES pH 6, 500 mM NaCl, 60 mM succinate. The Z1241145220-bound crystals grew in 30% PEG 300, 0.1 M MES pH 6, 210 mM ammonium phosphate. The Z4857158944-bound crystals grew in 30% PEG 300, 0.1 M MES pH 6, 560 mM ammonium phosphate. The cholesterol-bound crystals grew in 25% PEG300, 0.1 M MES pH 6, 400 mM sodium citrate, and 1% 1,2,3-heptanetriol. All crystals grew in the presence of 1 μM of ligand, except for the cholesterol structure, which had no ligand present during crystal growth. Crystals were harvested using either MicroLoops LD or mesh loops (MiTeGen) and stored in liquid nitrogen until data collection. Data collection was performed at Advanced Photon Source GM/CA beamlines 23ID-B and 23ID-D. Data collection used a 10 μm beam and diffraction images were collected in 0.2° oscillations at a wavelength of 1.254858 Å for the PB28-bound crystals and a wavelength of 1.033167 Å for all other crystals. A complete data set was obtained from a single crystal in each case.
Data reduction and refinement.
Diffraction data were processed in HKL200048 and in XDS49, and statistics are summarized in Table 1. The PB28-bound structure was solved using molecular replacement starting with a Rosetta50 homology model generated using the structure of EBP (Protein Data Bank accession 6OHT). Matthews probability predicted four copies in the asymmetric unit. Initially, a single copy of this model was placed using Phaser51 giving a marginally interpretable electron density map. This model did not fit well into density and was replaced with Idealized helices that were used as a search model for an additional copy. The resulting dimer was duplicated and manually placed into unmodeled density. The resulting structure was iteratively refined in Phenix52 and manually rebuilt in Coot53. Final refinement statistics are summarized in Extended Data Table 1. The PB28 structure was used as a model for molecular replacement for all other datasets. In the case of the structure modeled as cholesterol-bound, electron density for a sterol-shaped ligand was observed (Extended Data Fig. 1i) and tentatively modeled as cholesterol based on the high (millimolar) concentration of cholesterol in the crystallization conditions and the compatibility of cholesterol with the shape of the electron density in the binding pocket. The receptor was purified in the presence of ditolylguanidine (DTG), but no DTG was present in the precipitating solution, and electron density was clearly incompatible with bound DTG. We cannot exclude the possibility that some other compound structurally similar to cholesterol was carried through the purification and is the ligand observed in the binding pocket. Figures containing electron density or structures were prepared in PyMOL54 v2.5 or UCSF Chimera55 v1.15.
Preparation of membranes for radioligand binding.
The human σ2 receptor was cloned into pcDNA3.1 (Invitrogen) mammalian expression vector with an amino-terminal protein C tag followed with a 3C protease cleavage site. Mutations were introduced by Site-directed mutagenesis using HiFi HotStart DNA Polymerase (Kapa Biosystems). Expi293 cells were transfected using FectoPRO (Polyplus-transfection) according to manufacturer instruction. Cells were harvested by centrifugation and lysed by osmotic shock in a buffer containing 20 mM HEPES, pH 7.5, 2 mM MgCl2,1:100,000 (vol/vol) benzonase nuclease (Sigma Aldrich), and cOmplete Mini EDTA-free protease-inhibitor tablets (Sigma Aldrich). The lysates were homogenized with a glass dounce tissue homogenizer and then centrifuged at 20,000 × g for 20 min. After centrifugation, the membranes were resuspended in 50 mM Tris, pH 8.0, divided into 100 μL aliquots, flash frozen in liquid nitrogen, and stored at –80 °C until use.
Saturation and competition binding in Expi293 membranes.
Saturation binding was performed with a method similar to that of Chu and Ruoho56. Briefly, membrane samples from Expi293 cells (Thermo Fisher Scientific) expressing wild-type or mutant σ2 receptor, prepared as described above, were thawed, homogenized with a glass dounce, and diluted in 50 mM Tris, pH 8.0. Binding reactions were done in 100 μL, with 50 mM Tris pH 8.0, [3H]-DTG (PerkinElmer), and supplemented with 0.1% bovine serum albumin to minimize non-specific binding. To assay non-specific binding, equivalent reactions containing 10 μM haloperidol were performed in parallel. Competition assays were performed in a similar fashion with 10 nM [3H]-DTG and the indicated concentration of the competing ligand. Samples were shaken at 37 °C for 90 min. Afterward, the reaction was terminated by massive dilution and filtration over a glass microfiber filter with a Brandel harvester. Filters were soaked with 0.3% polyethyleneimine for at least 30 min before use. Radioactivity was measured by liquid scintillation counting. Data analysis was done in GraphPad Prism 9.0, with Ki values calculated by Cheng-Prusoff correction using the experimentally measured probe dissociation constant.
Circular dichroism.
Far-UV circular dichroism (CD) spectra (185–260 nm) were measured with a JASCO J-815 (JASCO Inc., Tokyo, Japan), with a Peltier temperature controller and single cuvette holder and Spectra Manager II software for data collection and analysis. Data was collected using 1 mm path length cuvette, bandwidth of 1 nm, data pitch of 0.5 nm, scanning speed of 50 nm/min, continuous scanning mode, and with 5 accumulations. Protein concentration was 0.25 mg/ml (10 μM) in 10 mM potassium phosphate pH 7.4, 250 mM potassium fluoride. Ligands were at 12 μM. Melt curves were measured at 222 nm between temperatures 20–95 °C, bandwidth of 1 nm, and a ramp rate of 1 °C/min with 10 s wait time. Calculation of Tm was done in Spectra Manager II by finding the peak of the first derivative of the melt curves, calculated using the Savitzky-Golay filter.
Size-exclusion chromatography with multi-angle light scattering (SEC-MALS).
The oligomeric state of σ2 receptor was assessed by SEC–MALS using a Wyatt Dawn Heleos II multi-angle light scattering detector and Optilab TrEX refractive index monitor with an Agilent isocratic HPLC system Infinity II 1260. Receptor was prepared as described above, but with no ligand added during purification. The ligand-free receptor was diluted to 1 mg/ml in SEC–MALS buffer (0.01% LMNG, 20 mM HEPES pH 7.5, 150 mM sodium chloride). Ligands were added to a final concentration of 1 μM and the sample was incubated with ligand for 2 h at room temperature (21 °C). Separation steps were performed in SEC–MALS buffer with a Tosoh G4SWxl column at a flow rate of 0.5 ml min−1. Data analysis used the Astra software package version 6.1.4.25 (Wyatt) using the protein conjugate method with a dn/dc value of 0.21 (mL/g) for detergent and 0.185 (mL/g) for protein.
Molecular docking.
The σ2 receptor bound to cholesterol (PDB ID: 7MFI) was used in the docking calculations. The structure was protonated at pH 7.0 by Epik and PROPKA in Maestro57 (2019 release). Based on the mutagenesis data14, E73 was modeled as a neutral residue. AMBER united atom charges were assigned to the structure. To model more realistic low protein dielectric boundary of this site, we embedded the receptor into a lipid-bilayer to capture its native environment in endoplasmic reticulum (ER) membrane, then followed by a 50 ns coarse-grained molecular dynamic (MD) simulation with a restricted receptor conformation. A more detailed protocol can be found on the DISI wiki page (http://wiki.docking.org/index.php/Membrane_Modeling). The volume of the low dielectric and the desolvation volume was extended out 2.2 Å and 1.2 Å, respectively, from the surface of protein and modelled lipid-bilayer using spheres calculated by SPHGEN. Energy grids were pre-generated with AMBER force fields using CHEMGRID for van der Waals potential58, QNIFFT59 for Poisson–Boltzmann-based electrostatic potentials, and SOLVMAP60 for ligand desolvation.
