Summary
By providing the scientific community with uniform and standardized resources of consistent quality, plasmid repositories play an important role in enabling scientific reproducibility. Plasmids containing insertion sequence elements (IS elements) represent a challenge from this perspective, as they can change the plasmid structure and function. In this study, we conducted a systematic analysis of a subset of plasmid stocks distributed by plasmid repositories (The Arabidopsis Biological Resource Center and Addgene) which carry unintended integrations of bacterial mobile genetic elements. The integration of insertion sequences was most often found in, but not limited to, pBR322‐derived vectors, and did not affect the function of the specific plasmids. In certain cases, the entire stock was affected, but the majority of the stocks tested contained a mixture of the wild‐type and the mutated plasmids, suggesting that the acquisition of IS elements likely occurred after the plasmids were acquired by the repositories. However, comparison of the sequencing results of the original samples revealed that some plasmids already carried insertion mutations at the time of donation. While an extensive BLAST analysis of 47 877 plasmids sequenced from the Addgene repository uncovered IS elements in only 1.12%, suggesting that IS contamination is not widespread, further tests showed that plasmid integration of IS elements can propagate in conventional Escherichia coli hosts over a few tens of generations. Use of IS‐free E. coli hosts prevented the emergence of IS insertions as well as that of small indels, suggesting that the use of IS‐free hosts by donors and repositories could help limit unexpected and unwanted IS integrations into plasmids.
Certain plasmids distributed by plasmid repositories were found to carry unintended insertion sequences. In some cases, these insertions provided a detectable fitness advantage to the Escherichia coli host. Some of these plasmids acquired the insertion prior to their deposition to the repository, others acquired it afterwards.

Introduction
Mobile genetic elements or transposable elements (TEs) are segments of DNA that can modify their genomic locus via the process of transposition. The smallest autonomous TEs, called insertion sequence (IS) elements, are found in prokaryotic cells, and consist largely of a transposase gene surrounded by inverted repeats (Mahillon and Chandler, 1998). Bacterial IS elements have a patchy distribution on the phylogenetic tree, and show dramatic variation in copy numbers even when comparing closely related strains (Sawyer et al., 1987; Wagner, 2006). At the upper extreme, there are examples of bacteria, such as Microcystis aeruginosa or Sitophilus oryzae primary endosymbiont which harbour more than 500 ISes within their genomes (Plague et al., 2008; Lin et al., 2011; Oakeson et al., 2014). A study, however, comparing 262 genomes originating from archaea and phylogenetically distant bacteria (representing Firmicutes, Actinobacteriae, Mollicutes, Spirochaetes, Cyanobacteriae and all five classes of Proteobacteriae and Chlamydiae), found the median number to be only 12, with a quarter of the sequences carrying none at all (Touchon and Rocha, 2007).
Despite their simplicity, multiple examples underline the role of IS elements in the adaptation of bacteria to environmental changes (Reynolds et al., 1981; Hall, 1999; Soto et al., 2004; Carlson et al., 2009; Kaleta et al., 2010; Zhang et al., 2017). Their contribution to the rate of mutations falls in the range 4–98%, depending on the genetic screen used for measurement (Hall, 1998, 1999; Halliday and Glickman, 1991; Feher et al., 2006). Among the ISes of Escherichia coli, IS1 seems to be the most active with an overall transposition rate of 2.79 × 10‐5 transposition/element/generation (Sousa et al., 2013). In addition, various forms of environmental stresses have been shown to induce IS transposition (Eichenbaum and Livneh, 1998; Drevinek et al., 2010; Pasternak et al., 2010; Umenhoffer et al., 2010). The fundamental properties of the IS types found in various strains of E. coli, along with their copy numbers, can be found in Appendix S1.
In parallel with the spread of gene cloning in molecular biology came reports describing sporadic IS integrations in the plasmid‐encoded transgenes (Blumenthal et al., 1985; Rawat et al., 2009). Further attention was garnered by the fact that in certain cases, IS transposition was found to be the primary mechanism causing inactivation of the cloned gene of interest (Rood et al., 1980; Nakamura and Inouye, 1981; Muller et al., 1989; Chen and Yeh, 1997; Valle‐Garcia et al., 2014; Rugbjerg et al., 2018; Fan et al., 2019). IS elements have also been shown to lead to instability of a cosmid library by integrating into the vector backbone and causing deletions or other rearrangements (Fernandez et al., 1986).
