Abstract
Successful in vitro amplification of fungal DNA in clinical specimens has been reported recently. In a collaboration among five European centers, the frequency and risk of contamination due to airborne spore inoculation or carryover contamination in fungal PCR were analyzed. The identities of all contaminants were specified by cycle sequencing and GenBank analysis. Twelve of 150 PCR assays that together included over 2,800 samples were found to be contaminated (3.3% of the negative controls were contaminated during the DNA extraction, and 4.7% of the PCR mixtures were contaminated during the amplification process). Contaminants were specified as Aspergillus fumigatus, Saccharomyces cerevisiae, and Acremonium spp. Further analysis showed that commercially available products like zymolyase powder or 10× PCR buffer may contain fungal DNA. In conclusion, the risk of contamination is not higher in fungal PCR assays than in other diagnostic PCR-based assays if general precautions are taken.
Invasive fungal infections have emerged as major infectious complications in immunocompromised hosts. As conventional diagnostic tests, such as culture systems or histopathology, show low sensitivity or specificity, numerous protocols for sensitive detection of fungal DNA by in vitro amplification with PCR have been established. Examples of primer target sequences are either the Candida-specific lanosterol demethylase gene (2), mitochondrial genes (13), aspartic proteinase genes (6), genes encoding the fungal heat shock proteins Cahsp 70 (1) or HSP 90 (3), or highly conserved ribosomal gene sequences (5, 18).
PCR assays show variable sensitivity with a detection limit of 10 fg of fungal DNA, corresponding to 1 to 10 fungal cells per ml of blood (5). Some of the assays are able to detect up to 40 different fungal species (18), including all clinically relevant fungal pathogens. In a typical PCR amplification, 1012 identical amplicons can be generated. As these amplicons may serve as targets for further reactions, carryover contaminations have to be excluded. Post-PCR treatment of the amplicons with UV light and 8-methoxypsoralen (8) or pre-PCR treatment with uracil-DNA-glycosylase digestion (7) has been proven to efficiently inactivate amplicons from previous PCRs.
Fungal spores, such as conidia from Aspergillus spp. and other molds, might be present in the air. Thus, airborne spore inoculation during the DNA extraction process could potentially lead to false-positive results, especially if panfungal primers are applied (5, 18).
The detection of fungal pathogens from blood by PCR has been performed independently in five different centers (Huddinge Hospital, Huddinge, Sweden; Hammersmith Hospital, London, Great Britain; Hôpital Saint Louis, Paris, France; Charité, Berlin, Germany; and Department of Internal Medicine, University of Tuebingen, Tuebingen, Germany) by using the same protocols for DNA extraction, PCR amplification, and hybridization. DNA was extracted under a biosafety hood as described previously (12) in a separate room with equipment exclusively used for DNA extraction. Amplicons were never processed in this area. Enzymatic lysis of the leukocytes was performed by proteinase K digestion (concentration, 200 μg/ml) (Boehringer, Mannheim, Germany), and spheroplasting of fungal cells was carried out by zymolyase incubation (0.3 mg/ml) (ICN, Costa Mesa, Calif.). DNA was precipitated and purified by the QIAmp tissue kit (Qiagen, Hilden, Germany).
In all laboratories PCR was performed in a separate room with equipment exclusively used for PCR. People pipetting PCR mixtures in this room were wearing one-way gowns, sterile gloves, and face masks. Standard PCR conditions were applied by using 100 pM of each primer (5′-ATT GGA GGG CAA GTC TGG TG and 5′-CCG ATC CCT AGT CGG CAT AG; Roth, Karlsruhe, Germany); these primers target a highly conserved region of the fungal 18S small-subunit rRNA gene. Thirty-four cycles of repeated denaturation (94°C, 30 s), annealing (62°C, 1 min), and extension (72°C, 2 min) were applied. Amplicons were detected by standard gel electrophoresis in a 2% Tris-acetate-EDTA (TAE) agarose gel (Sigma, Deissenhofen, Germany) using ethidium bromide staining, followed by Southern blot hybridization with a species-specific digoxigenin-labeled oligonucleotide (Roth) for Aspergillus fumigatus (5′-CAT GGC CTT CAC TGG CGT TGG GGG GAA CCA) and anti-digoxigenin antibodies conjugated with alkaline phosphatase (Boehringer Mannheim) (5).
In order to control naturally arising DNA from airborne sources (e.g., fungal spores) one negative control per five extracted samples was included during each DNA extraction procedure. Negative controls consisted of sterile water or blood from healthy individuals and were subjected to all preparation steps in parallel with the extracted samples.
