Abstract
Mutations of rpoB associated with rifampin resistance were studied in 37 multidrug-resistant (MDR) clinical strains of Mycobacterium tuberculosis isolated in Italy. At least one mutated codon was found in each MDR strain. It was always a single-base substitution leading to an amino acid change. Nine different rpoB alleles, three of which had not been reported before, were found. The relative frequencies of specific mutations in this sample were different from those previously reported from different geographical areas, since 22 strains (59.5%) carried the mutated codon TTG in position 531 (Ser→Leu) and 11 (29.7%) had GAC in position 526 (His→Asp).
The emergence of multidrug-resistant (MDR) strains of Mycobacterium tuberculosis poses a serious problem in tuberculosis control and stresses the need for the development of rapid and reliable diagnostic methods for drug susceptibility testing in clinical isolates. Recent advances in the understanding of the genetic basis of drug resistance have allowed for the development of DNA-based methods for the detection of resistance to antituberculosis drugs (for a review, see Musser [10]). These methods can be used, in conjunction with molecular typing (18), to elucidate the molecular epidemiology of drug resistance in M. tuberculosis. Epidemiological data are essential for the development of diagnostic strategies and, by providing information on the geographical distribution of resistant alleles, can help us understand whether mutated alleles arise independently or are attributable, in certain instances, to the spread of a genotype (6, 15). Mutations in rpoB, the gene coding for the β subunit of the RNA polymerase, are responsible for resistance to rifampin (RFM) (10), a fundamental drug for the therapy of tuberculosis (1). In this work we analyzed the mutations occurring in the rpoB gene of MDR strains of M. tuberculosis isolated in Italy.
RFM-resistant strains.
MDR isolates of M. tuberculosis from hospitals in northern and central Italy were collected from three clinical microbiology laboratories (Ospedale Umberto I, Ancona; Ospedale di Careggi, Florence; and Ospedale Forlanini, Rome). The 37 MDR strains that formed the object of this investigation (Table 1) were from different patients and were typed by DNA fingerprinting (IS6110) (17) in order to avoid the inclusion of identical strains responsible for outbreaks (data not shown). The MICs for RFM and rifabutin (RFB) were determined in 7H11 agar (4). Strains were considered resistant to RFM when MICs were >1 μg/ml and resistant to RFB when MICs were >0.5 μg/ml. All MDR strains were resistant to both RFM and RFB.
TABLE 1.
Resistance patterns of MDR M. tuberculosis strains isolated in Italya
| No. of strainsb | RFM | INH | STR | AMI | CIP | PZA | EMB | ETH |
|---|---|---|---|---|---|---|---|---|
| 9 | R | R | ||||||
| 5 | R | R | R | |||||
| 5 | R | R | R | R | ||||
| 4 | R | R | R | R | ||||
| 3 | R | R | R | R | ||||
| 2 | R | R | R | R | R | |||
| 2 | R | R | R | R | R | |||
| 1 | R | R | R | R | R | |||
| 1 | R | R | R | R | R | R | ||
| 1 | R | R | R | R | R | R | R | |
| 1 | R | R | R | R | R | |||
| 1 | R | R | R | R | R | |||
| 1 | R | R | R | R | R | |||
| 1 | R | R | R | R | ||||
| Total | 37 | 37 | 28 | 11 | 11 | 9 | 4 | 2 |
INH, isoniazid; STR, streptomycin; AMI, amikacin; CIP, ciprofloxacin; PZA, pyrazinamide; EMB, ethambutol; ETH, ethionamide; R, resistant.
n = 37.
Sequencing of rpoB.