The resulting docking setup was evaluated for its ability to enrich known σ2 ligands over property-matched decoys. Decoys are unlikely to bind to the receptor because despite their similar physical properties to known ligands, they are topologically dissimilar. We extracted 10 known σ2 ligands from ChEMBL(https://www.ebi.ac.uk/chembl/) including PB28 and roluperidone whose crystallographic poses were report here. Five-hundred and forty-two property-matched decoys were generated by the DUDE-Z pipeline61. Docking performance was evaluated based on the ability to enrich the knowns over the decoys by docking rank, using log adjusted AUC values (logAUC). The docking setup described above was able to achieve a high logAUC of 39 and to recover the crystal poses of PB28 and roluperidone with RMSD values of 0.93 and 0.77 Å, respectively. This docking setup gave the best retrospective enrichment and pose reproduction among three ligand-bound σ2 structures (Supplementary Fig. 5). We also constructed an ‘extrema’ set61 of 61,687 molecules using the DUDE-Z web server (http://tldr.docking.org) to ensure that molecules with extreme physical properties were not enriched. The docking setup enriched close to 90% mono-cations among the top1000 ranking molecules. To check if the limited amounts of knowns and property-matched decoys over-trained the docking parameters, the enrichment test was run using 574 additional σ2 ligands from S2RSLDB42 (http://www.researchdsf.unict.it/S2RSLDB) against the ‘extrema’ set. The resulting high logAUC of 41 demonstrated the docking setup was still able to enrich knowns over decoys on a 112-fold larger test set, indicating the favorable docking parameters for launching an ultra-large-scale docking campaign.
Four-hundred and ninety million cations from ZINC15 (http://zinc15.docking.org), characterized by similar physical properties as σ1/2 known ligands (for instance, with calculated octanol-water partition coefficients (cLogP) <=5 and with 250 Da <molecular weight <=400 Da), was then docked against the σ2 ligand binding site using DOCK3.8. Of these, 469 million molecules were successfully docked. On average, 3,502 orientations were explored and for each orientation, 183 conformations were averagely sampled. In total, more than 314 trillion complexes were sampled and scored. The total calculation time was 177,087 hours, or 3.7 calendar days on a cluster of 2,000 cores.
The top-ranking 300,000 molecules were filtered for novelty using the ECFP4-based Tanimoto coefficient against 2,232 σ1/2 ligands in ChEMBL (https://www.ebi.ac.uk/chembl/) and 574 σ2 ligands from S2RSLDB (http://www.researchdsf.unict.it/S2RSLDB). Molecules with Tanimoto coefficient (Tc) ≥ 0.35 were eliminated. The remaining 196,170 molecules were clustered by ECFP4-based Tc of 0.5, resulting in 33,585 unique clusters. From the top 5,000 novel chemotypes, molecules with > 2 kcal/mol internal strains were filtered out using strain_rescore.py in Macromodel (2019 release). After filtering for novelty and diversity, the docked poses of the best-scoring members of each chemotype were manually inspected for favorable and diversified interactions with the σ2 site, such as the salt bridge with Asp29, the hydrogen bond with His21/Val146 and the π-π stacking with Tyr50/Trp49. Ultimately, 86 compounds were chosen for testing, 79 of which were successfully synthesized.
Hit-rate curve prediction.
To guide the design of scoring bins for the hit rate curve, 1,000 docked poses were sampled in bins every 2.5 kcal/mol from the best score of −65 kcal/mol up to −22.5 kcal/mol. We chose this 2.5 kcal/mol distance between the bins to span the range with enough points (bins) to define a potential hit-rate vs. docking score curve. At the top of what we expected to be the curve, we increased the bin sizes because the density of molecules at these very highest ranks was relatively low. Correspondingly, at the lowest scores we added several more bins, also at a larger spacing, to help us get a robust lower baseline. The estimated hit rate was calculated by the number of sensible docked poses divided by 1,000. The criteria to define a sensible docked pose contains 1) no unsatisfied hydrogen bond donors; 2) less than 3 unsatisfied hydrogen acceptors; 3) forms a salt bridge with Asp29; 4) total torsion strain energy < 8 units; 5) maximum strain energy per torsion angle < 3 units. The first three filters were implemented based on LUNA (https://github.com/keiserlab/LUNA), which calculated all the intra- and interactions of a docked pose with the receptor, then hashed them into a binary fingerprint. The strain energy was calculated by an in-house population-based method62. Based on the shape of the estimated prior curve (Supplementary Fig. 6), more scoring bins are selected in the higher estimated hit-rate region: −65, −59.73 and −57.5 kcal/mol. After that, every scoring bin was 2.5 kcal/mol from each other till −37.5. The last four bins were 5 kcal/mol from each other. 13,000 molecules sampled were from these 14 scoring bins were filtered by novelty and internal torsion strain described above. The remaining 9,216 novel and non-strained molecules were cluster by the LUNA 1024-length binary fingerprint of a Tc = 0.32, resulting in 6,681 clusters. The first 40 chemotypes were attempted to be purchased from each scoring bin. After the evaluation of synthesis availability from the vendors, 491 molecules were ordered (Supplementary Tables 1 and 3).
Hit-rate curve fitting.
To fit the Bayesian hit-rate models we used Stan63 (v2.21.2) via BRMS64 (v2.14.4), with generic parameters: iter=4000, and cores=4. Here are the model specific parameters. For both hit-picking prior and posterior Sigmoid models formula=bmrs::formula(hit ~ top * inv_logit(hill*4/top*(dock_energy - dock50)), top + hill + dock50 ~ 1, nl=TRUE), where hill is scaled by 4/top so it is the slope of the curve at the dock50 irrespective of the value of Top. For Prior Sigmoid model, prior=c(brms::prior(normal(.5, .2), lb=0, ub=1, nlpar=“top”), brms::prior(normal(−50, 10), nlpar=“dock50”), brms::prior(normal(−.1, .1), ub=−.001, nlpar=“hill”)), inits=function(){list(top=as.array(.5), dock50=as.array(−50), hill=as.array(−.1))}, family=gaussian(). Updating the Prior sigmoid model with the mean expected hit-rate for each computationally analyzed tranche yielded an estimate and 95% credible interval for the sigma parameter for the Gaussian response of 20 [15, 30]%, but did not significantly adjust the distributions for Top, Hill, or Dock50 (Supplementary Fig. 7). Therefore, to estimate the posterior sigmoid model, we transferred the per-parameter prior distributions and initial values and used the family=bernoulli(“identity”). To compare models, we used the loo package to add the Pareto smoothed importance sampling leave-one-out (PSIS-LOO) and Bayesian version of the R265 (loo_R2) information criteria. Figures were generated using tidybayes66, ggplot267, and tidyverse68 packages in R69.
Analoging within the make-on-demand library.