PCR-grade water was used as a DNA-free negative control (one per 10 amplified specimens) to exclude any carryover contamination during the PCR process. These controls contained all necessary components for PCR except template DNA.
Fifteen contaminated negative control samples from four of the European centers (five samples from Huddinge Hospital, five from Hammersmith, three from Hôpital Saint Louis, and two from Charité) were shipped on dry ice to Tuebingen for cycle sequencing with an ABI 373A sequencer (Applied Biosystems, Dreieich, Germany) by using the following protocol. All amplicons were purified with the QIAquick purification kit (Qiagen) as described in the manufacturer’s protocol. Cycle PCR was performed in a GeneAmp 2400 PCR cycler (Perkin Elmer, Dreieich, Germany) by applying 25 cycles (10 s at 96°C, 5 s at 56°C, and 4 min at 60°C). After purification of the PCR product with CentriSep columns (Perkin Elmer), samples were sequenced by using the BigDye Terminator kit (Applied Biosystems). Sequence data were collected and compared with known sequences from fungal, mammalian, human, viral, bacterial, and organelle DNA databases kept at the European Bioinformatic Institute, Cambridge, United Kingdom (BLAST database program).
At Medical Hospital Tuebingen, 180 DNA extractions (each with 16 samples on average) comprising a total of 2,800 specimens (including 450 negative controls) followed by 150 PCR assays (each with 24 samples, including controls) were performed between June 1996 and May 1998.
Eight percent of the PCR assays were found to be invalid because of contaminations: negative controls from five independent DNA extractions showed a false-positive result after amplification (3.3%), and in addition seven PCR mixtures (4.7%) were contaminated.
The five DNA extraction controls contained DNA from Aspergillus spp. When this DNA was reamplified with internally transcribed spacer DNA (ITS) primers (5′-TCC GTA GGT GAA CCT GCG G and 5′-TCC TCC GCT TAT TGA TAT GC), two additional bands formed in the gel, indicating the presence of genomic DNA. Thus, we presume that this contamination occurred due to airborne spore inoculation.
Negative controls of the PCR mixtures from seven assays were false positive because of reamplification of target DNA (A. fumigatus), most probably because of carryover contaminations. When ITS primers were used no additional band was formed, indicating that no genomic DNA was present. This contamination was never detected when new PCR reagents were applied.
Fifteen contaminated negative controls from the other four centers were sequenced. All these negative controls showed positive results after gel electrophoresis. Three samples contained DNA amplicons from A. fumigatus, and 12 control samples contained DNA from Saccharomyces cerevisiae.
In order to check for the presence of fungal DNA in lysing enzymes (zymolyase, lyticase, and lysing enzyme extracted from Trichoderma harzianum), three different protocols of extraction of serial fungal dilutions (105 to 100 CFU of A. fumigatus/ml) were run in parallel. Zymolyase (ICN) is a β-1,3-laminaripentaohydrolase prepared from a submerged culture of Arthrobacter luteus and partially purified by affinity chromatography (9) in 50 mM Tris–10 mM EDTA–28 mM mercaptoethanol–0.3 mg of zymolyase/ml. Zymolyase was first isolated from brewery sewage at Kirin Brewery, Takasaki, Japan. Lyticase (Sigma) is a partially purified gamma-irradiated powder isolated from a culture of A. luteus containing 20% protein (16) and dissolved in 50 mM Tris–1 mM EDTA–20% mercaptoethanol–5 U of lyticase (18) or in 50 mM potassium phosphate–10 mM mercaptoethanol–5 U of lyticase (16). Lysing enzyme (Sigma) from T. harzianum (14) is a lyophilized powder extracted from a mold containing 80% protein with cellulase, protease, and chitinase activity and is aseptically filled (2 mg/ml, in PCR-grade water) (Fresenius, Bad Homburg, Germany). All enzymes were incubated at 37°C for 1 h.
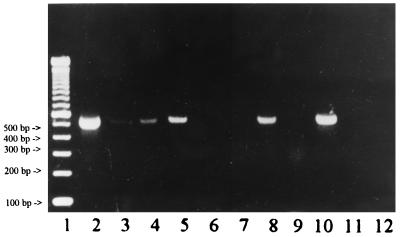
We could demonstrate that zymolyase powder from specific lot numbers in different dilutions contained fungal DNA (Fig. 1). Sequencing and GenBank sequence homology analysis showed that the lot for which data are shown in Fig. 1 contained DNA from S. cerevisiae (100% identity in a 368-bp overlap in the variable region of the 18S rRNA gene of S. cerevisiae). Identical lots were also used independently in three other laboratories (Huddinge Hospital, Huddinge, Sweden; Hôpital Saint Louis, Paris, France; and Charité, Berlin, Germany). GenBank sequence homology analysis showed a 100% identity of the amplicons from all four centers.
