Abstract
Background
Certain Fusarium exometabolites have been reported to inhibit seed germination of the cereal-parasitizing witchweed, Striga hermonthica, in vitro. However, it is unknown if these exometabolites will consistently prevent S. hermonthica incidence in planta. The study screened a selection of known, highly phytotoxic Fusarium exometabolites, in identifying the most potent/efficient candidate (i.e., having the greatest effect at minimal concentration) to completely hinder S. hermonthica seed germination in vitro and incidence in planta, without affecting the host crop development and yield.
Results
In vitro germination assays of the tested Fusarium exometabolites (i.e., 1,4-naphthoquinone, equisetin, fusaric acid, hymeglusin, neosolaniol (Neo), T-2 toxin (T-2) and diacetoxyscirpenol (DAS)) as pre-Striga seed conditioning treatments at 1, 5, 10, 20, 50 and 100 µM, revealed that only DAS, out of all tested exometabolites, completely inhibited S. hermonthica seed germination at each concentration. It was followed by T-2 and Neo, as from 10 to 20 µM respectively. The remaining exometabolites reduced S. hermonthica seed germination as from 20 µM (P < 0. 0001). In planta assessment (in a S. hermonthica-sorghum parasitic system) of the exometabolites at 20 µM showed that, although, none of the tested exometabolites affected sorghum aboveground dry biomass (P > 0.05), only DAS completely prevented S. hermonthica incidence. Following a 14-d incubation of DAS in the planting soil substrate, bacterial 16S ribosomal RNA (rRNA) and fungal 18S rRNA gene copy numbers of the soil microbial community were enhanced; which coincided with complete degradation of DAS in the substrate. Metabolic footprinting revealed that the S. hermonthica mycoherbicidal agent, Fusarium oxysporum f. sp. strigae (isolates Foxy-2, FK3), did not produce DAS; a discovery that corresponded with underexpression of key genes (Tri5, Tri4) necessary for Fusarium trichothecene biosynthesis (P < 0.0001).
Conclusions
Among the tested Fusarium exometabolites, DAS exhibited the most promising herbicidal potential against S. hermonthica. Thus, it could serve as a new biocontrol agent for efficient S. hermonthica management. Further examination of DAS specific mode of action against the target weed S. hermonthica at low concentrations (≤ 20 µM), as opposed to non-target soil organisms, is required.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-022-03471-6.
Keywords: Striga hermonthica, Fusarium oxysporum f. sp. strigae, Fusarium exometabolites, Diacetoxyscirpenol, Biopesticides, Targeted metabolomics, Metabolic footprinting, Trichothecene gene expression
Background
Cereal production in sub-Saharan Africa (SSA), like many crops in tropical/subtropical agroecosystems, is challenged by various biotic, abiotic, socio-cultural and socio-economic factors [4, 59]. In terms of biotic constraints, the obligate hemiparasitic witchweed, Striga spp., especially Striga hermonthica (Delile) Benth., is a major factor [19, 54]. According to Gressel et al. [23], out of the 26.23 million ha of all crop fields that were infested by Striga spp. in SSA, sorghum fields alone accounted for roughly 20 million ha, thereby causing an estimated yield loss of 6.5 to 6.9 million t per annum. Infestation by S. hermonthica begins after the release of organic stimulants (i.e., strigolactones) by the host cereal plants, inducing germination of preconditioned S. hermonthica seeds. Then, the elongating radicle of the S. hermonthica seedling develops into a haustorium, which is used to attach and penetrate the host cereal root [60]. Successful parasitism of the host plant by S. hermonthica is ensured by vascular continuity between the parasite and the host. Through this channel, S. hermonthica accesses the host resources to support its life cycle, while the host plant health deteriorates (until death in serious cases), owing mainly to water and nutrient deficiency [66].
Fusarium oxysporum f. sp. strigae (Fos) is a well-known bioherbicide (mycoherbicide), that has proven effective for controlling parasitic Striga spp., under both natural and artificial environments [52, 58, 64]. Although, various Fos isolates with significant pathogenicity towards S. hermonthica have been reported [82], in some cases, the bioherbicidal effectiveness of Fos isolates against S. hermonthica has shown to be inconsistent [6, 7, 76]. The inconsistent effectiveness of Fos isolates against differing S. hermonthica populations is, amongst others, a major challenge that deters the widespread application of any bioherbicide against its target weed [18, 26]. Like every functioning ecosystem (natural or artificial), the Fos–S. hermonthica–cereal crop pathosystem is made up of complex networks of bipartite, tripartite or multipartite interactions that occur both within and between the biotic (cereal crop, S. hermonthica, microorganisms) and abiotic (climate, water, soil physico-chemistry) components [48, 53]. These ecologically complex interactions may eventually reduce or in some cases enhance the efficacy of mycoherbicides against their target weeds [38]. However, with respect to the bipartite interaction between Fos and S. hermonthica, the role of genetic diversity as an underlying factor for this inconsistent effectiveness has been well studied. For instance, the classification of genetically contrasting Fos isolates (e.g., Foxy-2 and FK3) into separate vegetative compatibility groups (VCG) and mating types [13], and population genomic structure in S. hermonthica populations with varying susceptibility response to Foxy-2 and FK3 [5]. As a result, variation in the metabolome between genetically structured S. hermonthica groups were revealed through the sequenced genomic regions of the associated loci to show the differences in the protein-coding nucleotide sequences [5]. Meanwhile, in Fos, metabolomic studies have been mainly limited to the detection of particular secondary metabolites that are produced by the fungi and released into culture media e.g., fusaric acid and dehydrofusaric acid [3, 61], and/or into the infected S. hermonthica shoot to cause phytotoxicity, e.g., beauvericin [51]. Screening of the differential exometabolome of genetically contrasting Fos isolates to identify specific secondary metabolites with proven biopesticidal efficacy against S. hermonthica incidence in planta is lacking. This knowledge would bring awareness on specific bioactive metabolites that constitute the Fos exometabolomic arsenal for preventing S. hermonthica incidence.
The ability of several fungal secondary metabolites, especially Fusarium toxins, to hamper early growth stages (germination, germ tube development/attachment), and later stages (post-attachment/aboveground development) of the life cycle of parasitic weeds have been mentioned in numerous studies. With this, they have a considerable potential as biological tools for the parasitic weed management [10, 71, 77]. In the context of S. hermonthica, some specific Fusarium extracellular metabolites (exometabolites) have shown complete inhibition of the seed germination in vitro, when used as post-Striga seed conditioning treatments at very low concentrations (≤ 1 mM). For example, T-2 toxin at 10 μM [83], neosolaniol at 240 μM [67], and fusaric acid at 1 mM [28]. However, it is unknown if these highly bioactive Fusarium exometabolites will maintain their potency against S. hermonthica incidence in planta. Also, in non-parasitic plants, some Fusarium exometabolites have similarly shown to cause complete germination inhibition at very low concentrations, for instance; 1,4-naphthoquinone at 63 μM against Pinus thunbergii Parl. pollen germination [36], and hymeglusin at 50 μM against Brassica juncea (L.) Czern. seed germination [40]. For other Fusarium exometabolites, such as equisetin, the concentration causing complete inhibition of germination in plants has not been determined [78]. Unfortunately, the potential of these latter Fusarium exometabolites to inhibit S. hermonthica seed germination or incidence is not known. Thus, the identification of highly bioactive Fusarium exometabolites with broadscale efficacy against diverse S. hermonthica populations has been suggested as a promising direction for overcoming the limitations of the inconsistent effectiveness of Fos isolates against S. hermonthica [6]. In fact, fungal exometabolites, especially mycotoxins, play important roles in shaping the soil microbial community dynamics. This could impact the microbial abundance positively e.g., by serving as carbon source to support soil microbial proliferation; or adversely (antibiosis) e.g., by acting as antimicrobials to suppress soil microbial growth [44, 75]. Hence, in the latter case, microbial elimination (degradation) of the mycotoxin from the environment becomes a crucial ecological concern, as soil microorganisms are an integral part of any balanced ecosystem.
In spite of the progress made in identifying some particular Fusarium exometabolites that are effective at very low concentrations against S. hermonthica seed germination, there are still major knowledge gaps, especially regarding the exometabolite consistency in preventing S. hermonthica incidence in planta. Therefore, the main research questions of this study were: Among a set of known highly phytotoxic, germination-inhibiting Fusarium exometabolites [36, 40, 77], which is the most potent/efficient candidate (i.e., having the greatest effect at minimal concentration) against S. hermonthica seed germination? Is the potency of this candidate exometabolite consistent against S. hermonthica incidence in planta, whilst unaffecting the host crop development/yield? These questions serve as fundamental determiner for drawing greater attention into this strategy and encourage the use of fungal exometabolites as biopesticides to better combat the S. hermonthica menace. The study hypothesized that among the tested Fusarium exometabolites, the most potent/efficient candidate exometabolite against S. hermonthica, is part of the Fos exometabolome composition which is used for attacking S. hermonthica. Thus, the main objectives of the study were to: (i) examine the performance of a set of highly phytotoxic Fusarium exometabolites against S. hermonthica seed germination (in vitro) and incidence (in planta), towards identifying the most potent/efficient candidate exometabolite; and (ii) screen the exometabolome of contrasting Fos isolates (Foxy-2 and FK3), in determining if these agents produce the candidate exometabolite.
Methods
Preparatory activities: seed materials, seed sterilization, microbial samples, and exometabolites
Striga hermonthica (Delile) Benth. seeds were sampled from various locations in SSA (Table 1). Sorghum (Sorghum bicolor L. Moench) seeds (cultivar PI563294) were sampled from Maradi, Niger. Our previous research studies confirmed the susceptibility of this sorghum cultivar to S. hermonthica [5, 6]. The S. hermonthica and sorghum seeds were separately surface-sterilized using sodium hypochlorite, Tween 20® and double distilled water (ddH2O), then air-dried, according to the procedure of Anteyi and Rasche [5, 6]. Fusarium oxysporum f. sp. strigae (Fos) isolates Foxy-2 and FK3 were obtained from the University of Hohenheim, Stuttgart, Germany. Fusarium venenatum strain O86 (DSM number 23361) was obtained from the Leibniz Institute DSMZ (German Collection of Microorganisms and Cell Cultures) GmbH, Braunschweig, Germany. The following Fusarium exometabolites were purchased as isolated and purified forms from their suppliers; 1,4-naphthoquinone (1,4-Nq) (Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany), diacetoxyscirpenol (DAS) (Romer Labs Deutschland GmbH, Butzbach, Germany), equisetin (Equi) (Sigma-Aldrich Chemie GmbH), fusaric acid (FuA) (Sigma-Aldrich Chemie GmbH), hymeglusin (Hym) (Sigma-Aldrich Chemie GmbH), neosolaniol (Neo) (VWR International GmbH, Bruchsal, Germany) and T-2 toxin (T-2) (BIOMOL GmbH, Hamburg, Germany).
