Summary
Fungal infections have emerged as a major global threat to human health because of the increasing incidence and mortality rates every year. The emergence of drug resistance and limited arsenal of antifungal agents further aggravates the current situation resulting in a growing challenge in medical mycology. Here, we identified that ponatinib, an FDA‐approved antitumour drug, significantly enhanced the activity of the azole fluconazole, the most widely used antifungal drug. Further detailed investigation of ponatinib revealed that its combination with fluconazole displayed broad‐spectrum synergistic interactions against a variety of human fungal pathogens such as Candida albicans, Saccharomyces cerevisiae and Cryptococcus neoformans. Mechanistic insights into the mode of action unravelled that ponatinib reduced the efflux of fluconazole via Pdr5 and suppressed the expression of the proton pump, Pma1. Taken together, our study identifies ponatinib as a novel antifungal that enhances drug activity of fluconazole against diverse fungal pathogens.
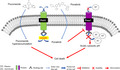
Model depiction of the role of ponatinib in potentiation of the antifungal efficacy of fluconazole by inhibition of fluconazole efflux via Pdr5 and suppression of Pma1 expression. Ponatinib (PubChem CID: 24826799) promotes intracellular fluconazole (PubChem CID:3365) accumulation by binding to Pdr5. On the other hand, ponatinib perturbs the cytosolic pH homeostasis by inhibiting the proton pump Pma1 located at the plasma membrane. Eventually, the dual action of ponatinib through these two pathways leads to cell death.
Introduction
Combinatorial therapy with commercially available drugs has been becoming an effective strategy to address the rising concern of the limited arsenal of antifungals coupled with the growing number of drug‐resistant clinical isolates and to satisfy the ever‐increasing clinical requirements (Mishra et al., 2007; Ruggero and Topal, 2014). Currently available antifungals could be usually classified into following three major categories: polyenes, echinocandins and azoles (Shapiro et al., 2011). Polyenes with antifungal activities were discovered and developed more than 60 years ago, and the molecules were found to inhibit fungal growth by binding to the ergosterol components in the plasma membrane (Anderson et al., 2014). However, translational research has shown that polyenes, which are mainly excreted from urine and bile, are able to cause severe nephrotoxicity and hepatotoxicity, and thus greatly limit their clinical applications (Gray et al., 2012). Echinocandins were introduced over 10 years ago, and the compounds were appreciated by its ability to inhibit the synthesis of (1, 3)‐β‐glucan, a major component of fungal cell wall (Shekhar‐Guturja et al., 2016b). Although echinocandins have been recognized as one of today’s best‐studied non‐ribosomal peptide natural product families, problems still exist, especially a very low level of oral bioavailability and a short half‐life. Azoles, known as the most widely used antifungal agents, have been used in clinic for more than 40 years (Shapiro et al., 2011). The azole‐based drugs were identified to effectively inactivate the ergosterol synthase (cytochrome P450) and block ergosterol synthesis, resulting in the accumulation of C‐14 methyl sterol (Shekhar‐Guturja et al., 2016b). As for the potential side‐effect of this drug family, studies have shown that metabolized azoles in the liver are mainly excreted from bile which easily results in hepatotoxicity.
Notably, the development of new antifungal turns out to be expensive, laborious and time‐consuming and cannot be relied on during the recurrent and emerging challenge of fungal infections. For example, it took nearly 30 years for echinocandins, the most recent class of antifungals, to develop and obtain success from bench to bedside (Basso et al., 2020). There is a pressing need to develop new antifungal therapeutic agents, and recent studies have strongly suggested that drug repurposing provides an attractive solution for antifungal development, with apparent advantages including the validated information about the knowledge base for the pharmacokinetics and pharmacodynamics, a dramatic shortening of the Research & Development (R&D) cycle and the huge cut of the R&D costs to achieve maximal utilization of the medical resources. There have been several examples of development of new antifungals based on this strategy. For instance, a previous study showed that sertraline, which is a selective inhibitor of central serotonin reuptake with well‐established antidepressant and anxiolytic activity, exhibits a synergistic effect with fluconazole (FLC) against Cryptococcus neoformans in a Galleria mellonella model (Spitzer et al., 2011). More recently, clofazimine, a lipophilic riminophenazine antibiotic compound which has been in clinical use for almost 40 years, but almost nothing was known about its mechanism of action, acts synergistically with fluconazole against diverse fungal species (Robbins et al., 2015). In addition, the natural product beauvericin, also a cyclohexadepsipeptide mycotoxin, was found to effectively potentiate the activity of fluconazole against some major human fungal pathogens (Shekhar‐Guturja et al., 2016b). These studies will boost the development of more systemic approaches to repurposing compounds for tackling the rising risk of fungal infection.
In this study, we provided strong evidence that ponatinib significantly potentiated fluconazole efficacy and exhibited a broad‐spectrum antifungal effect against diverse fungal pathogens, including C. albicans, S. cerevisiae and C. neoformans. More importantly, synergy testing of ponatinib and fluconazole in resistant C. albicans strains resulted in a reversal of fluconazole resistance. Following the elucidation of the mechanism of action, we finally concluded that ponatinib potentiates the antifungal efficacy of fluconazole, providing clues for developing new therapeutic strategies against fungal infections.
Results
The combination of ponatinib with fluconazole exerts a broad‐spectrum synergistic and fungicidal activity
Ponatinib, a multitargeted receptor tyrosine kinase (RTK) inhibitor, was identified through screening an FDA‐approved drug library (L4200, TargetMol) (Mei et al., 2020). We observed that a combination of ponatinib (Pona) and fluconazole (FLC) exhibited a much stronger inhibitory effect on the growth of the wild type C. albicans strain SC5314, when compared with ponatinib or fluconazole alone (Fig. 1A–D, Fig. S1). To test whether the function of ponatinib is conserved, we examined its ability to potentiate the activity of fluconazole against a variety of other fungal pathogens. As expected, ponatinib significantly enhanced the antifungal activity of fluconazole against S. cerevisiae (Fig. 1E–H) and the major human fungal pathogen C. neoformans (Fig. 1I–L). The synergistic activity of ponatinib with fluconazole was further confirmed by the checkerboard assay (FICI < 0.5), showing a powerful antifungal combination with broad‐spectrum activity against diverse human fungal pathogens (Odds, 2003; Jansen et al., 2009; Spitzer et al., 2011; Robbins et al., 2015; Shekhar‐Guturja et al., 2016b) (Table 1). Moreover, we found through a lactate dehydrogenase (LDH) assay that a concentration of ponatinib at 16 μM or lower was considered non‐toxic after incubation with the endothelial cells (Fig. S2).
Fig. 1.
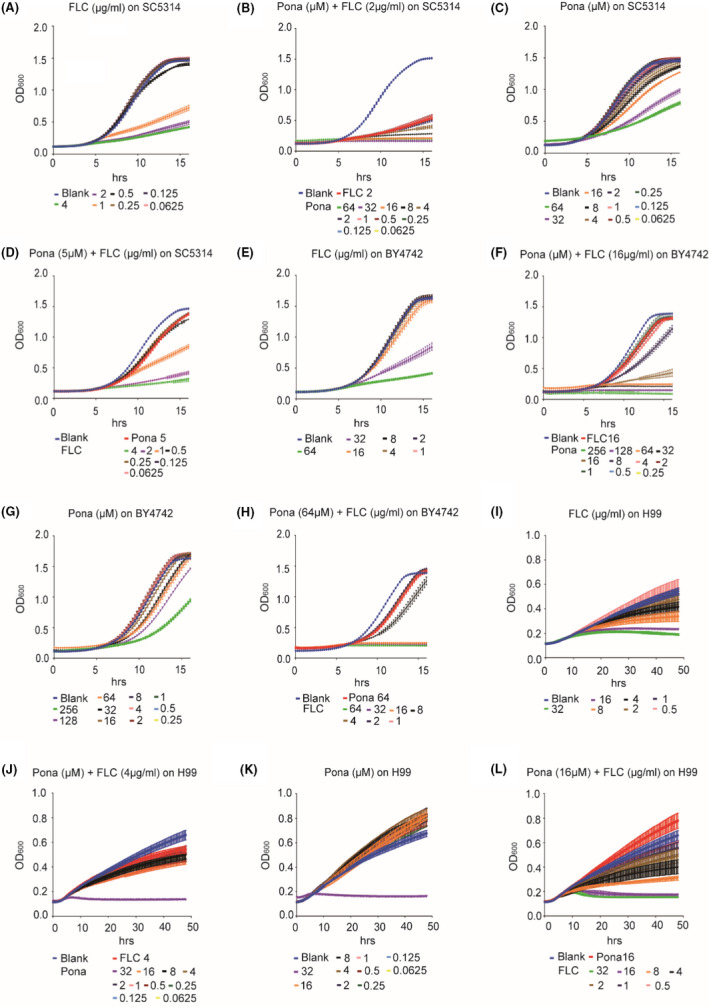
Combination of ponatinib and fluconazole synergistically inhibits growth of C. albicans, S. cerevisiae and C. neoformans. C. albicans (SC5314) (A–D), S. cerevisiae (BY4742) (E–H) and C. neoformans (H99) (I–L) isolates were subjected to twofold serial dilutions of fluconazole, ponatinib or both in YPD medium. OD600 was measured every 15 min at 30°C, and FICI was calculated using the reference guidelines for CLSI broth microdilution method (M38‐A).
Table 1.
Interaction between fluconazole and ponatinib against diverse human fungal pathogens by checkerboard microdilution assay.
| Strain | MIC alone | MIC combination | FICI for combination | Mode of interaction | ||
|---|---|---|---|---|---|---|
| FLC (μg ml‐1) | Pona (μM) | FLC (μg ml‐1) | Pona (μM) | |||
| SC5314 | 0.5 | 16 | 0.0625 | 0.0625 | 0.13 | Syn |
| BY4742 | 16 | 128 | 4 | 0.25 | 0.25 | Syn |
| H99 | 4 | 32 | 0.5 | 0.125 | 0.13 | Syn |
| CCC49 | 64 | 16 | 2 | 0.0625 | 0.04 | Syn |
| CCC80 | 32 | 8 | 2 | 0.0625 | 0.07 | Syn |
Syn, Synergistic.