The following study analyses sequences from plasmids stored in two research repositories, the Arabidopsis Biological Resource Center (ABRC; https://abrc.osu.edu/) at The Ohio State University and Addgene (https://www.addgene.org/), the non‐profit plasmid repository. Both repositories rely on depositions from the scientific community, prior to or after publication of data involving the deposited plasmids. Depositing in repositories accelerates science by enabling timely access to new research material and supports scientific reproducibility by providing authenticated and high‐quality material. As part of its plasmid authentication and quality control process, Addgene obtains full plasmid sequences and annotates IS insertions detected in the sequence.
Here, we describe the type and the extent of IS insertions in deposited plasmids and discuss potential solutions to prevent IS integrations into plasmids.
Results
Identification of IS5 elements in ABRC stocks
As part of ABRC’s quality control efforts, we investigated a number of complaints about incorrect restriction patterns in plasmids from a series of plant binary vectors based on the pCambia backbone (plasmids 1–25). Most researchers reported an incorrect EcoRI restriction digest pattern reflecting the presence of an extra 1.2 kb of sequence. ABRC’s quality control process involves analysis of two or three colonies derived from the ‘distribution’ glycerol stock from which samples are prepared for distribution, as well as two or three colonies derived from an original glycerol stock received from the donor of the plasmid. Restriction digest with EcoRI of ABRC stocks with the pCambia backbone confirmed the presence of the additional EcoRI site in a number of samples and revealed that single colonies, derived from individual original and distribution stocks, had different restriction patterns. In the example shown in Fig. 1A, one colony has the expected restriction pattern based on the known sequence of the plasmid, while the other shows the presence of the unexpected EcoRI fragment. The incorrect restriction pattern was identical for all pCambia plasmid colonies giving unexpected restriction digest results. We located the putative insertion in plasmid 8 based on diagnostic restriction digests and have shown by Sanger sequencing of the region between the Kanamycin resistance gene and the ColEI origin of replication that it represents a bacterial insertion element IS5, which provides the extra EcoRI restriction site appearing in the plasmid (Fig. 1B). The IS5 insertion is located on the plasmid backbone outside of the left border and does not transfer to the plant as part of the T‐DNA.
Fig. 1.
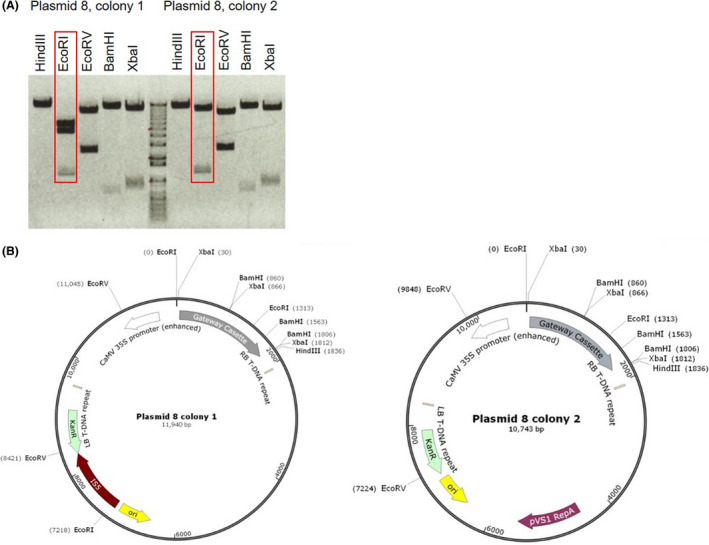
Identification of IS5 in a plant binary vector by restriction digest.
A. Restriction digest of plasmid 8 isolated from two individual colonies derived from an original glycerol stock received from the donor of the plasmid with five restriction enzymes. EcoRI digestions are marked, showing an additional EcoRI site in colony 1. The centre lane shows the 1 Kb Plus Ladder (Invitrogen) as a marker.
B. Schematic representation of plasmid 8, showing the location of the restriction sites for the enzymes used to digest colonies 1 and 2 as shown in A. The location of the IS insertion with an additional EcoRI site is shown in colony 1.
