Abstract
The eukaryotic transcription is orchestrated from a chunk of the DNA region stated as the core promoter. Multifarious and punctilious core promoter signals, viz., TATA-box, Inr, BREs, and Pause Button, are associated with a subset of genes and regulate their spatiotemporal expression. However, the core promoter architecture linked with these signals has not been investigated exhaustively for several species. In this study, we attempted to envisage the adaptive binding landscape of the transcription initiation machinery as a function of DNA structure. To this end, we deployed a set of k-mer based DNA structural estimates and regular expression models derived from experiments, molecular dynamic simulations, and theoretical frameworks, and high-throughout promoter data sets retrieved from the eukaryotic promoter database. We categorized protein-coding gene core promoters based on characteristic motifs at precise locations and analyzed the B-DNA structural properties and non-B-DNA structural motifs for 15 different eukaryotic genomes. We observed that Inr, BREd, and no-motif classes display common patterns of DNA sequence and structural environment. TATA-containing, BREu, and Pause Button classes show a deviant behavior with the TATA class displaying varied axial and twisting flexibility while BREu and Pause Button leaned toward G-quadruplex motif enrichment. Intriguingly, DNA meltability and shape signals are conserved irrespective of the presence or absence of distinct core promoter motifs in the majority of species. Altogether, here we delineated the conserved DNA structural signals associated with several promoter classes that may contribute to the chromatin configuration, orchestration of transcription machinery, and DNA duplex melting during the transcription process.
1. Introduction
Gene expression is a quintessential process where genetic information is deciphered for phenotype creation, launched from the transcription stage. The transcription initiation in eukaryotes is an intricate process with several factors, viz., RNA polymerase, transcription factors, promoter regions, and chromatin configuration, modulating the mechanism. RNA polymerase II is greatly conserved and is the main catalytic enzyme in the preinitiation complex (PIC), which accounts for the transcription of all protein-coding genes.1,2 A set of general transcription factors (GTFs), namely, TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH, can carry out basal transcription. GTFs contribute to the promoter recognition, recruitment of polymerase, DNA unwinding, and transcription start site recognition.1 Experiments on DNA-protein cross-linking of human PIC showed that TBP, TFIIB, and TFIIF contact the promoter DNA at the upstream of the transcription start sites (TSS), while TFIIE overlaps with the TSS.2 Promoter regions are the genetic determinants, acting as cis regulators of transcription. In eukaryotes, promoters are broadly categorized into three kinds, namely, core, proximal, and distal regions.3 The core promoter region is a minimal segment of genomic DNA encompassing the TSS interplays with Pol-II and general transcription machinery for orchestration of the pre-initiation complex.4−7 Recent studies indicated that core promoters occupy the [−40 to +40] nucleotide regions relative to the TSS.8−10 They are marked by precisely positioned sequence motifs like TATA-box, Initiator element (Inr), TFIIB recognition element (BRE), DNA replication-related element (DRE), TCT motif (or polypyrimidine initiator), downstream promoter element (DPE), Pause Button, X core promoter element 1 (XCPE1), and motif-ten element (MTE).3,5−7,9,11,12 TATA-box is the best-characterized ancient core promoter element that binds to the TBP subunit of the TFIID multiprotein complex.13 It is observed in cellular organisms varying from archaea14 and yeast to metazoans and plants.12,15 The Initiator element (Inr) is regarded as the most frequent core promoter motif that is positioned across the TSS.7,9,16 Inr may interact with several GTFs with the highest correlation for TFIID binding.13 The two TFIIB recognition elements, BREu and BREd, are known to be present at upstream and downstream locations of TATA-boxes.7,17,18 These two elements may regulate basal transcription levels in conjunction with TATA-boxes.7,17 The TCT element or polypyrimidine Inr, another core promoter element, is observed at the TSS of Drosophila and human ribosomal protein genes.19−21 The DPE and MTE core promoter elements located downstream of TSS are conserved from Drosophila to humans, and they may act as TFIID binding sites.22−24 The Pause Button is another downstream promoter with a wider distribution around the downstream of TSS, which has been identified in the paused Pol II sites in Drosophila.25,26 The majority of the core promoter elements act as recognition sites for the GTFs. Proximal and distal promoters, which vary the rate of transcription, extend up to a few hundred to kilo-base pairs and contain multiple transcription factor binding sites, namely, enhancers, insulators, and silencers.27 The proximal and distal promoter elements like enhancers also can recruit RNAPII and may act as a regulatory checkpoint for RNAPII delivery to the core promoter.28 However, enhancers are linked to a single gene or a specific set of genes to precisely modulate spatiotemporal patterns of their expression.4,29 Of note, the promoter elements are noticed in only a subset of genes.6,11,12 The promoter elements also differ in the position, number, type, and combination of these cognate elements for a gene.4,30 Their distribution and conservation may be dependent on the type of organism. Thus, it is interesting to understand the architecture of promoter sequences underlying transcription initiation with the identity or absence of consensus elements. Furthermore, sequence examination of the promoter region alone furnishes limited information about its functionality. The events in the RNA polymerase II transcription initiation pathway include searching for a suitable promoter DNA, inducing bend to promoter DNA by TFIID or its TATA-box binding protein (TBP), opening of DNA, and subsequent transcription bubble formation,31 which cohere in the backdrop of the DNA’s three-dimensional configuration. Hence, it is crucial to investigate the promoter as DNA structural manifestations.
Genomes are regarded as continuous thermodynamic landscapes encoding three types of information, namely, genetic information, cis-regulatory code, and nucleosome positioning information. While genetic information can be attributed solely to the primary sequence, cis-regulatory and nucleosome positioning codes depend on DNA structural properties.32 Physiological DNA may exist in diverse structural configurations depending on the sequence context.33 It may completely polymorph from its double-helical structure to non-double-helical forms such as G-quadruplexes, intercalated motifs, cruciform DNA, triple helices, hairpins, and slipped structures.33,34 Non-B-DNA structures have been revealed to be key modulators of several physiological mechanisms.35−39 Within a double-helical form, DNA can conform to the B or Z forms. Furthermore, perturbations in rotational and translational dinucleotide parameters of B-DNA account for several DNA structural features such as intrinsic curvature, flexibility, duplex stability, and groove shape.33,40 Investigations on these signature features have been carried out to understand DNA transcription factor recognition,41−44 characterize origins of replication in eukaryotes,45−47 predict promoter regions in several lineages spanning from yeast to mammals,48−50 and reveal associations among the DNA structure and gene expression variability.33,51 In a recent study, we used DNA duplex stability, bendability, and curvature to delineate TATA-containing and TATA-less promoters in six eukaryotes.12 Another study also reported the bendability characteristics of TATA-less promoters.52 However, these studies did not report differences in several other classes of promoters and investigated a limited number of biologically relevant DNA structural features. In this study, we systematically investigate the links between DNA structural alterations and several eukaryotic promoter classes, focusing on DNA duplex stability (melting and mechanical), flexibility (axial, torsional, and stretching), DNA shape (propeller twist, minor groove width, and wedge), and different non-B DNA forming motifs corresponding to phased A-tracts, G-quadruplexes, cruciform DNA, H-DNA, slipped and hairpin DNA, and Z-DNA.
2. Results and Discussion
DNA displays several lines of polymorphism due to alterations in the backbone, intra-base pair parameters, inter-base pair parameters, and groove geometry to the global helical axis path. The variations can be non-B-DNA structures and B-DNA structural alterations, which together we call DNA sequence-dependent structural features. B-DNA alterations include DNA meltability, flexibility (bendability, twistability, and stretchability), and groove shape (minor groove width, propeller twist, and wedge), and non-B-DNA features include G-quadruplexes, cruciforms, triplex DNA, and hairpin DNA. In the current study, we have extensively examined the association of DNA structural alterations in 15 species as a function of core promoter categories. We presume that cis-regulatory structural alterations can act as proximal or local signatures. The results of structural analysis are presented in the manuscript as (i) a sequence analysis (GC composition, CpG island prevalence, and hexanucleotide motif computation for the −500 to +500 region); (ii) a non-B-DNA motif enrichment analysis for the −500 to +500 region; (iii) DNA shape signals in the vicinity of transcription start sites [−40 to +40 region]; (iv) DNA thermal and mechanical stability in the −100 to +50 region; and (v) three types of DNA flexibility—axial flexibility, torsional flexibility, and stretching stiffness—and an intrinsic curvature-related feature wedge in the −150 to −1 region.
