Abstract
Bile acids have been linked to pathomechanism and prognosis in various types of cancers. The present study aimed to investigate the effect of bile acids on the molecular change in gastric epithelial cancer cells and to evaluate gastric bile acid concentration in patients with early gastric cancer (EGC). Human gastric cancer cells (AGS and NCI-N87 cell lines) were treated with several bile acid types to determine their effect on molecular changes in the cells. Gastric levels of individual bile acids were measured (primary unconjugated or conjugated bile acids and secondary bile acids) in 39 participants (20 controls and 19 patients with EGC). Exposing gastric epithelial cancer cells to primary bile acids in vitro upregulated the expression of early growth response factor 1 (Egr-1) and the oncogenes including c-Jun, c-Myc and Snail, whereas a p42/44 MAPK inhibitor exposure reduced their expression. There was a significant difference in age and presence of atrophic gastritis with intestinal metaplasia in background mucosa between controls and patients with EGC. There were significant differences in the levels of unconjugated or conjugated primary bile acids between controls and EGC patients except lithocholic acid. After adjustment of age and presence of atrophic gastritis with intestinal metaplasia, the levels of cholic acid [adjusted odds ratio (aOR) 4.3; 95% confidence interval (CI): 1.2-16.2; P=0.029] and glycochenodeoxycholic acid [aOR 9.9; 95% CI: 1.3-75.3; P=0.027] were significantly higher in patients with EGC compared with controls. In conclusion, bile acids upregulate Egr-1 in gastric cancer cells via the MAPK signaling pathway, and higher gastric levels of primary bile acids are associated with EGC. Therefore, exposure of gastric cells to primary bile acids may play a role in gastric carcinogenesis.
Keywords: stomach, bile acids, early growth response factor 1, oncogene, gastritis, atrophic
Introduction
Gastric cancer is one of the most common cancers worldwide (1). Even though Helicobacter pylori (H. pylori) infection is known to be the major risk factor for the development of gastric cancer, H. pylori eradication does not completely prevent gastric cancer, and other genetic or environmental factors might influence gastric cancer development (1–3).
Bile acids are cholesterol derivate and are required for absorption and transport of fat (2). Bile acids exist in enterohepatic organs such as the liver, gall bladder, and intestine, which contain high levels of bile acids. We frequently observed considerable bile colored fluid in the stomach of patients who had undergone gastric surgery, cholecystectomy, or sphincterotomy, and healthy controls with bile reflux who had no history of gastric surgery. Recently, several clinical studies have suggested that bile reflux is associated with premalignant gastric lesions such as atrophic gastritis and intestinal metaplasia (3,4). Furthermore, bile acid receptors, including G-protein-coupled bile acid receptor 1 (TGR5) and farnesoid X receptor (FXR), have been known to be expressed in the mucosa of patients with Barrett's esophagus, esophageal adenocarcinoma, and advanced gastric cancer (5–7). Acidified bile acids also activated the transcription factor c-Myc, which is associated with tumor progression and telomerase activity (2). However, the association between intragastric bile acid and patients with early gastric cancer (EGC) remains unelucidated. In addition, there is limited information about the effects of bile acids on the molecular change in gastric epithelial cells. Early growth response factor-1 (Egr-1) is a transcription factor, which has been known to be implicated in biological process including tissue injury, immune response, and fibrosis. It is also related to the inflammation, cell proliferation, cell differentiation, and initiation and progression of cancer (8). Egr-1 can be activated through stimuli by growth factors, tumor necrosis factors, inflammatory factors, reactive oxygen species, and bacteria such as H. pylori (9). Until now, there is limited information whether bile acids activate Egr-1 in gastric epithelial cells and which molecular mechanism is involved in the activation of Egr-1 by bile acids.
In this study, we aimed to investigate the effect of bile acids on the activation of Egr-1 and the related molecular change in gastric epithelial cells and to evaluate the difference in gastric bile acid concentration between controls and patients with early gastric cancer.