Table 1.
Sampled S. hermonthica origin and phenotypic response to Fos isolates Foxy-2 and FK3
| S. hermonthica origin | Geographic coordinates (in decimal degrees) | Source | Sampled population phenotypic response to Fos isolates Foxy-2 and FK3 a | |
|---|---|---|---|---|
| Latitude | Longitude | |||
| Abuja (Nigeria) | 9.066667 | 7.483333 | IITA-Nigeria | Susceptible to Foxy-2 and FK3 |
| Wad-Medani (Sudan) | 14.4 | 33.533333 | UG-Sudan | Susceptible to Foxy-2 and FK3 |
| Kibos (Kenya) | −0.066667 | 34.816667 | CIMMYT-Kenya | Susceptible to FK3; non-susceptible to Foxy-2 |
| Sirinka (Ethiopia) | 11.75 | 39.6 | SARC-Ethiopia | Susceptible to FK3; non-susceptible to Foxy-2 |
IITA-Nigeria – International Institute of Tropical Agriculture, Ibadan, Nigeria
UG-Sudan – University of Gezira, Wad Medani, Sudan
CIMMYT-Kenya – International Maize and Wheat Improvement Centre, Kibos research facility, Kenya
SARC-Ethiopia – Sirinka Agricultural Research Centre, Sirinka, Ethiopia
a − according to [5]
Performance of the Fusarium exometabolites against S. hermonthica
In vitro seed germination assay
The germination inhibitory potential of the test Fusarium exometabolites (i.e., 1,4-Nq, DAS, Equi, FuA, Hym, Neo and T-2) against S. hermonthica seeds were examined at 1, 5, 10, 20, 50 and 100 μM concentrations. The exometabolites were initially diluted with acetone (Carl Roth GmbH) to 1 mM, then by serial dilutions with ddH2O to the respective study concentrations. 100–250 surface-sterilized S. hermonthica seeds (collected from Sirinka, Ethiopia) were laid on a sterilized 0.8 cm glass fibre filter paper (GFFP) punched disc. Four S. hermonthica seed-carrying punched discs (i.e., four replicates) were placed on a doubled layer of sterilized 9 cm GFFP, contained in a 9 cm petri dish. Then, 6 mL of the exometabolites (or ddH2O as control) were added as pre-Striga seed conditioning treatments to each petri dish. Thus, the treatments included (1): ddH2O (control); (2): Hym; (3): 1,4-NQ; (4): FuA; (5): Equi; (6): Neo; (7): T-2; (8): DAS. For S. hermonthica seed preconditioning, the petri dishes were covered, sealed with parafilm and incubated for 14 d in a dark chamber at 30 °C. Afterwards, the GFFP punched discs (carrying the preconditioned S. hermonthica seeds) were re-placed on a single layer of new, sterilized 9 cm GFFP, in a petri dish. 3 mL of a synthetic strigolactone analog, rac-GR24 (0.1 ppm) (Chiralix B.V., Nijmegen, Netherlands) was added to every petri dish, and they were re-incubated at 30 °C for 24 h. This was followed by counting the germinated S. hermonthica seeds using the Zeiss Stemi 2000-C Stereomicroscope (Carl Zeiss Microscopy GmbH, Jena, Germany), together with the Zeiss AxioCam HRc (Carl Zeiss Light Microscopy, Göttingen, Germany). The experiment was performed in two repetitions. The least study concentration where all the tested exometabolites significantly reduced S. hermonthica seed germination was utilized as working concentration for further investigations.
Planting experiments
Sixty-five g of planting soil substrate, made up of a blend of 80% modular seed substrate (Klasmann-Deilmann GmbH, Geeste, Germany) and 20% maize rhizosphere (collected from the top layer (0–20 cm) of a maize field in Filderstadt, Germany: Latitude 48.6483, Longitude 9.2475) were filled into 20 × 5 × 2 cm rhizoboxes, made from polyvinyl chloride. Then a plexiglass lid was used to cover each of the rhizoboxes. Sixty-five mg of S. hermonthica seeds (from Sirinka, Ethiopia) were sown in depth of 5 cm beneath the soil surface. Thereafter, using a 10 mL pipette, each of the rhizoboxes were treated with 65 mL of the test exometabolites at 20 μM working concentration (derived from the in vitro seed germination assay), by application on the soil surface. Thus, the treatments included (1): ddH2O without Striga (Striga-neg. control); (2): ddH2O + Striga (Striga-pos. control); (3): Hym + Striga; (4): 1,4-Nq + Striga; (5): FuA + Striga; (6): Equi + Striga; (7): Neo + Striga; (8): T-2 + Striga; (9): DAS + Striga. Each treatment was replicated thrice, and the set up was arranged in a randomized complete block design. For S. hermonthica seed preconditioning, the rhizoboxes were incubated at 30 °C for 14 d in a climate chamber without light (Percival Intellus Environmental Controller, EA-75HIL, Perry, Iowa, USA). This was followed by sowing a sorghum seed in each of the rhizoboxes, and maintaining the climate chamber at an alternating 12-h period of 31 °C with light (mean illuminance 46,000 lx) and 27 °C without light. The rhizoboxes were irrigated with nutrient solution twice weekly, which consisted of a mixture of 3 parts of 0.2% (v/v) Wuxal® universal liquid fertilizer (Aglukon Spezialdünger GmbH, Düsseldorf, Germany) and 2 parts of Yoshida nutrient solution [79]. The planting experiments lasted for 9 weeks after sowing sorghum, and the trials were performed in 2 experimental repetitions. Following a similar experimental setup, additional planting experiments were performed, but without including S. hermonthica in the design. At the end of the planting experiments, S. hermonthica incidence in a rhizobox was determined from the total amount of attached S. hermonthica belowground and aboveground. The sorghum aboveground biomass was harvested, dried in an oven at 65 °C for 10 d, then weighed to obtain the sorghum aboveground dry biomass (ABM) data.
Additional experiments focusing on DAS (the candidate exometabolite)
Consistency against diverse S. hermonthica populations
The germination inhibitory potential of DAS against seeds of S. hermonthica populations from Abuja (Nigeria), Wad-Medani (Sudan) and Kibos (Kenya) were also evaluated. The experiment followed similar technical procedures as described for the in vitro seed germination assay (see above). The treatments included (1): ddH2O (control); (2): DAS 1 μM; (3): DAS 20 μM. Each of the treatments were in 4 replications, and the experiment was done in two repetitions.
Effectiveness as a post-Striga seed conditioning treatment
The experiment followed similar technical procedures as described for the in vitro seed germination assay (see above). However, here, the 14-d S. hermonthica (from Sirinka, Ethiopia) seed preconditioning was performed with 6 mL ddH2O. Thereafter, 1.5 mL DAS (1 μM or 20 μM) was applied as a post-Striga seed conditioning treatment, together with 1.5 mL rac-GR24 (0.1 ppm). Hence, the treatments included (1): rac-GR24 (control); (2): DAS 1 μM + rac-GR24; (3): DAS 20 μM + rac-GR24. The treatments were in 3 replications each, and the experiment was done in two repetitions.
Impact on soil microbial community abundance
10 g of planting soil substrate were filled into 9 cm petri dishes, and supplied with either 10 mL of ddH2O, DAS 1 μM or DAS 20 μM. In a parallel setup, 10 mg unsterilized S. hermonthica seeds were mixed into the planting soil substrate in each of the petri dishes. Therefore, the treatments consisted of (1): ddH2O (Striga-neg. control); (2): ddH2O + Striga (Striga-pos. control); (3): DAS 1 μM; (4): DAS 1 μM + Striga; (5): DAS 20 μM; (6): DAS 20 μM + Striga. The treatments were in 3 replications each. Then, the petri dishes were covered and incubated in a dark chamber at 28 °C for 14 d. Thereafter, total DNA (deoxyribonucleic acid) was isolated from 500 mg of the incubated treatments using the Fast DNA® spin kit for soil (MP Biomedicals LLC, Solon, Ohio, USA). Effect of DAS on the soil microbial community abundance was evaluated from the major soil microbial groups i.e., bacteria and fungi, through the 16S ribosomal RNA (rRNA) and 18S rRNA gene copy numbers, respectively. This was done by real-time quantitative PCR (qPCR) on the StepOnePlus™ Real-Time PCR System (Applied Biosystems, Waltham, Massachusetts, USA). At a standardized DNA concentration of 10 ng μl− 1 for each treatment, a 20 μL qPCR reaction system, with 3-step cycling, was performed using the SensiFAST™ SYBR® Hi-ROX Kit (Bioline Meridian Bioscience, Paris, France), according to the manufacturer’s protocol. The primer information is provided in Table 2. qPCR runs for each treatment consisted of 3 biological replicates into 3 technical replicates.
Table 2.