IS element insertion is a widespread phenomenon
To test for the presence of an IS element by PCR, we designed primers flanking the putative insertion site, and used them to probe 25 plasmids from the ABRC collection with the same pCambia backbone. Plasmid preparations were not derived from a culture inoculated from a single colony, but PCR amplification using primers IS5‐flank‐ColE1‐F1 and IS5‐flank‐KanR‐R1 enabled us to distinguish between plasmid populations with no detectable IS element, which were expected to show the presence of a ~ 200 bp amplicon, and plasmids with an IS element, with expected amplicon size of ~ 1400 bp in a single culture. The results presented in Fig. 2A demonstrate that an IS element was present in half of the cultures of plasmids of the pCambia series, while the other half was IS free. Given an established PCR bias towards short amplicons, this result is most likely an underestimate of the representation of the IS element in a plasmid population and this bias should be taken into account for all subsequent PCR assay‐based results. We also tested other frequently ordered plasmids with similar vector backbones and histories of user complaints of incorrect digest patterns for the presence of IS insertions. We found the presence of IS elements in plasmids with pBIN20 and pFGC5941 vector backbones, although there were some plasmids that did not show the presence of an IS element (Fig. 2B). The cultures with IS element‐containing plasmids also contained plasmids without the IS insertion, indicating that they contained a mix of plasmids with and without the IS element. pCambia, pBIN20 and pFGC5941 are all derived from pBR322, which is itself susceptible to the IS transposition phenomenon (Amster and Zamir, 1986). The instance of IS element transposition into a position between Kanamycin resistance and the ColE1 origin of replication has also been described for the pGreenII vector and derivative clones (Watson et al., 2016). Given that pGreenII has no pBR322 ancestry, this finding suggests that the IS insertion occurs more frequently than recognized within the scientific community.
Fig. 2.
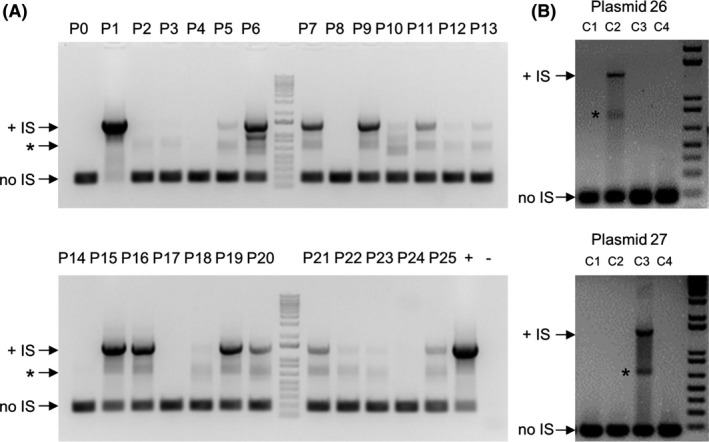
IS elements are present in plant binary vectors with different backbones.
A. PCR screening of a pCambia‐based binary vector series using primers flanking the putative IS insertion site. (‐) Represents negative control (water); (+) represents positive control (plasmid 1).
B. PCR screening of pFGC5941 (plasmid 26) and pBIN20 (plasmid 27) derivatives using primers flanking the putative IS insertion site. In both sections A and B, a PCR product of ~1400 bp indicates the presence of an IS insertion (+IS) while a product of ~200 bp indicates the absence of IS insertion (no IS). A non‐specific PCR product is marked with an asterisk.
Certain IS acquisitions occur prior to stock deposition, others occur afterwards
To test whether the insertion of IS elements occurs before or after a stock has been donated to the ABRC, we analysed a group of plant organellar markers in a pCambia backbone. An identical copy of these clones has been deposited to the Addgene plasmid repository, where they were sequenced at the whole genome level. PCR amplification from 16 different ABRC stocks from this group, using primers flanking the putative IS insertion site, showed that six of these clones contained an IS element, while 10 of them were IS free (Fig. 3). The next‐generation sequencing data on Addgene’s stocks were fully consistent with this result, with IS elements identified in the same six clones. Sequencing data additionally identified the insertion as an IS4 element which was annotated on the Addgene plasmid pages. While the sequence of these plasmids was affected, reports from donors and users suggested that the transposition of an IS element between the Kanamycin resistance and the origin of replication did not affect the functionality of these constructs. Most of the other ABRC plasmids representing expression vectors and constructs also showed correct/expected localization or function in plants, regardless of the IS element presence.