2.1. Sequence Characteristics of Eukaryotic Promoter Classes
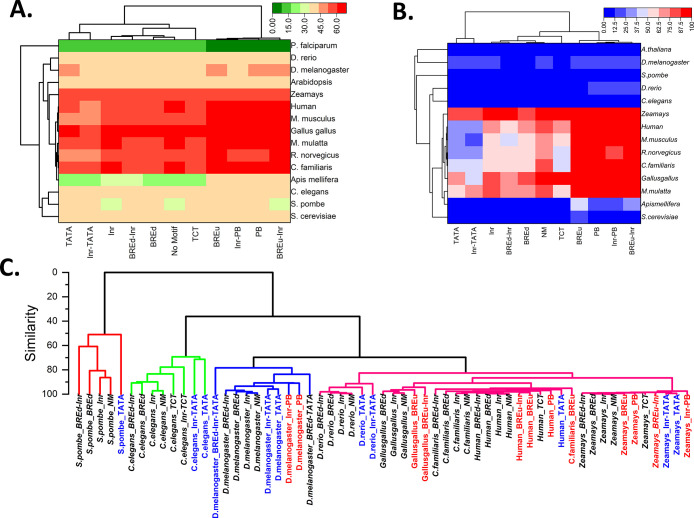
The eukaryotic core promoters are diversified in the context of sequence composition, and their functional activities are propelled by several sequence motifs.7 In this study, we sought to understand the sequence-dependent structural properties of eukaryotic promoter classes categorized based on the presence or absence of the cognate sequence motifs. Based on the literature survey10,11 and observed positional preference of promoter elements, we categorized them into 118 groups (Data Sets and Methods, Table S1). The percentages of notable promoter classes in 15 different eukaryotic species, viz., Saccharomyces cerevisiae,Schizosaccharomyces pombe,Plasmodium falciparum,Caenorhabditis. elegans,Apis mellifera,Drosophila melanogaster,Danio rerio,Gallus gallus,Canis familiaris,Rattus norvegicus,Mus musculus,Macaca mulatta,H. sapiens,Arabidopsis thaliana, and Zea mays are presented in Table 1. The Initiator element is the most prevalent in all species, and the result is in line with the previous literature.16 BREd and TATA-box classes represent the next most prevalent elements. BREu and TCT classes represent the next occurring classes. The remaining elements XCPE1, MTE, and DPE classes are underrepresented in all species. DRE is only relevant in fly species (Apis mellifera and Drosophila melanogaster). Interestingly, promoter sequences without any of these motifs represent the most prominent class. It is also observed that the percentages of promoters with two or more elements are less when compared to the total number of promoters in each promoter class category (Table S1). However, low-frequency elements such as TATA-Box, BREu, and Pause Button often exist along with Inr-containing promoter sequences. Hence, our categorization also included separate classes for a sequence constituting any two promoter elements in the above defined functional regions. Inr-TATA, BREu-Inr, BREd-Inr, and Inr-PB are observed to be notable classes with significant numbers in different species. Based on these data sets, we analyzed sequence composition as a function of GC percent distribution, CpG island occurrence, and k-mer word prevalence in various promoter classes. We have chosen the −500 to +500 region for composition analysis as the region might correspond to core, distal, and proximal promoter regions.48 The GC composition comparison of various classes of promoters was indicated as a heatmap in Figure 1A. The motif-less promoter sequences (no-motif class) may represent control sequences. To exclude contributions from promoter motifs, we removed the actual motifs and then calculated the GC percentage within the −500 to +500 region. It is observed that the Pause Button and BREu associated classes show higher GC than the no-motif class (p < 0.05, Wilcoxon signed-rank test). On the other hand, TATA and Inr-TATA promoter classes have low GC percentages when compared to motif-less promoter sequences in chickens, mammals, and plants, with more conspicuousness in mammals. Inr, TCT, BREd, and BREd-Inr classes did not indicate statistically significant differences in the GC composition in all 15 species. The clustergram in Figure 1A indicates the trends and relatedness of these promoter classes. We assumed that these differences may be attributed to the CpG island content in higher eukaryotes. Surprisingly, the result on the percentage of promoters with a CpG island is in line with our assumption (Figure 1B). The computation using the Takai–Jones algorithm53 revealed that yeasts, Plasmodium,C. elegans, and A. thaliana are devoid of CpG islands, whereas BREu, BREu-Inr, Pause Button, and Inr-PB in mammals, chickens, and plants showed a high preponderance of CpG islands compared to the no-motif category (Figure 1B).
Table 1. Percentage of Core Promoter Regions with Consensus Sequence Motifsa.
| organism | TATA | Inr | TCT | BREu | BREd | XCPE1 | DRE | DPE | PB | Inr-TATA | BREd-Inr | BREu-Inr | Inr-PB | no motif |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S. cerevisiae | 2.4 | 15.8 | 0.1 | 0.1 | 15.1 | 0.0 | 0.4 | 0.2 | 0.5 | 0.4 | 4.0 | 0.0 | 0.1 | 59.3 |
| S. pombe | 4.0 | 18.7 | 1.4 | 0.1 | 17.3 | 0.0 | 0.4 | 0.1 | 0.3 | 1.8 | 6.2 | 0.1 | 0.1 | 46.6 |
| P. falciparum | 17.9 | 2.4 | 1.7 | 0.0 | 31.5 | 0.0 | 0.0 | 0.0 | 0.0 | 1.7 | 4.7 | 0.0 | 0.0 | 21.5 |
| C.elegans | 3.3 | 31.1 | 2.8 | 0.6 | 10.3 | 0.0 | 0.4 | 0.1 | 0.7 | 2.3 | 8.5 | 0.7 | 0.7 | 30.7 |
| Apis mellifera | 4.4 | 24.1 | 0.4 | 0.3 | 13.8 | 0.0 | 1.7 | 0.5 | 1.3 | 1.8 | 7.0 | 0.6 | 1.5 | 36.0 |
| D. melanogaster | 3.8 | 28.4 | 0.5 | 0.3 | 8.0 | 0.0 | 4.8 | 0.3 | 1.2 | 3.6 | 9.6 | 0.3 | 2.2 | 25.5 |
| D. rerio | 1.7 | 28.9 | 0.9 | 0.7 | 11.4 | 0.1 | 0.0 | 0.4 | 0.7 | 1.2 | 6.9 | 0.5 | 0.5 | 43.6 |
| Gallus gallus | 1.2 | 19.6 | 1.5 | 8.7 | 5.4 | 0.5 | 0.0 | 0.3 | 2.3 | 0.7 | 2.6 | 3.1 | 0.7 | 49.9 |
| C. familiaris | 1.0 | 19.9 | 1.8 | 7.3 | 6.4 | 0.5 | 0.1 | 0.3 | 2.3 | 1.0 | 2.5 | 2.5 | 0.6 | 50.1 |
| R. norvegicus | 1.3 | 28.1 | 1.6 | 3.8 | 6.1 | 0.4 | 0.0 | 0.3 | 1.4 | 1.4 | 3.6 | 2.1 | 0.7 | 45.9 |
| M. musculus | 1.4 | 30.2 | 1.1 | 4.0 | 6.2 | 0.4 | 0.0 | 0.3 | 1.3 | 1.4 | 4.1 | 2.3 | 0.6 | 44.0 |
| M. mulatta | 0.8 | 26.1 | 1.4 | 6.0 | 6.1 | 0.5 | 0.0 | 0.4 | 1.8 | 0.6 | 3.0 | 3.3 | 0.7 | 45.7 |
| H. sapiens | 1.1 | 28.2 | 1.3 | 5.7 | 5.7 | 0.4 | 0.0 | 0.4 | 1.5 | 1.0 | 3.5 | 3.1 | 0.7 | 44.3 |
| A. thaliana | 9.6 | 18.4 | 2.3 | 0.1 | 10.4 | 0.0 | 0.4 | 0.1 | 0.6 | 4.6 | 4.2 | 0.1 | 0.2 | 43.4 |
| Zea mays | 4.4 | 23.4 | 1.2 | 2.8 | 5.5 | 0.1 | 0.1 | 0.2 | 1.5 | 2.6 | 1.8 | 1.2 | 0.6 | 51.3 |
The consensus motifs for upstream promoter elements for TATA-box, Initiator, TCT-element, TFIIB recognition element upstream (BREu), TFIIB recognition element downstream (BREd), XCPE1 (X core promoter element 1), and Drosophila DNA replication-related element (DRE) are TATAWAWR, BBCABW, YYCTTTYY, SSRCGCC, RTDKKKK, DSGYGGRASM, and WATCGATW, respectively. The consensus motifs for downstream promoter elements DPE and Pause Button are RGWCGTG and KCGRWCG, respectively.
Figure 1.
Sequence characteristics of eukaryotic promoter classes. (A) GC percentage trends in various promoter classes in 15 species. Actual consensus promoter elements were masked during GC percentage computation to avoid sequence biases. (B) CpG island distribution in various promoter classes. (C) Hierarchical clustering of hexamer motif distribution. Clustergrams were constructed based on the Pearson correlation clustering method for −500 to +500 regions.