Materials and methods
Cell culture
For our purposes, we required well-established, acid-stable gastric cancer cell lines with comparable levels of c-Myc expression (10). Accordingly, we purchased AGS (ATCC® CRL-1739™) and NCI-N87 (ATCC® CRL-5822™) cell lines from the American Type Culture Collection (Manassas, VA, USA). These gastric cancer cell lines were grown in Dulbecco's modified Eagle's medium (DMEM) (GIBCO Invitrogen) containing 4.5 mg/l glucose, 100 mg/l streptomycin, and 2 mM L-glutamine supplemented with 10% fetal bovine serum (FBS) (GIBCO Invitrogen). They were maintained at 37°C under a humidified 5% CO2 atmosphere in a CO2 incubator (Sanyo). Solutions of bile acids (Sigma-Aldrich) were prepared using appropriate solvents according to the manufacturer's protocols (Table SI). AGS and NCI-N87 cells were cultured in the growth medium for 24 h and then transferred to fresh, serum-free medium containing 100 µM of a bile acid for 48 h, with the bile acid being cholic acid (CA; Sigma-Aldrich, C9377), chenodeoxycholic acid (CDCA; Sigma-Aldrichl, C1129), taurocholic acid (TCA; Sigma-Aldrich, T4009), or glycochenodeoxycholic acid (GCDCA; Sigma-Aldrich, G0759). Afterward, we extracted the total RNA and total protein from the cells.
Western blotting
We extracted proteins from the gastric cells treated with bile acids using a radioimmunoprecipitation assay buffer (#R0278, Sigma-Aldrich) containing protease and phosphatase inhibitors (#p8340 and #p2850, respectively; Sigma-Aldrich). Proteins were separated by 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis, then blotted onto PVDF membranes (Millipore) according to the manufacturer's protocol. We then incubated the PVDF transfer membranes at 4°C overnight in diluted solutions of primary antibodies: anti-phospho-AKT (#9271), anti-AKT (#9272), anti-phospho-p42/44-MAPK (#9106), anti-p42/44-MAPK (#4695), anti-Egr-1 (#4154), anti-β-actin (#4967), anti-c-Jun (#9165), HRP-linked anti–rabbit IgG (#7074), and HRP-linked anti-mouse IgG (#7076) (Cell Signaling Technology, Inc.). We analyzed proteins using the Fujifilm LAS-3000 imaging system (Fujifilm). The fold change in protein expression was calculated by dividing the normalized signal intensity of the target band in the experimental sample by that of the target band in the control sample.
mRNA quantitation
Total RNA was extracted using TRIzol (Takara Bio Inc.). Briefly, 1 ml of Trizol solution was added into each well and the suspension was then moved to a 1.5 ml tube. After adding 200 µl of chloroform (Sigma-Aldrich Co. LLC) and vortex-mixing for 15 sec, the mixture was centrifuged at 20,000 × g for 20 min. The supernatant was then collected and mixed with equal amounts of 99.9% isopropyl alcohol (Merck), followed by centrifugation at 20,000 × g for 20 min. The pellet was washed with 1 ml of 70% ethyl alcohol (MERCK, Co. LLC), followed by centrifugation at 20,000 × g for 5 min. After removing the remaining ethyl alcohol, the RNA pellet was air dried at a 25°C. It was then resuspended in 50 µl of diethyl pyrocarbonate water. Total RNAs were converted to cDNAs using reverse transcription system (Promega Corporation). Real-time PCR was performed with Applied Biosystems StepOnePlus™ Real time PCR System (Life Technologies Corporation) according to the manufacturer's protocol. Glyceraldehyde 3-phosphate dehydrogenase was used as a control, and ΔΔCT values were calculated for cancer stem cell markers using the Taqman assay probes (Twist1, HS01675818_s1; c-Myc, HS00153408_m1; c-Jun, HS01103582_s1; Snail, HS00195591_m1; Thermo Fisher Scientific, Inc.).
Subjects
A total of 39 subjects [20 controls and 19 EGC patients] were enrolled in this study. We excluded patients with secondary bile reflux, defined as bile reflux after gastric surgery, patients with previous diagnosis of malignancy and patients with taking any medicines which might affect bile acid secretion or gastrointestinal motility such as steroid, prokinetics, lipid lowering agents, bile acid sequestrants, urodexoycholic acid and chenodexoycholic acid. This study was approved by our institutional review board (CNUH-2020-085). We explained the terms of participation in this study and obtained written informed consent from patients before endoscopic procedures.
Collection of gastric fluid
All endoscopic procedures were performed without foaming mucus remover or antispasmotics by an experienced endoscopist (SYP) in the early morning. Subjects were fasted for 12 h. First, we used distilled water to flush the endoscope clean; the gastric fluid in the fundus and greater curvature of the gastric body were aspirated into sterile collection traps immediately when the endoscope was introduced into the stomach. The collected fluid specimens were immediately cryopreserved at −80°C.