Fig. 3.
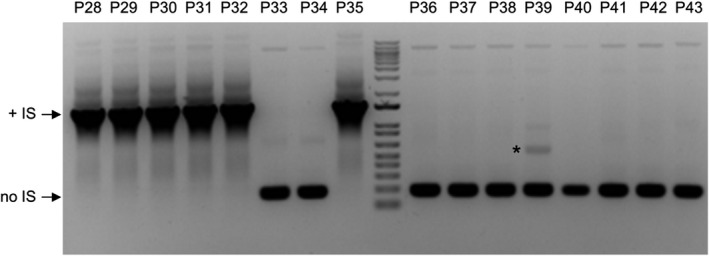
IS transposition occurs prior to stock deposition. PCR screening of a pCambia‐based binary vector series using primers flanking the putative IS insertion site. Plasmids isolated from 16 different stocks (P28–P43) were screened to detect the presence of an IS element. A PCR product of ~ 1400 bp indicates the presence of an IS insertion (+IS) while a product of ~ 200 bp indicates the absence of IS insertion (no IS). A non‐specific PCR product is marked with an asterisk.
The presence of shared integrated IS elements in plasmid preparations submitted to multiple plasmid repositories, however, does not exclude the possibility of IS acquisition during culturing at the repositories. Upon arrival of a plasmid preparation at Addgene, it is transformed into an E. coli host, and a single, sequenced clone is used to amplify the stock which is used to inoculate and grow distribution stocks. ABRC usually accepts plasmids as glycerol stocks in E. coli hosts, which are streaked on plates and several individual colonies are grown as liquid cultures for plasmid preparation and analysis (which may include restriction digestion, PCR and sequencing). If IS transposition did not occur during the growth of stock cultures, the stab cultures distributed by the repositories would be homogenous (especially if they had been transformed), and all cells sent to the recipient in a stab culture would be either with or without the insertion. To test whether this is the case, we obtained four pCambia‐derived stab cultures from two repositories: plasmids 27 and 44 from ABRC, and plasmids 45 and 46 from Addgene. Each stab culture was seeded onto LB+Km plates to obtain individual colonies. Ten colonies from each plating were inoculated and independently liquid grown to make small‐scale plasmid preparations. Restriction fragment analysis of the plasmid preparations revealed a variable restriction pattern among colonies for both plasmids 27 and 44 (Table 1, Fig. S1A and B), indicating that the obtained stab cultures were heterogeneous. These could be attributable either to IS transposition at the repository or the mixed nature of the glycerol stock submitted by the donors. Furthermore, all colonies of plasmid 45 carried various mutated versions of the plasmid (Table 1, Fig. S2A). Since Addgene inoculates a single transformant to generate cultures for storage, this is most likely the result of IS transposition that occurred after the founding of the seed cultures at the repository. In contrast, plasmid preparations of plasmid 46 were homogeneous and displayed only the expected restriction pattern (Table 1, Fig. S2B). The mixed content of some plasmid preparations shown on Fig. 2B also suggests the transposition of IS elements into certain plasmids during their propagation in conventional hosts, either at the repository or at the donor laboratory.
Table 1.
Measuring the purity of plasmids obtained from plasmid repositories.
| Plasmid | AbR | Size | Rep. origin | Copy no. | Ratio of correct restriction pattern a |
|---|---|---|---|---|---|
| Plasmid 27 | Km | 14.4 kb | IncP | High | 9:10 |
| Plasmid 44 | Km | 11.3 kb | ColE1 | High | 8:10 |
| Plasmid 45 | Km | 8.6 kb | ColE1 | High | 0:10 |
| Plasmid 46 | Km | 6.8 kb | ColE1 | High | 10:10 |
AbR, antibiotic resistance; Km, Kanamycin.
a
Restriction digestion of plasmid preparations made by culturing 10 colonies obtained after plating the respective stab culture.