To investigate the mutations associated to RFM resistance, a segment of the rpoB gene was sequenced. Genomic DNA from heat-inactivated suspensions of M. tuberculosis cells was obtained with the QIAamp Tissue kit (Qiagen). A 318-bp fragment of rpoB, from nucleotide 1807 to nucleotide 2124 (GenBank accession no. U12205), was amplified by PCR and sequenced by a nonradioactive manual method (Silver Sequence DNA sequencing system; Promega). The same primers (5′-CGA TCA CAC CGC AGA CGT TG-3′ and 5′-GGT ACG GCG TTT CGA TGA AC-3′) were used for PCR and DNA sequencing. Both DNA strands were sequenced, each using as a template the product of a different PCR. At least one mutated codon was found in each RFM-resistant strain; it was always a single-base substitution leading to an amino acid change. Nine different rpoB alleles were found, five with one mutation (33 strains [89.1%]), three with three mutations (3 strains [8.1%]), and one with two mutations (1 strain [2.7%]) (Table 2). The mutated allele carrying the TTG codon in position 531 was the most common (56.7%), together with the one carrying GAC in position 526 (24.3%). Three new alleles, those with three mutated codons, were recognized in this investigation (Table 2). Mutations already reported in the literature were found in codons 511, 512, 516, 526, and 531. In two strains we also found four new mutations: in codons 523 (TGG) and 525 (ATC) in one strain (GenBank accession no. AF055891) and in codons 541 (GAT) and 553 (CGC) in the other (GenBank accession no. AF055892). Twenty-two strains (59.5%) carried the mutated codon TTG in position 531 (Ser→Leu), and 11 strains (29.7%) had GAC in position 526 (His→Asp). A total of 34 of 37 strains (91.9%) had mutations in position 531 and/or position 526.
TABLE 2.
Relative frequencies of mutated rpoB alleles in Italian MDR isolates of M. tuberculosis
| Allelea | Amino acid change(s) | No. of strains (%) | MICb (μg/ml) of:
|
|
|---|---|---|---|---|
| RFM | RFB | |||
| TCG531TTG | Ser→Leu | 21 (56.7) | ≥128 (32–≥128) | ≥32 (4–≥32) |
| CAC526GAC | His→Asp | 9 (24.3) | ≥128 (16–≥128) | ≥32 (≥32) |
| CAC526TAC | His→Tyr | 1 (2.7) | ≥128 | 32 |
| CAC526CGC | His→Arg | 1 (2.7) | ≥128 | 32 |
| GAC516GTC | Asp→Val | 1 (2.7) | 64 | 1 |
| CAC526GAC TCG531TTG | His→Asp Ser→Leu | 1 (2.7) | ≥128 | 32 |
| CTG511CCG AGC512ACC GAC516GTCc | Leu→Pro Ser→Thr Asp→Val | 1 (2.7) | ≥128 | 32 |
| GAC516TAC GGG523TGG ACC525ATCd | Thr→Ile Gly→Trp Asp→Tyr | 1 (2.7) | ≥128 | 8 |
| CAC526GAC GAG541GAT TCG553GCGe | His→Asp Glu→Gly Ser→Ala | 1 (2.7) | ≥128 | 32 |
Codons are numbered according to the rpoB gene of Escherichia coli (14). The new base in each mutated codon is underlined.
Where there is more than one strain, the MIC is expressed as the mode and the range of MICs is given in parentheses.
New allele; GenBank accession no. AF055893.
New allele; GenBank accession no. AF055891.
New allele; GenBank accession no. AF055892.
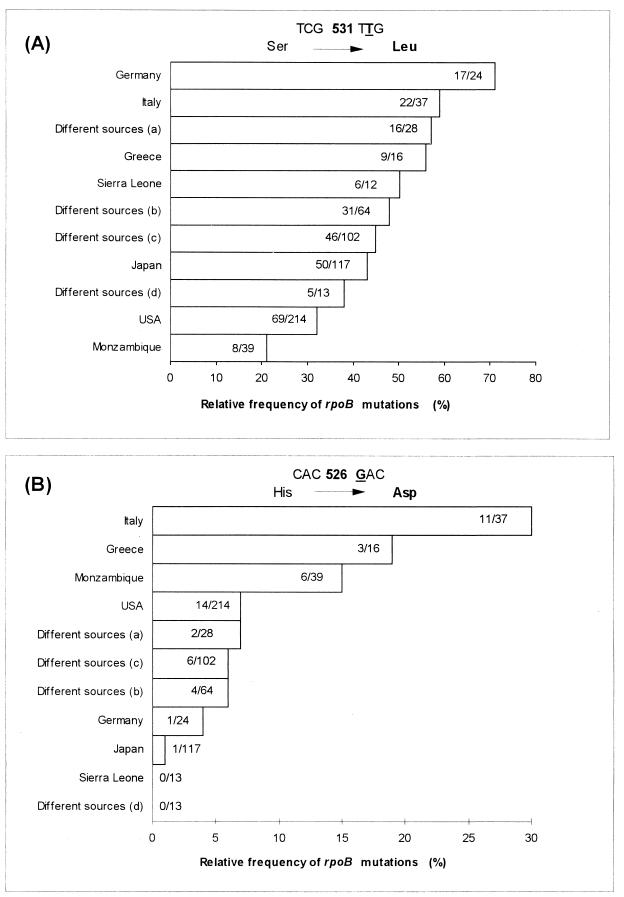
In this study on MDR strains of M. tuberculosis from northern and central Italy, RFM resistance was always found associated with RFB resistance, and in each strain we could detect the presence of a mutated rpoB allele. By comparing our data with the literature (Fig. 1), we observed that the two mutations occurring most frequently in Italian strains (531TTG and 526GAC) have different relative frequencies in strains from different countries. While 531TTG is absolutely the most common mutation worldwide, 526GAC appears to be prevalent in only a few countries, including Italy, Greece, and Mozambique. Among the possible explanations, transmission of MDR strains among patients could account for the disequilibrium in the geographical distributions of rpoB mutations. Of course, genetic-exchange mechanisms responsible for the spread of resistant alleles in M. tuberculosis cannot be completely ruled out, even if, due to the current understanding of mycobacterial genetics, they would be more difficult to postulate.