Next, we performed time‐kill curve analysis to determine if ponatinib renders fluconazole fungicidal. Candida albicans cells were subjected to fluconazole treatment with or without ponatinib in liquid medium, and the CFU ml‐1 of the suspension from each treatment was counted after plating onto the YPD medium. With the increment of ponatinib concentration in the combination group, we observed that the number of survival colonies decreased gradually with time, revealing that ponatinib can transform fluconazole from fungistatic into fungicidal (Fig. 2A). The ability to switch between yeast and hyphal morphology is critical for C. albicans pathogenicity (Witchley et al., 2019). To examine whether the combinatorial antifungal effect could be attributed to an impact on filamentation, we supplemented the hyphae‐inducing media with fluconazole, ponatinib or both and evaluated the hyphal growth of C. albicans. The media we used in the assay include M199, Spider or YPD medium with 10% serum, which are all traditional hyphae‐inducing media and have been widely used to assess the yeast–hyphae morphological change in C. albicans (Toenjes et al., 2005; Liu et al., 2017; Lim et al., 2020; Yang et al., 2020). As shown in Fig. 2B, the inhibitory effect was much more significant in the combinatorial group. Thus, ponatinib potentiates fluconazole activity against diverse fungal pathogens and exerts a fungicidal activity with fluconazole.
Fig. 2.
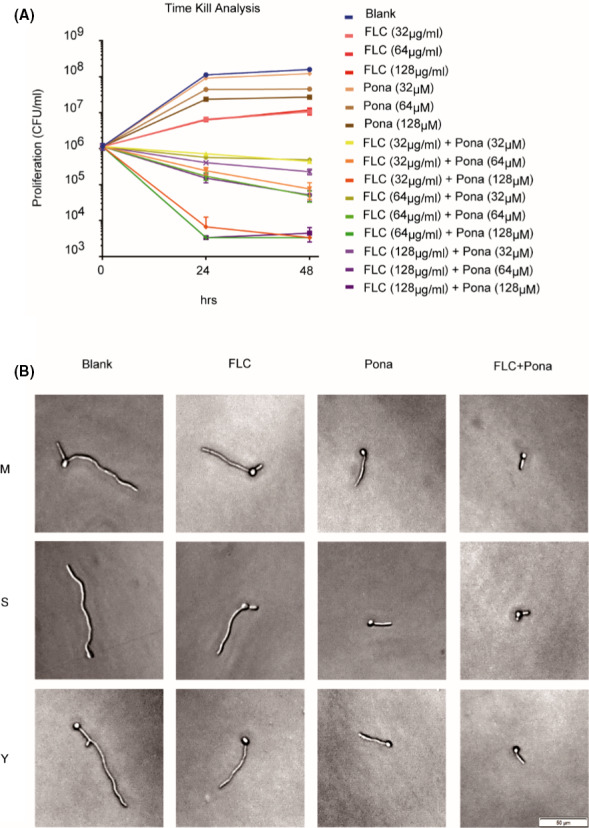
Ponatinib transforms fluconazole from fungistatic into fungicidal and suppresses hyphal formation.
A. C. albicans (SC5314) cells were inoculated into YPD liquid medium supplemented with or without indicated compounds (DMSO, 32‐128 μg/ml fluconazole, 32‐128 μM ponatinib or both). The survival colony‐forming units (CFUs) of SC5314 were counted after incubation at indicated periods of time (0, 24 and 48 h).
B. C. albicans (SC5314) cells were revived and resuspended in each of three different hyphae‐inducing media (M199 buffered to pH8 with 50 mM MOPS; Spider; and YPD with 10% serum) containing DMSO, fluconazole (2 μg ml‐1), ponatinib (5 μM) or the combination. Cells were incubated at 37 °C, and hyphal morphology was checked under light microscope after 4 h of incubation.
Ponatinib prevents the emergence of azole resistance in C. albicans
Fluconazole resistance in the pathogenic yeast C. albicans poses significant challenges for the treatment and prevention of Candida infections frequently confronted by patients in clinic. We therefore investigated whether the combination of fluconazole and ponatinib can also be effective against the fluconazole‐resistant clinical isolates of C. albicans. The minimal inhibitory concentration (MIC) values of fluconazole against two resistant isolates were 64 and 32 μg ml‐1, respectively, which were about 128‐ and 64‐fold higher than that of the standard laboratory strain SC5314 (Fig. S3). Ponatinib combined with fluconazole was very efficient in the inhibition of the resistant strains, as we documented that ponatinib increased the susceptibility of resistant strains to fluconazole by up to eightfold and the effective concentration of fluconazole decreased from 64 to 8 μg ml‐1 (Fig. 3B and F). In order to better evaluate the drug–drug interaction between ponatinib and fluconazole, we used the checkerboard assay as previously described (Rand et al., 1993) and calculated the FICI for the combination using a formula described in Experimental procedures. As shown in Table 1, ponatinib was highly synergistic with fluconazole with FICI = 0.04 for CCC49 and 0.07 for CCC80, highlighting its clinical applicability against fluconazole‐resistant C. albicans isolates.
Fig. 3.
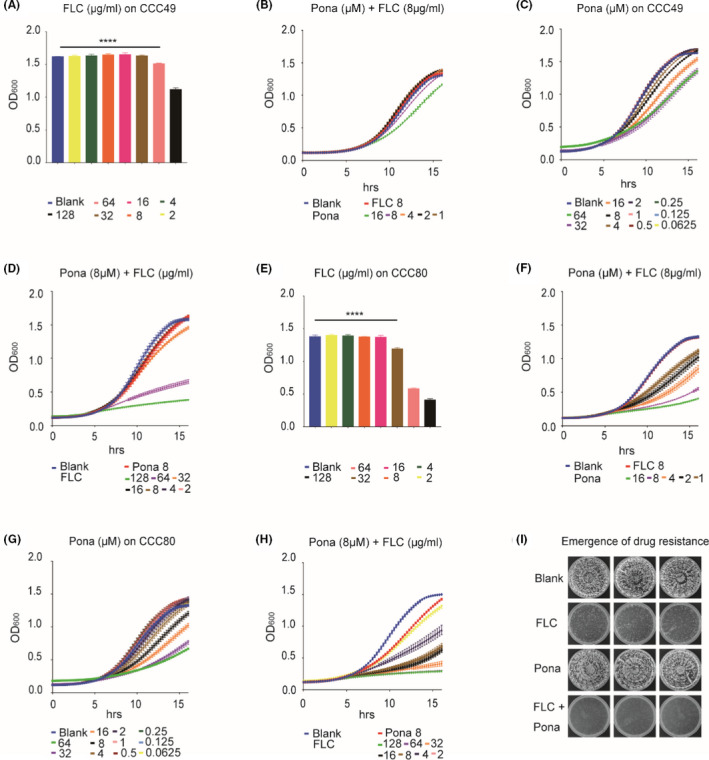
Ponatinib acts synergistically with fluconazole against azole‐resistant clinical isolates and prevents the emergence of resistance. CCC49 (A–D) and CCC80 (E–H) were subjected to twofold serial dilutions of fluconazole, ponatinib or both in YPD medium. OD600 was measured every 15 min at 30°C for 16 h, and FICI was calculated by standard CLSI broth microdilution method (M38‐A).
I. C. albicans (SC5314) cells were inoculated into YPD plates containing indicated dosages of compounds (DMSO, 32 μg ml‐1 fluconazole, 32 μM ponatinib, combination). The cell growth of resistant strains was observed after 3 days. ****P < 0.0001 vs. DMSO (the one‐way ANOVA and Tukey multiple comparisons).
The above results prompted us to ask whether ponatinib could prevent the emergence of drug resistance, given that this event is often considered as a canonical case of evolution by natural selection (Shekhar‐Guturja et al., 2016b; Vincent et al., 2016; Fransen et al., 2017). We plated 3 × 103 C. albicans cells onto YPD plates supplemented with 32 μg ml‐1 fluconazole, 32 μM ponatinib or both. A plate without supplementation was used as a control. As shown in Fig. 3I, the emergence of resistance was significantly induced by fluconazole alone. However, the induction was potently suppressed by the combination of ponatinib. Our data further indicated that the desired final concentrations of fluconazole and ponatinib, which confer synergistic activity against drug‐resistant C. albicans isolates, are 104.48 μM (32 μg ml‐1) and 32 μM respectively. Collectively, our results demonstrated that ponatinib not only enhances the antifungal activity against fluconazole‐resistant isolates of C. albicans but also prevents the emergence of fluconazole resistance.
Ponatinib reduces efflux of fluconazole via Pdr5 and promotes the intracellular hyperaccumulation of fluconazole
Drug efflux is one of the most intensive investigated mechanisms for fluconazole resistance (Hampe et al., 2017). To test whether ponatinib might affect drug efflux to enhance the antifungal activity of fluconazole, we first screened the sensitivity of each of 16 S. cerevisiae mutants lacking individual ATP‐binding cassette (ABC) transporter to ponatinib (Suzuki et al., 2011). The results showed that only the pdr5Δ mutant was hypersensitive to ponatinib treatment, indicating that ponatinib might interact with Pdr5 (Fig. 4A and B).
Pdr5 is an ABC transporter which pumps out a series of structurally unrelated compounds, including azoles and rhodamine (Prasad and Goffeau, 2012; Shekhar‐Guturja et al., 2016b; de Moraes et al., 2020). Consistent with previous studies (Lamping et al., 2007; Prasad and Goffeau, 2012), we also confirmed that fluconazole is a substrate of Pdr5. First, we compared the vegetative growth of wild‐type and pdr5Δ mutant cells in the absence and presence of fluconazole and the results showed that the mutant lacking PDR5 exhibited hypersensitivity to fluconazole when compared to the wild type (WT). Next, we treated the WT with increasing concentrations of fluconazole and the cells were stained with Rhodamine 6G, which is a commonly used fluorescent dye for mimicking fluconazole efflux as both molecules share the same transporters in yeast (Maesaki et al., 1999). The assay measured the efflux activities in the yeast cells being incubated with fluconazole and ponatinib at increasing concentrations showing synergism based on the intensity of intracellular fluorescence (red colour). Beauvericin, known to effectively potentiate the activity of fluconazole against yeasts (Shekhar‐Guturja et al., 2016a; Shekhar‐Guturja et al., 2016b), was used as a control. Interestingly, the intracellular accumulation of Rhodamine 6G was found to correlate with the level of fluconazole (Fig. 4C, E). To assess the effect of ponatinib on Pdr5 activity, we examined rhodamine‐6G efflux in wild‐type S. cerevisiae strain BY4742 in incubation with beauvericin or fluconazole/ponatinib at the concentrations of ponatinib MIC showing synergism. We observed in Fig. 4D that the combinatorial use of fluconazole and ponatinib resulted in a dose‐dependent accumulation of red fluorescence, suggestive of an effective inhibition of the efflux of rhodamine‐6G. The results also supported a close association of the antifungal effects of the fluconazole/ponatinib combination with the function of efflux pumps in the yeast. Finally, qRT‐PCR analysis revealed that ponatinib treatment had no effect on the transcript levels of PDR5 in S. cerevisiae, suggesting that ponatinib may only perturb the activity of Pdr5 instead of its transcription (Fig. S4). Taken together, our data clearly demonstrate that ponatinib improves the efficacy of fluconazole by stimulating intracellular fluconazole accumulation through inhibition of Pdr5.