List of primers sets used for qPCR and RT-PCR analyses
| Primer id | Nucleotide sequence (5′ → 3′) | Target gene | Target group | Reaction | Reference |
|---|---|---|---|---|---|
| Eub338 (F) | ACCTACGGGAGGCAGCAG | 16S rRNA | Bacteria | qPCR | [39] |
| Eub518 (R) | ATTACCGCGGCTGCTGG | 16S rRNA | Bacteria | qPCR | [50] |
| FF390 (F) | CGATAACGAACGAGACCT | 18S rRNA | Fungi | qPCR | [73] |
| FR1 (R) | AICCATTCAATCGGTAIT | 18S rRNA | Fungi | qPCR | [73] |
| FsTri5 (F) | TGGAGAACTGGATGGTCTGG | Tri5 | TpFs | RT-qPCR | [2] |
| FsTri5 (R) | GACATAGCCGTGCATGAAGC | Tri5 | TpFs | RT-qPCR | [2] |
| FsTri4 (F) | GCCACTGCTGCTACTGTTGA | Tri4 | TpFs | RT-qPCR | [2] |
| FsTri4 (R) | GGTCGTTGTCCAGATGTTCTTG | Tri4 | TpFs | RT-qPCR | [2] |
| EF1A (F) | GGCTTTCACCGACTACCCTCCTCT | EF1A | Eukaryotes | RT-qPCR | [32] |
| EF1A (R) | ACTTCTCGACGGCCTTGATGACAC | EF1A | Eukaryotes | RT-qPCR | [32] |
F – Forward primer
R – Reverse primer
TpFs – Trichothecene producing Fusarium species (especially characterized in F. graminearum and F. sporotrichioides)
Degradation of DAS in the planting soil substrate (targeted metabolomics)
For sample isolation, the left-overs of the soil microbial cultures (see above) were transferred to sterile 50 mL falcon tubes, and 10 mL ethyl acetate (Merck KGaA, Darmstadt, Germany) were added to each tube. The tubes were incubated in a dark shaker overnight, at 100 rpm and 27 °C. Thereafter, the tubes were centrifuged for 20 min, at 4750 rpm and 4 °C, using the Allegra® X-15R centrifuge (Beckman Coulter GmbH, Krefeld, Germany). The ethyl acetate phase was isolated and utilized for targeted metabolomic analysis by liquid chromatography–mass spectrometry (LC-MS) system. For this, 500 μL of the ethyl acetate phase were completely dried under N2, and resuspended with 150 μL methanol (VWR, Radnor, Pennsylvania, USA). Five μL of the solution were injected into the Agilent 1290 Infinity LC, coupled with the HPLC column ZORBAX Eclipse Plus C18, 95 Å, 1.8 μm, 2.1 × 50 mm, at 40 °C (Agilent Technologies GmbH, Waldbronn, Germany). Mobile phases included 0.2% formic acid (VWR) (solvent A), and acetonitrile (VWR) with 0.2% (v/v) formic acid (solvent B). At a flow rate of 0.3 mL min− 1, the ‘time – composition’ gradient program was set at 0.00(min) – B = 0.0%, 1.50(min) – B = 0.0%, 7.00(min) – B = 15.0%, 19.00(min) – B = 75.0%, 22.00(min) – B = 100.0%, 22.10(min) – B = 0.0%, and 23.10(min) – B = 0.0%. All solvents used were LC-MS grade. The purified fraction, and standard DAS, were measured by electrospray ionization and fourier transform mass spectrometry, using the Q Exactive™ Plus Orbitrap™ System (Thermo Fisher Scientific, Waltham, Massachusetts, USA). It made use of positive polarity mode, with 4.2 kV spray capillary. A desolvation temperature of 380 °C and stepped collision energy of 20, 60, and 110 V were maintained. The data dependent acquisition method was adopted, with a full MS scan of 70,000 resolution, followed by dd-MS2 (TOP 5) 140 to 1200 m/z of 17,500 resolution. From the obtained [M + H] +, [M + Na] + and [M + NH4] + adducts, the more intense [M + NH4] + adduct was selected for further analysis. For the standard DAS, at a retention time of 11.17 min, it was measured at 384.201 m/z.
Screening of Fos exometabolome for DAS production
Fungal culturing and metabolic footprinting
1.6 cm agar-mycelial plug from actively growing potato dextrose agar (PDA) (Carl Roth GmbH) cultures of Fos isolates (Foxy-2 and FK3), including F. venenatum (as control), were separately transferred to sterile 50 mL falcon tubes containing 35 mL of 25% potato dextrose broth (PDB) (Carl Roth GmbH). Fusarium venenatum was used as control because of its ability to produce DAS [46]. In a parallel setup, 35 mg unsterilized S. hermonthica seeds were added to each tube. Thus, the treatments included (1): Foxy-2; (2): Foxy-2 + Striga; (3): FK3; (4): FK3 + Striga; (5): F. venenatum; (6): F. venenatum + Striga. The treatments were in 3 replications each. Then, the tubes were incubated in a dark shaker for 2 weeks, at 65 rpm and 28 °C. Following a similar experimental design, Foxy-2, FK3 and F. venenatum were also cultured using Czapek-Dox broth containing 2% peptone (Carl Roth GmbH), with static incubation for 3 weeks at 22 °C, under 16 h light and 8 h dark conditions [63]. Thereafter, 10 mL ethyl acetate were added to every tube, and they were re-incubated overnight. Then, the tubes were centrifuged for 20 min, at 4750 rpm and 4 °C. The ethyl acetate phase of the treatments was isolated and used for exometabolomic analysis by LC-MS system as described above.
RNA isolation and gene expression
Total RNA (ribonucleic acid) was isolated from 100 mg hyphal biomass of actively growing PDA cultures of Foxy-2, FK3 and F. venenatum (ISOLATE II RNA Plant Kit, Bioline GmbH, Luckenwalde, Germany). Trichothecene gene expression by the fungal isolates were determined by quantitative reverse transcription PCR (RT-qPCR), using the StepOnePlus™ Real-Time PCR System. At a standardized RNA concentration of 10 ng μl− 1 for each fungal sample, a 20 μl RT-qPCR reaction system, with 3-step cycling, was performed using the SensiFAST™ SYBR® Hi-ROX One-Step Kit (Bioline Meridian Bioscience), according to the manufacturer’s protocol. Trichothecene genes, Tri5 and Tri4, were the genes of interest. These are the first genes involved in the Fusarium trichothecene biosynthetic pathway, from the starting compound farnesyl pyrophosphate. Tri5 encodes trichodiene synthase (which converts farnesyl pyrophosphate to trichodiene), while Tri4 encodes a cytochrome P450 monooxygenase (which converts trichodiene to isotrichotriol). Isotrichotriol is converted to isotrichodermol (i.e., the first trichothecene in the pathway) through a non-enzymatic process. In presence of other relevant Tri-genes (i.e., Tri101, Tri11, Tri3, Tri13 and Tri7, and Tri8), isotrichodermol is eventually converted to more complex trichothecene chemical species e.g., DAS [29, 35]. Thus, Fusarium strains lacking trichodiene synthase do not produce trichothecenes [46]. A housekeeping gene, Eukaryotic translation elongation factor 1 alpha (EF1A), was used as the reference gene [32]. The primer information is given in Table 2. RT-qPCR runs for each fungal sample consisted of 3 biological replicates into 3 technical replicates. Relative gene expression was calculated by the Pfaffl equation [56].
Biostatistical analysis
Quantitative data from all the experiments in this study were tested by the analysis of variance (ANOVA), and Tukey’s range test (post hoc analysis), using SAS 9.4 (SAS Institute, Cary, North Carolina, USA). In presenting the Tukey’s range test of the S. hermonthica incidence and sorghum ABM from the planting experiments, the mean data were standardized (as percentage) in relation to the respective control treatments, without making alterations to the analyses output. Thus, the S. hermonthica incidence data were standardized relative to the ‘Striga-pos. Control’, and the sorghum ABM data were standardized relative to the ‘Striga-neg. Control’.
Results
The most potent/efficient candidate exometabolite against S. hermonthica
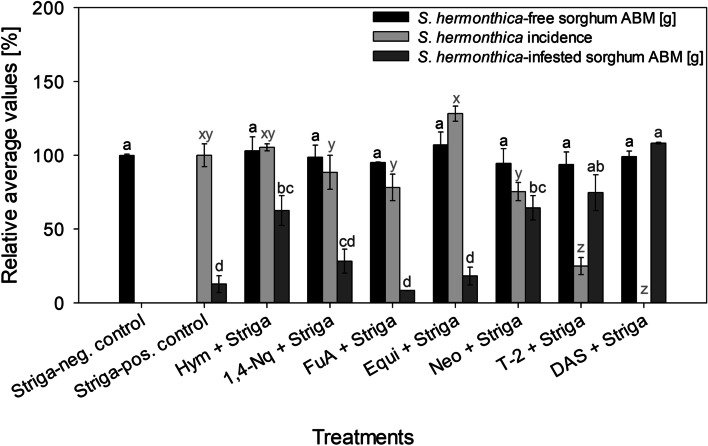
ANOVA indicated that at the various study concentrations, the treatments caused different effect on S. hermonthica seed germination (P < 0.0001). Tukey’s range test further revealed that at 1–10 μM, some exometabolites, particularly DAS, T-2 and Neo, exhibited stronger S. hermonthica seed germination inhibition compared to the others (Table 3). This concentration range (1–10 μM) was also characterised by wide variability (standard error (SE) > 1) in most of the treatments effect, including high instability of the response (S. hermonthica seed germination inhibition) in relation to increasing treatment concentration. Nevertheless, as from 10 μM, inhibition of S. hermonthica seed germination followed a concentration-dependent trend in relation to increasing treatment concentration. At ≥20 μM, all the exometabolites significantly reduced S. hermonthica seed germination, and they displayed more accurate/stable treatment effects (SE < 1). Hence, 20 μM was utilized as the exometabolite treatments working concentration in the planting experiments. All exometabolites completely inhibited S. hermonthica seed germination at 100 μM. Out of all tested exometabolites, only DAS completely inhibited S. hermonthica seed germination at every concentration (1–100 μM). The planting experiments revealed that at 20 μM, none of the exometabolites affected the ABM of S. hermonthica-free sorghum (P > 0.05). However, only DAS and T-2 supressed S. hermonthica incidence (P < 0.0001) (Fig. 1). Correspondingly, except for DAS and T-2, the ABM of S. hermonthica-infested sorghum for the rest exometabolites was lower than the Striga-neg. Control (P < 0.0001). DAS, nonetheless, was outstanding compared to T-2, because DAS completely hindered S. hermonthica incidence. This resulted in a larger ABM of S. hermonthica-infested sorghum. Further evaluation of DAS and T-2 showed that their ability to prevent S. hermonthica incidence was lower at 1 μM (Fig. 2). Nevertheless, DAS still suppressed S. hermonthica incidence (P < 0.05), while S. hermonthica incidence for T-2 was not different from the Striga-pos. Control (P > 0.05). The ABM of S. hermonthica-infested sorghum for both exometabolites were not different to the Striga-neg. Control (P > 0.05).
Table 3.
Tukey’s range test for S. hermonthica germination percentages (average) at different exometabolite concentrations
| 1 μM | 5 μM | 10 μM | 20 μM | 50 μM | 100 μM | |
|---|---|---|---|---|---|---|
| ddH2O | 55.000 (1.414) a | 55.000 (1.414) a | 55.000 (1.414) ab | 55.000 (1.414) a | 55.000 (1.414) a | 55.000 (1.414) a |
| Hym | 37.958 (4.495) ab | 40.798 (1.366) a | 11.362 (1.862) de | 0.551 (0.225) c | 0.689 (0.264) b | 0.000 (0.000) b |
| 1,4-NQ | 32.148 (3.309) bc | 34.859 (6.083) ab | 37.958 (8.161) bc | 4.135 (0.574) b | 0.965 (0.347) b | 0.000 (0.000) b |
| FuA | 28.662 (2.569) bc | 35.117 (5.164) ab | 16.268 (8.521) de | 1.516 (0.823) bc | 0.000 (0.000) b | 0.000 (0.000) b |
| Equi | 15.106 (6.939) cd | 54.225 (6.512) a | 26.338 (0.894) cd | 0.551 (0.225) c | 0.000 (0.000) b | 0.000 (0.000) b |
| Neo | 3.486 (2.480) d | 5.423 (0.775) c | 0.775 (0.447) e | 0.000 (0.000) c | 0.000 (0.000) b | 0.000 (0.000) b |
| T-2 | 4.648 (1.096) d | 0.775 (0.775) c | 0.000 (0.000) e | 0.000 (0.000) c | 0.000 (0.000) b | 0.000 (0.000) b |
| DAS | 0.000 (0.000) d | 0.000 (0.000) c | 0.000 (0.000) e | 0.000 (0.000) c | 0.000 (0.000) b | 0.000 (0.000) b |
Means having at least a letter in common are not significantly different (α = 0.05). Standard error (SE) is in parenthesis
Fig. 1.