Using 4 primary docking hits (ZINC450573233, ZINC533478938, ZINC548355486 and ZINC895657866) as queries in SmalWorld (https://sw.docking.org/) from the 28B make-on-demand library, a subset of Enamine REAL space, 20,005 analogues were selected by its default settings, then docked into the σ2 site for potential favorable interactions with His21, Tyr50, Gln77, and Val146.
Make-on-demand synthesis.
79 molecules that were prioritized by human inspection were delivered within 7 weeks with a 93% fulfilment rate, and 412 molecules by docking score alone were delivered within 4 weeks with an 82% fulfilment rate after a single synthesis attempt (Supplementary Tables 1 and 3–4). Most of the make-on-demand molecules were derived from Enamine REAL database (https://enamine.net/compound-collections/real-compounds). See Supplementary Information for synthesis procedure and characterization of compounds.
Yeast isomerase complementation assay.
The human σ2 receptor, ERG2, and EBP were subcloned into the URA3 shuttle vector p416GPD. The plasmids were transformed into the Erg2-deficient Saccharomyces cerevisiae strain Y17700 (BY4742; MATα; ura3Δ0; leu2Δ0; his3Δ1; lys2Δ0; YMR202w::kanMX4) (Euroscarf) by the lithium acetate/single-stranded carrier DNA/polyethylene glycol method. A single colony was picked from a URA-selective plate and grown in suspension. Yeast were diluted in sterile water in a five-fold serial dilution starting from O.D. 0.1. Two microliters of the yeast dilutions were spotted on a URA−selective plate either in the absence or the presence of sub-inhibitory concentrations of cycloheximide (50 ng/ml) and grown at 30°C for 24–48 h before imaging.
Sterol isomerization enzymatic assay.
EBP and σ2 were cloned into pcDNA3.1 (Invitrogen) mammalian expression vector with FLAG and protein C affinity tag, respectively. Proteins were purified as described for crystallography preparations, except no ligand was present during purification. Following size exclusion chromatography proteins were flash frozen in liquid nitrogen and kept at −80 °C until use. Zymostenol (CAS #566–97-2) and lathosterol (CAS #80–99-9) were purchased from Avanti Polar Lipids. For each sterol, a 2x solution was prepared by first dissolving DDM in isopropanol to 1% (w/v) and dissolving sterols in chloroform to a concentration of 1 mg/ml, followed by transferring 500 μM of the sterols to a new vial, evaporating under argon, and dissolving with DDM in a 1:20 (w/w) detergent to sterol ratio and a final 0.2% detergent in HEPES buffered saline (HBS; 20 mM HEPES pH 7.5, 150 mM NaCl). Proteins were diluted in HBS to 5 μM. Individual sterol standards were prepared by mixing each sterol 1:1 with HBS. A mixed sterol standard was prepared by mixing both sterols in a 1:1 ratio. For the enzymatic reactions, sterols were mixed in 1:1 ratio with the protein sample to give a final protein concentration of 2.5 μM, sterol concentration of 250 μM, and detergent concentration of 0.1%, in HBS. Reactions were incubated for 1 hour at 37 °C and then diluted 1:10 in methanol and kept at −20 °C until analysis by LC-MS. Samples were analyzed on a QE-plus mass spectrometer coupled to an Ultimate 3000 LC (Thermo fisher) in a method modified from Skubic et al.70. Five microliters were injected on a Force PFPP column coupled with an Allure PFPP column (both 2mm × 150 mm, Restek) maintained at 40°C. The mobile phases were A: methanol:isopropyl alcohol:water:formic acid (80:10:10:0.02) 5 mM ammonium formate, and B: isopropyl alcohol. The gradient was as follows: 0% B for 15 min, then 100% B in 1 second, maintained at 100% B for 5 min, followed by 5 min re-equilibration at 0% B. The flow rate was 0.15 mL min-1. The mass spectrometer was acquiring in t-SIM mode for the [M-H2O+H]+ ion (369.35158) with 70,000 resolution, and 0.5 m/z isolation. Standard samples for each compound were run first separately to obtain the retention time of each of the two isobaric compounds.
μOR activation assay.
To measure μOR Gi/o-mediated cAMP inhibition, 2.5 million HEK-293T cells (ATCC) were seeded in 10-cm plates. Eighteen to 24 hours later, upon reaching 85–90% confluency, cells were transfected using a 1:3 ratio of human μOR and a split-luciferase based cAMP biosensor (pGloSensorTM-22F; Promega). TransIT 2020 (Mirus Biosciences) was used to complex the DNA at a ratio of 3 μL TransIT per μg DNA, in OptiMEM (Gibco-ThermoFisher) at a concentration of 10 ng DNA per μL OptiMEM. Twenty-four hours later, cells were harvested from the plate using Versene (PBS + 0.5 mM EDTA, pH 7.4) and plated in poly-D-lysine-coated white, clear-bottom 96-well assay plates (Corning Costar #3917) at a density of 35,000 cells per well and incubated at 37 °C with 5% CO2 overnight. The next day, after aspiration of the culture medium, cells were incubated for 2 hours covered, at room temperature, with 40 μL assay buffer (CO2-independent medium, 10% FBS) supplemented with 2% (v/v) GloSensor™ reagent (Promega). To stimulate endogenous cAMP via β adrenergic-Gs activation, 5x drugs were prepared in 10x isoproterenol containing assay buffer (200 nM final concentration). For naloxone competition experiments, 5x naloxone (1 μM final concentration) was also added to each well. Luminescence was immediately quantified using a BMG Clariostar microplate reader. Data were analyzed using nonlinear regression in GraphPad Prism 9.0 (Graphpad Software Inc., San Diego, CA).
Off-target counterscreens.
Screening of compounds in the PRESTO-Tango GPCRome was accomplished as previously described41 with several modifications. First, HTLA cells were plated in DMEM with 10% FBS and 10 U ml−1 penicillin–streptomycin. Next, the cells were transfected using an in-plate PEI method71. PRESTO-Tango receptor DNAs were resuspended in OptiMEM and hybridized with PEI before dilution and distribution into 384-well plates and subsequent addition to cells. After overnight incubation, drugs were added to cells at 10 μM final concentration without replacement of the medium. The remaining steps of the PRESTO-Tango protocol were followed as previously described. For those six receptors for which activity was reduced to less than 0.5-fold of basal levels of relative luminescence units or for the one receptor for which basal signaling was increased greater than 3-fold of basal levels, assays were repeated as a full dose–response assay. Activity for none of the seven could be confirmed, and we discount the apparent activity seen in the single-point assay.
Radioligand binding screen of off-targets was performed by the National Institutes of Mental Health Psychoactive Drug Screen Program (PDSP)72. Detailed experimental protocols are available on the NIMH PDSP website at https://pdsp.unc.edu/pdspweb/content/PDSP%20Protocols%20II%202013–03-28.pdf.
Cell lines
All cell lines in this study were not authenticated. All cells used in this study are commercial and were obtained from vendors as indicated. Cells were confirmed to be mycoplasma free.
