Abstract
This study compared levels of human immunodeficiency virus type 1 RNA in plasma as measured by the Quantiplex branched-DNA and NucliSens nucleic acid sequence-based amplification assays. RNA was detectable in 118 of 184 samples (64.13%) by the Quantiplex assay and in 171 of 184 samples (92.94%) by the NucliSens assay. Regression analysis indicated that a linear relationship existed between the two sets of values (P < 0.0001), although the Quantiplex and NucliSens values were significantly different (P < 0.001), with the NucliSens values being approximately 0.323 log higher. Spearman correlation analysis indicated that the overall changes in patient viral load patterns were highly correlative between the two assays: r = 0.912, P < 0.0001. The lower limits of sensitivity were determined to be approximately 100 copies/ml and 1,200 to 1,400 copies/ml for the NucliSens and Quantiplex assays, respectively.
The measurement of human immunodeficiency virus type 1 (HIV-1) RNA levels in plasma (viral load) is presently one of the most valuable clinical tools for predicting HIV disease progression (3, 4, 6, 9, 12, 14, 17, 20), for determining the need to initiate or change antiretroviral therapy (2, 10, 11, 17, 20, 21), and for evaluating the efficacy of newly developed antiretroviral drugs (21). Several commercial assays, employing different molecular technologies, for measuring plasma HIV-1 RNA levels are available. These methods include reverse transcriptase PCR, which is used in the Amplicor HIV-1 Monitor assay (Roche Diagnostic Systems, Branchburg, N.J.) (16), branched-DNA (bDNA) techniques, which are used in the Quantiplex HIV RNA assay (Chiron Diagnostics, Emeryville, Calif.) (18, 23), and nucleic acid sequence-based amplification, which is used in the NucliSens HIV-1 RNA QT assay (Organon Teknika, Durham, N.C.) (13, 25, 26).
In addition to differences in the molecular bases of the assays, certain critical test parameters, including the comparability of the actual HIV-1 RNA copy numbers detected and the lower limits of assay sensitivity, can also vary significantly (13, 16, 18, 19, 23, 25, 26). The comparison of HIV-1 RNA values obtained from identical specimens by the various methods is important for two reasons. First, clinicians must often try to correlate viral load results from new patients to previous viral load results obtained by a different or less sensitive methodology. Second, clinical trial studies of new antiretroviral agents use different assays to measure responses to therapy. Equivalence of the RNA measurements would allow a more accurate comparison of the overall responses to new therapy regimens. Differences in HIV-1 RNA values could affect the total log reduction in RNA levels, the percentage of patients who achieve “undetectable” levels, and the determination of the potential success or failure of new experimental treatment regimens. To address this issue, we evaluated the NucliSens assay performance characteristics, including assay sensitivity, reproducibility, and accuracy, and examined the relationship between NucliSens and Quantiplex viral load values.
The NucliSens assay performance characteristics were determined by using HIV-1 RNA standards obtained from the AIDS Clinical Trials Group Virology Laboratories Quality Assurance Program (VQA). The VQA standards were serially diluted to yield the following standards (numbers in parentheses denote the number of times each standard was assayed over several independent test runs): 80 copies/ml (19), 100 copies/ml (20), 500 copies/ml (10), 1,000 copies/ml (8), 1,500 copies/ml (8), 3,000 copies/ml (20), 15,000 copies/ml (8), and 150,000 copies/ml (8). All standards were quantitated with the NucliSens assay according to the manufacturer’s instructions.