A recent study reported the evolutionary relationship in genomes based on the motif composition and showed that DNA sequence composition may be an effective methodology to understand the functional and evolutionary relationships.54 Here, we implemented a similar hexamer composition analysis to understand the relatedness between promoter classes in several genomes. We have carried out a hierarchical horizontal clustering analysis for hexamer words. The cluster analysis based on promoter regions for different species is in line with the ″tree of life″ inferred from the TimeTree database.55 Moreover, it revealed the relatedness between various promoter classes. Generally, TATA-containing promoters (TATA and TATA-Inr classes) and Pause Button classes (PB and Inr-PB) form the outlier groups in all species indicating their divergence. Inr, BREd, and no-motif classes are often clustered into the same group. Expectedly, the promoter classes within species have more motif relevance compared to other species. Altogether, the composition and CpG island analysis revealed similarities and differences between various promoter classes with TATA and Pause Button (or BREu) associated classes representing the two ends of the GC composition spectrum.
2.2. Non-B-DNA Enrichment May Be Linked with Promoter Architecture
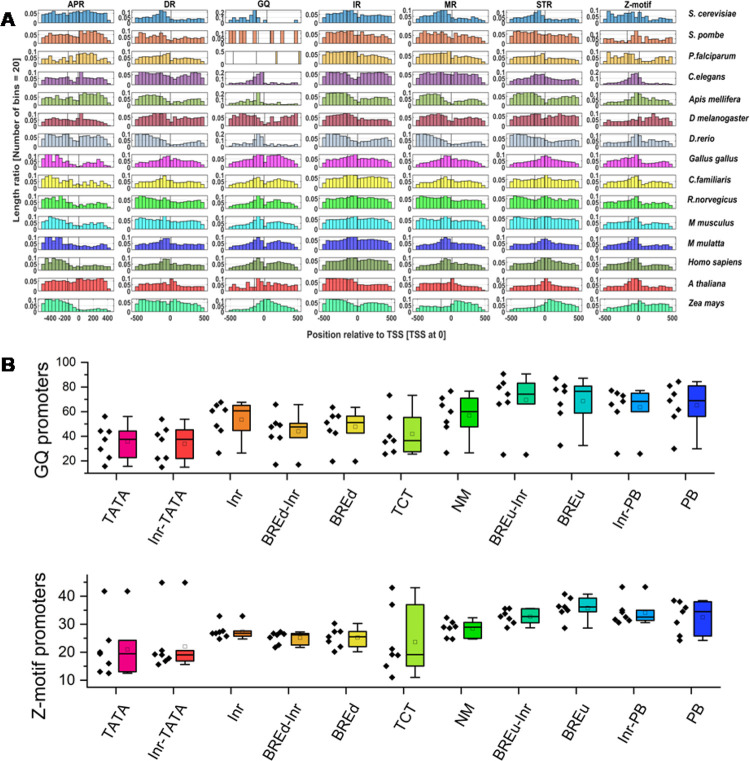
Now it is widely accepted that in vivo canonical B-DNA can morph into several non-B-DNA configurations based on the contextual sequences.34,36 Specifically, A-phased repeats, direct repeats, mirror repeats, inverted repeats, G-rich sequences, and YR repeats may form bent DNA, slipped structures, triplexes, cruciforms, G-quadruplexes, and Z-DNA structures, respectively.34,56 Here, we investigated these structurally constrained non-B-DNA prone motifs in the promoter classes of 15 species using the non-B-DNA tool.56 We delved into promoter classes using the positional distribution, length ratio of non-B-DNA motifs, and percentage of promoter sequences with at least one motif in the −500 to +500 region. The length ratio of the non-B-DNA motif is defined as ″the ratio of the length of the non-B-DNA motif to the total promoter sequence length″.37 Initially, we have investigated the general distribution of non-B-DNA motifs in all promoter sequences of different species with a major focus on the −100 to +50 region (core promoter region). The motif distributions indicate variable enrichment for different species with selective preponderances near transcription start sites (Figure 2A). The phased A-tracts do not indicate any prevalence in the core promoter regions compared to upstream or downstream regions. The direct repeats that can form slipped DNA are enriched in the immediate upstream of the core promoter regions (bin corresponding to the −100 to −50 region). G-quadruplex sequences seem to be the most enriched non-B-DNA type. They are enriched in the core promoter regions of all species except S. pombe, Plasmodium, and Drosophila. Inverted repeats do not reveal conspicuous signals in the core promoter regions for these species. Mirror repeats are observed in −300 to −100 regions but depleted in the vicinity of TSS in the fly and fish. They are also enriched in downstream regions in plants and immediate upstream regions in mammals. Short tandem repeats are observed in the vicinity of all species except Apis mellifera. The Z-DNA forming motifs have conspicuously enriched core promoter regions in all species. Overall, non-B-DNA computation indicated that the promoter DNA displays unique non-B-DNA structural motifs with a certain conservation in closely related species. Of note, positional enrichment of any non-B-DNA forming motif should not be regarded for functional evidence and evolutionary conservation in eukaryotes. However, several lines of experimental evidence indicated that the G-quadruplex and Z-DNA are associated with active transcriptional regions in genomes.34,37,38,57−60 A recent study indicated that direct repeats, mirror repeats, and triplex DNA repeats are moderately enriched in low-complexity repetitive sequences like heterochromatin, while inverted repeats and short tandem repeats displayed uniform distribution among gene-rich and gene-poor sequences.35 Another study based on immunofluorescence experiments and computational studies revealed the evolutionary pattern of G-quadruplex motifs. The density, length, and number of G-quadruplexes generally increased during evolution with higher stability in mammals.37 Relevant to the context, the analysis on ″length ratio of the non-B-DNA motif″ indicated that extreme enrichment of DR, IR, MR, and STRs was observed for P. falciparum (Figure S1A), whose genome has a very low GC content. G-quadruplexes and Z-DNA are highly preponderant at promoter regions with GC richness, i.e., mammals (Figure S1A).
Figure 2.
Non-B-DNA enrichment characteristics of eukaryotic promoter classes. (A) The positional distribution of seven non-B-DNA prone motifs is indicated as a normalized histogram for the −500 to +500 regions. (B) Boxplots for comparison of G-quadruplex motifs and Z-DNA motifs for 11 promoter classes. Dots indicate the percentage of promoters associated with G-quadruplex or Z-DNA-prone motifs for different promoter classes in chickens, mammals, and Zea mays. Code: APR: A-phased repeats, DR: direct repeats, MR: mirror repeats, IR: inverted repeats, GQ: G-quadruplex motifs, and STR: short tandem repeats.
Next, we analyzed the non-B-DNA-motif enrichment analysis for 11 promoter classes with a major focus on G-quadruplexes and Z-motifs (Figure 2B and Figure S1B). Based on our observation, the TATA, Inr-TATA, BREu, BREu-Inr, Pause Button, and Inr-PB classes display interesting signals. TATA and Inr-TATA classes possess fewer G-quadruplexes and Z-motifs in mammals compared to other classes (Figure 2B and Figure S1B). Contrastingly, the BREu and Pause Button associated classes seem to be more enriched with G-quadruplexes and Z-DNA (Figure 2B and Figure S1B). Here, we assume that G-quadruplex enrichment is an interesting observation. It is well known that the G-quadruplex motifs regulate transcription acting as an obstacle to the RNA polymerase or docking regions for specialized transcription factors.58 The Pause Button is known to be involved in the stalling of RNA polymerase.61,62 Furthermore, the high GC content and alternative DNA sequences in promoter sequences can contribute to Pol II pausing by recruitment of associated transcription factors.63
An interesting conjecture can be derived that the G-quadruplex and metazoan promoter element Pause Button may be linked together for pausing of RNA polymerase during the elongation process. The association of G-quadruplex structures with promoter motifs could generate specific patterns in transcription initiation or gene expression regulation.