IS‐containing plasmids have selective advantage over non‐IS plasmids
It was previously shown that the IS5 transposition into pGreenII significantly increased the bacterial growth rate and that the mutated pGreenII rapidly outcompeted the original plasmid with no insertion (Watson et al., 2016). Plasmid instability was proposed to be the result of natural selection favouring mutations that relieve the host cell from the burden of propagating the wild‐type construct. To test the stability of an ABRC plasmid stock without an IS insertion, we cultivated a stock of plasmid 1 derived from a single colony (previously shown by sequencing to be IS free), over multiple generations, and tested it using PCR amplification with primers flanking the putative IS insertion site. While the initial culture showed that no IS element was present in plasmids isolated either from donated originals (O1‐O2) or their distribution copies (D1‐D2), subsequent subculturing led to an increase in the proportion of the IS element‐containing plasmids (Fig. 4A). There was a clear difference in the rate at which the IS element was acquired when the original stocks were compared with the distribution copies, with the IS element readily detectable in D1 and D2 after the first subculture (Fig. 4A). This difference no longer existed after one more round of subculturing. By the third subculture, there were more plasmids with an IS element than without (Fig. 4A). Similarly, we observed the emergence of extra restriction bands caused by IS5 transposition when propagating two further pCambia‐derived vectors, plasmids 27 and 44 in E. coli DB3.1 for 70 generations (Fig. 4B Left, Centre). We screened the resulting plasmids with multiple PCRs (described in the Experimental Procedures) and found IS5 insertions in both. Sequencing verified this result: for plasmid 27, insertion of IS5 was detectable either at position 7187 or at 6336, which corresponds to the two regions flanking the KmR gene. No plasmid carried both insertions. (For a potential hypothesis explaining the selective advantage of IS5 insertions, see Appendix S2) Analysis of plasmid 44 verified an insertion of IS5 at position 10 185, directly downstream of the KmR gene. IS integration into the pCambia series, however, is not obligatory: plasmid 46 did not show a detectable change in its restriction pattern during 70 generations of growth in DB3.1, despite having a backbone nearly identical to that of plasmid 44 (Fig. 4B, Right).
Fig. 4.
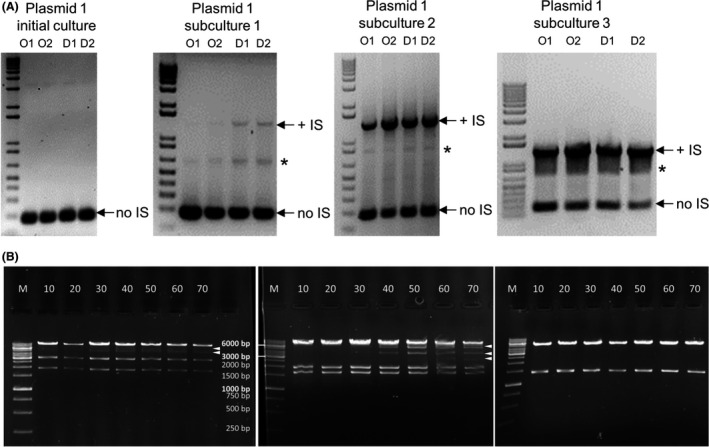
The presence of IS elements provides selective advantage to certain plasmids in E. coli DB3.1.
A. Time‐series PCR screening of plasmid 1 using primers flanking the putative IS insertion site. Single colonies of the ABRC original (O1–O2) and distribution stocks (D1–D2) were subjected to three rounds of subculturing, with appropriate aliquots taken out and used for the analysis. A PCR product of ~1400 bp indicates the presence of an IS insertion (+IS) while a product of ~200 bp indicates the absence of IS insertion (no IS). A non‐specific PCR product is marked with an asterisk.
B. Changes in the restriction pattern of various plasmids propagated in E. coli DB3.1. The numbers on top of each gel photo represent the number of generations the culture had gone through at the time of sampling. Left: plasmid 27, digested with EcoRI + NheI. Expected bands: 9491 bp, 3050 bp and 1862 bp. An extra band appears between 3 kbp and 6 kbp beginning at generation 30, marked by an arrowhead. Centre: plasmid 44, digested with EcoRI + HindIII. Expected bands: 7823 bp, 1946 bp and 1637 bp. Extra bands appear beginning at generation 40, marked by arrowheads. Right: plasmid 46, digested with EcoRI + EcoRV. Expected bands: 5447 bp and 1399 bp. Only expected bands are visible in all lanes. M: GeneRuler 1 kbp DNA Ladder (Thermo Scientific).