FIG. 1.
Comparison of the relative frequencies of rpoB mutations in RFM-resistant M. tuberculosis strains from different countries. The mutated codons TTG in position 531 (A) and GAC in position 526 (B) represent the two most frequent mutations in Italian strains. Data reported here are from the present study (for Italy) and from the literature for Germany (15), Greece (7), Sierra Leone (15), Japan (12, 13, 16), the United States (3, 6, 8, 11), Mozambique (2), and “different sources” a (9), b (17), c (19), and d (4). The number of strains in which the mutation was found/total number of strains studied is given in each bar.
Nucleotide sequence accession numbers.
The new alleles found in this study have been deposited in GenBank under accession no. AF055891, AF055892, and AF055893.
Acknowledgments
We thank P. Chiaradonna, C. Piersimoni, E. Tortoli, and M. Tronci for providing clinical strains and valuable advice.
This study was supported by grants from the Istituto Superiore di Sanità (Progetto Nazionale Tubercolosi, no. 96/D/T55).
REFERENCES
- 1.Bass J B, Jr, Farer L S, Hopewell P C, O’Brien R, Jacobs R F, Ruben F, Snider D E, Jr, Thornton G., Jr Treatment of tuberculosis and tuberculosis infection in adults and children. American Thoracic Society and The Centers for Disease Control and Prevention. Am J Respir Crit Care Med. 1994;149:1359–1374. doi: 10.1164/ajrccm.149.5.8173779. [DOI] [PubMed] [Google Scholar]
- 2.Caugant D A, Sandven P, Eng J, Jeque J T, Tonium T. Detection of rifampin resistance among isolates of Mycobacterium tuberculosis from Mozambique. Microb Drug Resist. 1995;1:321–326. doi: 10.1089/mdr.1995.1.321. [DOI] [PubMed] [Google Scholar]
- 3.Cooksey R C, Morlock G P, Glickman S, Crawford J T. Evaluation of a line probe assay kit for characterization of rpoB mutations in rifampin-resistant Mycobacterium tuberculosis isolates from New York City. J Clin Microbiol. 1997;35:1281–1283. doi: 10.1128/jcm.35.5.1281-1283.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heifets L B. Antituberculosis drugs: antimicrobial activity in vitro. In: Heifets L B, editor. Drug susceptibility in the chemotherapy of mycobacterial infections. Boca Raton, Fla: CRC Press Inc.; 1991. pp. 14–49. [Google Scholar]
- 5.Heym B, Honore N, Pernot-Truffot C, Banerjee A, Schurra C, Jacobs W R, Jr, van Embden J D A, Grosset J H, Cole S T. Implications of multidrug resistance for the future of short-course chemotherapy of tuberculosis: a molecular study. Lancet. 1998;344:293–298. doi: 10.1016/s0140-6736(94)91338-2. [DOI] [PubMed] [Google Scholar]
- 6.Kapur V, Li L-L, Iordanescu S, Hamrick M R, Wanger A, Kreiswirth B N, Musser J M. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase b subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J Clin Microbiol. 1994;32:1095–1098. doi: 10.1128/jcm.32.4.1095-1098.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsiota-Bernard P, Vrioni G, Marinis E. Characterization of rpoB mutations in rifampin-resistant clinical Mycobacterium tuberculosis isolates from Greece. J Clin Microbiol. 1998;36:20–23. doi: 10.1128/jcm.36.1.20-23.