2.3. Eukaryotic Promoter Classes Are Characterized by Unique Shape Features
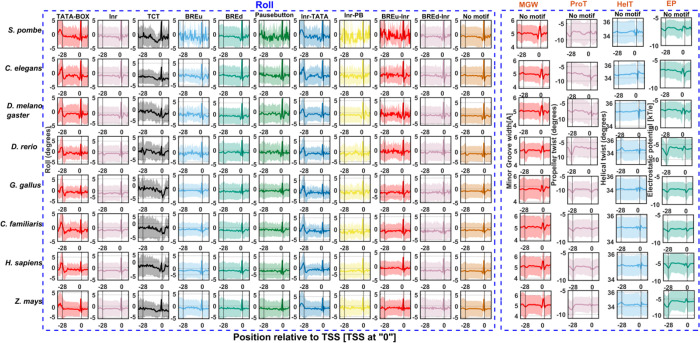
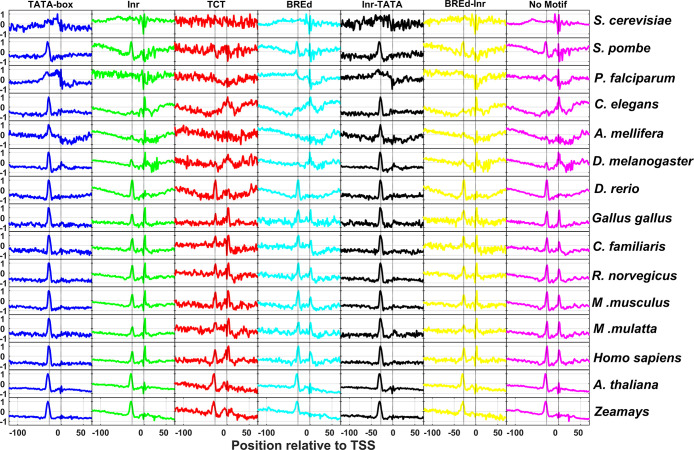
Irrespective of the identity of consensus signals, core promoters act as docking stations for orchestration of the preinitiation machinery of transcription. However, the prevalence of DNA shape signals in the vicinity of transcription start sites has not been reported in eukaryotes. For the first time, we exhaustively inspected five DNA shape features, namely, minor groove width, propeller twist, helical twist, roll, and electrostatic potential near −40 to +40 regions for several promoter classes. Figure 3 indicates the most prominent shape feature roll for the 11 promoter classes of 8 organisms. For brevity, we have shown roll angles for TATA-box, Inr, TCT, BREu, BREd, Pause Button, Inr-TATA, Inr-PB, BREu-Inr, BREd-Inr, and no-motif classes and other properties for the no-motif class only. Minor groove width, propeller twist, helical twist, and electrostatic potential profiles are shown in the Supplementary Information (Figures S2–S5). It is expected that AT-rich sequences of the TATA-box display strong signals. Surprisingly, all promoter classes, irrespective of species type, indicate similar but less conspicuous shape signals near the position corresponding to TSS. At the local regions of the transcription start site, all promoter classes and at the −28 region of the TATA class is characterized by the increased roll, reduced helical twist, widening of the minor groove, more negative propeller twist, and more negative electrostatic potential. The local rise in roll angles indicates a preponderance of YR nucleotide steps [CA, TA, CG, TG] that are known to have weak stacking interactions. Meanwhile, more negative propeller twist angles weaken hydrogen bonding patterns between base pairs.42 Furthermore, the reduction in helical twist favors unwinding that may lead to the bending of DNA.64 It is well known that TFIIB recognizes the upstream region of the TBP binding site through major groove interactions and the downstream region with minor groove interactions.17,65 However, neither the upstream nor downstream regions corresponding to TATA sites did not indicate any signatures. These results are comparable to a recent study on the role of DNA shape in prokaryotic transcription.66 The authors demonstrated that the α subunit C-terminal domain of Escherichia coli RNA polymerase recognizes the upstream promoter DNA element by virtue of its unique shape, i.e., larger negative electrostatic potential and narrow minor groove width.66 Furthermore, a recent study by the Yanhui Xu group resolved the longstanding argument of preinitiation complex formation in the absence of TATA-box using cryoelectron microscopy structural studies.31 They reported that a shared TFIID-binding pattern and loading of TBP to TATA-box-containing and TATA-less promoters with TBP also induce bending of TATA-less promoter sequences in PIC.31 Based on our observations and recent reports, we surmise that the shape of DNA has a crucial role in TFIID DNA recognition. Shape signatures in the vicinity of TSS enhance energetically less costlier DNA deformation required for TFIID recognition, DNA duplex separation, and transcription initiation. Next, we analyzed the physicochemical property of the DNA duplex stability in the core promoter regions.
Figure 3.
DNA shape signals in promoter classes of eukaryotes. The property roll has been shown for 11 promoter classes of 8 eukaryotes. Minor groove width, propeller twist, helical twist, and electrostatic potential are indicated for the no-motif class only.
2.4. DNA Duplex Stability Is a Characteristic Feature of All Eukaryotic Promoter Classes
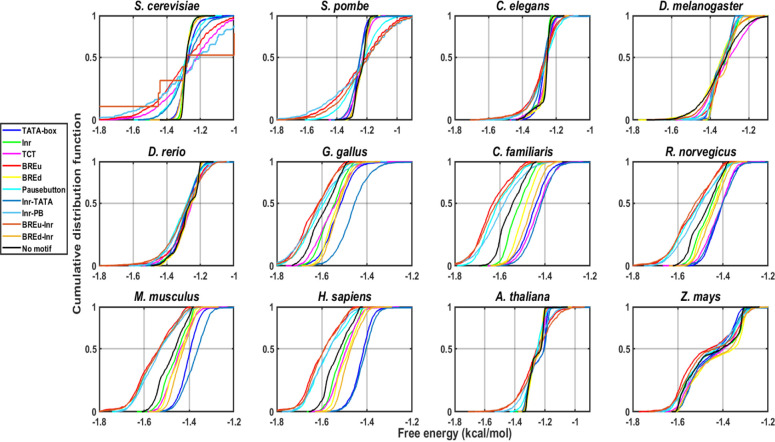
Genetic information ingrained in the hydrophobic core of the double-helical DNA is deciphered during transcription. The mechanism requires a local denaturation of double-helical DNA in the vicinity of the transcription start site. Earlier studies showed that the local DNA separation signals or low DNA duplex stability regions that facilitate the melting of DNA in core promoter regions are conserved from prokaryotes to humans.12,33,48 Here, we sought to understand the conservancy of low stability signals in several promoter classes for 15 species. To this end, we employed two models of DNA duplex stability: one based on melting studies67 and another based on the mechanistic strength of base stacking.68 The previous literature revealed that the stability profiles of different species can be broadly categorized into two groups: AT-rich and GC-rich.33,48,50 The first class represents lower eukaryotes (yeasts, P. falciparum,D. melanogaster, and C. elegans) where stability steeply reduces in the vicinity of promoters. Another kind of promoter region, where stability is higher at a longer range, is displayed for the case of mammals and birds.33,48 Of note, genome size is also linked to the type of profiles. In general, genomes with smaller sizes have a sharp low stability region, while genomes with high GC have higher stability.48,50 Our results for average promoter behavior are in line with previous results for longer range sequences [−500 to +500 regions] (not shown). However, within promoter classes, distinctions and commonalities exist. TATA-containing and TATA-Inr promoter classes have lower stability compared to the no-motif class, and the Pause Button, Inr-PB, BREu, and BREu-Inr classes have higher stability. We confirmed these differences using a two-sample KS test and plotted them as a cumulative distribution function (Figure 4). Next, we focused on the signals at shorter scales, i.e., the core promoter region. The position-specific mean feature value was plotted for the region −100 to +50 for melting stability (Figure 5) and mechanical stability (Figure S6) in all different promoter classes of 15 species, and we detected two local peaks corresponding to regions −1 and −28. The TATA-box displays the most conspicuous low melting stability signals at the −28 region. However, the mechanical stability property reveals a more prominent signal in the vicinity of TSS including TATA-class (Figure S6). Here, for the first time, we attempted to understand DNA duplex stability using a mechanical stability model that indicated similar signatures. The result affirms that not only the transcription machinery but the DNA low stability signatures in different promoter classes are conserved. The TA step of the TATA-box may provide the required support for inducing conformational change through kinking. Of note, according to the Santalucia melting stability model, the TA step shows the least stability in comparison to other steps. The Initiator element is known for the higher tendency of DNA melting for transcriptional bubble formation indicated by sharp low stability signatures in this study. Surprisingly, all promoter classes belonging to 15 species indicated a similar trend, revealing the conservation of low stability signals in promoter regions. Our results are in line with a recent study where the authors employed a modified Ising-type DNA instability model for human promoter characterization.69 The promoter classes in different species have extreme differences in sequence composition for various species yet indicate signature profiles. Furthermore, we argue that the propensity for localized low stability signatures may be independent of any protein interactions or DNA manipulation in the core promoter is a conserved structural property due to the conserved arrangement of the sequence itself. Our results surmise that the lower DNA duplex stability encoded by the DNA sequence itself is a crucial factor for the transcription process.
Figure 4.
Cumulative distribution function plots for different promoter classes. The DNA duplex stability values for −500 to +500 regions of different promoter classes like TATA-box, Inr, TCT, BREd, Inr-TATA, BREd-Inr, and no-motif for 12 eukaryotic organisms are shown.
Figure 5.