Relative average values of S. hermonthica-free sorghum ABM (aboveground dry biomass), S. hermonthica incidence and S. hermonthica-infested sorghum ABM to the chemical treatments at 20 μM concentration. Striga-neg. control = ddH2O without S. hermonthica. Striga-pos. control = ddH2O with S. hermonthica. Bars having at least a letter in common represent treatments means that are not significantly different (α = 0.05). Error bars indicate standard error (SE)
Fig. 2.
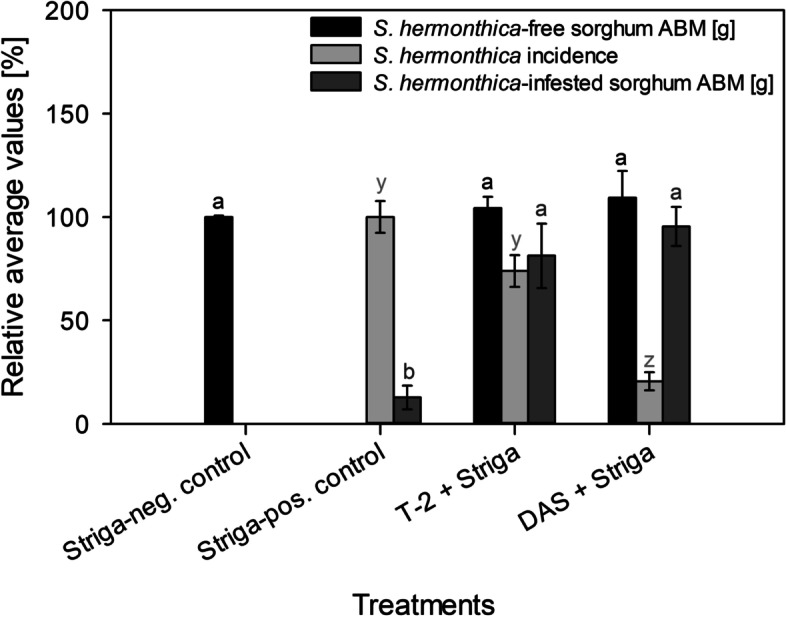
Relative average values of S. hermonthica-free sorghum ABM (aboveground dry biomass), S. hermonthica incidence and S. hermonthica-infested sorghum ABM to T-2 and DAS at 1 μM concentration. Striga-neg. control = ddH2O without S. hermonthica. Striga-pos. control = ddH2O with S. hermonthica. Bars having at least a letter in common represent treatments means that are not significantly different (α = 0.05). Error bars indicate standard error (SE)
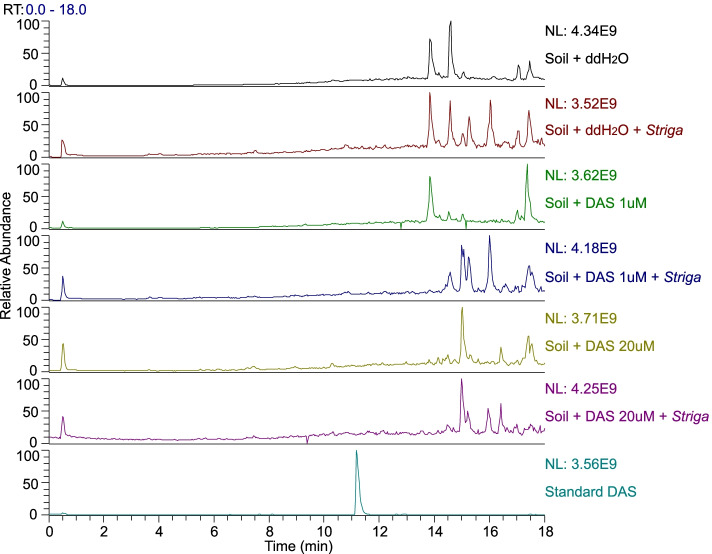
At 20 μM, DAS consistently inhibited the seed germination of diverse S. hermonthica populations from Abuja (Nigeria), Wad-Medani (Sudan) and Kibos (Kenya) completely (Table 4). Complete germination inhibition of the diverse S. hermonthica populations was also achieved at 1 μM DAS, except for the population from Kibos, which was almost completely inhibited (P < 0.0001). Similarly, application of DAS as a post-Striga seed conditioning treatment resulted in the complete inhibition of S. hermonthica seed germination at 20 μM (Fig. 3). This potential was reduced at 1 μM DAS, nevertheless, it was effective (P < 0.0001). Incubation of the planting soil substrate with 20 μM DAS for 14 days, strikingly increased the copy numbers of the 16S rRNA (bacterial community) (P < 0.0001) and the 18S rRNA (fungal community) genes (P < 0.01) (Fig. 4). The difference between the Striga-pos. and Striga-neg. control treatments indicated that addition of the S. hermonthica seeds moderately promoted the microbial abundance in the setup. This could have occurred either by contamination from the unsterilized S. hermonthica seeds, or the S. hermonthica seeds serving as nutrient substrate for microbial growth. However, comparing the DAS-treated samples to the control treatments revealed that DAS application stimulated proliferation of the bacterial and fungal communities in a concentration-dependent manner (i.e., 20 μM > 1 μM > control). Whereby, the fungal community multiplied faster than the bacterial community, as DAS concentration increased. LC-MS analysis revealed that DAS was eventually degraded in the planting soil substrate after the 14-d incubation. This was indicated by the absence of DAS signal in the LC-MS data of the DAS-treated samples (Fig. 5).
Table 4.
Tukey’s range test for germination percentages (average) of diverse S. hermonthica populations at different DAS concentrations
| Abuja (Nigeria) | Wad-Medani (Sudan) | Kibos (Kenya) | |
|---|---|---|---|
| ddH2O (control) | 27.375 (2.625) a | 31.250 (1.750) a | 34.500 (0.833) a |
| DAS 1 μM | 0.000 (0.000) b | 0.000 (0.000) b | 0.583 (0.468) b |
| DAS 20 μM | 0.000 (0.000) b | 0.000 (0.000) b | 0.000 (0.000) b |
Means having at least a letter in common are not significantly different (α = 0.05). Standard error (SE) is in parenthesis
Fig. 3.
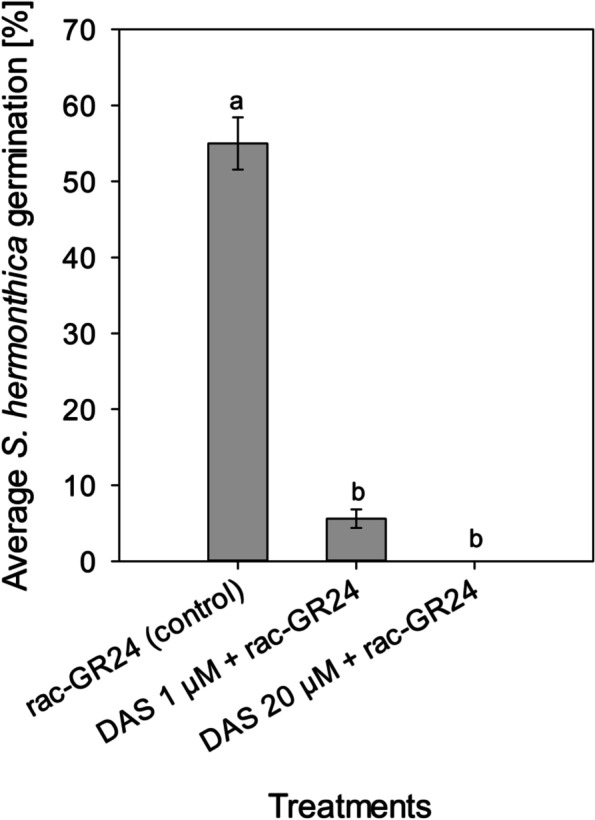
Average S. hermonthica germination response to post-Striga seed conditioning treatment with DAS. Bars having at least a letter in common represent treatments means that are not significantly different (α = 0.05). Error bars indicate standard error (SE)
Fig. 4.
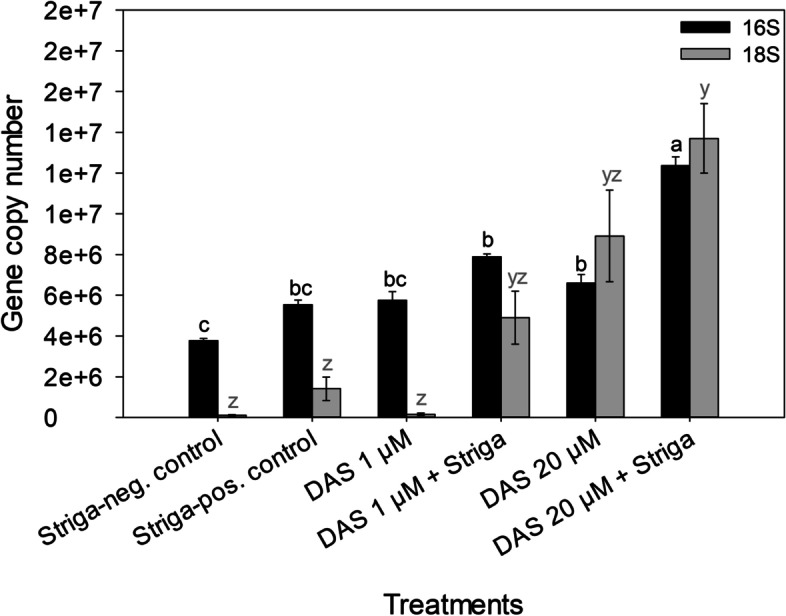
16S rRNA and 18S rRNA gene copy numbers in planting soil substrate after a 14-d incubation period. Striga-neg. control = soil + ddH2O without S. hermonthica seeds. Striga-pos. control = soil + ddH2O + S. hermonthica seeds. Bars having at least a letter in common represent treatments means that are not significantly different (α = 0.05). Error bars indicate standard error (SE)
Fig. 5.