FIG. 1.
Gel electrophoresis (2% TAE-agarose gel) showing amplified DNA of potentially contaminated samples (specific bands of approximately 500-bp length). Lanes: 1, 100-bp marker; 2, zymolyase, contaminated lot; 3, zymolyase, contaminated lot, 1:10 dilution; 4, 10× buffer from digoxigenin (Dig)-labeling kit (Boehringer Mannheim), contaminated lot; 5, 10× buffer from Dig-labeling kit (Boehringer Mannheim), contaminated lot; 6, zymolyase, regular lot, not contaminated; 7, proteinase K, regular lot, not contaminated; 8, lyticase, contaminated lot; 9, lysing enzyme extracted from T. harzianum; 10, positive control, A. fumigatus DNA, extracted from 103 CFU; 11, negative control of the extraction; and 12, negative control of the PCR.
After treatment of the zymolyase powder with UV light (30 min at 312 nm) PCR amplification was found to be negative. After the zymolyase was dissolved in 0.9% sodium chloride and seeded out onto Sabouraud glucose agar (incubation at 37°C for 48 h) no fungal growth could be observed. It was concluded that fungal DNA but not yeast was the cause of contamination (10).
Further tests showed a specific signal of 500 bp in the agarose gel when Tris buffer containing lyticase was amplified (Fig. 1). Conventional lyticase is an only partially purified powder. Recombinant lyticase expressed in Escherichia coli is available from Sigma; however, the use of recombinant enzyme increases the costs for DNA extraction (3 U are US$0.60, and zymolyase for one extraction is US$0.12). A faint band of 300 bp could be observed after amplification of buffer containing lysing enzyme, which is extracted from the mold T. harzianum. Despite repeated cycle sequencing this amplicon could not be further specified. No band could be observed when PCR was performed on solutions used to redissolve the enzymes. All buffers separately analyzed by PCR and gel electrophoresis yielded negative results.
Amplification of serial dilutions of Aspergillus conidia showed an identical detection limit of 20 CFU with zymolyase or lyticase in Tris buffer but a limit of 1,000 CFU with lyticase in potassium phosphate buffer.
Another set of amplification reactions showed contaminations in one component of the PCR mixture. Detailed and repeated serial analysis of all components of the PCR mixture (nucleotides, primers, water, magnesium chloride, 10× buffer, and Taq polymerase) showed a positive specific band only in the 10× PCR buffer supplied by Boehringer Mannheim. This result could be reproduced when different tubes of the same lot were used. When a new lot was used no signal was detectable. Sequencing of the amplicon followed by GenBank sequence homology analysis showed a homology of 99% with DNA of Acremonium spp.
Another enzyme which is commonly included in DNA extraction protocols is proteinase K, an endopeptidase used for leukocyte lysis (4). According to the manufacturer, Boehringer Mannheim, it is a purified powder from a culture of a fungus, Tritirachium album. However, contaminating DNA in the proteinase K fraction could not be observed over a 3-year period (Fig. 1).
In conclusion, PCR is a powerful technique with applications in many fields, e.g., medical diagnostics, forensic analysis, and population genetics. Since 1990, a large number of protocols for the detection of fungal pathogens by the PCR methodology have been published.
We could demonstrate that there are only a very limited number of fungal species potentially contaminating samples during DNA extraction and amplification procedures and that these originate from airborne spore inoculation or carryover contamination. Commercially available reagents like lysis enzymes also contained fungal DNA. This shows that many companies seem not to be aware of the fact that their products might be used for fungal PCR.
Although fungal spores are ubiquitous in the air, our collective experience shows that the frequency of contamination does not seem to be higher in fungal PCR than in diagnostic PCR-based techniques targeting nonfungal pathogens. Precautions to prevent airborne and carryover contaminations as well as the use of sufficient negative controls have to be applied carefully (11, 15).
Acknowledgments
We thank L. Klingspor, Huddinge Hospital, Huddinge, Sweden, T. Rogers, Hammersmith Hospital, London, Great Britain, and C. Lacroix, Hôpital Saint Louis, Paris, France, for their collaboration.
This work was supported by the Deutsche Krebshilfe (grant 70/2199/KaI).