Fig. 4.
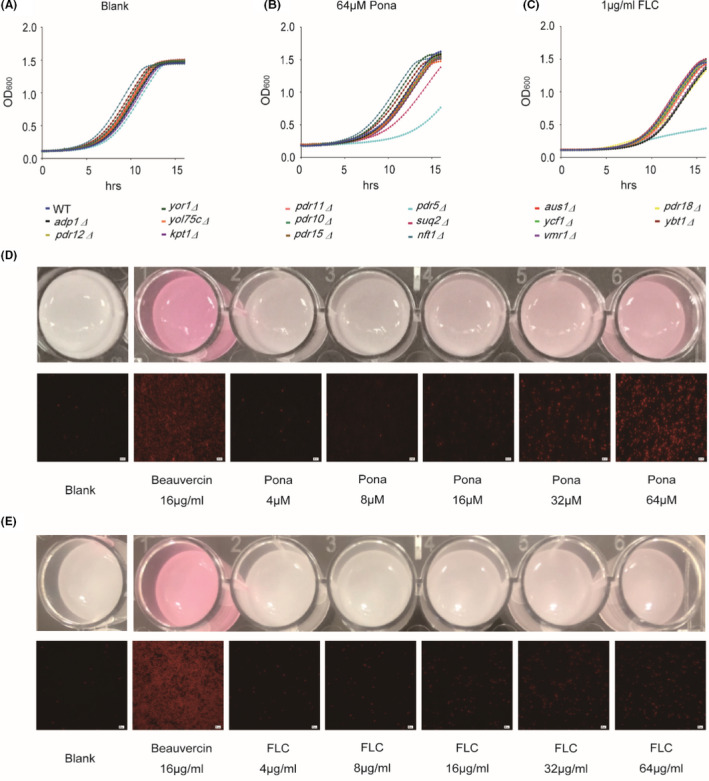
Ponatinib enhances intracellular hyperaccumulation of fluconazole via Pdr5.
A–C. The S. cerevisiae WT and 16 ABC mutants were separately treated with Ponatinib or fluconazole at indicated concentrations.
D–E. BY4742 was pretreated with beauvericin, ponatinib or fluconazole, following by addition of the same concentration of rhodamine‐6G. The intracellular accumulation of rhodamine‐6G was observed under fluorescence microscopy.
Whole‐genome sequencing identifies Pma1 as the putative target of ponatinib
Whole‐genome sequencing is a technology widely exploited to identify the targets of antifungal agents in the model yeast Saccharomyces cerevisiae (Shekhar‐Guturja et al., 2016b). Our results in Fig. 1 showed that Ponatinib alone also harbours the inhibitory activity against the growth of yeast cells, implying the presence of additional targets. We therefore sought to identify the target of ponatinib using the method of whole‐genome sequencing. To do that, the predominant efflux effect of 16 ABC transporters, which are highly expressed in the yeast cells, has to be minimized or excluded firstly. We therefore conducted a screen for ponatinib‐resistant isolates in the background of a specific S. cerevisiae mutant lacking all 16 ABC transporter‐encoding genes (ABC16 strain; strain 2336) (Suzuki et al., 2011; Shekhar‐Guturja et al., 2016b). The parental strain RY0566 was used as WT control. Specifically, about 3 × 107 cells ml‐1 of ABC16 strain were plated onto YPD medium supplemented with 64 μM ponatinib and we randomly isolated and sequenced ten ponatinib‐resistant colonies (Fig. 5A–D). Our whole‐genome sequencing of the 10 isolates revealed that nine out of 10 mutants harboured the same missense mutation, T558S, in Pma1 (Fig. 5E and F). Interestingly, it has been reported that the P‐type H+‐ATPase Pma1 is an essential proton pump able to regulate cytosolic pH homeostasis (Zhang et al., 2010), partly validating our screening results.
Fig. 5.
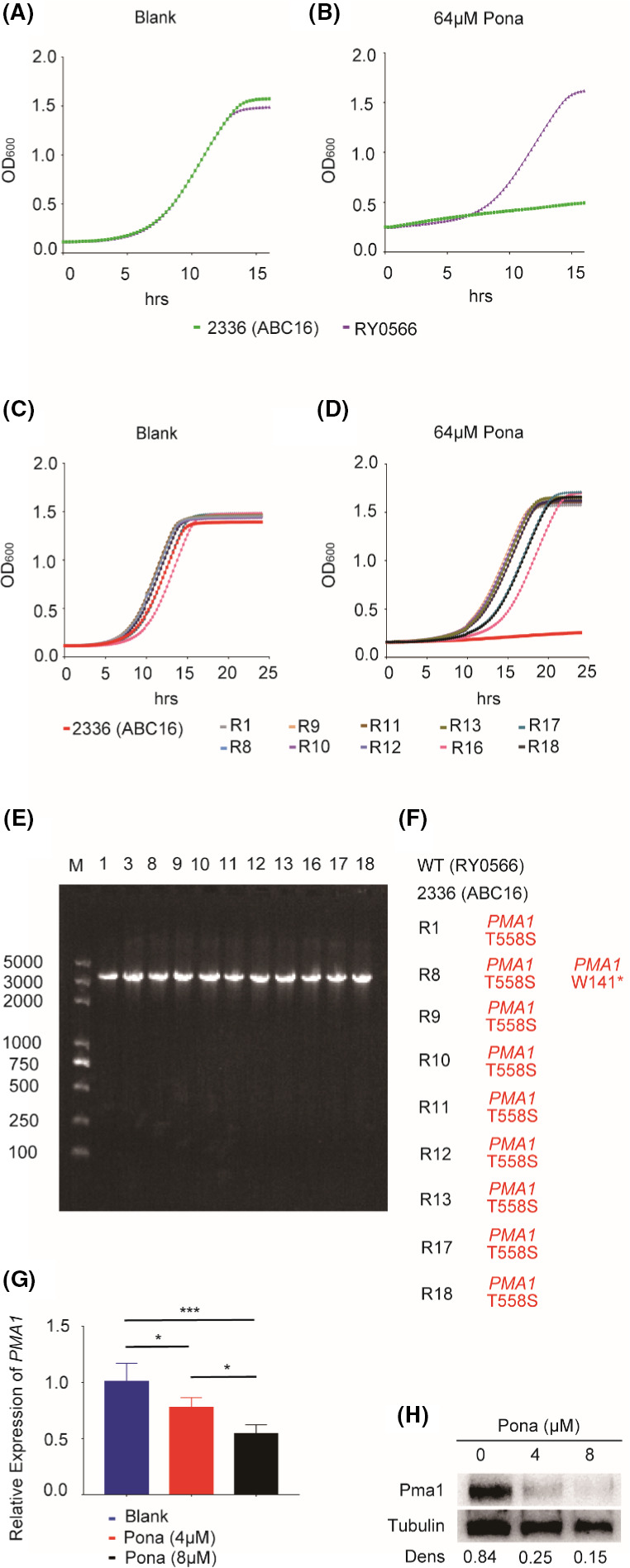
Identification of the ponatinib target by whole‐genome sequencing.
A–B. Strains RY0566 (wild type) and 2336 (the 16ABC mutant lacking all 16 efflux pump‐encoding genes) were treated with ponatinib.
C–D. Strain 2336 and ponatinib‐resistant mutants were treated with ponatinib.
E. Gel electrophoresis results of PCR products. DNA was extracted from drug‐resistant mutant cells and was amplified by PCR. (F) Sequencing results of PCR products. Mutations identified by whole‐genome sequencing of resistant mutants were indicated as amino acid changes. All resistant mutants harbour identical mutations in Pma1.
G. The total RNAs were extracted from the ABC16 mutant strain 2336 treated with ponatinib and prepared for qRT‐PCR. Error bars represent SDs of three biological triplicates.
H. The protein extracts were prepared from the ABC16 mutant cells treated with ponatinib and subject to western analysis, using antibodies against Pma1 and tubulin respectively. Blots were quantified using Image J. Tubulin was used as a loading control. *P < 0.05; ***P < 0.001 or ****P < 0.0001 (the one‐way ANOVA and Tukey multiple comparison).
To further evaluate the relationship between ponatinib compound and Pma1 expression, we first quantified and compared the transcript levels of PMA1 in the absence or presence of ponatinib. As shown in Fig. 5G, ponatinib significantly repressed the transcriptional expression of PMA1 in a dose‐dependent manner. Moreover, we found that Pma1 protein expression could also be modulated by ponatinib, as the immunoblot analysis clearly showed that the Pma1 protein levels were decreased dramatically by treating the cells with ponatinib, and the reduction is also dose‐dependent (Fig. 5H). In microbial organisms, the extrusion of protons by the electron transport chain causes an electrochemical gradient of protons, known as the proton motive force (PMF), which is generated across the cell membrane (Mitchell, 2011). Given the role of ponatinib as an essential proton pump in regulation of cytosolic pH, we ask there may exist a possible relationship between PMF and the accumulation of intracellular fluconazole by ponatinib, in other words, the proton motive force (PMF) and drug efflux pumps may play a central role in the drug combination effect. Our results showed that the enhancement of proton transport by increasing the proton motive force (PMF) after treatment with fluconazole, could be suppressed by supplementation of ponatinib (Fig. S5), further supporting the notion that ponatinib shows synergistic interaction when combined with fluconazole. Taken together, the results shown here highly support that Pma1 is the potential target of ponatinib (Fig. 6).