Mass chromatogram of planting soil substrate samples after a 14-d incubation. Standard DAS signal at 11.17 min
Determination of DAS production by Fos
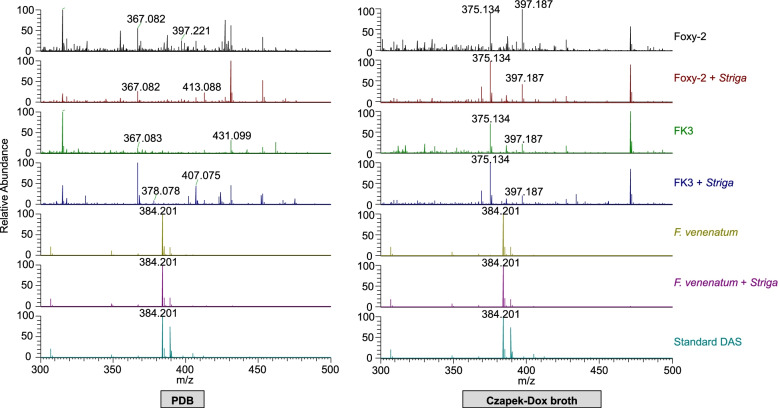
Exometabolomic investigation revealed that F. venenatum produced DAS in both the PDB and Czapek-Dox broth media cultures. This was regardless of the addition or non-addition of S. hermonthica seeds to the culturing media. Contrarily, both Fos isolates (Foxy-2, FK3) did not produce DAS under similar culturing conditions (Fig. 6). RT-qPCR analysis further revealed that for both Foxy-2 and FK3, the trichothecene genes Tri5 and Tri4 were underexpressed (P < 0.0001) (Fig. 7). Fusarium venenatum expressed Tri5 gene at 349.8-fold higher than Foxy-2, and 7951.3-fold higher than FK3. Also, F. venenatum expressed Tri4 gene at 17.7-fold higher than Foxy-2, and at 23.8-fold higher than FK3.
Fig. 6.
Mass spectrum of fungal samples cultured in PDB and Czapek-Dox broth media
Fig. 7.
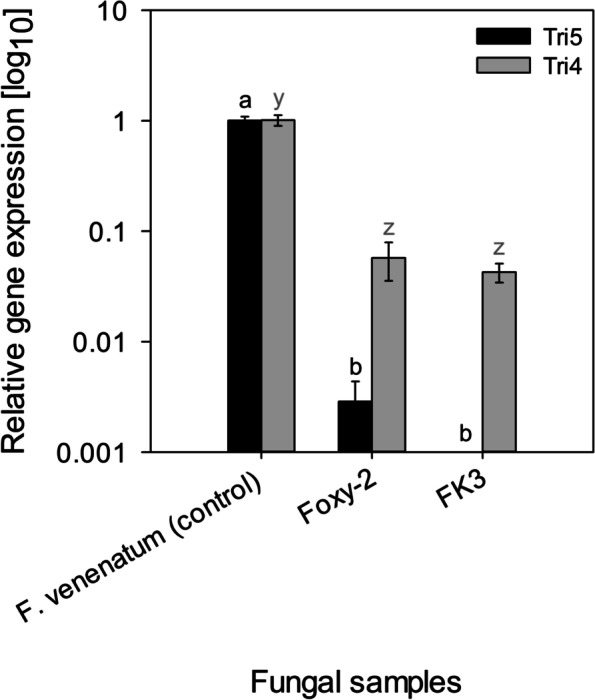
Relative expression of trichothecene genes Tri5 and Tri4 by F. venenatum and Fos isolates (Foxy-2 and FK3). Bars having at least a letter in common represent treatments means that are not significantly different (α = 0.05). Error bars indicate standard error (SE)
Discussion
DAS revealed outstanding suppression of S. hermonthica
It was the fundamental finding of this study that out of all tested Fusarium exometabolites, DAS exhibited the greatest potential to control S. hermonthica. DAS presented a clearly larger advantage than the other tested exometabolites in terms of its ability to completely inhibit S. hermonthica seed germination in vitro, at all studied concentrations (1–100 μM). Comparably, in a previous study on Orobanche ramosa (also a parasitic weed), DAS was the most promising Fusarium toxin tested as well, where it caused complete inhibition of the seed germination at 10 and 100 μM, but at 1 and 0.1 μM, it suppressed the germination to 2 and 7%, respectively [84]. With non-parasitic plants, on the other hand, 1 μM DAS failed to inhibit Arabidopsis seed germination, but at 10 μM, the germination was reduced from about 95 to 40% [42]. Furthermore, in Nicotiana sylvestris, pollen germination inhibition started from 20 ng mL− 1 (0.055 μM) DAS at 26.7%, but at 200 ng mL− 1 (0.55 μM), it was completely inhibited [65]. Because the variable susceptibility of diverse S. hermonthica populations to Fos isolates has been a major drawback in biocontrol of S. hermonthica [5, 7], the ability of DAS to consistently inhibit seed germination of diverse S. hermonthica populations in our study is a very interesting discovery. After DAS, T-2 was the second best-performing exometabolite tested in our study, followed by Neo. DAS, T-2 and Neo are all type A trichothecenes. Trichothecenes are sesquiterpene mycotoxins that are made up of a core ring structure, which contains an epoxide (tricyclic ether) at the 12,13 carbon positions, including a double bond at the 9, 10 carbon positions (see Supplementary File). These functional groups (particularly the former) are mainly responsible for their toxicity [14]. Also, type A trichothecenes differ from other trichothecene types i.e., type B (e.g., deoxynivalenol), type C (e.g., crotocin) and type D (e.g., satratoxin H), primarily because of the substitution at the carbon 8 of the core ring structure. This could be a hydroxyl group (e.g., Neo), an ester function (e.g., T-2), or no functional group/oxygen substitution (e.g., DAS) [45]. Hence, this group sameness may explain the moderately close biopesticidal potential of the type A trichothecenes against S. hermonthica in our study, particularly DAS and T-2. Nevertheless, T-2 performance was limited by several factors. For instance, it started causing complete germination inhibition of S. hermonthica seeds in vitro as from 10 μM (this was in agreement with Zonno and Vurro [83]), it was unable to completely hinder S. hermonthica incidence at 20 μM in planta, and it was unable to suppress S. hermonthica incidence at 1 μM. Moreover, due to loss of acetyl side groups on carbons 4 and 15, and an isovaleryl group at carbon 8, toxicity of DAS is generally lower [20, 42, 69]. We assume that the unique chemical structure of DAS may be the basis for its outstanding biopesticidal performance against S. hermonthica, compared to all other exometabolites tested in this study.
Preconditioning (i.e., dark stratification over a given period, in presence of moisture and suitably warm-temperature), is an indispensable, preliminary treatment, that is required for the release of parasitic plant seeds (Orobanche sp., Striga sp.) from primary dormancy, in preparation for germination [43]. During the seed preconditioning stage, essential metabolic pathways in the seed, such as respiration, synthesis of DNA, protein and hormones, are activated [17, 30]. Previous studies had mainly focused on using fungal exometabolites as post-Striga seed conditioning treatment against a single S. hermonthica population, mostly sampled from Sudan [1, 28, 67, 83]. However, as shown from our study, pre-Striga seed conditioning treatment with DAS consistently inhibited seed germination of diverse S. hermonthica populations, and DAS was effective both as a pre- and post-Striga seed conditioning treatment. It therefore implied that the mode by which DAS specifically inhibited S. hermonthica seed germination is not limited to the general disruption of vital cytological pathways or processes that occurs during S. hermonthica seed preconditioning for which trichothecenes are widely known, such as disruption of protein/DNA synthesis, mitochondrial function and mitotic division [9, 15, 24]. Perhaps, it could be connected to the disruption of other post-Striga seed conditioning pathways that are essential for S. hermonthica seed germination e.g., strigolactone perception. Because the strigolactone signal, which triggers S. hermonthica seed germination is perceived by strigolactone receptor, hence disruption of the strigolactone receptor (e.g., by mutation, or physical/chemical inhibitors) could lead to the inability of the receptor to perceive/transduce strigolactone signal [60, 62]. For instance, certain chemical inhibitors of the strigolactone receptors in some flowering plants (i.e., the α/β-hydrolase family proteins e.g., decreased apical dominance 2 in petunia (DAD2), and DWARF14 (D14) in Arabidopsis (AtD14) and rice (OsD14)), were recently identified. Examples of these inhibitors include, 2MN [41], N-Ph [25] and DL1 [80]. Thus, this major variation in the S. hermonthica seed germination pathway may serve as an important point of action for DAS. However, the exact mode of DAS inhibitory action against S. hermonthica seed germination remained elusive.
DAS application in planta did not affect sorghum biomass and soil microbial abundance
Treatment of the planting soil substrate with 20 μM DAS did not affect the ABM of S. hermonthica-free sorghum. This observation was the same for all tested exometabolites. It was previously reported that shoot morphogenesis was severely inhibited in Nicotiana tabacum grown on DAS-treated MS agar as from 5 μM, but was completely blocked at 20 μM DAS [49]. Also, Arabidopsis shoot growth was severely inhibited and root elongation completely blocked, when grown on 10 μM DAS-treated MS agar [42]. Thus, the extent of toxicity of DAS and other trichothecenes varies with the test plant [42, 55]. This, therefore, corroborates the need to ascertain an exometabolite working concentration on S. hermonthica host plants, before the exometabolite is adopted for parasitic weed management.