Animals
Animal experiments were approved by the UCSF Institutional Animal Care and Use Committee and were conducted in accordance with the NIH Guide for the Care and Use of Laboratory animals. Adult (8–10 weeks old) male C56BL/6 mice (strain #664) were purchased from the Jackson Laboratory. Mice were housed in cages on a standard 12:12 hour light/dark cycle with food and water ad libitum. We did not perform sample-size calculations. We modeled our sample sizes for behavioral studies on previous studies using a similar approach to our own, which have been demonstrated to be capable of detecting significant changes73,74. The animals were randomly assigned to the treatment group and control group. For behavioral experiments, animals were initially placed into one cage and allowed to free run for a few minutes. Next, each animal was randomly picked up, injected with the drug or vehicle control, and placed into a separate cylinder before the behavior test. All experiments were for animal behavior and followed this randomization protocol. For all behavioral testing the experimenter was always blind to treatment. All experiments were in animals and under blinding conditions.
Compounds
All ligands used in the animal studies were synthesized by Enamine (https://enamine.net/) (Supplementary Table 5) and dissolved 30 minutes prior testing. PB28 and Z1665845742 were resuspended in 0.9% NaCl. Z4857158944 and Z4446724338 were resuspended in 20% cyclodextrin. PD-144418 was resuspended in 20% Kolliphor.
Behavioral analyses
For all behavioral tests, animals were first habituated for 1 hour in Plexiglas cylinders. The experimenter was always blind to treatment. All tests were conducted 30 minutes after subcutaneous injection of the compounds. Hindpaw mechanical thresholds were determined with von Frey filaments using the up-down method75. For the ambulatory (rotarod) test, mice were first trained on an accelerating rotating rod, 3 times for 5 min, before testing with any compound.
Spared-nerve injury (SNI) model of neuropathic pain
Under isoflurane anesthesia, two of the three branches of the sciatic nerve were ligated and transected distally, leaving the sural nerve intact. Behavior was tested 7 to 14 days after injury and in situ hybridization was performed one week post-injury.
In situ hybridization
In situ hybridization was performed using fresh DRG tissue from adult mice (8–10 week old), following Advanced Cell Diagnostics’ protocol and as previously described76. All images were taken on an LSM 700 confocal microscope (Zeiss) and acquired with ZEN 2010 (Zeiss). Adjustment of brightness/contrast and changing of artificial colors (LUT) were done with Photoshop. The same imaging parameters and adjustments were used for all images within an experiment.
Statistical analyses of animal studies
All animal statistical analyses were performed with GraphPad Prism 8.0 (GraphPad Software Inc., San Diego, CA) unless otherwise noted. All data are reported as means ± SEM unless otherwise noted. Dose-response experiments were analyzed with one-way ANOVA and time-course experiments were analyzed with two-way ANOVA, and both experiments used Dunnett’s multiple comparison post-hoc test to determine differences between specific treatments and vehicle controls visualized in the figures. Rotarod experiments were analyzed using one-way ANOVA (saline, Z1665845742, and Z4857158944) or unpaired two-tailed Student’s t-test (kolliphor and Z4446724338). Details of analyses, including number of tested animals and groups, degrees of freedom, and p-values can be found in the figure legends.
Extended Data
Extended Data Figure 1 |. Characterization of σ2 receptor.
a, Size-exclusion chromatography with multi-angle light scattering of the human σ2 receptor. The σ2 receptor was run either without ligand or with 1 μM of the indicated ligand. Lines indicate calculated total mass (gray), detergent micelle (blue), and protein (purple). b, Sequence alignment between the human and bovine σ2 protein sequences performed using T-coffee77. Residues that line the binding pocket are marked in red. c, Circular dichroism analysis of the bovine σ2 receptor alone (black) or with the indicated ligand. Data is representative of multiple experiments. d, Circular dichroism melting curves of the bovine σ2 receptor. Temperature was raised from 20 °C to 90 °C and molar ellipticity was measured at 222 nm. Protein was incubated either with or without indicated ligand at 12 μM. Melting temperatures for each measurement are indicated with a circle. Data is representative of multiple experiments e, Size-exclusion chromatography (SEC) of the bovine σ2 receptor. Blue trace is after proteolytic tag removal. Red trace is protein applied on size exclusion after reapplying the tag-free protein on affinity resin to remove proteins with intact tags. The trace presented is representative of multiple purifications. f, Analysis of receptor purity after the second SEC using SDS-PAGE. Gray rectangle in e represents fractions chosen for analysis. The SDS-PAGE presented here is representative of multiple purifications. See Source Data for uncropped version. g, Crystals of bovine σ2 receptor in the lipidic cubic phase. h, Aspartate 56 (D56) is important for receptor structure but not for ligand binding. A tight network of hydrogen bonds that bridges extracellular loop 1 to TM helix 4 is depicted with black dashed lines. i, Electron density maps for the various ligands. Polder maps78 were calculated in Phenix. Maps are contoured at a level of 3 σ. j, View of cholesterol binding pose, showing contacts with other binding pocket residues. Hydrogen bonds are marked with black dashed lines. k, Yeast complementation assay. A ΔERG2 yeast strain was transformed with plasmids harboring the indicated genes. Yeast cells were grown to logarithmic phase and diluted to OD600 of 0.1, and then further diluted in a five-fold serial dilution series. Two microliters of each dilution were spotted on plates. Yeast cells were grown either in permissive conditions of no cycloheximide or in the restrictive conditions of 50 ng/ml cycloheximide, which requires functional Δ8–9 sterol isomerase activity for viability. ERG2 and EBP can act as sterol isomerases and rescue the growth of ΔERG2 Saccharomyces cerevisiae while the σ2 receptor, the σ1 receptor, or any other member of the EXPERA family cannot. l, EBP can catalyze the conversion of zymostenol to lathosterol while σ2 cannot. Standards are in dark gray. EBP converts zymostenol to lathosterol (apricot) but does not convert lathosterol to zymostenol (dark red). The σ2 receptor does not convert lathosterol to zymostenol (dark blue) or zymostenol to lathosterol (light purple). Structures of zymostenol and lathosterol are depicted below the traces.
Extended Data Figure 2 |. Comparisons of the distribution of docking scores.
a-d, The distribution of docking scores of tested molecules for hit rate curves against σ2 (left column) and D4 (right column) receptors. All tested molecules are grouped based on docking score bins. The distributions are shown in box plots for a, net electrostatic energy, b, ligand desolvation energy, c, van der Waals (vdW) energy and d, delta ligand desolvation energy after recalculating atomic desolvation energy based on the docked pose. e-h, Comparison of hit rates and affinities achieved by combined docking score and human inspection and these achieved by docking score alone. e, Overall hit rates for selecting compounds from the first 3 scoring bins by each strategy: human prioritization and docking score (orange), or docking score alone (blue). Hit rate is the ratio of active compounds/tested compounds; the raw numbers appear at the top of each bar. f, Hit rates for selecting compounds at different scoring ranges by each strategy: human prioritization and docking score (orange) or docking score alone (blue). g, Distribution of the binding affinity level among the hits from e (top panel). We measured competition binding curves for 14 docking hits from human prioritization and docking score, and 7 hits from the docking score alone. These are divided into three affinity ranges: <5 nM; 5 nM–50 nM; >50 nM; Distribution of the binding affinity level among the hits from all different scoring ranges (bottom panel). We measured competition binding curves for 14 docking hits from human prioritization and docking score, and 17 hits from the docking score alone. h. Hit-rate curve comparison with/without human picks. The hit rate without human picks at the top plateau is 39% and at the bottom plateau is 0%, and the docking score (dock50) and slope at the maximum (slope50) are −46.5 kcal mol−1 and −3.5% per kcal mol−1, respectively.