Measurement of bile acids by liquid chromatography with tandem mass spectrometry
We analyzed the gastric juice using a mass spectrometer API 4000Q TRAP (AB Sciex), First, we diluted the gastric juice 20- to 200-fold using distilled water. Then, diluted 100 µl of the gastric juice was mixed with an internal standard solution (CA-d5 ng/ml in 50% methanol). The mixed solution was then vortex-mixed for 3 sec, 200 µl of acetonitrile was added, and then centrifuged at 20,000 × g for 2 min. We injected 20 µl of the diluted supernatant that was diluted with 180 µl of 20 mM ammonium acetate. We used the standard component of Sigma-Aldrich C9377, G0759, C1129, T4009, L6250, D2510 for CA, CDCA, TCA, GCDCA, lithochholic acid (LCA), and deoxycholic acid (DCA) (Sigma-Aldrich Co. LLC). We analyzed LC-MS/MS data of each bile acid by the Analyst software version 1.6.3 (AB Sciex Pte. Ltd.).
Diagnosis of H. pylori infection and histology
Subjects were considered to be infected with H. pylori if the results of at least one of four diagnostic tests [rapid urease test, histologic results, H. pylori polymerase chain reaction (PCR), and (13C.)-urea breath test] were positive. All biopsy and resected specimens were evaluated for background histology and tumor histology based on the Vienna classification system by an expert pathologist (CYD) (11).
Statistical analysis
Statistical analysis was performed using SPSS version 23.0 (SPSS, Inc.). Continuous data are shown as mean ± standard deviation or median (interquartile range, IQR), while categorical data are shown as absolute and relative frequencies. Categorical data were examined using Fisher's exact test or the chi-squared test. Variables with a skewed distribution were performed using non-parametric tests (Mann-Whitney test, Kruskal Wallis test) and Spearman non-parametric test. For adjustment of variables, binary logistic regression models with enter were used to evaluate the association between the levels of each bile acid and EGC. The data in regression analysis were shown as adjusted odds ratios (aOR) with 95% confidence interval (CIs). P<0.05 was considered to indicate a statistically significant difference.
Results
Primary bile acids upregulate Egr-1 expression and oncogenes by modulating p42/44 MAPK signaling in human gastric cancer cells
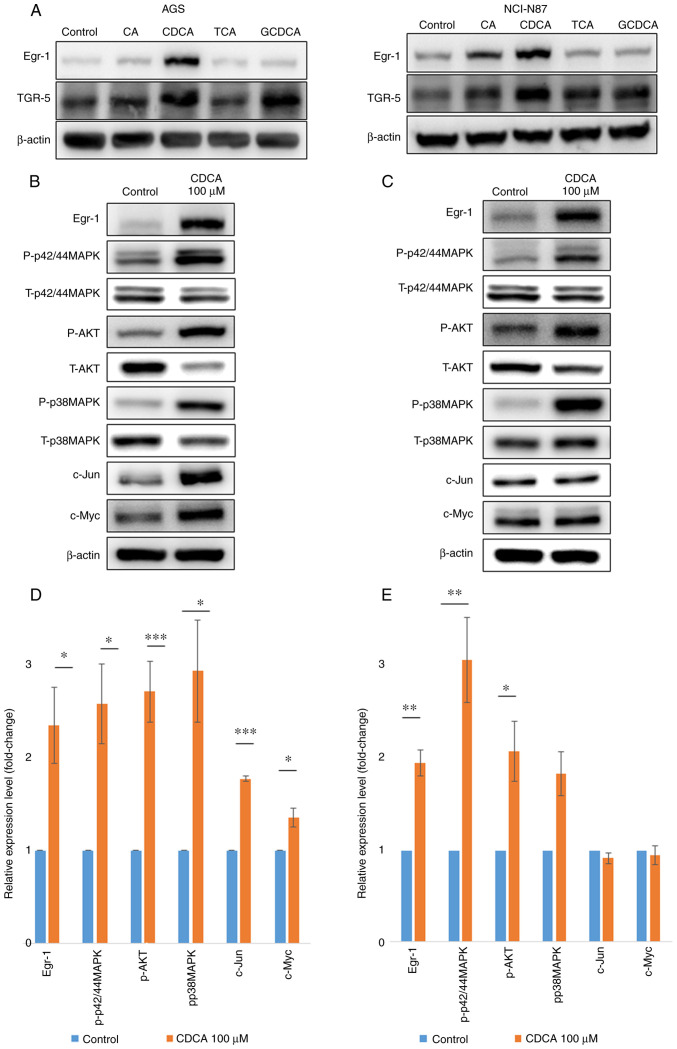
To investigate the effects of bile acids on the expression of the bile acid receptor TGR5 and the transcription factor Egr-1, human gastric cancer cells (AGS and NCI-N87) were treated for 48 h with 100-µM solutions of several unconjugated and conjugated primary bile acids, and the expression levels of TGR5 and Egr-1 were determined by western blotting. In AGS cells, Egr-1 expression was increased by both unconjugated and conjugated primary bile acids, but in NCI-N87 cells, it was increased only by unconjugated primary bile acids (CA and CDCA; Fig. 1A). In contrast, bile acids did not induce TGR5 overexpression in AGS or NCI-N87 cells. Of all the bile acids, CDCA was associated with the most significant increase in Egr-1 expression in both AGS and NCI-N87 cells. Therefore, CDCA was selected for further experiments, as presented in Figs. 1 and 2.