The soil bacterial and fungal community abundances were increased in relation to increasing DAS concentration applied to the planting soil substrate. Accordingly, this observation coincided with the complete degradation of DAS in the soil substrates after a 14-d incubation of the experimental setup. Numerous microbes, especially bacteria and fungi, possess enzymatic gene products to specifically and efficiently degrade the 12,13-epoxy ring that is important for trichothecene toxicity [16, 74]. For instance, bovine rumen microorganisms e.g., Eubacteria strain BBSH 797, selectively degrade the 12,13-epoxy ring of DAS, T-2 and deoxynivalenol by enzymatic reduction into de-epoxy and deacylated products [21, 68]. Also, widely diverse soil and freshwater bacteria species including, but not limited to, Curtobacterium sp., Bacillus sp., Pseudomonas sp., Arthrobacter sp. Agrobacterium sp., Brevibacterium sp., Blastobacter sp., Alcaligenes sp., Microbacterium sp. and Xanthomonas sp., have been reported to completely degrade T-2, DAS, Neo and type B trichothecenes (nivalenol and fusarenon-X); whereby, the degraded product is assimilated and utilized as a source of carbon and energy for cellular growth/multiplication [8, 72]. On the other hand, fungal degradation of trichothecenes is mainly by acetylation and deacetylation of the trichothecene molecule to produce significantly less-toxic acetoxy products, without necessarily affecting the trichothecene core [81]. For example, the degradation of DAS by resting cells of Mucor mucedo and growing cells of F. oxysporum f. sp. vasinfectum, to produce triacetoxyscirpene and 3-acetoxyscirpene-4,15-diol respectively [11, 12]. Thus, the ability of numerous environmental microbes to efficiently degrade trichothecenes, and assimilate the end products as carbon/energy source for cellular multiplication, may explain the correlation we observed between increasing DAS treatment concentration in the planting soil substrate and the soil microbial community abundance, whereby DAS was eventually degraded in the planting soil substrate.
Fos does not produce DAS
Fos, represented by Foxy-2 and FK3 isolates, did not produce DAS according to the exometabolomic (LC-MS) analysis. This observation corresponded to both Fos isolates underexpression of key genes (Tri5 and Tri4) that are necessary for initiating the Fusarium trichothecene biosynthesis pathway. It was on this note that the expression of Tri101, Tri11, Tri3, Tri13 and Tri7, and Tri8 genes by Fos were not further analysed in this study, since Foxy-2 and FK3 showed underexpression of Tri5 and Tri4 genes; an observation in line with the Fos exometabolomic analysis. This discovery gives a new insight that DAS (or essentially, trichothecenes) is not part of Fos exometabolome composition. This finding was in accordance with Proctor et al. [57] and Khatibi et al. [31], that categorized F. oxysporum as a non-producer of trichothecenes. However, it is worthy to note that some authors had reported the isolation of trichothecenes e.g., DAS and T-2, from F. oxysporum [22, 27, 47]. Although, Ghosal et al. [22] had clearly specified the forma specialis of the F. oxysporum they investigated i.e., F. oxysporum f. sp. carthami, which was different from ours. However, Mirocha et al. [47] and Hasan [27] did not further specify the forma specialis of F. oxysporum used in their experiment. It was revealed that F. oxysporum carry both non-functional (pseudo-Tri101) and functional (Tri201) trichothecene 3-O-acetyltransferase genes in their genome. The Tri201 gene acts like the Tri101 gene in known Fusarium trichothecene producers (e.g., F. sporotrichioides), which is responsible for C3 acetylation of 3-hydroxytrichothecenes (e.g., isotrichodermol) into a less-toxic product (e.g., isotrichodermin). This action is both a means of self-defence against toxic trichothecenes, and is also part of the Fusarium trichothecene biosynthesis pathway [33–35]. Therefore, it might be suggested that the ancestor of F. oxysporum was a trichothecene producer, before the divergence of trichothecene producers from non-producers in the evolution of Fusarium species [70]. Our finding, in contrast to Ghosal et al. [22], Mirocha et al. [47], Hasan [27], perhaps illustrates the proposed divergent evolution of trichothecene producers from non-producers in F. oxysporum.
Conclusion
Among the tested Fusarium exometabolites, DAS was unequalled for completely inhibiting S. hermonthica seed germination in vitro, and preventing S. hermonthica incidence in planta. Notwithstanding the promising attributes of DAS in this study, there is need to further investigate its specific mode of action against the germination of S. hermonthica seeds. This investigation would be a critical step, before performing in situ verification (field trials) of the S. hermonthica-biopesticidal efficacy of DAS. Through this, DAS specificity of action against the target weed (S. hermonthica) at very low concentrations (≤ 20 μM), as opposed to non-target soil organisms, will be clearly understood.
Also, our study revealed that contrary to F. venenatum, Fos is a non-producer of DAS. It therefore raises the question if F. venenatum could be a complementary bioherbicide for controlling S. hermonthica. In this regard, an additional positive feature of this ubiquitous soil-borne saprophytic fungus is its inability to produce some mycotoxins, including T-2, deoxynivalenol, nivalenol, zearalenone and sambucoin [46]. Furthermore, F. venenatum is phylogenetically closely related to the phytopathogenic fungi F. graminearum, which is globally notorious for causing Fusarium head blight in cereals and vascular wilt to non-cereal plants, however, the non-phytopathogenic status of F. venenatum was confirmed [37]. Thus, as part of future research directions, F. venenatum could be tested for its S. hermonthica incidence prevention in planta. This will reveal whether the quantity of DAS produced by F. venenatum in the soil will sufficiently prevent S. hermonthica germination, whilst unaffecting non-target organisms. Therefore, F. venenatum could serve as a sustainable, cheaper (compared to isolated/purified DAS), and proactive biocontrol agent for S. hermonthica eradication. Another option would be to test if the co-inoculation of F. venenatum and Fos isolate (with known pathogenicity towards the given S. hermonthica population) will better increase the overall S. hermonthica biocontrol efficiency through synergism. This is based on the assumption that DAS from F. venenatum will primarily attack S. hermonthica germination, while Fos will attack the incidence of germinated or attached S. hermonthica seedlings that escaped the reach of DAS in the soil.
Supplementary Information
Acknowledgements
The publication is an output of a PhD scholarship at the University of Hohenheim in the framework of the project German-Ethiopian SDG Graduate School: Climate Change Effects on Food Security (CLIFOOD) between the Food Security Centre, University of Hohenheim (Germany) and the Hawassa University (Ethiopia), supported by the German Academic Exchange Service (DAAD) with funds from the German Federal Ministry for Economic Cooperation and Development (BMZ).
We thank the following individuals for their kind assistance in this study: Dr. Charles I. Nwankwo (University of Hohenheim, Stuttgart, Germany) for sorghum seeds from Maradi, Niger; Mr. Nnanna N. Unachukwu (International Institute of Tropical Agriculture (IITA), Ibadan, Nigeria) for S. hermonthica seeds from Abuja, Nigeria; Mr. Tamirat Tesfaye Abegaz (Sirinka Agricultural Research Centre (SARC), Sirinka, Ethiopia) and Dr. Gashaw Meteke (Hawassa University, Hawassa, Ethiopia) for S. hermonthica seeds from Sirinka, Ethiopia; Dr. Dan Makumbi (International Maize and Wheat Improvement Centre (CIMMYT), Kenya) for S. hermonthica seeds from Kibos, Kenya; and Prof. Dr. Joachim Sauerborn (emeritus professor, University of Hohenheim, Stuttgart, Germany) for S. hermonthica seeds from Wad Medani, Sudan through Dr. Eldur Balla Zahran (University of Gezira, Wad Medani, Sudan); Dr. Jens Pfannstiel (University of Hohenheim, Stuttgart, Germany) for general advice on the design of the metabolomic experiments.
Abbreviations
- 1,4-Nq
1,4-naphthoquinone
- ABM
Aboveground dry biomass
- ANOVA
Analysis of variance
- CIMMYT-Kenya
International Maize and Wheat Improvement Centre, Kibos research facility, Kenya
- DAS
Diacetoxyscirpenol
- DNA
Deoxyribonucleic acid
- EF1A
Eukaryotic translation elongation factor 1 alpha
- Equi
Equisetin
- F
Forward primer
- Fos
Fusarium oxysporum f. sp. strigae
- FuA
Fusaric acid
- Hym
Hymeglusin
- IITA-Nigeria
International Institute of Tropical Agriculture, Ibadan, Nigeria
- LC-MS
Liquid chromatography-mass spectrometry
- Neo
Neosolaniol
- PDA
Potato dextrose agar
- PDB
Potato dextrose broth
- qPCR
Real-time quantitative Polymerase chain reaction
- R
Reverse primer
- RNA
Ribonucleic acid
- rRNA
Ribosomal ribonucleic acid
- RT-qPCR
Quantitative reverse transcription Polymerase chain reaction
- SARC-Ethiopia
Sirinka Agricultural Research Centre, Sirinka, Ethiopia
- SE
Standard error
- SSA
Sub-Saharan Africa
- T-2
T-2 toxin
- TpFs
Trichothecene producing Fusarium species
- UG-Sudan
University of Gezira, Wad Medani, Sudan
Authors’ contributions
WOA designed and performed the experiments, collected and analysed the data, and prepared the manuscript. IK performed the LC-MS analyses. FR conceived of the study, supervised the work, and critically reviewed the manuscript draft. All authors have read and approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. The study was funded by the German Federal Ministry for Economic Cooperation and Development (BMZ). Supported by the German Academic Exchange Service (DAAD). The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All data supporting the findings in this study are presented within the manuscript.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors have no competing interest to declare.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ahmed NE, Sugimoto Y, Babiker AGT, Mohamed OE, Ma Y, Inanaga S, Nakajima H. Effects of Fusarium solani isolates and metabolites on Striga germination. Weed Sci. 2001;49(3):354–358. [Google Scholar]
- 2.Alexander NJ, McCormick SP, Blackburn JA. Effects of xanthotoxin treatment on trichothecene production in Fusarium sporotrichioides. Can J Microbiol. 2008;54(12):1023–1031. doi: 10.1139/W08-100. [DOI] [PubMed] [Google Scholar]
- 3.Amalfitano C, Pengue R, Andolfi A, Vurro M, Zonno MC, Evidente A. HPLC analysis of fusaric acid, 9,10-dehydrofusaric acid and their methyl esters, toxic metabolites from weed pathogenic Fusarium species. Phytochem Anal. 2002;13(5):277–282. doi: 10.1002/pca.648. [DOI] [PubMed] [Google Scholar]
- 4.Amelework BA, Shimelis HA, Tongoona P, Mengistu F, Laing MD, Ayele DG. Sorghum production systems and constraints, and coping strategies under drought-prone agro-ecologies of Ethiopia. South African Journal of Plant and Soil. 2016;33(3):207–217. [Google Scholar]
- 5.Anteyi WO, Rasche F. Population genetic structure and marker-trait associations in east and west African Striga hermonthica with varying phenotypic response to Fusarium oxysporum f. sp. strigae isolates Foxy-2 and FK3. Plant J. 2020;104(2):391–402. doi: 10.1111/tpj.14930. [DOI] [PubMed] [Google Scholar]
- 6.Anteyi WO, Rasche F. Role and in vivo localization of Fusarium oxysporum f. sp. strigae and Bacillus subtilis in an integrated Striga hermonthica biocontrol system. PhytoFrontiers™. 2021;1(1):51–61. [Google Scholar]
- 7.Avedi EK, Ochieno DMW, Ajanga S, Wanyama C, Wainwright H, Elzein A, Beed FD. Fusarium oxysporum f. sp. strigae strain foxy 2 did not achieve biological control of Striga hermonthica parasitizing maize in Western Kenya. Biol Control. 2014;77:7–14. [Google Scholar]
- 8.Beeton S, Bull AT. Biotransformation and detoxification of T-2 toxin by soil and freshwater bacteria. Appl Environ Microbiol. 1989;55(1):190–197. doi: 10.1128/aem.55.1.190-197.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bin-Umer MA, McLaughlin JE, Basu D, McCormick S, Tumer NE. Trichothecene mycotoxins inhibit mitochondrial translation—implication for the mechanism of toxicity. Toxins. 2011;3(12):1484–1501. doi: 10.3390/toxins3121484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cimmino A, Masi M, Evidente M, Superchi S, Evidente A. Fungal phytotoxins with potential herbicidal activity: chemical and biological characterization. Nat Prod Rep. 2015;32(12):1629–1653. doi: 10.1039/c5np00081e. [DOI] [PubMed] [Google Scholar]
- 11.Claridge CA, Schmitz H. Microbial and chemical transformations of some 12,13-epoxytrichothec-9,10-enes. Appl Environ Microbiol. 1978;36(1):63–67. doi: 10.1128/aem.36.1.63-67.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claridge CA, Schmitz H. Production of 3-acetoxyscirpene-4,15-diol from anguidine (4,15-diacetoxyscirpene-3-ol) by Fusarium oxysporum f.sp. vasinfectum. Appl Environ Microbiol. 1979;37(4):693–696. doi: 10.1128/aem.37.4.693-696.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Klerk M. The characterization of populations of Fusarium oxysporum f. sp. strigae from Kenya and Nigeria. In: diversity and detection of Kenyan and Nigerian populations of Fusarium oxysporum f.sp. strigae. Master thesis, Stellenbosch University, Chapter. 2017;2:25–41. [Google Scholar]
- 14.de Oliveira CAF, Corassin CH, Corrêa B, Oswald IP. Animal health: Mycotoxins. In: Van Alfen NK, editor. Encyclopedia of agriculture and food systems. Pages: Academic Press; 2014. p. 358–77. isbn:9780080931395.