DNA melting stability profiles of TATA-box, Inr, TCT, BREd, Inr-TATA, BREd-Inr, and no-motif classes for 15 species. The x axis in all subplots ranges from −100 to +50 with TSS being positioned at ″0″. For comparative analysis, the y axis has been scaled from −1 to +1.
2.5. DNA Flexibility May Confer Additional Features for Chromosome Organization
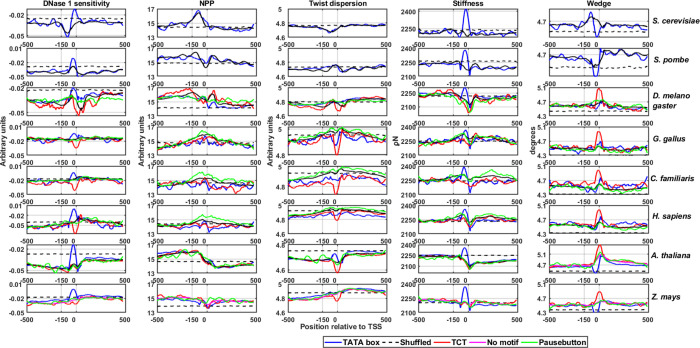
The transcription process occurs in the background of chromatin configuration, and the promoter DNA should be accessible to the transcription machinery. Previous studies indicated that promoter regions are characterized by nucleosome depletion regions70 and are characterized by DNA rigidity.33,52 However, few studies also reported that the TATA-containing promoters are found to be more flexible than TATA-less and neighboring sequences as well.12,52 Here, we extended the DNA flexibility computation for several promoter classes by incorporating novel models. We discretized DNA flexibility into axial, torsional, and stretching flexibility and characterized the promoter architecture. Axial flexibility was measured by DNase I sensitivity and the nucleosome positioning preference model as indicated in the previous literature.12 We adopted twisting flexibility or torsional flexibility from the Haran et al. model where a high dispersion in helical twist indicates torsional more torsional flexibility.71 The stretching flexibility has been characterized by a model based on the stiffness of the DNA under 0 N force.72 Furthermore, the wedge model that indicates intrinsic curvature was also employed. Figure 6 and Figure S7 indicate the flexibility signature profiles for different promoter classes derived from these five models. In all species, TATA-containing promoters (TATA and Inr-TATA) display more flexibility with the DNase I sensitivity model compared to other classes and randomized sequences near TSS. Surprisingly, the TCT class displays high rigidity in all species. The observation is in line with earlier research, which revealed that TCT promoters in Drosophila are associated with strongly positioned nucleosomes.19,73 The NPP model, however, indicates that all promoter classes are rigid compared to neighboring sequences. Promoter regions also display less torsional dispersion, indicating their rigidity. However, the TCT class is torsionally less flexible compared to other classes. The stiffness model indicated that promoters are high in a stiffer region at the upstream of TSS followed by less stiffer regions through the downstream locations. Further, the wedge model indicated that the promoter displays less wedge angles at upstream regions followed by a higher wedge at downstream regions around TSS sites. These models provide information about sequence-dependent flexibility settings in the vicinity of TSS. Here, the three types of DNA flexibility can be connected to the process of transcription initiation. The DNA should be less bendable or rigid to maintain nucleosome-free regions or anti-nucleosomal barriers. The rigid DNA may guide the easy sliding of the transcription machinery to determine the perfect transcription initiation regions.12,40 During transcription, the machinery has to escape from the promoter regions and the higher energetic cost required for DNA crookedness during the preinitiation complex formation may favor the process. Here, three types of flexibility—axial, stretching, and twisting flexibilities—in the core promoter DNA indirectly regulate the mechanism through conserved sequences. However, a clear exception is observed for TATA-containing promoters, particularly at TATA sites.
Figure 6.
DNA flexibility profiles of S. cerevisiae, S. pombe,D. melanogaster, Gallus gallus, C. familiaris, H. sapiens, A. thaliana, and Zea mays. Different structural properties, bending flexibility (DNase 1 sensitivity, nucleosome positioning preference model), twisting flexibility (twist dispersion model), stretching stiffness, and wedge were calculated for promoter classes TATA-box, TCT, no-motif, shuffled no-motif, and Pause Button.
To summarize the structural analysis, the promoter classes display local DNA melting signals for DNA opening, a flexible and optimal binding interface for the recognition of TFIID complexes through DNA shape preferences, a nucleosome depletion region for promoter access, and discrete preferences for non-B-DNA for higher-order modulation for transcriptional flexibility.
3. Conclusions
Remarkable progress has been made in the past four decades in discerning the promoter architecture, underpinning the precise positioning of a multitude of promoter elements and their role in the spatio-temporal gene expression. Although the common principles underlying the mechanism of transcription initiation of protein-coding genes have been broadly deciphered, a substantial number of structural studies are based on the TBP-based preinitiation complex on TATA-box-containing promoters. In this study, we carried out a deep computational investigation on promoter classes using a simple sequence-to-structure mapping strategy from structural models derived from experiments and theoretical studies. It has been observed that less frequent promoter motifs TATA-box, BREu, and Pause Button often co-occur with the Inr motif in eukaryotes. Our results indicated that in all species studied, TATA-containing promoters (TATA-only or Inr-TATA) are AT-rich with low CpG island preference, while the BREu and Pause Button associated classes displayed the opposite trend. The TATA class is characterized by fewer G-quadruplexes and Z-motifs in mammals compared to other classes. Contrastingly, the BREu and Pause Button classes seem to be enriched with direct repeats, G-quadruplexes, and Z-DNA compared to other promoter classes. For the first time, our study reported DNA shape signals in eukaryotic core promoter regions. All promoter classes of different species indicate higher positive roll values, lower values of helical twist, wider minor grooves, and high negative electrostatic potentials in the vicinity of TSS. They also displayed low stability signals and unique DNA flexibility features irrespective of the promoter class or species in the vicinity of TSS. These findings may explain the role of DNA structure in transcription initiation in the absence of TATA-boxes. A common theme that emerges from our study is that a plethora of DNA structural features that deviate from the ideal behavior fine-tune the process of transcription initiation. The DNA structural strategy implemented in this study complements experimental approaches of promoter biology and can provide mechanistic insights into transcription initiation. These structural signals can be integrated with epigenetic information and can be utilized for the enhanced prediction of promoter DNA-GTF recognition in genomes, refining promoter prediction methodologies, and modeling gene expression pathways. Nonetheless, there are certain limitations to our study. The current approach does not implement DNA structure under the influence of DNA methylation. The promoter classification may be dubious due to the lack of promoter activity definitions of majority of the promoter elements at a larger scale. However, our report provides a deep and expansive prospectus of the evolutionarily conserved conceptual framework for the DNA structure of sequences encompassing transcription initiation sites in eukaryotic genomes.
4. Data Sets and Methods
4.1. Promoter Sequence Data Set
The promoter sequence data sets of nucleotide sequences for 15 eukaryotic species were downloaded from the Eukaryotic Promoter Database (EPD) (http://epd.vital-it.ch).74 The EPD database provides the transcription start site information that has been identified through high-throughput experiments.74 We have selected the most representative promoter for a gene for every organism. The resulting sequences were further processed to remove redundant sequences or sequences with ″N″ nucleotides. Final numbers for different promoter data sets comprise 5114 for Saccharomyces cerevisiae, 4796 for Schizosaccharomyces pombe, 4028 for Plasmodium falciparum, 13,399 for Drosophila melanogaster, 5454 for Apis mellifera, 10,174 for Danio rerio, 6364 for Caenorhabditis elegans, 16,398 for H. sapiens, 8832 for Macaca mulatta, 20,190 for Mus musculus, 11,759 for Rattus norvegicus, 5627 for Gallus gallus, 6291 for Canis familiaris, 22,647 for Arabidopsis thaliana and 15,620 for Zea mays. We have extracted promoter sequences [−500 to +500] relative to the transcription start site for further processing.