Fig. 6.
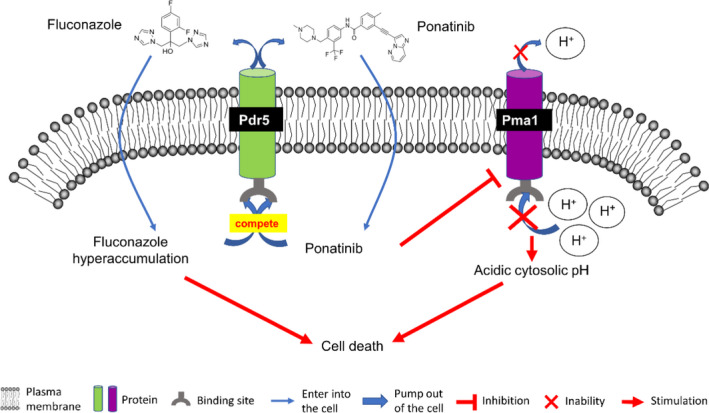
Model depiction of the role of ponatinib in potentiation of the antifungal efficacy of fluconazole by inhibition of fluconazole efflux via Pdr5 and suppression of Pma1 expression. Ponatinib (PubChem CID: 24826799) promotes intracellular fluconazole (PubChem CID:3365) accumulation by binding to Pdr5. On the other hand, ponatinib perturbs the cytosolic pH homeostasis by inhibiting the proton pump Pma1 located at the plasma membrane. Eventually, the dual action of ponatinib through these two pathways leads to cell death.
Discussion
The global incidence of fungal infection has increased over the last few decades due to the expanded ageing population, widespread azole resistance and the marked increase in the number of immunocompromised patients. Due to the fact that invasive fungal infections and fungal sepsis are rapidly increasing with considerable morbidity and mortality, especially in the intensive care unit (ICU), combinatorial antifungal therapy has been becoming a promising therapeutic strategy to enhance drug effectiveness and ameliorate the emergence of drug resistance and of course, has been gaining increasing attention within both industry and academia. Ponatinib is an FDA‐approved oral tyrosine kinase inhibitor (Mitchell et al., 2018) and has been evaluated to impede tumour progression, including tumours in thyroid, breast, ovary and lung, neuroblastoma, rhabdoid tumours, gastrointestinal stromal tumours, et al (Musumeci et al., 2018). In this study, we discovered a novel function of this compound, which acts as a potentiator of azole activity against diverse human fungal pathogens, as well as the highly fluconazole‐resistant isolates of C. albicans. Moreover, our work illustrated that the mechanism of the synergistic combinations that potentiate the antifungal fluconazole is associated with the inhibitory activity of ponatinib, as shown by inhibiting efflux of fluconazole via Pdr5 and suppressing the expression of the P‐type H+‐ATPase Pma1. We provided evidence that ponatinib may execute two levels of actions in the synergism: First, it reduces efflux of fluconazole by inhibiting Pdr5 and thus promotes the intracellular hyperaccumulation of fluconazole; Second, it may interfere with the cytoplasmic pH homeostasis by affecting the expression of the putative target Pma1, as the P‐type H+‐ATPase Pma1 has been reported to be an essential proton pump operating to regulate the cytosolic pH homeostasis (Zhang et al., 2010). Importantly, the observation that the effective concentration of ponatinib appears to be non‐toxic towards endothelial cells greatly assists in dealing with the concern of the potential drug side‐effect, which may limit its application in future translational studies. Of course, the interaction between ponatinib and fluconazole identified so far is certainly only the tip of the iceberg and awaits more systemic analysis.
Currently, the most urgent problem related to fungal infections in clinic is the emergence of multidrug resistance due to the fungistatic activity of antifungals like fluconazole. Identification of a novel antifungal candidate which can transform the fungistatic effect of fluconazole to fungicidal will be an effective strategy for clearance of fungal pathogens. Our results indicate that ponatinib not only inhibits the growth of a broad‐spectrum of fungal pathogens but also converts fluconazole from fungistatic to fungicidal which prevents the emergence of azole resistance, providing a promising therapeutic strategy for antifungal development in the future.
The ABC (ATP‐binding cassette) superfamily contains membrane proteins that transport a wide variety of substrates, such as metabolic products, lipids and sterols, and drugs, across extra‐ and intracellular membranes. In microbial organisms, the multidrug efflux pumps, which belong to members of the ABC superfamily, act to effectively pump drugs out of cells, significantly reduce the intracellular concentration of antifungals and prevent the emergence of drug resistance (Nakamura et al., 2001; Coste et al., 2006; Shapiro et al., 2011). Among them, Pdr5 is the most abundant ABC transporter in S. cerevisiae and shares sequence homology with Cdr1 in C. albicans (Kontoyiannis and Lewis, 2002). Studies have revealed that Pdr5 is able to squeeze out hundreds of structurally independent hydrophobic compounds to pass through the plasma membrane (Kolaczkowski et al., 1996; Egner et al., 1998; Golin et al., 2003). Intriguingly, Pdr5 was the only target that came out of the screen when we used the 16 ABC transporter mutants to evaluate the efflux effect of ponatinib, suggesting that Pdr5 could be the substrate of Ponatinib and by inactivating the activity of Pdr5, the compound prevents the efflux of fluconazole from the fungal cells. In the future, more evidence is required to support our hypothesis, especially those implicated in assaying direct interaction between Pdr5 and ponatinib.
The cytosolic pH homeostasis plays a key role in cell growth (Gillies et al., 1981, Casey et al., 2010, Dechant et al., 2010, Dechant et al., 2014, Martínez‐Muñoz and Kane, 2017; Saliba et al., 2018). The ability to regulate intracellular pH enables C. albicans to survive in both extremely acidic and alkaline microenvironments (Casey et al., 2010; Cyert and Philpott, 2013; Felcmanova et al., 2017; Martínez‐Muñoz and Kane, 2017; Saliba et al., 2018; Xu et al., 2019). The critical elements determining pH homeostasis are as follows: proton pumps, exchangers and buffers (Casey et al., 2010). Proton pumps, such as Pma1 in the plasma membrane, are required to establish certain pH gradient (Ferreira et al., 2001; Orij et al., 2011; Martínez‐Muñoz and Kane, 2017). Exchangers can transport ions and solutes against the gradient by using the energy stored in pH gradients or ion gradients to determine the final pH (Brett et al., 2005; Ohgaki et al., 2011; Kondapalli et al., 2014). Buffers protect the cells or organelles from the disturbance of short‐term pH fluctuation (Casey et al., 2010; Poznanski et al., 2013). Pma1, which is independent of ion pumps, is a structural component of the plasma membrane accounting for 20–40% of the total plasma membrane proteins (Monk et al., 1991). Strikingly, this P‐type H+‐ATPase plays a key role in cytosolic pH regulation (Serrano, 1988; van der Rest et al., 1995) by pumping the proton out of the cells and consuming an impressive amount of intracellular ATP which accounts for approximately a quarter of total ATP consumption (Giacomello et al., 2013), highlighting the potential of Pma1 as an antifungal target. Interestingly, harnessing whole‐genome sequencing and RT‐PCR analysis, we argued that ponatinib may target Pma1 to perturb the pH homeostasis in the cytoplasm, eventually leading to cell death. Supporting this proposition requires more evidence; however, the effective inhibitor of Pma1 has not been determined in clinic (Stewart et al., 1988; Monk et al., 1995; Perlin et al., 1997; Monk et al., 2005; Chan et al., 2007; Billack et al., 2009; Ottilie et al., 2018). Thus, alternative strategies have to be considered.
Numerous studies have demonstrated that fluconazole is widely used in clinic for the treatment of fungal infection by selectively interfering with the activity of cytochrome P‐450 and inhibiting the biosynthesis of ergosterol in the plasma membrane. However, targeting a single drug target is usually difficult to achieve the desired effect and prone to drug resistance. In our work, we found that ponatinib acts as a fluconazole potentiator through suppression of not only multidrug efflux via Pdr5 but also Pma1 expression in vitro, supporting that the multi‐target drug ponatinib could enhance the effectiveness of fluconazole via two action modes. Compared with the single drug administration, drug combinatorial therapy has demonstrated great advantages in overcoming drug resistance and improving therapeutic efficacy, since this therapy harbours multi‐target activities and confers high information‐processing capacity and functional diversity, especially the simultaneous regulation of multiple inputs into the signalling network. Moreover, the drug combinatorial therapy allows many parts of the network to be activated at once and has been applied in the treatment of many chronic diseases such as cancer and thus has drawn intensive attention from researchers and pharmaceutical enterprises. Of course, it has to be noted that the main problem faced by repurposing the existing multi‐target drugs is their potential adverse effects. In addition, the optimal drug concentration for each target is often different, and it is difficult for a multi‐target drug to achieve the optimal pharmacological effect for each target, which may affect its synergistic effect. Thus, testing clinically relevant pharmacodynamics and pharmacokinetics of the prioritized drug combinations needs to be seriously considered for identification, development and optimization of efficacious combinatorial drug treatments.
Experimental procedures
Strains and culture condition
The strains used in this study (Table 2) were stored at −80°C and routinely grown in yeast peptone dextrose (YPD) medium.
Table 2.