Our laboratory was able to detect HIV-1 RNA in the 80-copy/ml standard in 11 of 19 specimens tested (57.89%) and in the 100-copy/ml standard in 17 of 20 specimens (85%). RNA was detected 100% of the time at all other input levels tested. Assay reproducibility was evaluated by determining the standard deviation from the mean log value obtained for each of the standards after the multiple test runs. The standard deviations for the standards tested are as follows: 80 copies/ml, ±0.322 log; 100 copies/ml, ±0.359 log; 500 copies/ml, ±0.182 log; 1,000 copies/ml, ±0.113 log; 1,500 copies/ml, ±0.164 log; 3,000 copies/ml, ±0.096 log; 15,000 copies/ml, ±0.101 log; and 150,000 copies/ml, ±0.067 log. The overall assay variation was ±0.1755 log, which was within the expected variation of the assay in accordance with the published literature (25). Assay accuracy was evaluated by calculating the difference between the input log value and the mean of the actual log values obtained for each standard. The mean log differences from the expected log values are as follows: 80 copies/ml, +0.117 log; 100 copies/ml, −0.118 log; 500 copies/ml, −0.026 log; 1,000 copies/ml, +0.008 log; 1,500 copies/ml, −0.120 log; 3,000 copies/ml, +0.006 log; 15,000 copies/ml, −0.063 log; and 150,000 copies/ml, −0.228 log. The overall assay accuracy for the standards tested was −0.053 log. The recovery of the input copies ranged from 94.10% for 100 RNA copies/ml to 109.30% for 80 RNA copies/ml, with an overall mean recovery of 99.15% of the input RNA copies.
The relationship between NucliSens and Quantiplex HIV-1 RNA values was determined by comparing the viral loads measured by both assays in 184 serial plasma samples collected, with Institutional Review Board approval, from 14 patients participating in the protease inhibitor nelfinavir (Agouron Pharmaceuticals, Inc., La Jolla, Calif.) phase III clinical trials. In accordance with the nelfinavir clinical trial protocol, bDNA levels were analyzed in duplicate 1-ml volumes with a modified 1.0 version of the Quantiplex assay by Corning SciCor, Inc. (Indianapolis, Ind.). NucliSens HIV-1 RNA measurements were performed on site according to the manufacturer’s instructions with an initial 200-μl input volume (detection limit = 500 copies/ml). All samples undetectable at a 200-μl input volume were retested with a 1-ml input volume (detection limit = 100 copies/ml).
As shown in Table 1, the Quantiplex assay detected HIV-1 RNA in 118 of 184 specimens, and the NucliSens assay detected HIV-1 RNA in 171 of 184 specimens. With a 200-μl specimen input volume, the NucliSens assay identified 22 of 37 samples (59.46%) with HIV-1 RNA levels between 47 and 500 copies/ml. HIV-1 RNA was detected in 15 additional specimens after the specimen input volume was increased to 1 ml. All samples with RNA detectable by the Quantiplex assay were positive by the NucliSens assay. The results for 162 specimens (88.04%) were concordant based upon the detection limits of 100 copies/ml for the NucliSens assay and 500 copies/ml for the Quantiplex assay (manufacturer’s claim). There were 22 samples with discordant results that had NucliSens values of >500 copies/ml (mean value = 1,350 copies/ml), but were reported as having <500 copies/ml by the Quantiplex assay.
TABLE 1.
Comparison of HIV-1 RNA detection rates in serial plasma samples obtained with the Quantiplex and NucliSens assays
| Categorya | No. of specimens | % with RNA detected |
|---|---|---|
| bDNA positive, NucliSens positive | 118 | 64.13 |
| bDNA negative, NucliSens negative | 13 | 7.06 |
| bDNA negative, NucliSens positiveb | 31 | 16.85 |
| bDNA negative, NucliSens positivec | 22 | 11.96 |
| bDNA positive (total) | 118 | 64.13 |
| NucliSens positive (total) | 171 | 92.94 |
Lower limits of detection were 500 copies/ml for Quantiplex (bDNA) and 100 copies/ml for NucliSens.
Values of <500 copies/ml.
Values of ≥500 copies/ml.