4.2. Classification of Promoter Sequences
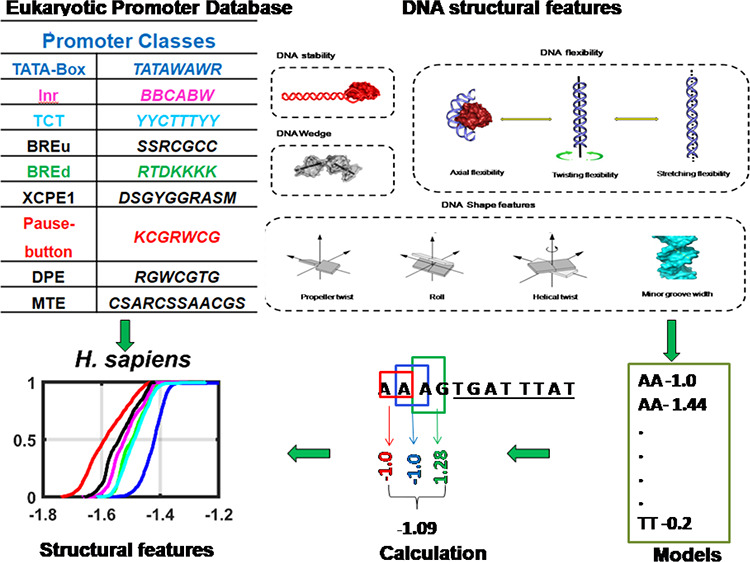
The key aspect of this study is categorizing promoter sequences into various classes based on the presence or absence of well-defined consensus core promoter motifs. Earlier studies defined the functionality of eukaryotic promoter elements based on certain benchmarks such as precise positioning, maximum motif length, minimal consensus motif, and conservation across orthologous regions.10,12,75 We utilized the criterion of the presence of minimal cognate motifs for seven upstream motifs and three downstream motifs at a specified location relative to the transcription start site to categorize them into various promoter classes. The consensus motifs for upstream promoter elements for TATA-box, Initiator, TCT-element, TFIIB recognition element upstream (BREu), TFIIB recognition element downstream (BREd), XCPE1 (X core promoter element 1), and Drosophila DNA replication-related element (DRE) are TATAWAWR, BBCABW, YYCTTTYY, SSRCGCC, RTDKKKK, DSGYGGRASM, and WATCGATW, respectively. The consensus motifs for downstream promoter elements MTE, DPE, and Pause Button are CSARCSSAACGS, RGWCGTG, and KCGRWCG, respectively.10 TATA-box-containing promoters and BREu-containing and BREd-containing motifs were defined by the presence of a consensus motif at −40 to −1 relative to TSS. Inr-containing, TCT-containing, and XCPE1-containing promoters are defined as sequences with cognate elements within the 10 nucleotide deviation from their precisely defined positions, i.e., −3 to +3, −2 to +6, and −8 to +2, respectively. DRE (DNA replication-related element)-containing promoters are defined with sequences having motifs within the −100 to −1 region relative to TSS. For downstream promoter classes, we have chosen +1 to +40 regions for MTE and DPE and +1 to +60 for Pause Button, respectively. Promoter regions overlapping with different motifs in functional regions are listed into separate classes. For example, a promoter sequence that contains a TATA-box and Inr element in the defined functional region is classified as Inr-TATA class. Hence, promoter classes include motif alone and overlapping categories. Overall, 118 categories were obtained based on 10 promoter motifs for 15 species (Table S1). A stringent criterion for all 15 species has been utilized, although certain discrepancies exist in our classification. For instance, a previous study utilized the −150 to −1 region to classify promoters into TATA-containing and TATA-less promoters in six eukaryotes.12
4.3. Computation of DNA Sequence Composition
Sequence characteristics of promoter regions [−500 to +500] such as GC percentage, hexanucleotide motifs, and CpG islands were computed. GC percentage was carried out using the ″infoseq″ tool of the EMBOSS package.76 Actual consensus promoter elements were masked during GC percentage computation. Hexamer motif analysis was done using the Cserhati et al. motif analysis method.54 For all promoter classes of 15 species, all possible hexamers were computed. The promoter classes where any hexamer is missing were omitted for further analysis. A clustergram was constructed based on the Pearson correlation coefficient as a distance metric. The noncanonical promoter signatures, the CpG islands are screened using the CpG searcher program, with cutoffs of (i) sequence length >500 nucleotides, (ii) GC percentage of the sequence >55, and (iii) observed/expected ratio of CpG > 0.65.53
4.4. Computation of B-DNA Structural Features and Non-B-DNA Prone Motifs
DNA structural features are broadly divided into two categories: B-DNA structural features and non-B-DNA forming structural motifs.
The B-DNA structure is an entanglement of various physicochemical and conformational properties functioning at local and global levels.40 Several secondary structural variations of DNA, namely, duplex stability, flexibility, wedge, and DNA shape, were analyzed in this study.
4.4.1. DNA Stability
The physicochemical property DNA stability has been analyzed in the context of thermal and mechanical stability. A dinucleotide model from unified parameters of melting studies on 108 oligonucleotides67 was utilized to predict the melting stability of a DNA fragment as in the previous literature.12,40,45,48,51 However, for the first time, we inferred mechanical stability based on single-stack free energy increments from tethered DNA origami beam experiments.68
4.4.2. DNA Flexibility
The flexibility of DNA molecules can be categorized into three directional components: axial flexibility (or bendability), twisting flexibility (twistability), and stretching flexibility.77 Bendability computation has been carried out by trinucleotide models detailed in the previous research.12,40,45,51 DNase I cutting preferences of trinucleotide steps are predicted by the DNase 1 sensitivity model78 and the nucleosome positioning preference (NPP) model.79 Twistability can be estimated as the variance or dispersion of helical twist based on X-ray crystal structural data.80 A higher variance indicates more torsional flexibility and vice versa. Earlier, Haran et al. and Resnick et al. utilized the approach to link p53 binding affinity to twisting flexibility.71,81 Stretching flexibility (springiness) can be inferred from the stiffness modulus of the two base-paired steps. The model has been deciphered from the reported results of 1 ms long molecular dynamic studies on benchmark DNA sequences of size 18 base pairs.72 A DNA wedge model from BMHT studies provides clues about local bending or intrinsic curvature.82 The dinucleotide structural features duplex stability, twist dispersion, and DNA stretching flexibility are predicted by a one-nucleotide moving window of 15 nt, while trinucleotide feature models (DNase 1 sensitivity and nucleosome positioning preference) are computed by a 30-nt window. For a given DNA sequence, the corresponding k-mer steps are replaced with numerical values of look-up tables of structural descriptors. Structural signatures are depicted using average profiles over all sequences in a class of promoters.
4.4.3. DNA Shape
The features minor groove width, propeller twist, helical twist, roll, and electrostatic potential are predicted using the DNAshapeR package.83 The tool uses a sliding pentamer window based on structural data of Monte Carlo simulations of 2121 DNA fragments.84
4.4.4. Non-B-DNA Prone Motifs
Different non-B-DNA forming motifs corresponding to phased A-tracts, G-quadruplexes, cruciform DNA, H-DNA, slipped DNA (STR), and Z-DNA were computed using the non-B-DNA tool.56
Acknowledgments
V.R.Y. is grateful to the Science and Engineering Research Board (SERB), Department of Science and Technology (DST), Government of India, for the Early-Career Research Grant (ECR/2017/001100/LS). A. V acknowledges JRF/SRF fellowship from the SERB-DST grant (ECR/2017/001100/LS) awarded to V.R.Y.
Glossary
Abbreviations
- TSS
transcription start site
- PIC
pre-initiation complex
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c04603.
Percentage of core promoter regions with consensus sequence motifs (Table S1); non-B-DNA enrichment characteristics of eukaryotic promoter classes (Figure S1); minor groove width signals in promoter classes of eukaryotes (Figure S2); propeller twist signals in promoter classes of eukaryotes (Figure S3); helical twist profiles of different promoter classes of eukaryotes (Figure S4); electrostatic potential profiles of promoter classes (Figure S5); DNA mechanical stability profiles of different promoter classes (Figure S6); and DNA flexibility profiles of different promoter classes (Figure S7) (PDF)
Author Contributions
V.R.Y. conceived the project. V.R.Y. and A.V. carried out data collection, processing, computation of features, interpretation of the results, and drafting. All authors read and approved the manuscript.
This work was funded by an Early-Career Research Grant (ECR/2017/001100/LS) from the Science and Engineering Research Board (SERB), Department of Science and Technology (DST), Government of India.
The authors declare no competing financial interest.
Notes
The data sets are retrieved from public web resources. Derived data supporting the findings of this study are available from the corresponding author (V.R.Y.) on request.