Strains used in this study.
| Strain name | Parent | Genotype |
Strain background /construction |
References |
|---|---|---|---|---|
| C. albicans | ||||
| SC5314 | Wild type | This work | ||
| CCC49 | Fluconazole‐resistant clinical isolates | This work | ||
| CCC80 | Fluconazole‐resistant clinical isolates | This work | ||
| S. cerevisiae | ||||
| BY4742 | Wild type | Suzuki et al. (2011) | ||
| adp1Δ | BY4742 | adp1Δ transformed with WT | Suzuki et al. (2011) | |
| snq2Δ | BY4742 | snq2Δ transformed with WT | Suzuki et al. (2011) | |
| ycf1Δ | BY4742 | ycf1Δ transformed with WT | Suzuki et al. (2011) | |
| pdr15Δ | BY4742 | pdr15Δ transformed with WT | Suzuki et al. (2011) | |
| yor1Δ | BY4742 | yor1Δ transformed with WT | Suzuki et al. (2011) | |
| vmr1Δ | BY4742 | vmr1Δ transformed with WT | Suzuki et al. (2011) | |
| pdr11Δ | BY4742 | pdr11Δ transformed with WT | Suzuki et al. (2011) | |
| nft1Δ | BY4742 | nft1Δ transformed with WT | Suzuki et al. (2011) | |
| kpt1Δ | BY4742 | kpt1Δ transformed with WT | Suzuki et al. (2011) | |
| ybt1Δ | BY4742 | ybt1Δ transformed with WT | Suzuki et al. (2011) | |
| pdr18Δ | BY4742 | pdr18Δ transformed with WT | Suzuki et al. (2011) | |
| yol075cΔ | BY4742 | yol075cΔ transformed with WT | Suzuki et al. (2011) | |
| aus1Δ | BY4742 | aus1Δ transformed with WT | Suzuki et al. (2011) | |
| pdr5Δ | BY4742 | pdr5Δ transformed with WT | Suzuki et al. (2011) | |
| pdr10Δ | BY4742 | pdr10Δ transformed with WT | Suzuki et al. (2011) | |
| pdr12Δ | BY4742 | pdr12Δ transformed with WT | Suzuki et al. (2011) | |
| BY4741 | Wild type | |||
|---|---|---|---|---|
| RY0566 | BY4741 |
Isogenic control MATa hΔ::tetO2‐GFP‐URA3 can1Δ::GMToolkit ‐a [CMVpr‐rtTA KANMX4 STE2pr‐Sp‐his5]lyp1Δ his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 |
Suzuki et al. (2011) | |
| 2336 | BY4741 | Green Monster MATa adp1Δ snq2Δ ycf1Δ pdr15Δ yor1Δ vmr1Δ pdr11Δ nft1Δ bpt1Δ ybt1Δ ynr070wΔ yol075Δ aus1Δ pdr5Δ pdr10Δ pdr12Δ can1Δ::GMT oolkit‐a (CMVpr‐rtTA KANMX4 STE2pr‐Sp‐his5) his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 Each ABC‐transporter deletion contains ADHterm‐tetO2pr‐GFP(S65T)‐CYC1termURA3. | Suzuki et al. (2011) | |
| R1 | 2336 | Ponatinib resistant isolate | This work | |
| R8 | 2336 | Ponatinib resistant isolate | This work | |
| R9 | 2336 | Ponatinib resistant isolate | This work | |
| R10 | 2336 | Ponatinib resistant isolate | This work | |
| R11 | 2336 | Ponatinib resistant isolate | This work | |
| R12 | 2336 | Ponatinib resistant isolate | This work | |
| R13 | 2336 | Ponatinib resistant isolate | This work | |
| R16 | 2336 | Ponatinib resistant isolate | This work | |
| R17 | 2336 | Ponatinib resistant isolate | This work | |
| R18 | 2336 | Ponatinib resistant isolate | This work | |
| C. neoformans | ||||
| H99 | Wild type | This work | ||
Minimal Inhibitory Concentration (MIC) Assay
MIC assay was evaluated in 96‐well microtitre plates using the broth dilution testing reference method M27‐A3/S4, as recommended by the Clinical and Laboratory Standards Institute (CLSI) (Rex, 2008). Twofold serial dilutions of fluconazole (HY‐B0101; MedChemExpress, Shanghai, China) or ponatinib (S1490; Selleck, Shanghai, China) were prepared along the columns or rows of a 96‐well plate. Overnight cultures were diluted to an OD600 of 1.0, followed by another 100‐fold dilution to reach a final OD600 of ~ 0.01 (Chen et al., 2011; Chen and Noble, 2012). The plates were incubated at 30°C and photographed by ChemiDoc MP (Bio‐Rad, Shanghai, China). The MIC was defined as the lowest drug concentration that caused a specified reduction in visible growth comparing with that of control. All strains were assessed in biological triplicates with three technical replicates.
LDH Assay
To test the cytotoxicity of ponatinib on mammalian cells, 2 × 104 endothelial cells (NCM460) were seeded overnight in Dulbecco's modified Eagle’s medium (DMEM) supplemented with 10% FBS and kept at 37°C in a CO2 incubator. Cells were then exposed to twofold serial dilutions of ponatinib for 1.5 h at 37°C before being lysed. Cytotoxicity was measured using Cytotoxicity LDH Assay Kit‐WST (NG715, DOJINDO, Shanghai, China).
Checkerboard assay
Checkerboard assay was evaluated in 96‐well microtitre plates using the reference guidelines for broth microdilution method (M38‐A2) from the Clinical and Laboratory Standards Institute (CLSI) (Alexander, 2008). The procedures were performed as described in a previous study (Hsieh et al., 1993). Briefly, each compound was serially diluted in two‐fold in a 96‐well plate, either across columns of the plates (fluconazole) or rows of the plates (ponatinib). Overnight cultures were diluted to an OD600 of 1.0, followed by another 100‐fold dilution to reach a final OD600 of ~ 0.01 (Chen et al., 2011; Chen and Noble, 2012). The plates were incubated at 30°C, and the OD600 was measured every 15min. The fractional inhibitory concentration index (FICI) of each drug combination was determined according to the standard CLSI protocol, and FICI < 0.5 was defined as synergy (Brown et al., 2014). The formula is calculated as follows: FICI = (MICA in combination/MICA alone) + (MICB in combination/MICB alone).
Filamentation assay
Optimal induction of filamentous growth in C. albicans was achieved by incubating the yeast cells on hyphal‐inducing medium, following the procedures described previously (Flanagan et al., 2017).
Emergence of drug resistance in vitro
For emergence of drug resistance in vitro, fresh overnight yeast cultures were washed twice in PBS and diluted into an optical density (OD600) of 0.5. After 10‐fold serial dilutions, cells were spread onto appropriate agar plates supplemented with 32 μg ml‐1 fluconazole, 32 μM ponatinib or both.
Time‐kill curve analysis
Overnight culture of C. albicans was diluted to a final OD600 of 0.05 in liquid YPD medium containing fluconazole (32, 64, 128 μg ml‐1), ponatinib (32, 64, 128 μM) or both in 96‐well plates with a final volume of 200 μl. The suspension was diluted at 1:250 000 and plated on YPD plates at indicated periods of time (0, 24 h or 48 h). The plates were incubated at 30°C for 24 h before determining the colony‐forming units (CFUs) by counting.
Rhodamine‐6G (R6G) staining assay
Cells grown overnight in YPD liquid medium at 30 °C were washed with ice‐cold glucose‐free phosphate‐buffered saline (PBS) and incubated at 30°C for 1 h under starvation to reduce the ATP‐binding cassette (ABC) efflux pumps activity. The collected cells were then washed and diluted to 108 cells ml‐1 in ice‐cold PBS, which were exposed to 4, 8, 16, 32, 64 μM ponatinib or 4, 8, 16, 32, 64 μg ml‐1 fluconazole respectively. Beauvericin at a concentration of 16 μg ml‐1 was added as a positive control with dimethyl sulfoxide (DMSO) as the negative control. All samples were incubated for another 2 h at 30°C. After treatment with 10 μM (final concentration) R6G, cells were incubated for another 1.5 h at 30°C. The external R6G was then removed by washing with PBS, and 2% glucose was added to the samples to reactivate the ABC efflux pumps. After incubation at 30oC for 1 h, the reactivated cells were washed and observed under fluorescent microscopy to monitor intracellular R6G accumulation.
Whole‐genome sequencing of ponatinib‐resistant isolates
The parental strain ABC16 and 10 selected ponatinib‐resistant isolates were separately cultured on solid YPD plates for DNA extraction. Then, the mutations in PMA1 of the above 11 isolates were validated by PCR and sequencing to identify the specific mutation (Heitman et al., 1991; Shekhar‐Guturja et al., 2016b; Vincent et al., 2016; Sukheja et al., 2017; Wang et al., 2020). The PCR reaction mixture is comprised of 10xHifi PCR buffer (2 μl), 2 mM dNTPs (1.2 μl), 50 mM MgSO4 (0.6 μl), 10 mM primers (0.4 μl), 5 μM Taq Enzyme (0.1 μl), DNA template (2 μl) and sterile water up to μl. For the PCR program, the conditions are followed by 95°C 5 min; 94°C 30 s, 55°C 30 s and 68°C 6 min and 40 s for 30 cycles; 68°C 10 min. The genomic sequences of all strains can be accessed through NCBI accession number PRJNA649097.
qRT‐PCR
Overnight cell cultures in YPD at 30°C were diluted to OD600 of 1.0. Cells were treated with DMSO, fluconazole, ponatinib or both and grown at 30°C for 3 h. The RNA was then extracted by hot phenol method and further treated with RT reagent with gDNA Eraser (Takara #RR047A, Beijing, China). The PCR was performed using Universal SYBR Green Supermix (Bio‐Rad #1725121) with the following program: 95°C for 30 s; 95°C for 5 s; and 60°C for 30 s, for 40 cycles. Primers are listed in Table 3. The data were analysed by the 2−ΔΔCt method, in which ΔΔC t = (C t value of target gene − C t value of reference gene)sample – (C t value of target gene − C t value of reference gene)control, as described previously (Livak and Schmittgen, 2001; Huang et al., 2007).
Table 3.
Primers used in this study.
| Primer name | Purpose | Sequence 5’ to 3’ |
|---|---|---|
| NL50 | Forward to amplify PMA1 | ACATTCAAAAGAAAGAAAAAAAATATACCCCAGCTAGTTAAAGAAAATCATTGAAAAGAATAAGAAGATAAGAAAGATTTAATTATCAAACAATATCAATCGGATCCCCGGGTTAATTAA |
| NL51 | Reverse to amplify PMA1 | TTGATAAAAAAATTAAAATTAAAATTAGAAAAATTAAACCAGAAAAATCAAGTTGATTAAAATGTGACAAAAATTATGATTAAATGCTACTTCAACAGGAGAATTCGAGCTCGTTTAAAC |
| NL220 | Forward for qRT‐PCR of PMA1 | GCCTGCTAAGACTTACGATGACGC |
| NL221 | Reverse for qRT‐PCR of PMA1 | TTCACCGGCGGCAACTGGAC |
| NL222 | Forward for qRT‐PCR of ACT1 | ATTATATGTTTAGAGGTTGCTGCTTTGG |
| NL223 | Reverse for qRT‐PCR of ACT1 | CAATTCGTTGTAGAAGGTATGATGCC |
Western blot
The cell lysis, protein extraction and Western blot procedures were performed as described in a previous study (Liu et al., 2017). The antibodies are listed in Table 4. For densitometry, Image J software (https://imagej.net/Downloads) was used as in a previous study (Flanagan et al., 2017).