Figure 1.
Upregulation of Egr-1 and oncogenes by primary bile acids. (A) Egr-1 and TGR-5 protein expression levels were measured in AGS and NCI-N87 cells using western blotting. CA and CDCA increased the Egr-1 expression but did not affect the TGR-5 expression in human gastric cancer cells. P-p42/44MAPK, AKT and p38MAPK protein expression levels were measured in (B) AGS and (C) NCI-N87 cells using western blotting. P-p42/44MAPK, AKT and p38MAPK protein expression levels were measured in (D) AGS and (E) NCI-N87 cells using quantitative reverse-transcription PCR. CDCA significantly increased (>2-fold) the phosphorylation of p42/44MAPK, AKT and p38MAPK in AGS and NCI-N87 cells. Fold change in protein expression was calculated by dividing the normalized signal intensity of the target band in the experimental sample by that of the target band in the control sample. *P<0.05, **P<0.01, and ***P<0.001. Egr-1, early growth response factor 1; TGR-5, G-protein coupled bile acid receptor 1; CA, cholic acid; CDCA, chenodeoxycholic acid; p-, phosphorylated; t-, total; TCA, taurocholic acid; GCDCA, glycochenodeoxycholic acid.
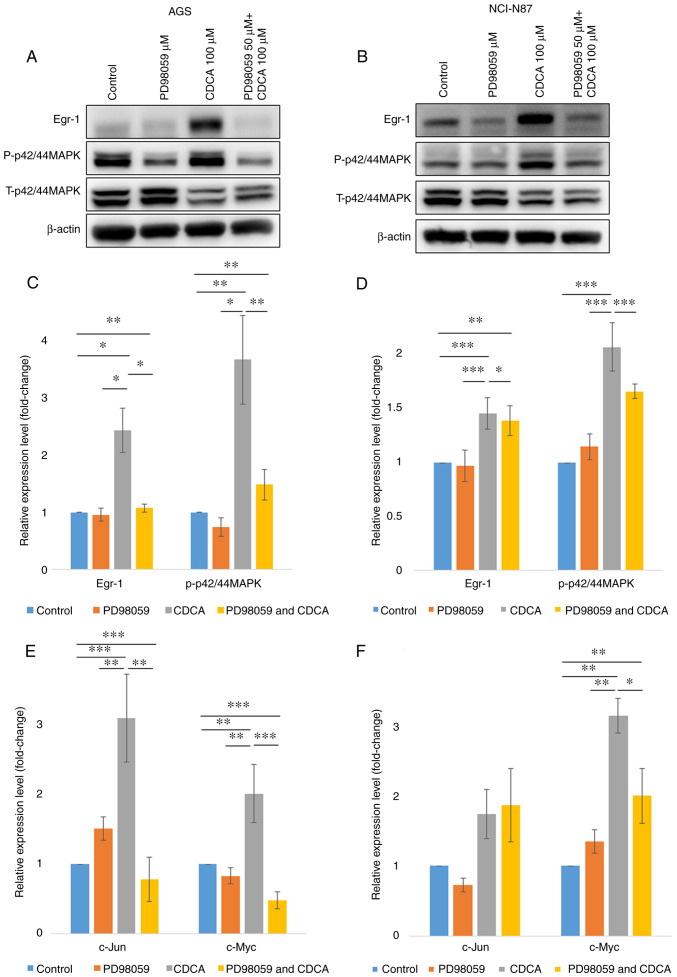
Figure 2.