Supplementary Material
References
- Grünberg S.; Hahn S. Structural insights into transcription initiation by RNA polymerase II. Trends Biochem. Sci. 2013, 38, 603–611. 10.1016/j.tibs.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury S.; Bernecky C.; Cramer P. Structural basis of transcription initiation by RNA polymerase II. Nat Rev Mol Cell Biol 2015, 16, 129–143. 10.1038/nrm3952. [DOI] [PubMed] [Google Scholar]
- Yella V. R.; Bansal M.. In silico Identification of Eukaryotic Promoters. In Systems and Synthetic Biology; 2015, pp. 63–75, 10.1007/978-94-017-9514-2_4. [DOI] [Google Scholar]
- Weingarten-Gabbay S.; Segal E. The grammar of transcriptional regulation. Hum. Genet. 2014, 133, 701–711. 10.1007/s00439-013-1413-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danino Y. M.; Even D.; Ideses D.; Juven-Gershon T. The core promoter: At the heart of gene expression. Biochim. Biophys. Acta 2015, 1849, 1116–1131. 10.1016/j.bbagrm.2015.04.003. [DOI] [PubMed] [Google Scholar]
- Roy A. L.; Singer D. S. Core promoters in transcription: old problem, new insights. Trends Biochem. Sci. 2015, 40, 165–171. 10.1016/j.tibs.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T.; Kadonaga J. T. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 2010, 339, 225–229. 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even D. Y.; Kedmi A.; Ideses D.; Juven-Gershon T. Functional Screening of Core Promoter Activity. Methods Mol. Biol. 2017, 1651, 77–91. 10.1007/978-1-4939-7223-4_7. [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T.; Hsu J.-Y.; Theisen J. W.; Kadonaga J. T. The RNA polymerase II core promoter—the gateway to transcription. Curr. Opin. Cell Biol. 2008, 20, 253–259. 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V.; Stark A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat Rev Mol Cell Biol 2018, 19, 621–637. 10.1038/s41580-018-0028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloutskin A.; Danino Y. M.; Orenstein Y.; Zehavi Y.; Doniger T.; Shamir R.; Juven-Gershon T. ElemeNT: a computational tool for detecting core promoter elements. Transcription 2015, 6, 41–50. 10.1080/21541264.2015.1067286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yella V. R.; Bansal M. DNA structural features of eukaryotic TATA-containing and TATA-less promoters. FEBS Open Bio 2017, 7, 324–334. 10.1002/2211-5463.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale S. T.; Kadonaga J. T. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003, 72, 449–479. 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- Reeve J. N. Archaeal chromatin and transcription. Mol. Microbiol. 2003, 48, 587–598. 10.1046/j.1365-2958.2003.03439.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y. Y.; Ichida H.; Abe T.; Suzuki Y.; Sugano S.; Obokata J. Differentiation of core promoter architecture between plants and mammals revealed by LDSS analysis. Nucleic Acids Res. 2007, 35, 6219–6226. 10.1093/nar/gkm685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.; Bolotin E.; Jiang T.; Sladek F. M.; Martinez E. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene 2007, 389, 52–65. 10.1016/j.gene.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W.; Roberts S. G. E. A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes Dev. 2005, 19, 2418–2423. 10.1101/gad.342405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W.; Roberts S. G. E. TFIIB and the regulation of transcription by RNA polymerase II. Chromosoma 2007, 116, 417–429. 10.1007/s00412-007-0113-9. [DOI] [PubMed] [Google Scholar]
- Vo Ngoc L.; Kassavetis G. A.; Kadonaga J. T. The RNA polymerase II core promoter in Drosophila. Genetics 2019, 212, 13–24. 10.1534/genetics.119.302021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. P. The architecture of mammalian ribosomal protein promoters. BMC evolutionary biology 2005, 5, 1–16. 10.1186/1471-2148-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry T. J.; Theisen J. W. M.; Hsu J.-Y.; Wang Y.-L.; Corcoran D. L.; Eustice M.; Ohler U.; Kadonaga J. T. The TCT motif, a key component of an RNA polymerase II transcription system for the translational machinery. Genes Dev. 2010, 24, 2013–2018. 10.1101/gad.1951110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke T. W.; Kadonaga J. T. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997, 11, 3020–3031. 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U.; Liao G.-c.; Niemann H.; Rubin G. M. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 2002, 3, 1–12. 10.1186/gb-2002-3-12-research0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke T. W.; Kadonaga J. T. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 1996, 10, 711–724. 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- Hendrix D. A.; Hong J.-W.; Zeitlinger J.; Rokhsar D. S.; Levine M. S. Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proceedings of the National Academy of Sciences 2008, 105, 7762–7767. 10.1073/pnas.0802406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. Paused RNA Polymerase II as a Developmental Checkpoint. Cell 2011, 145, 502–511. 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray G. A.; Hahn M. W.; Abouheif E.; Balhoff J. P.; Pizer M.; Rockman M. V.; Romano L. A. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 2003, 20, 1377–1419. 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- Kim T. K.; Shiekhattar R. Architectural and Functional Commonalities between Enhancers and Promoters. Cell 2015, 162, 948–959. 10.1016/j.cell.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindal G. A.; Farley E. K. Enhancer grammar in development, evolution, and disease: dependencies and interplay. Dev. Cell 2021, 56, 575–587. 10.1016/j.devcel.2021.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levo M.; Segal E. In pursuit of design principles of regulatory sequences. Nat Rev Genet 2014, 15, 453–468. 10.1038/nrg3684. [DOI] [PubMed] [Google Scholar]
- Chen X.; Qi Y.; Wu Z.; Wang X.; Li J.; Zhao D.; Hou H.; Li Y.; Yu Z.; Liu W.; Wang M.; Ren Y.; Li Z.; Yang H.; Xu Y.. Structural insights into preinitiation complex assembly on core promoters. Science 2021, 372 (). [DOI] [PubMed] [Google Scholar]
- Parker S. C.; Tullius T. D. DNA shape, genetic codes, and evolution. Curr. Opin. Struct. Biol. 2011, 21, 342–347. 10.1016/j.sbi.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal M.; Kumar A.; Yella V. R. Role of DNA sequence based structural features of promoters in transcription initiation and gene expression. Curr. Opin. Struct. Biol. 2014, 25, 77–85. 10.1016/j.sbi.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Kaushik M.; Kaushik S.; Roy K.; Singh A.; Mahendru S.; Kumar M.; Chaudhary S.; Ahmed S.; Kukreti S. A bouquet of DNA structures: Emerging diversity. Biochem. Biophys. Rep. 2016, 5, 388–395. 10.1016/j.bbrep.2016.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakopoulos-Soares I.; Morganella S.; Jain N.; Hemberg M.; Nik-Zainal S. Noncanonical secondary structures arising from non-B DNA motifs are determinants of mutagenesis. Genome Res. 2018, 28, 1264–1271. 10.1101/gr.231688.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiblet W. M.; Cremona M. A.; Cechova M.; Harris R. S.; Kejnovska I.; Kejnovsky E.; Eckert K.; Chiaromonte F.; Makova K. D. Long-read sequencing technology indicates genome-wide effects of non-B DNA on polymerization speed and error rate. Genome Res. 2018, 28, 1767–1778. 10.1101/gr.241257.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F.; Niu K.; Cui Y.; Li C.; Lyu M.; Ren Y.; Chen Y.; Deng H.; Huang L.; Zheng S.; Liu L.; Wang J.; Song Q.; Xiang H.; Feng Q. Genome-wide analysis of DNA G-quadruplex motifs across 37 species provides insights into G4 evolution. Commun Biol 2021, 4, 98. 10.1038/s42003-020-01643-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey U.; Sarkar S.; Teronpi V.; Yella V. R.; Kumar A. G-quadruplex motifs are functionally conserved in cis-regulatory regions of pathogenic bacteria: An in-silico evaluation. Biochimie 2021, 184, 40–51. 10.1016/j.biochi.2021.01.017. [DOI] [PubMed] [Google Scholar]
- Spiegel J.; Adhikari S.; Balasubramanian S. The Structure and Function of DNA G-Quadruplexes. Trends Chem 2020, 2, 123–136. 10.1016/j.trechm.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yella V. R.; Kumar A.; Bansal M., DNA Structure and Promoter Engineering. In Systems and Synthetic Biology; 2015, pp. 241–254, 10.1007/978-94-017-9514-2_13. [DOI] [Google Scholar]
- Yella V. R.; Bhimsaria D.; Ghoshdastidar D.; Rodriguez-Martinez J. A.; Ansari A. Z.; Bansal M. Flexibility and structure of flanking DNA impact transcription factor affinity for its core motif. Nucleic Acids Res. 2018, 46, 11883–11897. 10.1093/nar/gky1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R.; West S. M.; Sosinsky A.; Liu P.; Mann R. S.; Honig B. The role of DNA shape in protein–DNA recognition. Nature 2009, 461, 1248–1253. 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror I.; Golan T.; Levy C.; Rohs R.; Mandel-Gutfreund Y. A widespread role of the motif environment in transcription factor binding across diverse protein families. Genome Res. 2015, 25, 1268–1280. 10.1101/gr.184671.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.; Orenstein Y.; Jolma A.; Yin Y.; Taipale J.; Shamir R.; Rohs R. Transcription factor family-specific DNA shape readout revealed by quantitative specificity models. Mol. syst. biol. 2017, 13, 910. 10.15252/msb.20167238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yella V. R.; Vanaja A.; Kulandaivelu U.; Kumar A. Delving into Eukaryotic Origins of Replication Using DNA Structural Features. ACS Omega 2020, 5, 13601–13611. 10.1021/acsomega.0c00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X. Q.; Zeng J.; Yan H. Structural properties of replication origins in yeast DNA sequences. Physical biology 2008, 5, 036012 10.1088/1478-3975/5/3/036012. [DOI] [PubMed] [Google Scholar]