These time‐series results nevertheless demonstrate that due to the presence of these elements in the genomes of most commonly used E. coli strains, even a stock with no detectable IS‐mutated plasmid may eventually become a mix of IS‐mutated and normal plasmids, with the proportion of mutant plasmids increasing each time the stock is subcultured. Our findings are in line with earlier reports suggesting that insertion of an IS element can provide these plasmids a selective advantage over the wild type (Pósfai et al., 2006; Rugbjerg et al., 2018). The type of burden caused by the plasmid and the mechanism of release attained by the insertion, however, may be case specific.
IS acquisition of plasmids can be avoided with the use of IS‐free hosts
According to our hypothesis, the source of the IS elements identified in the plasmids above are the elements residing in the genomes of conventional E. coli hosts used for plasmid assembly and propagation. The correctness of newly assembled plasmids is surely verified by the constructing laboratories in most, if not all cases. The most commonly used methods of verification, restriction digestion and Sanger sequencing, however, may miss IS‐insertion mutants if the fraction of such mutants is low at the time of testing due to the low intensity of the unexpected bands or peaks on the electrophoresis readout of the two methods, respectively. This can result in the storage of mixed bacterial cultures as glycerol stocks and the deposition of mixed bacterial stocks or plasmid preparations. The small fraction of mutant‐carrying cells can expand due to their selective advantage at any later stage of use that involves bacterial growth. Similarly, at the repository, transposition of IS elements from the host bacteria can happen at any subsequent stage of culturing, even in the dividing cells of the stab culture. This can explain the distribution of mixed bacterial cultures, despite the availability of verified original stocks.
Therefore, a straightforward approach to avoid the emergence of insertion mutants would be to replace the currently used conventional E. coli hosts with IS‐free E. coli strains. Two such E. coli stains were chosen to test this strategy: MDS42 and BLK16 (Pósfai et al., 2006; Umenhoffer et al., 2017). They are derivatives of E. coli K12 MG1655 and E. coli BL21 (DE3), respectively, and have gone through systematic genome reduction processes which, besides removing many strain‐specific genomic islands, eliminated all active mobile genetic elements from their chromosomes. MDS42 is generally used as a cloning host due to its elevated transformability, while BLK16 is recommended for protein overexpression. IS elements have been completely deleted from the prior strain as opposed to the latter, where they were mostly inactivated by inserting premature stop codons (Pósfai et al., 2006; Umenhoffer et al., 2017).
As a starting point, strain MDS42 was transformed with the correct form (verified by restriction digestion) of two plasmids that were prone to acquire IS integrations: plasmids 27 and 44. Several colonies of each transformation were cultured (corresponding to 10 generations) to make small‐scale plasmid preparations and to choose a starting culture that displays the correct restriction pattern. Then, the respective cell lines were cultured for 60 more generations, making small‐scale plasmid preparations at every 10 generations for restriction analysis. Importantly, no change of any sort was observable in the restriction pattern of either plasmid (Fig. 5). Similarly, the correct restriction pattern was maintained for both plasmids when propagated for 70 generations in the IS‐free strain BLK16 (Fig. S3). These data support the notion that major reorganization of these plasmids, including the integration of an IS element, can be avoided if an IS‐free host strain is used for their propagation. It was not apparent at this stage, however, whether other types of mutations (single nucleotide exchanges, small insertions or deletions, etc.) had been acquired by the plasmids during propagation. This was possible considering the selective advantage of IS‐mutated plasmids described above, possibly relieving the host from some type of burden brought about by the wild‐type vectors. If this burden could be relieved by point mutations or small indels, a similar selection process would expand the mutant subpopulation within a relatively low number of generations.
Fig. 5.
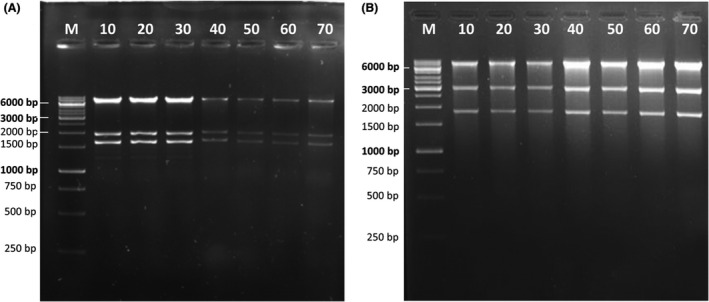
Stability of the restriction pattern of plasmids propagated in E. coli MDS42. The numbers on top of each gel photo represent the number of generations the culture had gone through at the time of sampling.