- 15.Desjardins AE, Hohn TM, McCormick SP. Trichothecene biosynthesis in Fusarium species: chemistry, genetics, and significance. Microbiol Mol Biol Rev. 1993;57(3):595–604. doi: 10.1128/mr.57.3.595-604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devreese M, De Backer P, Croubels S. Different methods to counteract mycotoxin production and its impact on animal health. Vlaams Diergeneeskundig Tijdschrift. 2013;82(4):181–190. [Google Scholar]
- 17.Ejeta G. (2005). Integrating biotechnology, breeding and agronomy in the control of the parasitic weed Striga spp. in sorghum. In proceedings of the international congress “in the wake of the double Helix: from the green revolution to the gene revolution” (Tuberosa R, Phillips RL and Gale M. (eds)). Bologna, Italy, pp. 239–251.
- 18.El-Sayed W. Biological control of weeds with pathogens: current status and future trends. Journal of Plant Diseases and Protection. 2005;112(3):209–221. [Google Scholar]
- 19.Emechebe AM, Ellis-Jones J, Schulz S, Chikoye D, Douthwaite B, Kureh I, Tarawali G, Hussaini MA, Kormawa P, Sanni A. Farmers' perception of the striga problem and its control in northern Nigeria. Exp Agric. 2004;40(2):215–232. [Google Scholar]
- 20.Eudes F, Comeau A, Rioux S, Collin J. Phytotoxicity of eight mycotoxins associated with the fusariosis of wheat spikelets. Can J Plant Pathol. 2000;22(3):286–292. [Google Scholar]
- 21.Fuchs E, Binder EM, Heidler D, Krska R. Structural characterization of metabolites after the microbial degradation of type a trichothecenes by the bacterial strain BBSH 797. Food Addit Contam. 2002;19(4):379–386. doi: 10.1080/02652030110091154. [DOI] [PubMed] [Google Scholar]
- 22.Ghosal S, Chakrabarti DK, Basu Chaudhury KC. Toxic substances produced by Fusarium I: Trichothecene derivatives from two strains of Fusarium oxysporum f. sp. carthami. J Pharm Sci. 1976;65(1):160–161. doi: 10.1002/jps.2600650145. [DOI] [PubMed] [Google Scholar]
- 23.Gressel J, Hanafi A, Head G, Marasas W, Obilana AB, Ochanda J, Souissi T, Tzotzos G. Major heretofore intractable biotic constraints to African food security that may be amenable to novel biotechnological solutions. Crop Prot. 2004;23(8):661–689. [Google Scholar]
- 24.Gurdaswani V and Ghag SB. (2020). Toxins from Fusarium species and their role in animal and plant diseases. In: new and future developments in microbial biotechnology and bioengineering. Recent advances in application of fungi and fungal metabolites: applications in healthcare. Eds. Singh J and Gehlot P. Elsevier B.V. Ch 2. Page 11. ISBN 978-0-12-821006-2.
- 25.Hamiaux C, Drummond RSM, Luo Z, Lee HW, Sharma P, Janssen BJ, Perry NB, Denny WA, Snowden KC. Inhibition of strigolactone receptors by N-phenylanthranilic acid derivatives: structural and functional insights. J Biol Chem. 2018;293(17):6530–6543. doi: 10.1074/jbc.RA117.001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harding DP, Raizada MN. Controlling weeds with fungi, bacteria and viruses: a review. Front Plant Sci. 2015;6:659. doi: 10.3389/fpls.2015.00659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hasan HAH. Phytotoxicity of pathogenic fungi and their mycotoxins to cereal seedling viability. Mycopathologia. 1999;148(3):149–155. doi: 10.1023/a:1007164617175. [DOI] [PubMed] [Google Scholar]
- 28.Idris AE, Abouzeid MA, Boari A, Vurro M, Evidente A. Identification of phytotoxic metabolites of a new Fusarium sp. inhibiting germination of Striga hermonthica seeds. Phytopathol Mediterr. 2003;42(1):65–70. [Google Scholar]
- 29.Ismail Y, McCormick S, Hijri M. A fungal symbiont of plant-roots modulates mycotoxin gene expression in the pathogen Fusarium sambucinum. PLoS One. 2011;6(3):e17990. doi: 10.1371/journal.pone.0017990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joel DM. The long-term approach to parasitic weeds control: manipulation of specific developmental mechanisms of the parasite. Crop Prot. 2000;19(8–10):753–758. [Google Scholar]
- 31.Khatibi PA, Newmister SA, Rayment I, McCormick SP, Alexander NJ, Schmale DG., 3rd Bioprospecting for trichothecene 3-O-acetyltransferases in the fungal genus Fusarium yields functional enzymes with different abilities to modify the mycotoxin deoxynivalenol. Appl Environ Microbiol. 2011;77(4):1162–1170. doi: 10.1128/AEM.01738-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim HK, Yun SH. Evaluation of potential reference genes for quantitative RT-PCR analysis in Fusarium graminearum under different culture conditions. The Plant Pathology Journal. 2011;27(4):301–309. [Google Scholar]
- 33.Kimura M, Shingu Y, Yoneyama K, Yamaguchi I. Features of Tri101, the trichothecene 3-O-acetyltransferase gene, related to the self-defense mechanism in Fusarium graminearum. Biosci Biotechnol Biochem. 1998;62(5):1033–1036. doi: 10.1271/bbb.62.1033. [DOI] [PubMed] [Google Scholar]
- 34.Kimura M, Tokai T, Matsumoto G, Fujimura M, Hamamoto H, Yoneyama K, Shibata T, Yamaguchi I. Trichothecene nonproducer Gibberella species have both functional and nonfunctional 3-O-acetyltransferase genes. Genetics. 2003;163(2):677–684. doi: 10.1093/genetics/163.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimura M, Tokai T, Takahashi-Ando N, Ohsato S, Fujimura M. Molecular and genetic studies of Fusarium trichothecene biosynthesis: pathways, genes, and evolution. Biosci Biotechnol Biochem. 2007;71(9):2105–2123. doi: 10.1271/bbb.70183. [DOI] [PubMed] [Google Scholar]
- 36.Kimura Y, Shimada A, Nakajima H, Hamasaki T. Structures of naphthoquinones produced by the fungus, Fusarium sp., and their biological activity toward pollen germination. Agric Biol Chem. 1988;52(5):1253–1259. [Google Scholar]
- 37.King R, Brown NA, Urban M, Hammond-Kosack KE. Inter-genome comparison of the Quorn fungus Fusarium venenatum and the closely related plant infecting pathogen Fusarium graminearum. BMC Genomics. 2018;19:269. doi: 10.1186/s12864-018-4612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knudsen GR, Dandurand L-MC. Ecological complexity and the success of fungal biological control agents. Advances in Agriculture. 2014;2014:542703. [Google Scholar]
- 39.Lane D. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. West Sussex, United Kingdom: John Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- 40.Liao P, Wang H, Hemmerlin A, Nagegowda DA, Bach TJ, Wang M, Chye ML. Past achievements, current status and future perspectives of studies on 3-hydroxy-3-methylglutaryl-CoA synthase (HMGS) in the mevalonate (MVA) pathway. Plant Cell Rep. 2014;33(7):1005–1022. doi: 10.1007/s00299-014-1592-9. [DOI] [PubMed] [Google Scholar]
- 41.Mashita O, Koishihara H, Fukui K, Nakamura H, Asami T. Discovery and identification of 2-methoxy-1-naphthaldehyde as a novel strigolactone-signaling inhibitor. J Pestic Sci. 2016;41(3):71–78. doi: 10.1584/jpestics.D16-028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masuda D, Ishida M, Yamaguchi K, Yamaguchi I, Kimura M, Nishiuchi T. Phytotoxic effects of trichothecenes on the growth and morphology of Arabidopsis thaliana. J Exp Bot. 2007;58(7):1617–1626. doi: 10.1093/jxb/erl298. [DOI] [PubMed] [Google Scholar]
- 43.Matusova R, van Mourik T, Bouwmeester HJ. Changes in the sensitivity of parasitic weed seeds to germination stimulants. Seed Sci Res. 2004;14(4):335–344. [Google Scholar]
- 44.McCormick SP. Microbial detoxification of mycotoxins. J Chem Ecol. 2013;39(7):907–918. doi: 10.1007/s10886-013-0321-0. [DOI] [PubMed] [Google Scholar]
- 45.McCormick SP, Stanley AM, Stover NA, Alexander NJ. Trichothecenes: from simple to complex mycotoxins. Toxins. 2011;3(7):802–814. doi: 10.3390/toxins3070802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller JD, MacKenzie S. Secondary metabolites of Fusarium venenatum strains with deletions in the Tri5 gene encoding trichodiene synthetase. Mycologia. 2000;92(4):764–771. [Google Scholar]
- 47.Mirocha CJ, Abbas HK, Kommedahl T, Jarvis BB. Mycotoxin production by Fusarium oxysporum and Fusarium sporotrichioides isolated from Baccharis spp. from Brazil. Appl Environ Microbiol. 1989;55(1):254–255. doi: 10.1128/aem.55.1.254-255.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mounde LG, Anteyi WO, Rasche F. Tripartite interaction between Striga spp., cereals, and plant root-associated microorganisms: a review. CAB Reviews. 2020;15(005):1–17. [Google Scholar]
- 49.Muhitch MJ, McCormick SP, Alexander NJ, Hohn TM. Transgenic expression of the TRI101 or PDR5 gene increases resistance of tobacco to the phytotoxic effects of the trichothecene 4,15-diacetoxyscirpenol. Plant Sci. 2000;157(2):201–207. doi: 10.1016/s0168-9452(00)00282-x. [DOI] [PubMed] [Google Scholar]
- 50.Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59(3):695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ndambi B. Investigating the mode of action of the mycoherbicide component Fusarium oxysporum f. sp. strigae on Striga parasitizing sorghum and its implication for Striga control in Africa. PhD thesis. Institute of Plant Production and Agroecology in the Tropics and Subtropics. 2011 University of Hohenheim, Germany: Ch. 5, pp 82–101.