Table 4.
Antibodies used in this study.
| Antibodies purpose | Antigen recognized | Species | Source or reference |
|---|---|---|---|
| Loading control | Tubulin | Rat | Abcam, #ab6161, Shanghai, China |
| Pma1 | Mouse | Gene Tex, #GTX24645, Alton Pkwy Irvine, CA, USA | |
| Secondary | Rat Ig | Goat | Cell Signaling TECHNOLOGY, #7077, Shanghai, China |
| Secondary | Mouse Ig | Goat | Arigo, #65350, Shanghai, China |
Proton pump in plasma membrane
The BY4741 strain was cultured overnight in YPD liquid medium at 30°C, and 5 × 108 cells were used for extraction of IOV (In‐side out), which was treated with fluconazole (16 μg ml‐1), ponatinib (8 μM) or both in 96 well black plates. The fluorescence intensity at excitation wavelength 490 nm and emission wavelength 530 nm was measured every 5 min for 1 h. The N‐ethylmaleimide (NEM) (10 μM) and orthovanadate (OV) (100 μM) were added as a positive control with dimethyl sulfoxide (DMSO) as the negative control (Van Dyke et al., 1985, Kaunitz and Sachs, 1986). IOV (In‐side out) was extracted using Fungus/Yeast Membrane Vesicle RSOV/IOV Prep Kit (GMS 10169.3 v.A; Shanghai CHENGONG Biotechnology Co., Ltd, Shanghai, China). The proton pump in the plasma membrane was measured using Proton Transport (P ATPase dependent) Assay Kit (GMS 10159.1.2 v.A; Shanghai CHENGONG Biotechnology Co., Ltd).
Statistics
All data were shown as the mean ± SDs in three independent experiments. All results were calculated from the means of three separate experiments. Statistical analysis was performed using GraphPad Prism 7, San Diego, CA, USA with the one‐way ANOVA and Tukey multiple comparison analysis at a *P < 0.05、***P < 0.001 or ****P < 0.0001 level of significance.
Funding Information
This study was supported by grants from the National Key R&D Program of China (2018YFC2000700; 2020YFA0907200), the National Nature Science Foundation (81630086, 81971993, 31900129, 81572053, 31870141, 31570140), the Key Research Program (ZDRW‐ZS‐2017‐1; KGFZD‐135‐19‐11;153831KYSB20170043) of the Chinese Academy of Sciences, the Major Science and Technology Innovation Program of Shanghai Municipal Education Commission (2019‐01‐07‐00‐01‐E00059), the Program for Young Eastern Scholar at Shanghai Institutions of Higher Learning (program QD2018016), Shanghai Pujiang Program (18PJ1406600), Innovative research team of high‐level local universities in Shanghai, the Innovation Capacity Building Project of Jiangsu Province (BM2020019).
Conflict of interests
The authors declare no conflicts of interest.
Author contribution
NNL, HW and CC conceived and designed the study; LL, NNL, HW and CC performed data analysis and wrote the manuscript; LL, TJ, YM, JZ, JL, JT, LW, JL and PY conducted all experiments and performed the statistical analysis of the data; LL, YP, HW, LZ, J.L.L.‐R., RSS, CC, NNL and HW discussed the experiments and results.
Supporting information
Fig. S1. Combination of ponatinib and fluconazole synergistically inhibits growth of C. albicans. C. albicans (SC5314) was subjected to 4 μM ponatinib combined with two‐fold serial dilutions of fluconazole in YPD medium. OD600 was measured every 15 min at 30°C.
Fig. S2. Ponatinib is non‐toxic at 16 μM or lower by lactate dehydrogenase assay. Endothelial cells (NCM460) were exposed to two‐fold serial dilutions of ponatinib in DMEM at 37°C for 1.5 h.
Fig. S3. The visible MIC of fluconazole or ponatinib on C. albicans, S. cerevisiae, C. neoformans and fluconazole‐resistant clinical C. albicans isolates. (A) Two‐fold serial dilutions of fluconazole or ponatinib in YPD without fungi. (B) C. albicans was exposed to two‐fold serial dilutions of fluconazole or ponatinib in YPD. (C) S. cerevisiae was exposed to two‐fold serial dilutions of fluconazole or ponatinib in YPD. (D) C. neoformans was exposed to two‐fold serial dilutions of fluconazole or ponatinib in YPD. (E) Fluconazole‐resistant clinical C. albicans isolates (CCC49) was exposed to two‐fold serial dilutions of fluconazole or ponatinib in YPD. (F) Fluconazole‐resistant clinical C. albicans isolates (CCC80) was exposed to two‐fold serial dilutions of fluconazole or ponatinib in YPD. The plates of C. albicans, S. cerevisiae and fluconazole‐resistant clinical C. albicans isolates were incubated at 30°C for 24h, the plate of C. neoformans was incubated at 30°C for 72h and all the plates were photographed by ChemiDoc MP (Bio‐Rad).
Fig. S4. Transcriptional expression of PDR5 in S. cerevisiae was quantified under treatment with ponatinib. Cells were subjected to DMSO or ponatinib in YPD at 30°C for 4 h. Experiments were performed in biological triplicates.
Fig. S5. The proton pump in the membrane was enhanced by fluconazole but inhibited when combined with ponatinib. IOV (In‐side out) of 4741 was subjected to DMSO, N‐ethylmaleimide (NEM) (10 μM) + Orthovanadate (OV) (100 μM), fluconazole (16 μg ml‐1), ponatinib (8 μM) or fluconazole (16 μg ml‐1) + ponatinib (8 μM) for 1 h. Experiments were performed in biological triplicates.
Acknowledgements
This study was supported by grants from the National Key R&D Program of China (2018YFC2000700; 2020YFA0907200), the National Nature Science Foundation (81630086, 81971993, 31900129, 81572053, 31870141, 31570140), the Key Research Program (ZDRW‐ZS‐2017‐1; KGFZD‐135‐19‐11; 153831KYSB20170043) of the Chinese Academy of Sciences, the Major Science and Technology Innovation Program of Shanghai Municipal Education Commission (2019‐01‐07‐00‐01‐E00059), the Program for Young Eastern Scholar at Shanghai Institutions of Higher Learning (program QD2018016), Shanghai Pujiang Program (18PJ1406600), Innovative research team of high‐level local universities in Shanghai, the Innovation Capacity Building Project of Jiangsu Province (BM2020019).
The authors thank Prof. Julia R. Koehler, Prof. Jinqiu Zhou and Prof. Ling Lu for kindly providing strains used in this work and all the lab members in Shanghai Jiao Tong University School of Medicine and at Institut Pasteur of Shanghai, Chinese Academy of Sciences, for their help in discussion and preparation of the manuscript.
Microb. Biotechnol. (2022) 15(2), 482–498
Contributor Information
Changbin Chen, Email: cbchen@ips.ac.cn.
Ning‐Ning Liu, Email: liuningning@shsmu.edu.cn.
Hui Wang, Email: huiwang@shsmu.edu.cn.
Data Availability Statement
The data sets generated from the current study have been deposited in the US National Center for Biotechnology Information (NCBI). The accession number for the gene expression profiling raw data reported in this paper is NCBI PRJNA: 649097. The Submission ID is SUB7844246. The link is provided in https://submit.ncbi.nlm.nih.gov/subs/bioproject/SUB7844246/overview.