A. Plasmid 44, digested with HindIII and EcoRI. Expected bands: 7823, 1946 and 1637 bp. Only expected bands are present in all lanes.
B. Plasmid 27, digested with NheI and EcoRI. Expected bands: 9491, 3050 and 1862 bp. All lanes display expected bands only. M: GeneRuler 1 Kbp DNA Ladder (Thermo Scientific).
To investigate this possibility, we carried out deep sequencing analysis of plasmid 27 preparations obtained from MDS42 after 10 and 70 generations of culturing respectively. Sequence analysis revealed 18 variants in the 70th generation sample relative to the reference sequence, listed in Table S2. All but one of these variants were present in the 10th generation sample as well, suggesting the majority of these variants are likely to have been present in the original sample, and did not arise as novel mutations during propagation in the IS‐free host strain. The exception was a C‐>A SNP at position 6929, scored as a heterozygous site, with read counts in the 70th generation sample of 4257 and 555 for the reference and variant alleles respectively. (For a potential hypothesis explaining the selective advantage of this mutation, which causes premature translation termination of the KmR gene, see Appendix S2) As expected, no signs of IS acquisitions were found in the sequencing reads, apart from the IS1 known to be present in the reference plasmid 27 sequence.
The frequency of E. coli ISes in Addgene sequences
We expanded our analysis by examining the type and frequency of IS elements in the Addgene plasmid collection. Addgene generated complete sequence data for 47 877 plasmids from their repository. We used these as part of a local BLAST+ (Camacho et al., 2009) analysis in which we created a custom BLAST database and searched this set of > 47 000 plasmids for evidence of each of 18 IS sequences of interest (Table 2; File S1). We limited our queries to transposable elements found in various E. coli strains, for we were interested not in mobile elements already residing in the insert DNA to be cloned into various vectors, but in those elements that have transposed during plasmid construction or storage, which is almost exclusively done in E. coli (Lodish et al., 2000). BLAST hits were filtered to help ensure hits reflected intact and viable IS sequences, as opposed to hits to partial IS sequences that might arise as a result of a valid local alignment (see Methods for filtering criteria). Of the 47 877 plasmids evaluated, 533 contained a single intact IS element, and an additional 2 each contained 2 unique intact IS elements, meaning that in total 535 (1.12%) of the plasmids contained at least 1 IS sequence. Within these 535 plasmids, 6 IS types were identified (Fig. 6). Of these, IS1A was most common (363 plasmids), followed by IS10R (127 plasmids), IS5 (40 plasmids) and Tn1000, IS2 and IS1F, which were each represented in five or fewer plasmids. This indicates that the plasmids identified to carry insertions in our restriction digestion screens described above are not the only ones found within repositories to harbour IS elements upon their deposition. In addition, IS5 is not the only E. coli‐derived mobile element to transpose into plasmids, and is by far neither the most common. A rapid analysis comparing the composition of the IS‐containing plasmid set to the entire collection revealed several significant alterations. In brief, the IS‐containing set displayed an enrichment of large plasmids (with sizes > 10 kbp), of low‐copy plasmids and of plasmids carrying a kanamycin resistance gene alone or in combination with a chloramphenicol resistance gene. Details of this analysis are described in Appendix S3.
Table 2.
IS elements used as queries.
| IS name | Accession No. |
|---|---|
| IS1A | X52534 |
| IS1B | X17345 |
| IS1D | X52536 |
| IS1F | X52538 |
| IS2 | M18426 |
| IS3 | X02311 |
| IS4 | J01733 |
| IS5 | J01735 |
| IS5B | U95365 |
| IS5D | X13668 |
| IS5Y | ECK0261 a |
| IS10R | J01829 |
| IS30D | X62680 |
| IS150 | X07037 |
| IS186A | M11300 |
| IS186B | X03123 |
| IS911 | X17613 |
| Tn1000 | X60200 |
The full IS5Y sequence is available in Supp_file_1.fasta of the Supplement.
a
Refers to insH5 transposase.
Fig. 6.