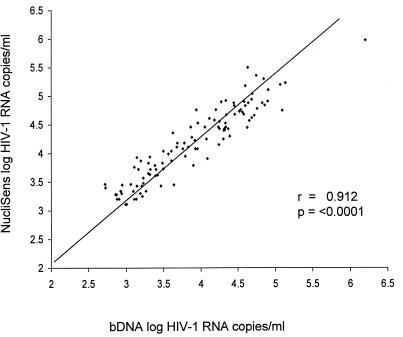
A comparison of the absolute and log-transformed values, obtained by the Wilcoxon signed-rank test, found a significant difference (P < 0.001) between the HIV-1 RNA values of the two tests. The mean ratio of Quantiplex absolute values to NucliSens absolute values was 1:2.441. The NucliSens RNA values were, on average, 0.323 log higher than the Quantiplex values. Regression analysis (Fig. 1) using the log-transformed values determined that there was a significant linear relationship between the Quantiplex and NucliSens values (r = 0.912, P < 0.0001). The intercept of 0.30 was not significantly different from 0 (95% confidence interval, −0.01 to 0.61). The slope of 0.87 was significantly different from 1 (95% confidence interval, 0.80 to 0.95). This means that although the two values correlate significantly, the NucliSens values are consistently higher than Quantiplex values. Spearman correlation coefficient analysis indicated that the overall changes in patient viral load patterns were highly correlative between the two assays (P < 0.001). These results were in agreement with several studies that found that the values obtained by the first-generation nucleic acid sequence-based amplification assay and the Amplicor reverse transcriptase PCR Monitor assay were most often comparable but significantly different from the values obtained with the Quantiplex assay (1, 5, 7, 8, 15, 19, 22, 24).
FIG. 1.
Linear regression plot of correlation between NucliSens and Quantiplex HIV-1 RNA values.
The NucliSens assay was significantly more sensitive than the Quantiplex assay in detecting HIV-1 RNA, due to two factors. First, the lower limit of detection of the NucliSens assay was determined to be approximately 100 copies/ml (with a 1-ml input) based upon validation studies using VQA standards. Second, although a direct evaluation of the Quantiplex assay sensitivity was not performed, the correlation data suggests that the detection limit is actually in the range of 1,200 to 1,400 copies/ml. This would explain the 22 discrepant samples, which had NucliSens values of >500 copies/ml but were undetectable by the Quantiplex assay. The differences observed in this study, related both to the values of the two assays and the manufacturer’s detection limits and to our estimated detection limit for the Quantiplex assay, may be related to methods of testing and quantitation. The NucliSens assay is based on target amplification, while the Quantiplex assay is based on signal amplification. The NucliSens results are extrapolated from the values obtained from three internal calibrators of known RNA copies coamplified with each test sample. Quantiplex results are based upon a calibration curve generated from external standards.
The successful use of highly active antiretroviral therapy in the treatment of HIV disease has demonstrated the need for accurate, ultrasensitive viral load assays to monitor the course of antiretroviral therapy. The ability to lower the detection limit by increasing the specimen input volume has proved to be a significant benefit of the NucliSens assay. The enhanced sensitivity of the NucliSens assay permitted the detection of a sustained rebound in viral load in 11 of the 14 patients on 15 occasions sooner than the Quantiplex assay. The rapid detection of increased viral replication could allow the earlier identification of patients on a failing regimen or indicate the need to supplement the regimen with an additional antiretroviral agent.
These studies clearly indicate that it is best to choose one viral load assay for monitoring patients and that the results from different assays should not be used interchangeably. One must be aware of the differences in the accuracy and sensitivity of the assays; all “undetectable” results are not equal. In addition, these differences must be taken into account when data from clinical trials of new antiretroviral agents is interpreted.
Acknowledgments
This study was funded in part by the Jane and Dayton Brown and Dayton Brown, Jr., Virology Laboratory and a grant from Organon Teknika Corp., Boxtel, The Netherlands.
We thank Dean Winslow of Agouron Pharmaceuticals for kindly providing Quantiplex viral load results, Joseph Romano for editorial review, and Nina Kohn for statistical analysis. We sincerely thank the nurses and volunteer patients from the North Shore University Hospital Center for AIDS Research and Treatment who faithfully participated in these studies.