References
- Alexander, B.D. (2008) Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts. Approved Standard M38‐A2, Clinical and Laboratory Standards Institute. [Google Scholar]
- Anderson, T.M. , Clay, M.C. , Cioffi, A.G. , Diaz, K.A. , Hisao, G.S. , Tuttle, M.D. , et al. (2014) Amphotericin forms an extramembranous and fungicidal sterol sponge. Nat Chem Biol 10: 400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso, V. , Tran, D.Q. , Ouellette, A.J. , and Selsted, M.E. (2020) Host defense peptides as templates for antifungal drug development. J Fungi 6: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billack, B. , Santoro, M. , and Lau‐Cam, C. (2009) Growth inhibitory action of ebselen on fluconazole‐resistant Candida albicans: role of the plasma membrane H+‐ATPase, Microbial drug resistance. (Larchmont N.Y.) 15: 77–83. [DOI] [PubMed] [Google Scholar]
- Brett, C.L. , Tukaye, D.N. , Mukherjee, S. , and Rao, R. (2005) The yeast endosomal Na+K+/H+ exchanger Nhx1 regulates cellular pH to control vesicle trafficking. Mol Biol Cell 16: 1396–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.C.S. , Nelson, J. , VanderSluis, B. , Deshpande, R. , Butts, A. , Kagan, S. , et al. (2014) Unraveling the biology of a fungal meningitis pathogen using chemical genetics. Cell 159: 1168–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey, J.R. , Grinstein, S. , and Orlowski, J. (2010) Sensors and regulators of intracellular pH. Nat Rev: Mol Cell Biol 11: 50–61. [DOI] [PubMed] [Google Scholar]
- Chan, G. , Hardej, D. , Santoro, M. , Lau‐Cam, C. , and Billack, B. (2007) Evaluation of the antimicrobial activity of ebselen: role of the yeast plasma membrane H+‐ATPase. J Biochem Mol Toxicol 21: 252–264. [DOI] [PubMed] [Google Scholar]
- Chen, C. , and Noble, S.M. (2012) Post‐transcriptional regulation of the Sef1 transcription factor controls the virulence of Candida albicans in its mammalian host. PLoS Pathog 8: e1002956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. , Pande, K. , French, S.D. , Tuch, B.B. , and Noble, S.M. (2011) An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell Host Microbe 10: 118–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste, A. , Turner, V. , Ischer, F. , Morschhäuser, J. , Forche, A. , Selmecki, A. , et al. (2006) A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans . Genetics 172: 2139–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyert, M.S. , and Philpott, C.C. (2013) Regulation of cation balance in Saccharomyces cerevisiae . Genetics 193: 677–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moraes, D.C. , Cardoso, K.M. , Domingos, L.T.S. , do Carmo Freire Ribeiro Pinto, M. , Monteiro, R.Q. , and Ferreira‐Pereira, A. (2020) β‐Lapachone enhances the antifungal activity of fluconazole against a Pdr5p‐mediated resistant Saccharomyces cerevisiae strain. Braz J Microbiol 51: 1051–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechant, R. , Binda, M. , Lee, S.S. , Pelet, S. , Winderickx, J. , and Peter, M. (2010) Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V‐ATPase. EMBO J 29: 2515–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechant, R. , Saad, S. , Ibáñez, A.J. , and Peter, M. (2014) Cytosolic pH regulates cell growth through distinct GTPases, Arf1 and Gtr1, to promote Ras/PKA and TORC1 activity. Mol Cell 55: 409–421. [DOI] [PubMed] [Google Scholar]
- Egner, R. , Rosenthal, F.E. , Kralli, A. , Sanglard, D. , and Kuchler, K. (1998) Genetic separation of FK506 susceptibility and drug transport in the yeast Pdr5 ATP‐binding cassette multidrug resistance transporter. Mol Biol Cell 9: 523–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felcmanova, K. , Neveceralova, P. , Sychrova, H. , and Zimmermannova, O. (2017) Yeast Kch1 and Kch2 membrane proteins play a pleiotropic role in membrane potential establishment and monovalent cation homeostasis regulation. FEMS Yeast Research 17: 10.1093/femsyr/fox053 [DOI] [PubMed] [Google Scholar]
- Ferreira, T. , Mason, A.B. , and Slayman, C.W. (2001) The yeast Pma1 proton pump: a model for understanding the biogenesis of plasma membrane proteins. J Biol Chem 276: 29613–29616. [DOI] [PubMed] [Google Scholar]
- Flanagan, P.R. , Liu, N.‐N. , Fitzpatrick, D.J. , Hokamp, K. , Köhler, J.R. , and Moran, G.P. (2017) The Candida albicans TOR‐activating GTPases Gtr1 and Rhb1 Coregulate starvation responses and biofilm formation. mSphere 2: e00477–17. 10.1128/mSphere.00477-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen, F. , Hermans, K. , Melchers, M.J.B. , Lagarde, C.C.M. , Meletiadis, J. , and Mouton, J.W. (2017) Pharmacodynamics of fosfomycin against ESBL‐ and/or carbapenemase‐producing Enterobacteriaceae. J Antimicrob Chemother 72: 3374–3381. [DOI] [PubMed] [Google Scholar]
- Giacomello, M. , De Mario, A. , Scarlatti, C. , Primerano, S. , and Carafoli, E. (2013) Plasma membrane calcium ATPases and related disorders. Int J Biochem Cell Biol 45: 753–762. [DOI] [PubMed] [Google Scholar]
- Gillies, R.J. , Ugurbil, K. , den Hollander, J.A. , and Shulman, R.G. (1981) 31P NMR studies of intracellular pH and phosphate metabolism during cell division cycle of Saccharomyces cerevisiae . Proc Natl Acad Sci USA 78: 2125–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golin, J. , Ambudkar, S.V. , Gottesman, M.M. , Habib, A.D. , Sczepanski, J. , Ziccardi, W. , and May, L. (2003) Studies with novel Pdr5p substrates demonstrate a strong size dependence for xenobiotic efflux. J Biol Chem 278: 5963–5969. [DOI] [PubMed] [Google Scholar]
- Gray, K.C. , Palacios, D.S. , Dailey, I. , Endo, M.M. , Uno, B.E. , Wilcock, B.C. , and Burke, M.D. (2012) Amphotericin primarily kills yeast by simply binding ergosterol. Proc Natl Acad Sci USA 109: 2234–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampe, I.A.I. , Friedman, J. , Edgerton, M. , and Morschhäuser, J. (2017) An acquired mechanism of antifungal drug resistance simultaneously enables Candida albicans to escape from intrinsic host defenses. PLoS Pathog 13: e1006655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman, J. , Movva, N.R. , and Hall, M.N. (1991) Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science (New York N.Y.) 253: 905–909. [DOI] [PubMed] [Google Scholar]
- Hsieh, M.H. , Yu, C.M. , Yu, V.L. , and Chow, J.W. (1993) Synergy assessed by checkerboard a critical analysis. Diag Microbiol Infect Dis 16: 343–349. [DOI] [PubMed] [Google Scholar]
- Huang, C. , Yang, L. , Li, Z. , Yang, J. , Zhao, J. , Dehui, X.u. , et al. (2007) Detection of CCND1 amplification using laser capture microdissection coupled with real‐time polymerase chain reaction in human esophageal squamous cell carcinoma. Cancer Genet Cytogenet 175: 19–25. [DOI] [PubMed] [Google Scholar]
- Jansen, G. , Lee, A.Y. , Epp, E. , Fredette, A. , Surprenant, J. , Harcus, D. , et al. (2009) Chemogenomic profiling predicts antifungal synergies. Mol Syst Biol 5: 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaunitz, J.D. , and Sachs, G. (1986) Identification of a vanadate‐sensitive potassium‐dependent proton pump from rabbit colon. J Biol Chem 261: 14005–14010. [PubMed] [Google Scholar]
- Kolaczkowski, M. , van der Rest, M. , Cybularz‐Kolaczkowska, A. , Soumillion, J.P. , Konings, W.N. , and Goffeau, A. (1996) Anticancer drugs, ionophoric peptides, and steroids as substrates of the yeast multidrug transporter Pdr5p. J Biol Chem 271: 31543–31548. [DOI] [PubMed] [Google Scholar]
- Kondapalli, K.C. , Prasad, H. , and Rao, R. (2014) An inside job: how endosomal Na(+)/H(+) exchangers link to autism and neurological disease. Front Cell Neurosci 8: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontoyiannis, D.P. , and Lewis, R.E. (2002) Antifungal drug resistance of pathogenic fungi. Lancet (London, England) 359: 1135–1144. [DOI] [PubMed] [Google Scholar]
- Lamping, E. , Monk, B.C. , Niimi, K. , Holmes, A.R. , Tsao, S. , Tanabe, K. , et al. (2007) Characterization of three classes of membrane proteins involved in fungal azole resistance by functional hyperexpression in Saccharomyces cerevisiae . Eukaryot Cell 6: 1150–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, J.Y. , Park, Y.H. , Pyon, Y.H. , Yang, J.M. , Yoon, J.Y. , Park, S.J. , et al. (2020) The LAMMER kinase is involved in morphogenesis and response to cell wall‐ and DNA‐damaging stresses in Candida albicans . Med Mycol 58: 240–247. [DOI] [PubMed] [Google Scholar]
- Liu, N.N. , Flanagan, P.R. , Zeng, J. , Jani, N.M. , Cardenas, M.E. , Moran, G.P. , and Köhler, J.R. (2017) Phosphate is the third nutrient monitored by TOR in Candida albicans and provides a target for fungal‐specific indirect TOR inhibition. Proc Natl Acad Sci USA 114: 6346–6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. , and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Method. Methods (San Diego, Calif.) 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Maesaki, S. , Marichal, P. , Vanden Bossche, H. , Sanglard, D. , and Kohno, S. (1999) Rhodamine 6G efflux for the detection of CDR1‐overexpressing azole‐resistant Candida albicans strains. J Antimicrob Chemother 44: 27–31. [DOI] [PubMed] [Google Scholar]
- Martínez‐Muñoz, G.A. , and Kane, P. (2017) Vacuolar and plasma membrane proton pumps collaborate to achieve cytosolic pH homeostasis in yeast. J Biol Chem 292: 7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei, Y. , Jiang, T. , Zou, Y. , Wang, Y. , Zhou, J. , Li, J. , et al. (2020) FDA approved drug library screening identifies robenidine as a repositionable antifungal. Front Microbiol 11: 996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, N.N. , Prasad, T. , Sharma, N. , Payasi, A. , Prasad, R. , Gupta, D.K. , and Singh, R. (2007) Pathogenicity and drug resistance in Candida albicans and other yeast species. A review. Acta microbiologica et immunologica Hungarica 54: 201–235. [DOI] [PubMed] [Google Scholar]
- Mitchell, P. (2011) Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. 1966. Biochem Biophys Acta 1807: 1507–1538. [DOI] [PubMed] [Google Scholar]
- Mitchell, R. , Hopcroft, L.E.M. , Baquero, P. , Allan, E.K. , Hewit, K. , James, D. , et al. (2018) Targeting BCR‐ABL‐independent TKI resistance in chronic myeloid leukemia by mTOR and autophagy inhibition. J Natl Cancer Inst 110: 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk, B.C. , Kurtz, M.B. , Marrinan, J.A. , and Perlin, D.S. (1991) Cloning and characterization of the plasma membrane H(+)‐ATPase from Candida albicans . J Bacteriol 173: 6826–6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk, B.C. , Mason, A.B. , Kardos, T.B. , and Perlin, D.S. (1995) Targeting the fungal plasma membrane proton pump. Acta Biochim Pol 42: 481–496. [PubMed] [Google Scholar]