- 52.Nzioki HS, Oyosi F, Morris CE, Kaya E, Pilgeram AL, Baker CS, Sands DC. Striga biocontrol on a toothpick: a readily deployable and inexpensive method for smallholder farmers. Front Plant Sci. 2016;7:1121. doi: 10.3389/fpls.2016.01121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olff H, Alonso D, Berg MP, Eriksson BK, Loreau M, Piersma T, Rooney N. Parallel ecological networks in ecosystems. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2009;364(1524):1755–1779. doi: 10.1098/rstb.2008.0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ouedraogo N, Sanou J, Kam H, Traore H, Adam M, Gracen V, Danquah EY. Farmers' perception on impact of drought and their preference for sorghum cultivars in Burkina Faso. Agricultural Science Research Journal. 2017;7(9):277–284. [Google Scholar]
- 55.Perincherry L, Lalak-Kańczugowska J, Stępień Ł. Fusarium-produced mycotoxins in plant-pathogen interactions. Toxins. 2019;11(11):664. doi: 10.3390/toxins11110664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Proctor RH, McCormick SP, Alexander NJ, Desjardins AE. Evidence that a secondary metabolic biosynthetic gene cluster has grown by gene relocation during evolution of the filamentous fungus Fusarium. Mol Microbiol. 2009;74(5):1128–1142. doi: 10.1111/j.1365-2958.2009.06927.x. [DOI] [PubMed] [Google Scholar]
- 58.Rebeka GT. Integrating sorghum [Sorghum bicolor (L.) Moench] breeding and biological control using Fusarium oxysporum against Striga hermonthica in Ethiopia. PhD thesis. African Center for Crop Improvement, School of Agricultural, Earth and Environmental Sciences, College of Agriculture, Engineering and Science. 2013 University of KwaZulu-Natal, Republic of South Africa: Ch. 3&4, pp 66–107.
- 59.Reynolds TW, Waddington SR, Anderson CL, Chew A, True Z, Cullen A. Environmental impacts and constraints associated with the production of major food crops in sub-Saharan Africa and South Asia. Food Security. 2015;7(4):795–822. [Google Scholar]
- 60.Saucet SB, Shirasu K. Molecular parasitic plant-host interactions. PLoS Pathog. 2016;12(12):e1005978. doi: 10.1371/journal.ppat.1005978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Savard ME, Miller JD, Ciotola M, Watson AK. Secondary metabolites produced by a strain of Fusarium oxysporum used for Striga control in West Africa. Biocontrol Sci Tech. 1997;7(1):61–64. [Google Scholar]
- 62.Seto Y, Yasui R, Kameoka H, Tamiru M, Cao M, Terauchi R, Sakurada A, Hirano R, Kisugi T, Hanada A, Umehara M, Seo E, Akiyama K, Burke J, Takeda-Kamiya N, Li W, Hirano Y, Hakoshima T, Mashiguchi K, Noel JP, Kyozuka J, Yamaguchi S. Strigolactone perception and deactivation by a hydrolase receptor DWARF14. Nat Commun. 2019;10:191. doi: 10.1038/s41467-018-08124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shams M, Mitterbauer R, Corradini R, Wiesenberger G, Dall'Asta C, Schuhmacher R, Krska R, Adam G, Berthiller F. Isolation and characterization of a new less-toxic derivative of the Fusarium mycotoxin diacetoxyscirpenol after thermal treatment. J Agric Food Chem. 2011;59(17):9709–9714. doi: 10.1021/jf2022176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shayanowako AIT, Shimelis H, Laing MD, Mwadzingeni L. Variance components and heritability of traits related to Striga asiatica resistance and compatibility to Fusarium oxysporum f.sp. strigae in maize. Maydica. 2018;638(1):1–8. [Google Scholar]
- 65.Siriwardana TM, Lafont P. New sensitive biological assay for 12,13-epoxytrichothecenes. Appl Environ Microbiol. 1978;35(1):206–207. doi: 10.1128/aem.35.1.206-207.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spallek T, Mutuku JM, Shirasu K. The genus Striga: a witch profile. Mol Plant Pathol. 2013;14(9):861–869. doi: 10.1111/mpp.12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sugimoto Y, Ahmed NE, Yasuda N, Inanaga S. Trichothecene inhibitors of Striga hermonthica germination produced by Fusarium solani. Weed Sci. 2002;50(5):658–661. [Google Scholar]
- 68.Swanson SP, Nicoletti J, Rood HD, Jr, Buck WB, Cote LM, Yoshizawa T. Metabolism of three trichothecene mycotoxins, T-2 toxin, diacetoxyscirpenol and deoxynivalenol, by bovine rumen microorganisms. J Chromatogr. 1987;414(2):335–342. doi: 10.1016/0378-4347(87)80058-0. [DOI] [PubMed] [Google Scholar]
- 69.Thompson WL, Wannemacher RW. Structure - function relationships of 12,13-epoxytrichothecene mycotoxins in cell culture: comparison to whole animal lethality. Toxicon. 1986;24(10):985–994. doi: 10.1016/0041-0101(86)90004-8. [DOI] [PubMed] [Google Scholar]
- 70.Tokai T, Fujimura M, Inoue H, Aoki T, Ohta K, Shibata T, Yamaguchi I, Kimura M. Concordant evolution of trichothecene 3-O-acetyltransferase and an rDNA species phylogeny of trichothecene-producing and non-producing fusaria and other ascomycetous fungi. Microbiology. 2005;151(2):509–519. doi: 10.1099/mic.0.27435-0. [DOI] [PubMed] [Google Scholar]
- 71.Triolet M, Guillemin J-P, Andre O, Steinberg C. Fungal-based bioherbicides for weed control: a myth or a reality? Weed Res. 2020;60(1):60–77. [Google Scholar]
- 72.Ueno Y, Nakayama K, Ishii K, Tashiro F, Minoda Y, Omori T, Komagata K. Metabolism of T-2 toxin in Curtobacterium sp. strain 114-2. Appl Environ Microbiol. 1983;46(1):120–127. doi: 10.1128/aem.46.1.120-127.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vainio EJ, Hantula J. Direct analysis of wood-inhabiting fungi using denaturing gradient gel electrophoresis of amplified ribosomal DNA. Mycol Res. 2000;104(8):927–936. [Google Scholar]
- 74.Vanhoutte I, Audenaert K, De Gelder L. Biodegradation of mycotoxins: tales from known and unexplored worlds. Front Microbiol. 2016;7:561. doi: 10.3389/fmicb.2016.00561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Venkatesh N, Keller NP. Mycotoxins in conversation with bacteria and fungi. Front Microbiol. 2019;10:403. doi: 10.3389/fmicb.2019.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Venne J, Beed F, Avocanh A, Watson A. Integrating Fusarium oxysporum f. sp. strigae into cereal cropping systems in Africa. Pest Manag Sci. 2009;65(5):572–580. doi: 10.1002/ps.1741. [DOI] [PubMed] [Google Scholar]
- 77.Vurro M, Boari A, Evidente A, Andolfi A, Zermane N. Natural metabolites for parasitic weed management. Pest Manag Sci. 2009;65(5):566–571. doi: 10.1002/ps.1742. [DOI] [PubMed] [Google Scholar]
- 78.Wheeler MH, Stipanovic RD, Puckhaber LS. Phytotoxicity of equisetin and epi-equisetin isolated from Fusarium equiseti and F. pallidoroseum. Mycol Res. 1999;103(8):967–973. [Google Scholar]
- 79.Yoshida S, Forno DA, Cook JH, and Gomez KA. Laboratory manual for physiological studies of rice (3rd ed.), pp 61–66. The international rice research institute, Laguna, Philippines 1976.
- 80.Yoshimura M, Sato A, Kuwata K, Inukai Y, Kinoshita T, Itami K, Tsuchiya Y, Hagihara S. Discovery of shoot branching regulator targeting strigolactone receptor DWARF14. ACS Central Science. 2018;4(2):230–234. doi: 10.1021/acscentsci.7b00554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yoshizawa T, Onomoto C, Morooka N. Microbial acetyl conjugation of T-2 toxin and its derivatives. Appl Environ Microbiol. 1980;39(5):962–966. doi: 10.1128/aem.39.5.962-966.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zimmermann J, Klerk MD, Musyoki MK, Viljoen A, Watson AK, Beed F, Gorfer M, Cadisch G, Rasche F. An explicit AFLP-based marker for monitoring Fusarium oxysporum f.sp. strigae in tropical soils. Biol Control. 2015;89(4):42–52. [Google Scholar]
- 83.Zonno MC, Vurro M. Effect of fungal toxins on germination of Striga hermonthica seeds. Weed Res. 1999;39(1):15–20. [Google Scholar]
- 84.Zonno MC, Vurro M. Inhibition of germination of Orobanche ramosa seeds by Fusarium toxins. Phytoparasitica. 2002;30(5):519–524. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings in this study are presented within the manuscript.