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moghazeh S L, Pan X, Arain T, Stover C K, Musser J M, Kreiswirth B N. Comparative antimycobacterial activities of rifampin, rifampentine, and KRM-1648 against a collection of rifampin-resistant Mycobacterium tuberculosis isolates with known rpoB mutations. Antimicrob Agents Chemother. 1996;40:2655–2657. doi: 10.1128/aac.40.11.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morris S, Bai G H, Suffys P, Gomez-Portillo L, Fairchok M, Rouse D. Molecular mechanisms of multiple drug resistance in clinical isolates of Mycobacterium tuberculosis. J Infect Dis. 1995;171:954–960. doi: 10.1093/infdis/171.4.954. [DOI] [PubMed] [Google Scholar]
- 10.Musser J M. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin Microbiol Rev. 1995;8:496–514. doi: 10.1128/cmr.8.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nachamkin I, Kang C, Weinstein M P. Detection of resistance to isoniazid, rifampin, and streptomycin in clinical isolates of Mycobacterium tuberculosis by molecular methods. Clin Infect Dis. 1997;24:894–900. doi: 10.1093/clinids/24.5.894. [DOI] [PubMed] [Google Scholar]
- 12.Ohno H, Koga H, Kohno S, Tashiro T, Hara K. Relationship between rifampin MICs for rpoB mutations of Mycobacterium tuberculosis strains isolated in Japan. Antimicrob Agents Chemother. 1996;40:1053–1056. doi: 10.1128/aac.40.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohno H, Koga H, Kuroita T, Tomono K, Ogawa K, Yanagihara K, Yamamoto Y, Miyamoto J, Tashiro T, Kohno S. Rapid prediction of rifampin susceptibility of Mycobacterium tuberculosis. Am J Respir Crit Care Med. 1997;155:2057–2063. doi: 10.1164/ajrccm.155.6.9196115. [DOI] [PubMed] [Google Scholar]
- 14.Ovchinnikov Y A, Monastyrskaya G S, Gubanov V V, Guryev S O, Chertov O Y, Modyanov N N, Grinkevich V A, Makarova I A, Marchenko T V, Polovnikova I N, et al. The primary structure of Escherichia coli RNA polymerase. Nucleotide sequence of the rpoB gene and amino-acid sequence of the beta-subunit. Eur J Biochem. 1981;116:621–629. doi: 10.1111/j.1432-1033.1981.tb05381.x. [DOI] [PubMed] [Google Scholar]
- 15.Rinder H, Dobner P, Feldmann K, Rifai M, Bretzel G, Rusch-Gerdes S, Loscher T. Disequilibria in the distribution of rpoB alleles in rifampicin-resistant M. tuberculosis isolates from Germany and Sierra Leone. Microb Drug Resist. 1997;3:195–197. doi: 10.1089/mdr.1997.3.195. [DOI] [PubMed] [Google Scholar]
- 16.Taniguchi H, Aramaki H, Nikaido Y, Mizuguchi Y, Nakamura M, Koga T, Yoshida S-I. Rifampicin resistance and mutation of rpoB in Mycobacterium tuberculosis. FEMS Microbiol Lett. 1996;144:103–108. doi: 10.1111/j.1574-6968.1996.tb08515.x. [DOI] [PubMed] [Google Scholar]
- 17.Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston M J, Matter L, Schopfer K, Bodmer T. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 18.van Soolingen D, de Haas P E W, Hermans P W M, van Embden J D A. DNA fingerprinting of Mycobacterium tuberculosis. Methods Enzymol. 1994;235:196–205. doi: 10.1016/0076-6879(94)35141-4. [DOI] [PubMed] [Google Scholar]
- 19.Williams D L, Waguespack C, Eisenach K, Crawford J T, Portaels F, Salfinger M, Nolan C M, Abe C, Sticht-Groh V, Gillis T P. Characterization of rifampin resistance in pathogenic mycobacteria. Antimicrob Agents Chemother. 1994;38:2380–2386. doi: 10.1128/aac.38.10.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]