REFERENCES
- 1.Arancia S, Sandini S, Cassone A, De Bernardis F, La Valle R. Construction and use of PCR primers from a 70 kDa heat shock protein gene for identification of Candida albicans. Mol Cell Probes. 1997;11:329–336. doi: 10.1006/mcpr.1997.0125. [DOI] [PubMed] [Google Scholar]
- 2.Buchman T G, Rossier M, Merz W G, Charache P. Detection of surgical pathogens by in vitro DNA amplification, part 1: rapid identification of Candida albicans by in vitro amplification of a fungus-specific gene. Surgery. 1990;108:338–347. [PubMed] [Google Scholar]
- 3.Crampin A C, Matthews R C. Application of the polymerase chain reaction to the diagnosis of candidosis by amplification of an HSP 90 gene fragment. J Med Microbiol. 1993;39:233–238. doi: 10.1099/00222615-39-3-233. [DOI] [PubMed] [Google Scholar]
- 4.Ebeling W, Hennrich N, Klockow M, Metz H, Orth H D, Lang H. Proteinase K from Tritirachium album. Eur J Biochem. 1974;47:91–97. doi: 10.1111/j.1432-1033.1974.tb03671.x. [DOI] [PubMed] [Google Scholar]
- 5.Einsele H, Hebart H, Roller G, Löffler J, Rothenhöfer I, Müller C A, Bowden R A, van Burik J-A, Engelhard D, Kanz L, Schumacher U. Detection and identification of fungal pathogens in blood by using molecular probes. J Clin Microbiol. 1997;35:1353–1360. doi: 10.1128/jcm.35.6.1353-1360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flahaut M, Sanglard D, Monod M, Bille J, Rossier M. Rapid detection of Candida albicans in clinical samples by DNA amplification of common regions from C. albicans-secreted aspartic proteinase genes. J Clin Microbiol. 1998;36:395–401. doi: 10.1128/jcm.36.2.395-401.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gobet P, Buisson J C, Vagner O, Naciri M, Grappin M, Comparot S, Harly G, Aubert D, Varga I, Camerlynck P, Bonnin A. Detection of Cryptosporidium parvum DNA in formed human feces by a sensitive PCR-based assay including uracil-N-glycosylase inactivation. J Clin Microbiol. 1997;35:254–256. doi: 10.1128/jcm.35.1.254-256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jinno Y, Yoshiura K, Niikawa N. Use of psoralen as extinguisher of contaminated DNA in PCR. Nucleic Acids Res. 1990;18:6739. doi: 10.1093/nar/18.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kitamura K, Kaneko T, Yamamoto Y. Lysis of viable yeast cells by enzymes of Arthrobacter luteus. J Gen Appl Microbiol. 1972;18:57–71. doi: 10.1016/0003-9861(71)90053-1. [DOI] [PubMed] [Google Scholar]
- 10.Kitchin P A, Szotyori Z, Fromholc C, Almond N. Avoidance of false positives. Nature. 1990;344:201. doi: 10.1038/344201a0. [DOI] [PubMed] [Google Scholar]
- 11.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 12.Löffler J, Hebart H, Schumacher U, Reitze H, Einsele H. Comparison of different methods for fungal DNA extraction from cultures and blood. J Clin Microbiol. 1997;35:3311–3312. doi: 10.1128/jcm.35.12.3311-3312.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyakawa Y, Mabuchi T, Kagaya K, Fukazawa Y. Isolation and characterization of a species-specific DNA fragment for detection of Candida albicans by polymerase chain reaction. J Clin Microbiol. 1992;30:894–900. doi: 10.1128/jcm.30.4.894-900.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Müller F M, Werner K E, Kasai M, Francesconi A, Chanock S J, Walsh T J. Rapid extraction of genomic DNA from medically important yeasts and filamentous fungi by high-speed cell disruption. J Clin Microbiol. 1998;36:1625–1629. doi: 10.1128/jcm.36.6.1625-1629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarkar G, Sommer S. Shedding light on PCR contamination. Nature. 1990;343:27. doi: 10.1038/343027a0. [DOI] [PubMed] [Google Scholar]
- 16.Scott J H, Schekman R. Lyticase: endoglucanase and protease activities that act together in yeast cell lysis. J Bacteriol. 1980;142:414–423. doi: 10.1128/jb.142.2.414-423.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang C M, Holden D W, Aufauvre-Brown A, Cohen J. The detection of Aspergillus spp. by the polymerase chain reaction and its evaluation in bronchoalveolar fluid. Am Rev Respir Dis. 1993;148:1313–1317. doi: 10.1164/ajrccm/148.5.1313. [DOI] [PubMed] [Google Scholar]
- 18.Van Burik J-A, Myerson D, Schreckhise R W, Bowden R A. Panfungal PCR assay for detection of fungal infection in human blood specimens. J Clin Microbiol. 1998;36:1169–1175. doi: 10.1128/jcm.36.5.1169-1175.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]