REFERENCES
- 1.Alaeus A, Lidman K, Sonnerberg A, Albert J. Subtype-specific problems with quantification of plasma HIV-1 RNA. AIDS. 1997;11:859–865. doi: 10.1097/00002030-199707000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Bagnarelli P, Menzo S, Valenza A, Paolucci S, Petroni S, Scalise G, Sampaolesi R, Manzin A, Varaldo P E, Clementi M. Quantitative molecular monitoring of human immunodeficiency virus type 1 activity during therapy with specific antiretroviral compounds. J Clin Microbiol. 1995;33:16–23. doi: 10.1128/jcm.33.1.16-23.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coombs R W, Welles S L, Hooper C, Reichelderfer P S, D’Aquila R T, Japoyr A J, Johnson V A, Kuritzkes D R, Richman D D, Kwok S, Todd J, Jackson J B, De Gruttola V, Crumpacker C S, Kahn J. Association of plasma human immunodeficiency virus type 1 RNA level with risk of clinical progression in patients with advanced infection. J Infect Dis. 1996;174:704–712. doi: 10.1093/infdis/174.4.704. [DOI] [PubMed] [Google Scholar]
- 4.Coombs R W. HIV-1 burden as a marker of disease progression and clinical response to therapy in AIDS. Clin Lab Med. 1994;14:301–311. [PubMed] [Google Scholar]
- 5.Coste J, Montes B, Reynes J, Peeters M, Segarra C, Vendrell J P, Delaporte E, Segondy M. Comparative evaluation of three assays for the quantitation of human immunodeficiency virus type 1 RNA in plasma. J Med Virol. 1996;50:293–302. doi: 10.1002/(SICI)1096-9071(199612)50:4<293::AID-JMV3>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 6.Cunningham A L, Dwyer D E, Dowton D N. Viral markers in HIV infection and AIDS. J Acquired Immune Defic Syndr. 1993;6(Suppl. 1):S32–S35. [PubMed] [Google Scholar]
- 7.Dyer J R, Gilliam B L, Eron J J, Grosso L, Cohen M S, Fiscus S A. Quantitation of human immunodeficiency virus type 1 RNA in cell free seminal plasma: comparison of NASBA™ with Amplicor™ reverse transcription-PCR amplification and correlation with quantitative culture. J Virol Methods. 1996;60:161–170. doi: 10.1016/0166-0934(96)02063-0. [DOI] [PubMed] [Google Scholar]
- 8.Griffith B P, Rigsby M O, Garner R B, Gordon M M, Chacko T M. Comparison of the Amplicor HIV-1 Monitor test and the nucleic acid sequence-based amplification assay for quantitation of human immunodeficiency virus RNA in plasma, serum, and plasma subjected to freeze-thaw cycles. J Clin Microbiol. 1997;35:3288–3291. doi: 10.1128/jcm.35.12.3288-3291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hogervost E, Jurriaans S, de Wolf F, van Wijk A, Wiersma A, Valk M, Roos M, van Gemen B, Coutinho R, Miedema F, Goudsmit J. Predictors for non and slow progression in human immunodeficiency virus (HIV) type 1 infection: low viral RNA copy numbers in serum and maintenance of HIV-1 p24-specific but not V3-specific antibody levels. J Infect Dis. 1995;171:811–821. doi: 10.1093/infdis/171.4.811. [DOI] [PubMed] [Google Scholar]
- 10.Holodniy M, Katzenstein D A, Israelski D M, Merigan T C. Reduction in plasma human immunodeficiency virus ribonucleic acid after dideoxynucleoside therapy as determined by the polymerase chain reaction. J Clin Investig. 1991;88:1755–1759. doi: 10.1172/JCI115494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holodniy M, Katzenstein D A, Winters M A, Montoya J, Shafer R, Kozal M, Ragni M, Merigan T C. Gene amplification techniques to measure viral load and genotypic resistance in asymptomatic subjects treated with combination therapy. J Acquired Immune Defic Syndr. 1993;6:366–369. [PubMed] [Google Scholar]
- 12.Katzenstein T L, Pederson C, Nielsen C, Lundgren J D, Jakobsen P H, Gerstoft J. Longitudinal serum HIV RNA quantification: correlation to viral phenotype at seroconversion and clinical outcome. AIDS. 1996;10:167–173. doi: 10.1097/00002030-199602000-00006. [DOI] [PubMed] [Google Scholar]