Extended Data Figure 3 |. Analogs of σ2 receptor ligands and the effect of a structural water molecule.
a-c, Initial hits and selected analogs of σ2 receptor ligands. Competition binding curves on the top panel, 2D drawings of compounds are on the bottom panel. Parent compound is indicated by gray background. Points shown as mean ± SEM from three technical replicates. a, Parent compound ZINC548355486 and its three potent analogues. b, Parent compound ZINC895657866 and its three potent analogues. c, Parent compound ZINC450573233 and its three potent analogues. d-f, The binding site of the σ2 receptor contains a structural water. d, Water coordination at the binding site of the σ2 receptor. Water molecule is depicted as a red sphere. Hydrogen bonds are indicated by black dashed lines. e, Saturation binding curve to measure the dissociation constant (Kd) of [3H]-DTG for the various mutants of σ2 receptor meant to disrupt water coordination. Residues proximal to the structural water were chosen for mutation. Residues were mutated to the indicated amino acid. Points shown as mean ± SEM from three technical replicates. f, Competition binding measurement of affinity of Z1241145220 in various mutants of σ2. Points shown as mean ± SEM from three technical replicates.
Extended Data Figure 4 |. Effect of systemic σ receptor ligands on motor behavior.
a, Response of mice to a von Frey filament after spared nerve injury (SNI). All five ligands are compared to their respective vehicles (PD-144418 10 mg/kg (n = 5) and 30 mg/kg (n = 5) vs. kolliphor (n = 5), one-way ANOVA, F(2, 12) = 7.49, p = 0.008; Z4446724338 10 mg/kg (n = 10) and 20 mg/kg (n = 5) vs cyclodextrin (n = 10), one-way ANOVA, F(2, 22) = 25.12, p < 0.001; Z4857158944 10 mg/kg (n = 5) and 20 mg/kg (n = 5) vs cyclodextrin (n = 10), one-way ANOVA, F(2, 17) = 5.10, p = 0.02; Z1665845742 10 mg/kg (n = 10) and 20 mg/kg (n = 5) and PB28 30 mg/kg (n = 10) vs saline (n = 10), one-way ANOVA, F(3, 31) = 6.18, p = 0.002; asterisks define individual group differences to respective vehicle control using Dunnett’s multiple comparisons Post-hoc test; ns = not significant, * p < 0.05, ** p < 0.01, *** p < 0.001). Data shown are mean ± SEM. Data for higher doses and vehicles is replotted from Fig. 4. b, No sedation or motor impairment on the rotarod was observed after drug treatments compared to vehicle at 1 hour (Z1665845742 10 mg/kg (n = 5) and Z4857158944 20 mg/kg (n = 5) vs saline (n = 5), one-way ANOVA, F(2, 12) = 1.04, p = 0.38; Z4446724338 10 mg/kg (n = 5) vs kolliphor (n = 5), unpaired two-tailed Student’s t-test, t(8) = 0.47, p = 0.65) or 24 hours post-injection (Z1665845742 10 mg/kg (n = 5) and Z4857158944 20 mg/kg (n = 5) vs saline (n = 5), one-way ANOVA, F(2, 12) = 0.45, p = 0.65; Z4446724338 10 mg/kg (n = 5) vs kolliphor (n = 5), unpaired two-tailed Student’s t-test, t(8) = 0.72, p = 0.49); ns = not significant. Data shown are means ± SEM. c, Response of SNI mice to a von Frey filament after repeated injections of Z4446724338 10 mg/kg (n = 5). Mechanical thresholds were assessed 1 hour and 24 hours after four separate injections. Data shown are means ± SEM normalized to each mouse’s SNI baseline. d, Response of SNI mice to a von Frey filament after repeated injections of Z4857158944 10 mg/kg (n = 5). Mechanical thresholds were assessed 1 hour and 24 hours after four separate injections. Data shown are means ± SEM normalized to each mouse’s SNI baseline. e. Quantification of the expression levels of Sigmar1 (σ1) and Tmem97 (σ2) in wildtype (WT) and SNI mice detected by in situ hybridization (n = 3 mice per group). Representative images can be found in panel f. Data shown are mean ± SEM; unpaired two-tailed Student’s t-test— Sigmar1: t(4) = 0.5, p = 0.64; Tmem97: t(4) = 1.0, p = 0.37; ns = not significant. AU = arbitrary units. f, in situ hybridization of mouse dorsal root ganglion (DRG) sections for Sigmar1 (σ1) and Tmem97 (σ2) genes illustrates expression in myelinated (Nefh-positive; blue) and unmyelinated (Acpp-positive; red) subsets of sensory neurons and no change after SNI.
Extended Data Figure 5 |. Off-target profiling of Z4446724338, Z1665845742, and Z4857158944.
a-c, TANGO screens against a panel of 320 GPCRs for the indicated σ2 ligand. a, Z4446724338, b, Z1665845742, c, Z4857158944. d, GloSensor μOR-mediated cAMP inhibition (Gi activation) by DAMGO, Z4446724338, Z1665845742, and Z4857158944. e-f, Follow-up does-response curves for pain-related receptors that showed activation in a-c. e, Z4446724338 and Z1665845742 against 5HT1A. f, Z4857158944 against κOR. Data shown are means ± SEM.
Extended Data Figure 6 |. Paw withdrawal thresholds and in situ intensity measurements.
a, Paw withdrawal thresholds (PWT) before (blue bar) and after (red bar) spared nerve injury (SNI), as well as after SNI + treatment (purple bar). For easier visualization of individual data points, data was also plotted without the pre-SNI baseline. Data are the same as in Figure 4b and Extended Data Figure 4a, but without the normalization to the individual post-SNI baselines and are expressed as mean ± SEM; mice per group: saline (n = 10); cyclodextrin (n = 10); kolliphor (n = 5); PB28 30 mg/kg (n = 10); PD-144418 10 mg/kg (n = 5) and 30 mg/kg (n = 5); Z4446724338 10 mg/kg (n = 10) and 20 mg/kg (n = 5); Z1665845742 10 mg/kg (n = 5) and 20 mg/kg (n = 5); Z4857158944 10 mg/kg (n = 5) and 20 mg/kg (n = 5); unpaired two-tailed Student’s t-test. b, PWTs 1 hour, 24 hours, and 48 hours after saline or drug treatment. Data are the same as in Figure 4c, but without the normalization to the individual post-SNI baselines, and are expressed as mean ± SEM. Significance levels determined using Dunnett’s multiple comparisons Post-hoc test reflect the difference between Z4446724338 and saline for simplicity (two-way ANOVA; time × treatment interaction: F(8, 80) = 2.4, p = 0.02; time: F(2, 74) = 5.2, p = 0.009; treatment: F(4, 40) = 3.3, p = 0.02; four treatment groups (n = 10) except PD-144418 (n = 5); ns = not significant. c, Response of SNI mice to a von Frey filament after repeated injections of Z4446724338 10 mg/kg (n = 5). Mechanical thresholds were assessed 1 hour and 24 hours after four separate injections. Data shown are paw withdrawal thresholds in grams, expressed as mean ± SEM. d, Response of SNI mice to a von Frey filament after repeated injections of Z4857158944 10 mg/kg (n = 5). Mechanical thresholds were assessed 1 hour and 24 hours after four separate injections. Data shown are paw withdrawal thresholds in grams, expressed as mean ± SEM.