Primary bile acid increases the expression levels of Egr-1 and oncogenes in human gastric cancer cells by modulating p42/44 MAPK signaling in human gastric cancer cells. To determine the signaling pathway by which CDCA induced the expression of oncogenes, 50 µM of p42/44 MAPK (PD98509) inhibitors were used. The expression level of Egr-1 was decreased by p42/44 MAPK inhibitors in (A) AGS and (B) NCI-N87. Egr-1 and p-p42/44MAPK protein expression levels were measured in (C) AGS and (D) NCI-N87 cells using western blotting. The expression of oncogenes were measured in (E) AGS and (F) NCI-N87 using quantitative reverse-transcription PCR. *P<0.05, **P<0.01 and ***P<0.001. Egr-1, early growth response factor 1; CDCA, chenodeoxycholic acid; p-, phosphorylated; t-, total.
To investigate the effect of bile acids on the AKT-MAPK signaling activation, AGS and NCI-N87 were treated with 100 µM of CDCA for 48 h. Treatment with CDCA stimulated phosphorylation of p42/44 MAPK, AKT, and p38 MAPK in both gastric cancer cell lines. We also identified the upregulation of c-Jun and c-Myc in AGS with 100 µM of CDCA for 48 h (Fig. 1B-E).
To determine the signaling pathway by which CDCA induced Egr-1 expression and upregulation of c-Jun and c-Myc, signaling inhibitors of p42/44 MAPK (PD98509) were used. As shown in Fig. 2, the expression of Egr-1 was decreased by inhibitors of p42/44 MAPK, while the inhibitor of p38 MAPK did not affect the expression of Egr-1 (data not shown). These results suggest that the CDCA-induced upregulation of Egr-1 was mediated through the p42/44MAPK signaling pathway. Likewise, p42/44 MAPK inhibitors in AGS cells downregulated CDCA-induced expression of c-Jun and c-Myc (all P<0.05). In NCI-N87 cells, the expression of c-Myc was decreased by p42/44 MAPK inhibitors (P<0.05, Fig. 2E and F).
Bile acids in gastric fluid
Baseline characteristics and measurement of bile acids of subjects
A total of 39 subjects were enrolled in this study. The purposes of endoscopic procedures were as follows: endoscopic surveillance or evaluation of dyspepsia in 20 controls and endoscopic resection, such as endoscopic mucosal resection or endoscopic submucosal dissection for EGC in 19 patients. The median age was 65 years (range, 24–85), with 26 males. H. pylori infection was in 43.6% (17/39) patients. There was a significant difference in age and histologic background of underlying gastric mucosa (presence of atrophic gastritis with intestinal metaplasia) among controls and patients with EGC (both P<0.05). However, there were no difference in gender, hypertension, diabetes and H. pylori status between 2 groups (Table SII).
We measured the concentration of bile acids from the gastric fluid. The levels of conjugated primary bile acids were higher than those of unconjugated primary bile acids (Fig. S1). The TCA level (median 1.71 µg/ml, IQR 0.00 µg/ml ~17.70 µg/ml) was higher than that of CA (median 0.02 µg/ml, IQR 0.0 µg/ml ~0.17 µg/ml, P<0.001), and the GCDCA level (median 2.36 µg/ml, IQR 0.00 µg/ml ~47.96 µg/ml) was higher than that of CDCA (median 0.00 µg/ml, IQR 0.00 µg/ml ~0.22 µg/ml, P<0.001, Fig. S1). The levels of secondary bile acids were lower than those of primary bile acids (P=0.001 for CA and DCA, P<0.001 for CDCA and LCA). The CA level correlated with CDCA level (rho=0.831, P<0.001) and the TCA level correlated with GCDCA level (rho=0.967, P<0.001).
Difference in bile acids from the gastric fluid in controls and patients with early gastric cancer
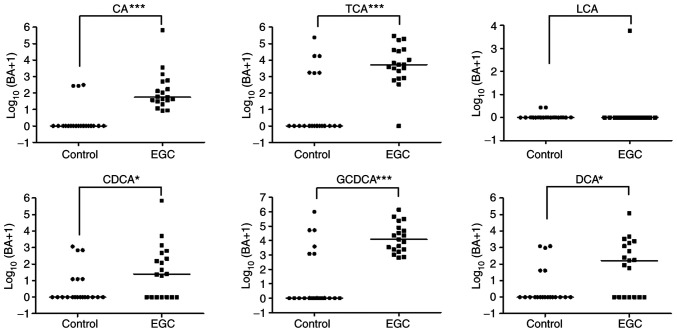
There were significant differences in CA, TCA, CDCA, GCDCA, and DCA levels between controls and patients with EGC (all P<0.05, Fig. 3). After adjustment for age and background histology (presence of atrophic gastritis with intestinal metaplasia), the levels of CA (aOR 4.3, 95% CI: 1.2~16.2, P=0.029) and GCDCA (aOR 9.9, 95% CI: 1.3~75.3, P=0.027) were significantly higher in patients with EGC than in controls.