- Monk, B.C. , Niimi, K. , Lin, S. , Knight, A. , Kardos, T.B. , Cannon, R.D. , et al. (2005) Surface‐active fungicidal D‐peptide inhibitors of the plasma membrane proton pump that block azole resistance. Antimicrob Agents Chemother 49: 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musumeci, F. , Greco, C. , Grossi, G. , Molinari, A. , and Schenone, S. (2018) Recent studies on ponatinib in cancers other than chronic myeloid leukemia. Cancers 10: 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, K. , Niimi, M. , Niimi, K. , Holmes, A.R. , Yates, J.E. , Decottignies, A. , et al. (2001) Functional expression of Candida albicans drug efflux pump Cdr1p in a Saccharomyces cerevisiae strain deficient in membrane transporters. Antimicrob Agents Chemother 45: 3366–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds, F.C. (2003) Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother 52: 1. [DOI] [PubMed] [Google Scholar]
- Ohgaki, R. , van, I. S. C. Matsushita, M. , Hoekstra, D. , and Kanazawa, H. (2011) Organellar Na+/H+ exchangers: novel players in organelle pH regulation and their emerging functions. Biochemistry 50: 443–450. [DOI] [PubMed] [Google Scholar]
- Orij, R. , Brul, S. , and Smits, G.J. (2011) Intracellular pH is a tightly controlled signal in yeast. Biochem Biophys Acta 1810: 933–944. [DOI] [PubMed] [Google Scholar]
- Ottilie, S. , Goldgof, G.M. , Cheung, A.L. , Walker, J.L. , Vigil, E. , Allen, K.E. , et al. (2018) Two inhibitors of yeast plasma membrane ATPase 1 (ScPma1p): toward the development of novel antifungal therapies. J Cheminform 10: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlin, D.S. , Seto‐Young, D. , and Monk, B.C. (1997) The plasma membrane H(+)‐ATPase of fungi. A candidate drug target? Ann N Y Acad Sci 834: 609–617. [DOI] [PubMed] [Google Scholar]
- Poznanski, J. , Szczesny, P. , Ruszczyńska, K. , Zielenkiewicz, P. , and Paczek, L. (2013) Proteins contribute insignificantly to the intrinsic buffering capacity of yeast cytoplasm. Biochem Biophys Res Comm 430: 741–744. [DOI] [PubMed] [Google Scholar]
- Prasad, R. , and Goffeau, A. (2012) Yeast ATP‐binding cassette transporters conferring multidrug resistance. Annu Rev Microbiol 66: 39–63. [DOI] [PubMed] [Google Scholar]
- Rand, K.H. , Houck, H.J. , Brown, P. , and Bennett, D. (1993) Reproducibility of the microdilution checkerboard method for antibiotic synergy. Antimicrob Agents Chemother 37: 613–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex, J.H. (2008) Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts. Approved Standard M27‐A3, Clinical and Laboratory Standards Institute. [Google Scholar]
- Robbins, N. , Spitzer, M. , Yu, T. , Cerone, R.P. , Averette, A.K. , Bahn, Y.‐S. , et al. (2015) An antifungal combination matrix identifies a rich pool of adjuvant molecules that enhance drug activity against diverse fungal pathogens. Cell Rep 13: 1481–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero, M.A. , and Topal, J.E. (2014) Development of echinocandin‐resistant Candida albicans candidemia following brief prophylactic exposure to micafungin therapy. Transpl infect Dis 16: 469–472. [DOI] [PubMed] [Google Scholar]
- Saliba, E. , Evangelinos, M. , Gournas, C. , Corrillon, F. , Georis, I. , and André, B. (2018) The yeast H+‐ATPase Pma1 promotes Rag/Gtr‐dependent TORC1 activation in response to H+‐coupled nutrient uptake. eLife 7: e31981. 10.7554/eLife.31981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano, R. (1988) Structure and function of proton translocating ATPase in plasma membranes of plants and fungi. Biochem Biophys Acta 947: 1–28. [DOI] [PubMed] [Google Scholar]
- Shapiro, R.S. , Robbins, N. , and Cowen, L.E. (2011) Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol Mol Biol Rev 75: 213–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar‐Guturja, T. , Gunaherath, G.M.K.B. , Wijeratne, E.M.K. , Lambert, J.‐P. , Averette, A.F. , Lee, S.C. , et al. (2016a) Dual action antifungal small molecule modulates multidrug efflux and TOR signaling. Nat Chem Biol 12: 867–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar‐Guturja, T. , Tebung, W.A. , Mount, H. , Liu, N. , Köhler, J.R. , Whiteway, M. , and Cowen, L.E. (2016b) Beauvericin potentiates azole activity via inhibition of multidrug efflux, blocks Candida albicans Morphogenesis, and is effluxed via Yor1 and circuitry controlled by Zcf29. Antimicrob Agents Chemother 60: 7468–7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer, M. , Griffiths, E. , Blakely, K.M. , Wildenhain, J. , Ejim, L. , Rossi, L. , et al. (2011) Cross‐species discovery of syncretic drug combinations that potentiate the antifungal fluconazole. Mol Syst Biol 7: 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, E. , Gow, N.A. , and Bowen, D.V. (1988) Cytoplasmic alkalinization during germ tube formation in Candida albicans . J Gen Microbiol 134: 1079–1087. [DOI] [PubMed] [Google Scholar]
- Sukheja, P. , Kumar, P. , Mittal, N. , Li, S. G. , Singleton, E. , and Russo, R. , et al. (2017) A novel small‐molecule inhibitor of the mycobacterium tuberculosis demethylmenaquinone methyltransferase MenG is bactericidal to both growing and nutritionally deprived persister cells. mBio 8: e02022–16. 10.1128/mBio.02022-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, Y.o. , Onge, R.P.S. , Mani, R. , King, O.D. , Heilbut, A. , Labunskyy, V.M. , et al. (2011) Knocking out multigene redundancies via cycles of sexual assortment and fluorescence selection. Nat Methods 8: 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toenjes, K.A. , Munsee, S.M. , Ibrahim, A.S. , Jeffrey, R. , Edwards, J.E. Jr , and Johnson, D.I. (2005) Small‐molecule inhibitors of the budded‐to‐hyphal‐form transition in the pathogenic yeast Candida albicans . Antimicrob Agents Chemother 49: 963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Rest, M.E. , Kamminga, A.H. , Nakano, A. , Anraku, Y. , Poolman, B. , and Konings, W.N. (1995) The plasma membrane of Saccharomyces cerevisiae: structure, function, and biogenesis. Microbiol Rev 59: 304–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke, R.W. , Hornick, C.A. , Belcher, J. , Scharschmidt, B.F. , and Havel, R.J. (1985) Identification and characterization of ATP‐dependent proton transport by rat liver multivesicular bodies. J Biol Chem 260: 11021–11026. [PubMed] [Google Scholar]
- Vincent, B.M. , Langlois, J.B. , Srinivas, R. , Lancaster, A.K. , Scherz‐Shouval, R. , Whitesell, L. , et al. (2016) A fungal‐selective cytochrome bc(1) inhibitor impairs virulence and prevents the evolution of drug resistance. Cell Chem Biol 23: 978–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Ye, X. , Yang, X. , Cai, Y. , Wang, S. , Tang, J. , et al. (2020) Discovery of novel antibiotics as covalent inhibitors of fatty acid synthesis. ACS Chem Biol 15: 1826–1834. [DOI] [PubMed] [Google Scholar]
- Witchley, J.N. , Penumetcha, P. , Abon, N.V. , Woolford, C.A. , Mitchell, A.P. , and Noble, S.M. (2019) Candida albicans morphogenesis programs control the balance between gut commensalism and invasive infection. Cell Host Microbe 25: 432–443.e436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H. , Whiteway, M. , and Jiang, L. (2019) The tricarboxylic acid cycle, cell wall integrity pathway, cytokinesis and intracellular pH homeostasis are involved in the sensitivity of Candida albicans cells to high levels of extracellular calcium. Genomics 111: 1226–1230. [DOI] [PubMed] [Google Scholar]
- Yang, H. , Tsang, P.C.S. , Pow, E.H.N. , Lam, O.L.T. , and Tsang, P.W. (2020) Potential role of Candida albicans secreted aspartic protease 9 in serum induced‐hyphal formation and interaction with oral epithelial cells. Microb Pathog 139: 103896. [DOI] [PubMed] [Google Scholar]
- Zhang, Y.Q. , Gamarra, S. , Garcia‐Effron, G. , Park, S. , Perlin, D.S. , and Rao, R. (2010) Requirement for ergosterol in V‐ATPase function underlies antifungal activity of azole drugs. PLoS Pathog 6: e1000939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Combination of ponatinib and fluconazole synergistically inhibits growth of C. albicans. C. albicans (SC5314) was subjected to 4 μM ponatinib combined with two‐fold serial dilutions of fluconazole in YPD medium. OD600 was measured every 15 min at 30°C.
Fig. S2. Ponatinib is non‐toxic at 16 μM or lower by lactate dehydrogenase assay. Endothelial cells (NCM460) were exposed to two‐fold serial dilutions of ponatinib in DMEM at 37°C for 1.5 h.
Fig. S3. The visible MIC of fluconazole or ponatinib on C. albicans, S. cerevisiae, C. neoformans and fluconazole‐resistant clinical C. albicans isolates. (A) Two‐fold serial dilutions of fluconazole or ponatinib in YPD without fungi. (B) C. albicans was exposed to two‐fold serial dilutions of fluconazole or ponatinib in YPD. (C) S. cerevisiae was exposed to two‐fold serial dilutions of fluconazole or ponatinib in YPD. (D) C. neoformans was exposed to two‐fold serial dilutions of fluconazole or ponatinib in YPD. (E) Fluconazole‐resistant clinical C. albicans isolates (CCC49) was exposed to two‐fold serial dilutions of fluconazole or ponatinib in YPD. (F) Fluconazole‐resistant clinical C. albicans isolates (CCC80) was exposed to two‐fold serial dilutions of fluconazole or ponatinib in YPD. The plates of C. albicans, S. cerevisiae and fluconazole‐resistant clinical C. albicans isolates were incubated at 30°C for 24h, the plate of C. neoformans was incubated at 30°C for 72h and all the plates were photographed by ChemiDoc MP (Bio‐Rad).
Fig. S4. Transcriptional expression of PDR5 in S. cerevisiae was quantified under treatment with ponatinib. Cells were subjected to DMSO or ponatinib in YPD at 30°C for 4 h. Experiments were performed in biological triplicates.
Fig. S5. The proton pump in the membrane was enhanced by fluconazole but inhibited when combined with ponatinib. IOV (In‐side out) of 4741 was subjected to DMSO, N‐ethylmaleimide (NEM) (10 μM) + Orthovanadate (OV) (100 μM), fluconazole (16 μg ml‐1), ponatinib (8 μM) or fluconazole (16 μg ml‐1) + ponatinib (8 μM) for 1 h. Experiments were performed in biological triplicates.
Data Availability Statement
The data sets generated from the current study have been deposited in the US National Center for Biotechnology Information (NCBI). The accession number for the gene expression profiling raw data reported in this paper is NCBI PRJNA: 649097. The Submission ID is SUB7844246. The link is provided in https://submit.ncbi.nlm.nih.gov/subs/bioproject/SUB7844246/overview.