Extended Data Table 1 |.
Data collection and refinement statistics
| PB28-bound (Se-labeled) | Roluperidone-bound (Native) | Z1241145220-bound (Native) | Z4857158944-bound (Native) | Cholesterol-bound (Native) | |
|---|---|---|---|---|---|
|
| |||||
| Data collection | |||||
| Space group | P21 | P21 | P212121 | P21 | P21 |
| Number of crystals | 1 | 1 | 1 | 1 | 1 |
| Cell dimensions | |||||
| a, b, c (Å) | 70.6, 55.2, 93.0 | 69.1, 54.2, 99.7 | 55.4, 61.5, 110.4 | 70.7, 55.4, 93.0 | 70.9, 54.9, 93.0 |
| α, β, γ (°) | 90, 95.0, 90 | 90, 91.1, 90 | 90, 90, 90 | 90, 94.5, 90 | 90, 94.3, 90 |
| Wavelength (Å) | 1.255 | 1.03320 | 1.03321 | 1.033167 | 1.03320 |
| Resolution (Å) | 33.88 – 2.942 (3.047 – 2.942) | 42.61 – 2.71 (2.81 – 2.71) | 49.5 – 2.41 (2.55 – 2.41) | 40.2 – 2.41 (2.55 – 2.41) | 47.24 – 2.8 (2.9 –2.8) |
| Rsym | 24.75 (88.16) | 26.11 (205.9) | 18.4 (177.4) | 19.67 (227.6) | 32.2 (220.2) |
| I/σI | 5.73 (0.93) | 5.90 (0.71) | 7.50 (0.7) | 5.18 (0.56) | 5.35 (0.94) |
| Completeness (%) | 98.67 (90.76) | 99.54 (99.87) | 99.55 (99.36) | 97.9 (88.3) | 99.7 (98.5) |
| Redundancy | 4.0 (3.4) | 6.8 (6.5) | 6.2 (4.4) | 4.4 (4.5) | 6.7 (6.9) |
| CC1/2 | 98.7 (49.5) | 99.4 (36.6) | 99.6 (26.9) | 99.5 (28.3) | 99.2 (42.6) |
| Refinement | |||||
| Resolution (Å) | 2.94 | 2.71 | 2.41 | 2.41 | 2.8 |
| No. reflections | 15228 | 20340 | 15165 | 27448 | 17720 |
| No. reflection used for Rfree | 1524 (10%) | 2004 (9.85%) | 1063 (7%) | 1370 (5%) | 1752 (9.89%) |
| Rwork / Rfree | 20.39 / 24.26 | 22.18 / 25.2 | 21.36 / 24.6 | 25.0 / 28.8 | 24.06 / 27.81 |
| No. atoms | |||||
| Protein | 5490 | 5472 | 2761 | 5393 | 5292 |
| Lipid/ion | 231 | 250 | 148 | 231 | 223 |
| Ligand | 108 | 108 | 48 | 100 | 112 |
| Water | 27 | 7 | 46 | 37 | 21 |
| B-factors (Å2) | |||||
| Protein | 49.68 | 67.89 | 50.19 | 57.24 | 64.29 |
| Lipid/ion | 52.48 | 65.57 | 59.49 | 62.32 | 62.33 |
| Ligand | 56.89 | 79.39 | 49.32 | 66.07 | 71.96 |
| Water | 45.66 | 62.72 | 57.08 | 57.77 | 61.02 |
| R.m.s. deviations | |||||
| Bond lengths (Å) | 0.003 | 0.003 | 0.005 | 0.003 | 0.003 |
| Bond angles (°) | 0.61 | 0.61 | 1.04 | 0.58 | 0.59 |
Extended Data Table 2 |.
14 of the highest-affinity direct docking hits for the σ2 receptor
| 2D drawing | ZINC ID | Rank | DOCK score (kcal/mol) | TC* | Ki (nM) |
Selectivity (σ1/σ2) | |
|---|---|---|---|---|---|---|---|
| σ2 | σ1 | ||||||
|
| |||||||
|
ZINC000450573233 | 4429 | −57.25 | 0.32 | 4.3 | 128 | 30 |
|
|
ZINC000895657866 | 19047 | −55.35 | 0.31 | 21.4 | 989.6 | 46 |
|
|
ZINCO01170548029 | 4945 | −57.11 | 0.35 | 22.6 | 727.2 | 32 |
|
|
ZINC000533478938 | 18545 | −55.38 | 0.30 | 34.5 | 1470 | 43 |
|
ZINC000921927365 | 983 | −59.01 | 0.31 | 67.3 | 1186 | 18 |
|
|
ZINC000548355486 | 7007 | −56.68 | 0.29 | 2.4 | 4.9 | 2 |
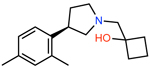
|
ZINC000348332392 | 931 | −59.07 | 0.28 | 33.7 | 2.9 | 0.1 |
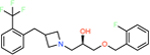
|
ZINC001254761628 | 16059 | −55.58 | 0.27 | 4.7 | 53 | 11.3 |
|
|
ZINC000544117725 | 3522 | −57.52 | 0.28 | 10 | 16.25 | 1.6 |
|
ZINC000170908795 | 13281 | −55.84 | 0.29 | 6.7 | 32.7 | 4.9 |
|
|
ZINC001196519317 | 9290 | −56.3 | 0.29 | 2.4 | 13.4 | 5.6 |
|
|
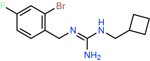
ZINC000656714762 | 1276 | −58.68 | 0.26 | 67.8 | 4.6 | 0.1 |
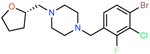
|
ZINC001237901728 | 11409 | −56.03 | 0.30 | 27 | 1.6 | 0.1 |
|
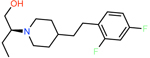
ZINC001460312963 | 11817 | −55.99 | 0.29 | 5.2 | 1.7 | 0.3 |
See Supplementary Table 1 for all 484 compounds tested.
TC, Tanimoto coefficient to sigma ligands from ChEMBL.
Extended Data Table 3 |.
Measured pharmacokinetic parameters for PB28, Z1665845742, Z4446724338 and Z4857158944 in male CD-1 mice by 10 mg/kg subcutaneous administration
| Pharmacokinetic Parameters | |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Type | Name | Tmax min | Cmax ng/ml (g) | AUC0→t (AUClast) ng*min/ml (g) | AUC0→∞ (AUCinf_obs) ng*min/ml (g) | T1/2 (HL_Lambda_z), min | Kel (Lambda_z), min−1 |
|
| |||||||
| Plasma | Z1665845742 | 20 | 968 | 99000 | 112000 | 185 | 0.00374 |
| Z4446724338 | 20 | 449 | 58300 | 60500 | 47.4 | 0.0146 | |
| Z4857158944 | 20 | 228 | 13300 | 14200 | 27.8 | 0.0249 | |
| PB28 | 60 | 42 | 8640 | 45900 | 740 | 0.000937 | |
|
| |||||||
| Brain | Z1665845742 | 20 | 3150 | 436000 | 509000 | 747 | 0.000928 |
| Z4446724338 | 20 | 7390 | 1140000 | 1150000 | 69.7 | 0.00995 | |
| Z4857158944 | 20 | 2960 | 247000 | 327000 | 452 | 0.00153 | |
| PB28 | 60 | 948 | 229000 | 240000 | 98.1 | 0.00706 | |
Supplementary Material
Acknowledgements
Funding to support this research was provided by NIH grant R01GM119185, The Vallee Foundation, and the Sanofi iAwards program (ACK), by DARPA grant HR0011–19-2–0020 and NIH grant R35GM122481 (BKS), and by grant GM133836 (JJI). GM/CA@APS has been funded by the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006, P30GM138396). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02–06CH11357. The Eiger 16M detector at GM/CA-XSD was funded by NIH grant S10 OD012289. We thank Dr. Kelly Arnett and the Harvard Center for Macromolecular Interactions for excellent support of biophysical experiments including circular dichroism and SEC-MALS. We also thank Charles Vidoudez and The Harvard Center for Mass Spectrometry for performing mass spectrometry analysis of sterols. Molecular graphics and analyses were performed with UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311.