Figure 3.
Gastric bile acids concentration in controls and patients with EGC. Concentrations of CA, TCA, CDCA, GCDCA and DCA in controls and patients with EGC measured using liquid chromatography with tandem mass spectrometry. *P<0.05 and ***P<0.001. BA, bile acids; CA, cholic acid; TCA, taurocholic acid; LCA, lithocholic acid; CDCA, chenodeoxycholic acid; GCDCA, glycochenodeoxycholic acid; DCA, deoxycholic acid; LGD, low grade dysplasia; EGC, early gastric cancer.
Discussion
In this study, we found that bile acid induced the upregulation of Egr-1 and oncogenes through MAPK signaling in gastric cancer cells. We also identified the presence of several primary and secondary bile acids in human gastric fluid, with gastric levels of primary bile acids (both conjugated and unconjugated) being higher in patients with EGC than in controls.
Bile acid has been known to be associated with the pathomechanism of gastric carcinogenesis. Several studies suggested the involvement of bile acid receptors in gastric carcinogenesis. Cao et al showed that TGR5 was overexpressed in gastric intestinal-type adenocarcinomas and was associated with poor prognosis in gastric cancer patients (5,12). Yu et al suggested that FXR was associated with Caudal type homeobox 2 (CDX2) and Mucin 2 (MUC2) expression, leading to gastric intestinal metaplasia (7). We identified the expression of TGR5 in gastric cancer cells in vitro. However, bile acids did not promote the expression of TGR5 in gastric cancer cell lines. Instead, we identified the overexpression of Egr-1 by the bile acid in gastric cancer cells. Until now, there is little information about the involvement of Egr-1 induced by bile acids in gastric carcinogenesis. A previous study demonstrated that Egr-1 was overexpressed in precancerous lesions of the stomach (13). Egr-1 has been implicated in biological processes, including inflammation, cell proliferation, cell differentiation, and cancer progression (8). In our study, the upregulation of Egr-1 in gastric cancer cells in vitro by bile acids require MAPK signaling, not the activation of TGR5. Allen et al also demonstrated that the activation of MAPK signaling is required for the upregulation of Egr-1 by bile acids in cholestasis liver injury models (14). This study also showed that primary bile acids increased the expression levels of the c-Myc and c-Jun genes through MAPK signaling in gastric cancer cell lines, which were involved in the initiation and development of gastric cancer (15,16). Our results suggest that continuous exposure of gastric epithelial cells to primary bile acids may be a factor in gastric carcinogenesis. Future research are needed to know the roles of bile acids in other gastric cancer cell lines with variable characteristics.
Recent studies used the concentration of total bile acids or each bile acid component to evaluate bile reflux status (3,17,18). We measured the levels of variable bile acids in the stomach. The levels of primary bile acids were higher than those of secondary bile acids. Among primary bile acids, conjugated bile acids were more abundant than unconjugated bile acids, consistent with a recent study (17). In addition, the levels of bile acids were correlated to each other.
Several studies reported that bile reflux was associated with reflux esophagitis, the proliferation of esophageal squamous cells, Barrett's adenocarcinoma, and intestinal metaplasia in the cardia (19,20). Moreover, recent studies suggested the association between bile acids and the risk of precancerous gastric lesions such as atrophic gastritis and intestinal metaplasia (3,7,21). Matsuhisa et al demonstrated that the total bile acid concentration was correlated with the grade of glandular atrophy and intestinal metaplasia of the stomach (3,22). Li et al showed that endoscopic bile reflux grading in patients with chronic gastritis and precancerous lesions was lower than that in patients with gastric cancer (4). Xu et al showed that DCA-induced macrophage-derived exosomes increased the expression of spasmolytic polypeptide expressing metaplasia markers in gastric organoids leading to intestinal metaplasia of gastric mucosa (23,24). In our study, the levels of primary bile acid (conjugated and unconjugated) in the gastric fluid were still higher in patients with EGC than in controls after adjustment of age and background gastric mucosa status. Previous studies also demonstrated that the levels of total bile acids in the gastric fluid was higher in patients with precancerous lesion such as intestinal metaplasia (3,22). Suggesting that a high concentration of bile acid may be involved in early steps of gastric carcinogenesis.