- 13.Kievits T, van Gemen B, van Strijp D, Schukkink R, Dircks M, Adriaanse H, Malek L, Sooknanan R, Lens P. NASBA isothermal enzymatic in vitro nucleic acid amplification optimized for the diagnosis of HIV-1 infection. J Virol Methods. 1991;35:273–286. doi: 10.1016/0166-0934(91)90069-c. [DOI] [PubMed] [Google Scholar]
- 14.Mellors J W, Kingsley L A, Rinaldo C R, Todd J A, Hoo B S, Kokka R, Gupta P. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann Intern Med. 1995;122:573–579. doi: 10.7326/0003-4819-122-8-199504150-00003. [DOI] [PubMed] [Google Scholar]
- 15.Mellors J W, Munoz A, Giorgio J V, Margolick J B, Tassoni C J, Gupta P, Kingsley L A, Todd J A, Saah A J, Detels R, Phair J P, Rinaldo C., Jr Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann Intern Med. 1997;126:946–954. doi: 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 16.Mulder J, McKinney N, Christopherson C, Sninsky J, Greenfield L, Kwok S. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J Clin Microbiol. 1994;32:292–300. doi: 10.1128/jcm.32.2.292-300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Brien W A, Hartigan P M, Martin D, Esinhart J, Hill A, Benoit S, Rubin M, Lahart C, Wray N, et al. Changes in plasma HIV-1 RNA and CD4+ lymphocyte counts and the risk of progression to AIDS. N Engl J Med. 1996;334:426–431. doi: 10.1056/NEJM199602153340703. [DOI] [PubMed] [Google Scholar]
- 18.Pachl C, Todd J A, Kern D G, Sheridan P J, Fong S J, Stempien M, Hoo B, Besemer D, Yeghiazarian T, Irvine B, Kolberg J, Kokka R, Neuwald P, Urdea M S. Rapid and precise quantification of HIV-1 RNA in plasma using a branched DNA signal amplification assay. J Acquired Immune Defic Syndr. 1995;8:446–454. doi: 10.1097/00042560-199504120-00003. [DOI] [PubMed] [Google Scholar]
- 19.Revets H, Marissens D, De Wit S, Lacor P, Clumeck N, Lauwers S, Zissis G. Comparative evaluation of NASBA HIV-1 RNA QT, AMPLICOR-HIV Monitor, and QUANTIPLEX HIV RNA assay, three methods for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1996;34:1058–1064. doi: 10.1128/jcm.34.5.1058-1064.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saag M S, Holodniy M, Kuritzkes D R, O’Brien W A, Coombs R, Poscher M E, Jacobsen D M, Shaw G M, Richman D D, Volberding P A. HIV viral load markers in clinical practice. Nat Med. 1996;2:625–629. doi: 10.1038/nm0696-625. [DOI] [PubMed] [Google Scholar]
- 21.Schooley R T. Correlation between viral load measurements and outcome in clinical trials of antiviral drugs. AIDS. 1995;9(Suppl. 2):S15–S19. [PubMed] [Google Scholar]
- 22.Schuurman R, Descamps D, Weverling G J, Kaye S, Tijnagel J, Williams I, van Leeuwen R, Tedder R, Boucher C A, Brun-Vezinet F, Loveday C. Multicenter comparison of three commercial methods for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1996;34:3016–3022. doi: 10.1128/jcm.34.12.3016-3022.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Urdea M. Synthesis and characterization of branched DNA (bDNA) for the direct and quantitative detection of CMV, HBV, HCV, and HIV. Clin Chem. 1993;39:725–726. [Google Scholar]
- 24.Vandamme A M, Schmit J C, Van Dooren S, Van Laethem K, Gobbers E, Kok W, Goubu P, Witvrouw M, Peetermans W, De Clercq E, Desmyter J. Quantification of HIV-1 RNA in plasma: comparable results with the NASBA HIV-1 RNA QT and the AMPLICOR HIV Monitor test. J Acquired Immune Defic Syndr. 1996;13:127–139. doi: 10.1097/00042560-199610010-00003. [DOI] [PubMed] [Google Scholar]
- 25.van Gemen B, Wiel P V D, van Beuningen R. The one-tube quantitative HIV-1 RNA NASBA: precision, accuracy, and application. PCR Methods Appl. 1995;4:S177–S184. doi: 10.1101/gr.4.4.s177. [DOI] [PubMed] [Google Scholar]
- 26.van Gemen B, van Beuningen R, Nabbe A, van Strijp D, Jurriaans S, Lens P, Kievits T. A one-tube quantitative HIV-1 RNA NASBA nucleic acid amplification assay using electrochemiluminescence (ECL) labeled probes. J Virol Methods. 1994;49:157–168. doi: 10.1016/0166-0934(94)90040-x. [DOI] [PubMed] [Google Scholar]