- Chen W.; Feng P.; Lin H. Prediction of replication origins by calculating DNA structural properties. FEBS Lett. 2012, 586, 934–938. 10.1016/j.febslet.2012.02.034. [DOI] [PubMed] [Google Scholar]
- Yella V. R.; Kumar A.; Bansal M. Identification of putative promoters in 48 eukaryotic genomes on the basis of DNA free energy. Sci. Rep. 2018, 8, 4520. 10.1038/s41598-018-22129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goñi J. R.; Pérez A.; Torrents D.; Orozco M. Determining promoter location based on DNA structure first-principles calculations. Genome Biol. 2007, 8, R263. 10.1186/gb-2007-8-12-r263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeel T.; Saeys Y.; Bonnet E.; Rouzé P.; Van de Peer Y. Generic eukaryotic core promoter prediction using structural features of DNA. Genome Res. 2008, 18, 310–323. 10.1101/gr.6991408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yella V. R.; Bansal M. DNA structural features and architecture of promoter regions play a role in gene responsiveness of S. cerevisiae. J. Bioinform. Comput. Biol. 2013, 11, 1343001. 10.1142/S0219720013430014. [DOI] [PubMed] [Google Scholar]
- Tirosh I.; Berman J.; Barkai N. The pattern and evolution of yeast promoter bendability. Trends Genet. 2007, 23, 318–321. 10.1016/j.tig.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Takai D.; Jones P. A. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 3740–3745. 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cserhati M.; Xiao P.; Guda C. K-mer-Based Motif Analysis in Insect Species across Anopheles, Drosophila, and Glossina Genera and Its Application to Species Classification. Comput. Math. Methods Med. 2019, 2019, 4259479. 10.1155/2019/4259479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.; Stecher G.; Suleski M.; Hedges S. B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017, 34, 1812–1819. 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- Cer R. Z.; Donohue D. E.; Mudunuri U. S.; Temiz N. A.; Loss M. A.; Starner N. J.; Halusa G. N.; Volfovsky N.; Yi M.; Luke B. T.; Bacolla A.; Collins J. R.; Stephens R. M. Non-B DB v2.0: a database of predicted non-B DNA-forming motifs and its associated tools. Nucleic Acids Res. 2012, 41, D94–D100. 10.1093/nar/gks955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S. I.; Ham S.; Park J.; Seo S. H.; Lim C. H.; Jeon H.; Huh J.; Roh T. Y. Z-DNA-forming sites identified by ChIP-Seq are associated with actively transcribed regions in the human genome. DNA Res. 2016, 23, 477–486. 10.1093/dnares/dsw031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N. The interplay between G-quadruplex and transcription. Curr. Med. Chem. 2019, 26, 2898–2917. 10.2174/0929867325666171229132619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig Lombardi E.; Londoño-Vallejo A. A guide to computational methods for G-quadruplex prediction. Nucleic Acids Res. 2020, 48, 1–15. 10.1093/nar/gkz1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong B.; Chen S.; Kwon J. A.; Rich A. Characterization of Z-DNA as a nucleosome-boundary element in yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 2229–2234. 10.1073/pnas.0611447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertner B.; Zeitlinger J. RNA polymerase II pausing during development. Development 2014, 141, 1179–1183. 10.1242/dev.088492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist D. A.; Dos Santos G.; Fargo D. C.; Xie B.; Gao Y.; Li L.; Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 2010, 143, 540–551. 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szlachta K.; Thys R. G.; Atkin N. D.; Pierce L. C. T.; Bekiranov S.; Wang Y. H. Alternative DNA secondary structure formation affects RNA polymerase II promoter-proximal pausing in human. Genome Biol. 2018, 19, 89. 10.1186/s13059-018-1463-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Bates D. L.; Dey R.; Chen P. H.; Machado A. C. D.; Laird-Offringa I. A.; Rohs R.; Chen L. DNA binding by GATA transcription factor suggests mechanisms of DNA looping and long-range gene regulation. Cell Rep. 2012, 2, 1197–1206. 10.1016/j.celrep.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolov D. B.; Chen H.; Halay E. D.; Usheva A. A.; Hisatake K.; Lee D. K.; Roeder R. G.; Burley S. K. Crystal structure of a TFIIB–TBP–TATA-element ternary complex. Nature 1995, 377, 119–128. 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- Lara-Gonzalez S.; Dantas Machado A. C.; Rao S.; Napoli A. A.; Birktoft J.; Di Felice R.; Rohs R.; Lawson C. L. The RNA Polymerase α Subunit Recognizes the DNA Shape of the Upstream Promoter Element. Biochemistry 2020, 59, 4523–4532. 10.1021/acs.biochem.0c00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SantaLucia J. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proceedings of the National Academy of Sciences 1998, 95, 1460–1465. 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilchherr F.; Wachauf C.; Pelz B.; Rief M.; Zacharias M.; Dietz H. Single-molecule dissection of stacking forces in DNA. Science 2012, 2, 1197. 10.1016/j.celrep.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Kantorovitz M. R.; Rapti Z.; Gelev V.; Usheva A. Computing DNA duplex instability profiles efficiently with a two-state model: trends of promoters and binding sites. BMC Bioinformatics 2010, 11, 604. 10.1186/1471-2105-11-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C.; Pugh B. F. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 2009, 10, 161–172. 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beno I.; Rosenthal K.; Levitine M.; Shaulov L.; Haran T. E. Sequence-dependent cooperative binding of p53 to DNA targets and its relationship to the structural properties of the DNA targets. Nucleic Acids Res. 2011, 39, 1919–1932. 10.1093/nar/gkq1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Gonzalez A.; Vilhena J. G.; Moreno-Herrero F.; Perez R. DNA Crookedness Regulates DNA Mechanical Properties at Short Length Scales. Phys. Rev. Lett. 2019, 122, 048102 10.1103/PhysRevLett.122.048102. [DOI] [PubMed] [Google Scholar]
- Rach E. A.; Winter D. R.; Benjamin A. M.; Corcoran D. L.; Ni T.; Zhu J.; Ohler U. Transcription initiation patterns indicate divergent strategies for gene regulation at the chromatin level. PLoS Genet. 2011, 7, e1001274 10.1371/journal.pgen.1001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreos R.; Ambrosini G.; Périer R. C.; Bucher P. The Eukaryotic Promoter Database: expansion of EPDnew and new promoter analysis tools. Nucleic Acids Res. 2015, 43, D92–D96. 10.1093/nar/gku1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basehoar A. D.; Zanton S. J.; Pugh B. F. Identification and distinct regulation of yeast TATA box-containing genes. Cell 2004, 116, 699–709. 10.1016/S0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- Rice P.; Longden I.; Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000, 16, 276–277. 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Bao L.; Zhang X.; Jin L.; Tan Z.-J. Flexibility of nucleic acids: From DNA to RNA. Chin. Phys. B 2016, 25, 018703 10.1088/1674-1056/25/1/018703. [DOI] [Google Scholar]
- Brukner I.; Sanchez R.; Suck D.; Pongor S. Sequence-dependent bending propensity of DNA as revealed by DNase I: parameters for trinucleotides. EMBO J. 1995, 14, 1812–1818. 10.1002/j.1460-2075.1995.tb07169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchwell S. C.; Drew H. R.; Travers A. A. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 1986, 191, 659–675. 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- Olson W. K.; Gorin A. A.; Lu X. J.; Hock L. M.; Zhurkin V. B. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl. Acad. Sci. U. S. A. 1998, 95, 11163–11168. 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inga A.; Storici F.; Darden T. A.; Resnick M. A. Differential Transactivation by the p53 Transcription Factor Is Highly Dependent on p53 Level and Promoter Target Sequence. Mol. Cell. Biol. 2002, 22, 8612–8625. 10.1128/MCB.22.24.8612-8625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolshoy A.; McNamara P.; Harrington R. E.; Trifonov E. N. Curved DNA without A-A: experimental estimation of all 16 DNA wedge angles. Proc. Natl. Acad. Sci. U. S. A. 1991, 88, 2312–2316. 10.1073/pnas.88.6.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu T.-P.; Comoglio F.; Zhou T.; Yang L.; Paro R.; Rohs R. DNAshapeR: an R/Bioconductor package for DNA shape prediction and feature encoding. Bioinformatics 2016, 32, 1211–1213. 10.1093/bioinformatics/btv735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T.; Yang L.; Lu Y.; Dror I.; Dantas Machado A. C.; Ghane T.; Di Felice R.; Rohs R. DNAshape: a method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic Acids Res. 2013, 41, W56–W62. 10.1093/nar/gkt437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.