In conclusion, bile acids activated Egr-1 expression in gastric cancer cells through the MAPK signaling pathway, and higher gastric concentrations of primary bile acids were associated with EGC. These findings suggest that exposure of gastric cells to primary bile acids may play a role in gastric carcinogenesis.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
This research was supported by The National Research Foundation of Korea (NRF) grant funded by the Korea government (grant nos. 2018R1C1B5043483 and 2020R1I1A1A01068428), The Chonnam National University Hospital Research Institute of Clinical Medicine (grant nos. CRI 18018-1 and BCRI19258) and Korean College of Helicobacter and Upper Gastrointestinal Research Foundation Grant.
Availability of data and materials
The datasets used and/or analyzed in the current study are available from the corresponding author on reasonable request.
Authors' contributions
All authors have read and approved this manuscript. SYP conceptualized, designed and supervised the study, analyzed and interpreted data and drafted and finalized the manuscript. SYP and SML confirm the authenticity of all the raw data. SML and MSP conducted the study, collected and interpreted data and drafted the manuscript. YDC analyzed and reviewed the pathological findings. JOC analyzed and interpreted the collected clinical data and drafted and reviewed the manuscript. YDJ analyzed and interpreted the experimental data. DHK and HSK analyzed electronic medical records about clinical data including demographic factors, endoscopic findings from enrolled patients.
Ethics approval and consent to participate
This study was approved by our institutional review board (approval no. CNUH-2020-085). The terms of participation in this study were explained and written informed consent was obtained from patients before endoscopic procedures.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, Znaor A, Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 2.Di Ciaula A, Wang DQ, Molina-Molina E, Lunardi Baccetto R, Calamita G, Palmieri VO, Portincasa P. Bile acids and cancer: Direct and environmental-dependent effects. Ann Hepatol. 2017;16((Suppl 1: s3-e105)):S87–S105. doi: 10.5604/01.3001.0010.5501. [DOI] [PubMed] [Google Scholar]
- 3.Matsuhisa T, Arakawa T, Watanabe T, Tokutomi T, Sakurai K, Okamura S, Chono S, Kamada T, Sugiyama A, Fujimura Y, et al. Relation between bile acid reflux into the stomach and the risk of atrophic gastritis and intestinal metaplasia: A multicenter study of 2283 cases. Dig Endosc. 2013;25:519–525. doi: 10.1111/den.12030. [DOI] [PubMed] [Google Scholar]
- 4.Li D, Zhang J, Yao WZ, Zhang DL, Feng CC, He Q, Lv HH, Cao YP, Wang J, Qi Y, et al. The relationship between gastric cancer, its precancerous lesions and bile reflux: A retrospective study. J Dig Dis. 2020;21:222–229. doi: 10.1111/1751-2980.12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao W, Tian W, Hong J, Li D, Tavares R, Noble L, Moss SF, Resnick MB. Expression of bile acid receptor TGR5 in gastric adenocarcinoma. Am J Physiol Gastrointest Liver Physiol. 2013;304:G322–G327. doi: 10.1152/ajpgi.00263.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong J, Behar J, Wands J, Resnick M, Wang LJ, DeLellis RA, Lambeth D, Souza RF, Spechler SJ, Cao W. Role of a novel bile acid receptor TGR5 in the development of oesophageal adenocarcinoma. Gut. 2010;59:170–180. doi: 10.1136/gut.2009.188375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu JH, Zheng JB, Qi J, Yang K, Wu YH, Wang K, Wang CB, Sun XJ. Bile acids promote gastric intestinal metaplasia by upregulating CDX2 and MUC2 expression via the FXR/NF-κB signalling pathway. Int J Oncol. 2019;54:879–892. doi: 10.3892/ijo.2019.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thiel G, Cibelli G. Regulation of life and death by the zinc finger transcription factor Egr-1. J Cell Physiol. 2002;193:287–292. doi: 10.1002/jcp.10178. [DOI] [PubMed] [Google Scholar]