Competing interest
A.C.K. is a founder and consultant for biotechnology companies Tectonic Therapeutic, Inc., and Seismic Therapeutic, Inc., as well as the Institute for Protein Innovation, a non-profit research institute. B.K.S. is a founder of Epiodyne, a company active in analgesia, and of BlueDolphin, which undertakes fee-for-service ligand-discovery.
Footnotes
Code availability
DOCK3.7 is freely available for non-commercial research http://dock.compbio.ucsf.edu/DOCK3.7/. A web-based version is available at http://blaster.docking.org/.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Ethical compliance
All animal experiments were approved by the Institutional Animal Care and Use Committee at UCSF and were conducted in accordance with the NIH Guide for the Care and Use of Laboratory animals.
Data availability
The coordinates and structure factors for PB28-bound σ2, roluperidone-bound σ2, Z1241145220-bound σ2, Z4857158944-bound σ2, and cholesterol-bound σ2 have been deposited in the PDB with accession codes 7M93, 7M94, 7M95, 7M96, and 7MFI respectively. The identities of the compounds docked in this study are freely available from the ZINC database (http://zinc15.docking.org) and active compounds may be purchased from Enamine. Any other data relating to this study are available from the corresponding authors on reasonable request. Source data are provided with this paper.
References
- 1.Waarde A van et al. Potential applications for sigma receptor ligands in cancer diagnosis and therapy. Biochimica Et Biophysica Acta Bba - Biomembr 1848, 2703–2714 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Harvey PD et al. Effects of Roluperidone (MIN-101) on two dimensions of the negative symptoms factor score: Reduced emotional experience and reduced emotional expression. Schizophr Res 215, 352–356 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Sahn JJ, Mejia GL, Ray PR, Martin SF & Price TJ Sigma 2 Receptor/Tmem97 Agonists Produce Long Lasting Antineuropathic Pain Effects in Mice. Acs Chem Neurosci 8, 1801–1811 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Intagliata S et al. Discovery of a Highly Selective Sigma-2 Receptor Ligand, 1-(4-(6,7-Dimethoxy-3,4-dihydroisoquinolin-2(1H)-yl)butyl)-3-methyl-1H-benzo[d]imidazol-2(3H)-one (CM398), with Drug-Like Properties and Antinociceptive Effects In Vivo. Aaps J 22, 94 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Quadir SG et al. The Sigma-2 receptor / transmembrane protein 97 (σ2R/TMEM97) modulator JVW-1034 reduces heavy alcohol drinking and associated pain states in male mice. Neuropharmacology 184, 108409 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grundman M et al. A phase 1 clinical trial of the sigma-2 receptor complex allosteric antagonist CT1812, a novel therapeutic candidate for Alzheimer’s disease. Alzheimer’s Dementia Transl Res Clin Interventions 5, 20–26 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riad A et al. Sigma-2 Receptor/TMEM97 and PGRMC-1 Increase the Rate of Internalization of LDL by LDL Receptor through the Formation of a Ternary Complex. Sci Rep-uk 8, 16845 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abate C et al. PB28, the Sigma-1 and Sigma-2 Receptors Modulator With Potent Anti–SARS-CoV-2 Activity: A Review About Its Pharmacological Properties and Structure Affinity Relationships. Front Pharmacol 11, 589810 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shields SD, Eckert WA & Basbaum AI Spared nerve injury model of neuropathic pain in the mouse: a behavioral and anatomic analysis. J Pain 4, 465–470 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Hellewell SB et al. Rat liver and kidney contain high densities of σ1 and σ2 receptors: characterization by ligand binding and photoaffinity labeling. European J Pharmacol Mol Pharmacol 268, 9–18 (1994). [DOI] [PubMed] [Google Scholar]
- 11.Hellewell SB & Bowen WD A sigma-like binding site in rat pheochromocytoma (PC12) cells: decreased affinity for (+)-benzomorphans and lower molecular weight suggest a different sigma receptor form from that of guinea pig brain. Brain Res 527, 244–253 (1990). [DOI] [PubMed] [Google Scholar]
- 12.Hanner M et al. Purification, molecular cloning, and expression of the mammalian sigma1-binding site. Proc National Acad Sci 93, 8072–8077 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langa F et al. Generation and phenotypic analysis of sigma receptor type I (sigma1) knockout mice. Eur J Neurosci 18, 2188–2196 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Alon A et al. Identification of the gene that codes for the σ2 receptor. Proc National Acad Sci 114, 7160–7165 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebrahimi-Fakhari D et al. Reduction of TMEM97 increases NPC1 protein levels and restores cholesterol trafficking in Niemann-pick type C1 disease cells. Hum Mol Genet 25, 3588–3599 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartz F et al. Identification of cholesterol-regulating genes by targeted RNAi screening. Cell Metab 10, 63–75 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Sanchez-Pulido L & Ponting CP TM6SF2 and MAC30, new enzyme homologs in sterol metabolism and common metabolic disease. Frontiers Genetics 5, 439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahdessian H et al. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc National Acad Sci 111, 8913–8918 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vilner BJ, John CS & Bowen WD Sigma-1 and sigma-2 receptors are expressed in a wide variety of human and rodent tumor cell lines. Cancer Res 55, 408–13 (1995). [PubMed] [Google Scholar]
- 20.Scott LL et al. Small molecule modulators of σ2R/Tmem97 reduce alcohol withdrawal-induced behaviors. Neuropsychopharmacol 43, 1867–1875 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vázquez-Rosa E et al. Neuroprotective Efficacy of a Sigma 2 Receptor/TMEM97 Modulator (DKR-1677) after Traumatic Brain Injury. Acs Chem Neurosci 10, 1595–1602 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stein RM et al. Virtual discovery of melatonin receptor ligands to modulate circadian rhythms. Nature 579, 609–614 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuller M et al. Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking. Sci Adv 7, eabf8711 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt HR & Kruse AC The Molecular Function of σ Receptors: Past, Present, and Future. Trends Pharmacol Sci 40, 636–654 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caffrey M & Cherezov V Crystallizing membrane proteins using lipidic mesophases. Nat Protoc 4, 706–731 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long T et al. Structural basis for human sterol isomerase in cholesterol biosynthesis and multidrug recognition. Nat Commun 10, 2452 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zimmermann L et al. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol 430, 2237–2243 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Audet M & Stevens RC Emerging structural biology of lipid G protein-coupled receptors. Protein Sci 28, 292–304 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt HR et al. Crystal structure of the human σ1 receptor. Nature 532, 527–530 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hubler Z et al. Accumulation of 8,9-unsaturated sterols drives oligodendrocyte formation and remyelination. Nature 560, 372–376 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lomize MA, Pogozheva ID, Joo H, Mosberg HI & Lomize AL OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res 40, D370–D376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyu J et al. Ultra-large library docking for discovering new chemotypes. Nature 566, 224–229 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischer A, Smieško M, Sellner M & Lill MA Decision Making in Structure-Based Drug Discovery: Visual Inspection of Docking Results. J Med Chem 64, 2489–2500 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Cendán CM, Pujalte JM, Portillo-Salido E, Montoliu L & Baeyens JM Formalin-induced pain is reduced in σ1 receptor knockout mice. Eur J Pharmacol 511, 73–74 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Puente B de la et al. Sigma-1 receptors regulate activity-induced spinal sensitization and neuropathic pain after peripheral nerve injury. Pain 145, 294–303 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Cendán CM, Pujalte JM, Portillo-Salido E & Baeyens JM Antinociceptive effects of haloperidol and its metabolites in the formalin test in mice. Psychopharmacology 182, 485–493 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Romero L et al. Pharmacological properties of S1RA, a new sigma-1 receptor antagonist that inhibits neuropathic pain and activity-induced spinal sensitization. Brit J Pharmacol 166, 2289–2306 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruna J et al. Efficacy of a Novel Sigma-1 Receptor Antagonist for Oxaliplatin-Induced Neuropathy: A Randomized, Double-Blind, Placebo-Controlled Phase IIa Clinical Trial. Neurotherapeutics 15, 178–189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vela JM, Merlos M & Almansa C Investigational sigma-1 receptor antagonists for the treatment of pain. Expert Opin Inv Drug 24, 883–896 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Kooistra AJ et al. GPCRdb in 2021: integrating GPCR sequence, structure and function. Nucleic Acids Res 49, gkaa1080- (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hauser AS et al. Pharmacogenomics of GPCR Drug Targets. Cell 172, 41–54.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kroeze WK et al. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat Struct Mol Biol 22, 362–369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nastasi G et al. S2RSLDB: a comprehensive manually curated, internet-accessible database of the sigma-2 receptor selective ligands. J Cheminformatics 9, 3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang X-P et al. Allosteric ligands for the pharmacologically dark receptors GPR68 and GPR65. Nature 527, 477–483 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang S et al. Structure of the D2 dopamine receptor bound to the atypical antipsychotic drug risperidone. Nature 555, 269–273 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jumper J et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gordon DE et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Otwinowski Z & Minor W Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 49.Kabsch W XDS. Acta Crystallogr Sect D Biological Crystallogr 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alford RF et al. An Integrated Framework Advancing Membrane Protein Modeling and Design. Plos Comput Biol 11, e1004398 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mccoy AJ et al. Phaser crystallographic software. J Appl Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liebschner D et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr Sect D Struct Biology 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr Sect D Biological Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schrödinger L The PyMOL Molecular Graphics System, Version 2.5.
- 55.Pettersen EF et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Chu UB & Ruoho AE Sigma Receptor Binding Assays. Curr Protoc Pharmacol Éditor Board S J Enna Ed Et Al 71, 1.34.1–21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weiner SJ et al. A new force field for molecular mechanical simulation of nucleic acids and proteins. J Am Chem Soc 106, 765–784 (1984). [Google Scholar]
- 58.Meng EC, Shoichet BK & Kuntz ID Automated docking with grid-based energy evaluation. J Comput Chem 13, 505–524 (1992). [Google Scholar]
- 59.Gallagher K & Sharp K Electrostatic Contributions to Heat Capacity Changes of DNA-Ligand Binding. Biophys J 75, 769–776 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mysinger MM & Shoichet BK Rapid Context-Dependent Ligand Desolvation in Molecular Docking. J Chem Inf Model 50, 1561–1573 (2010). [DOI] [PubMed] [Google Scholar]
- 61.Stein RM et al. Property-Unmatched Decoys in Docking Benchmarks. J Chem Inf Model 61, 699–714 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gu S, Smith MS, Yang Y, Irwin JJ & Shoichet BK Ligand Strain Energy in Large Library Docking. Biorxiv 2021.04.06.438722 (2021) doi: 10.1101/2021.04.06.438722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carpenter B et al. Stan : A Probabilistic Programming Language. J Stat Softw 76, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bürkner P-C brms : An R Package for Bayesian Multilevel Models Using Stan. J Stat Softw 80, (2017). [Google Scholar]
- 65.Gelman A, Goodrich B, Gabry J & Vehtari A R-squared for Bayesian Regression Models. Am Statistician 73, 1–6 (2018). [Google Scholar]
- 66.Kay M tidybayes: Tidy Data and Geoms for Bayesian Models. (2020). doi: 10.5281/zenodo.1308151. [DOI] [Google Scholar]
- 67.Wickham H ggplot2: Elegant Graphics for Data Analysis. (Springer-Verlag; New York, 2016). [Google Scholar]
- 68.Wickham H et al. Welcome to the tidyverse. J Open Source Softw 4, 1686 (2019). [Google Scholar]
- 69.Team, R. C. R: A Language and Environment for Statistical Computing. (2018).
- 70.Skubic C, Vovk I, Rozman D & Križman M Simplified LC-MS Method for Analysis of Sterols in Biological Samples. Molecules 25, 4116 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Longo PA, Kavran JM, Kim M-S & Leahy DJ Chapter Eighteen Transient Mammalian Cell Transfection with Polyethylenimine (PEI). Methods Enzymol 529, 227–240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Besnard J et al. Automated design of ligands to polypharmacological profiles. Nature 492, 215–220 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scherrer G et al. Dissociation of the Opioid Receptor Mechanisms that Control Mechanical and Heat Pain. Cell 137, 1148–1159 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Muralidharan A et al. Identification and characterization of novel candidate compounds targeting 6- and 7-transmembrane μ-opioid receptor isoforms. Brit J Pharmacol 178, 2709–2726 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chaplan SR, Bach FW, Pogrel JW, Chung JM & Yaksh TL Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Meth 53, 55–63 (1994). [DOI] [PubMed] [Google Scholar]
- 76.Solorzano C et al. Primary Afferent and Spinal Cord Expression of Gastrin-Releasing Peptide: Message, Protein, and Antibody Concerns. J Neurosci 35, 648–657 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Notredame C, Higgins DG & Heringa J T-coffee: a novel method for fast and accurate multiple sequence alignment11Edited by J. Thornton. J Mol Biol 302, 205–217 (2000). [DOI] [PubMed] [Google Scholar]
- 78.Liebschner D et al. Polder maps: improving OMIT maps by excluding bulk solvent. Acta Crystallogr Sect D Struct Biology 73, 148–157 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structure factors for PB28-bound σ2, roluperidone-bound σ2, Z1241145220-bound σ2, Z4857158944-bound σ2, and cholesterol-bound σ2 have been deposited in the PDB with accession codes 7M93, 7M94, 7M95, 7M96, and 7MFI respectively. The identities of the compounds docked in this study are freely available from the ZINC database (http://zinc15.docking.org) and active compounds may be purchased from Enamine. Any other data relating to this study are available from the corresponding authors on reasonable request. Source data are provided with this paper.