- 9.Wang B, Guo H, Yu H, Chen Y, Xu H, Zhao G. The role of the transcription factor EGR1 in cancer. Front Oncol. 2021;11:642547. doi: 10.3389/fonc.2021.642547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steele NG, Chakrabarti J, Wang J, Biesiada J, Holokai L, Chang J, Nowacki LM, Hawkins J, Mahe M, Sundaram N, et al. An organoid-based preclinical model of human gastric cancer. Cell Mol Gastroenterol Hepatol. 2019;7:161–184. doi: 10.1016/j.jcmgh.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlemper RJ, Riddell RH, Kato Y, Borchard F, Cooper HS, Dawsey SM, Dixon MF, Fenoglio-Preiser CM, Fléjou JF, Geboes K, et al. The Vienna classification of gastrointestinal epithelial neoplasia. Gut. 2000;47:251–255. doi: 10.1136/gut.47.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carino A, Graziosi L, D'Amore C, Cipriani S, Marchianò S, Marino E, Zampella A, Rende M, Mosci P, Distrutti E, et al. The bile acid receptor GPBAR1 (TGR5) is expressed in human gastric cancers and promotes epithelial-mesenchymal transition in gastric cancer cell lines. Oncotarget. 2016;7:61021–61035. doi: 10.18632/oncotarget.10477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park SY, Kim JY, Lee SM, Chung JO, Lee KH, Jun CH, Park CH, Kim HS, Choi SK, Rew JS, et al. Expression of early growth response gene-1 in precancerous lesions of gastric cancer. Oncol Lett. 2016;12:2710–2715. doi: 10.3892/ol.2016.4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allen K, Kim ND, Moon JO, Copple BL. Upregulation of early growth response factor-1 by bile acids requires mitogen-activated protein kinase signaling. Toxicol Appl Pharmacol. 2010;243:63–67. doi: 10.1016/j.taap.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shibata W, Maeda S, Hikiba Y, Yanai A, Sakamoto K, Nakagawa H, Ogura K, Karin M, Omata M. c-Jun NH2-terminal kinase 1 is a critical regulator for the development of gastric cancer in mice. Cancer Res. 2008;68:5031–5039. doi: 10.1158/0008-5472.CAN-07-6332. [DOI] [PubMed] [Google Scholar]
- 16.Chen R, Masuo K, Yogo A, Yokoyama S, Sugiyama A, Seno H, Yoshizawa A, Takaishi S. SNAIL regulates gastric carcinogenesis through CCN3 and NEFL. Carcinogenesis. 2021;42:190–201. doi: 10.1093/carcin/bgaa133. [DOI] [PubMed] [Google Scholar]
- 17.Zhao A, Wang S, Chen W, Zheng X, Huang F, Han X, Ge K, Rajani C, Huang Y, Yu H, et al. Increased levels of conjugated bile acids are associated with human bile reflux gastritis. Sci Rep. 2020;10:11601. doi: 10.1038/s41598-020-68393-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee W, Um J, Hwang B, Lee YC, Chung BC, Hong J. Assessing the progression of gastric cancer via profiling of histamine, histidine, and bile acids in gastric juice using LC-MS/MS. J Steroid Biochem Mol Biol. 2020;197:105539. doi: 10.1016/j.jsbmb.2019.105539. [DOI] [PubMed] [Google Scholar]
- 19.Goldman A, Shahidullah M, Goldman D, Khailova L, Watts G, Delamere N, Dvorak K. A novel mechanism of acid and bile acid-induced DNA damage involving Na+/H+ exchanger: Implication for Barrett's oesophagus. Gut. 2010;59:1606–1616. doi: 10.1136/gut.2010.213686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Souza RF. The role of acid and bile reflux in oesophagitis and Barrett's metaplasia. Biochem Soc Trans. 2010;38:348–352. doi: 10.1042/BST0380348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyun JJ, Yeom SK, Shim E, Cha J, Choi I, Lee SH, Chung HH, Cha SH, Lee CH. Correlation between bile reflux gastritis and biliary excreted contrast media in the stomach. J Comput Assist Tomogr. 2017;41:696–701. doi: 10.1097/RCT.0000000000000585. [DOI] [PubMed] [Google Scholar]
- 22.Matsuhisa T, Tsukui T. Relation between reflux of bile acids into the stomach and gastric mucosal atrophy, intestinal metaplasia in biopsy specimens. J Clin Biochem Nutr. 2012;50:217–221. doi: 10.3164/jcbn.11-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu X, Cheng J, Luo S, Gong X, Huang D, Xu J, Qian Y, Wan X, Zhou H. Deoxycholic acid-stimulated macrophage-derived exosomes promote spasmolytic polypeptide-expressing metaplasia in the stomach. Biochem Biophys Res Commun. 2020;524:649–655. doi: 10.1016/j.bbrc.2020.01.159. [DOI] [PubMed] [Google Scholar]
- 24.Weis VG, Sousa JF, LaFleur BJ, Nam KT, Weis JA, Finke PE, Ameen NA, Fox JG, Goldenring JR. Heterogeneity in mouse spasmolytic polypeptide-expressing metaplasia lineages identifies markers of metaplastic progression. Gut. 2013;62:1270–1279. doi: 10.1136/gutjnl-2012-302401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed in the current study are available from the corresponding author on reasonable request.