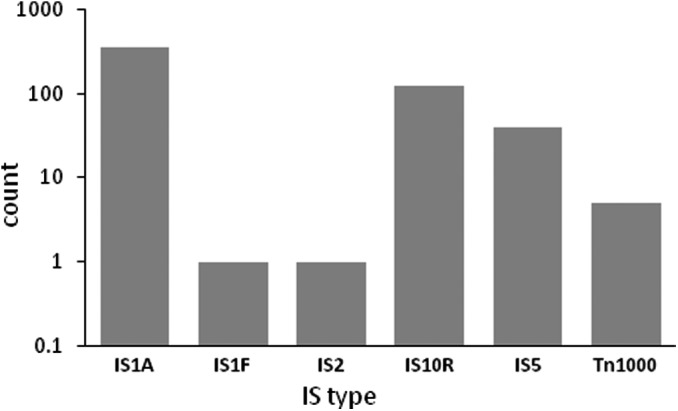
The distribution of IS elements found in the Addgene plasmid collection.
Discussion
This study was initiated by the observation that certain plasmids distributed by repositories gave unexpected restriction patterns. These pattern changes were found to be caused by the integration of IS elements into the respective plasmids. The phenomenon of IS transposition from the host chromosome to a plasmid has been known for decades, and in certain cases, has been shown to be the primary mutational mechanism to inactivate the cloned gene of interest (see Introduction). We nevertheless investigated this process in more detail for at least three reasons: (i) to explore the timing and the source of the transposition events, (ii) to measure how widespread this phenomenon is within a plasmid repository and (iii) to offer potential solutions to evade it, if necessary.
Our analysis revealed that in many cases, the plasmids submitted to the repositories already carried the IS elements. This was especially apparent when the same insertion mutation was detected in a plasmid that had been submitted to multiple repositories, and was sequenced upon arrival. In most cases, however, plasmids are only rejected if the insertions are unexpected, i.e. the ISes are not present in the sequence submitted by the donors along with the DNA. In addition, not all repositories apply routine sequencing of deposited plasmids, further explaining the entry of IS‐bearing plasmids. In addition, IS elements also enter the plasmids by transposing from the host cell’s chromosome during their storage and handling at the repositories. This is indicated by the mixed nature of certain distributed stab cultures, consisting of cells harbouring the wild type and cells harbouring the insertion mutant plasmids as well. We have also demonstrated that even when starting with pure cultures carrying wild‐type plasmids, insertion mutant forms can arise and become dominant in a cell culture within a few tens of generations when using conventional E. coli hosts for their propagation.
To explore how widespread the presence of IS elements is in a repository, we ran a BLAST analysis on the 47,877 plasmids available from and sequenced by Addgene, using as queries the 18 mobile elements of E. coli most often observed to transpose into plasmids. We identified an IS insertion in 1.12% of the plasmids, mostly resulting from IS1A and IS10R transpositions. We note, however, that this is likely to be an underestimation of the phenomenon for two reasons: (i) plasmids carrying unexpected IS insertions upon their submission are rejected by repositories that sequence the deposits, and (ii) plasmids found to be IS free upon their arrival to the repository may acquire insertions later, if grown in conventional E. coli hosts.
As a result of these observations, it is certainly relevant to ask if this phenomenon can be avoided. Earlier works have shown that the systematic deletion, inactivation and silencing of TEs were all capable of increasing genetic stability of the host at chromosomal and plasmid‐based loci alike (Csorgo et al., 2012; Umenhoffer et al., 2017; Geng et al., 2019; Nyerges et al., 2019). IS‐free strains have been engineered for E. coli (Pósfai et al., 2006; Park et al., 2014), Corynebacterium glutamicum (Choi et al., 2015) and Acinetobacter baylyi (Suarez et al., 2017) in the course of systematic genome reduction projects. We tested the feasibility of this solution using two of our IS‐free E. coli strains, MDS42 and BLK16. We showed here that two plasmids, which acquire IS insertions in a conventional host, maintained their correct restriction patterns during 70 generations of culturing in both MDS42 and BLK16. Deep sequencing of one of these plasmids, plasmid 27 propagated in MDS42, confirmed its IS‐free nature in the 70‐generation sample, albeit an SNP variant appeared as a minor fraction of the plasmid population. This is in line with earlier observations describing that by limiting the mutational repertoire of a cell, one can delay (but not completely avoid) the emergence of mutants that release the growth burden imposed by transgenes (Csorgo et al., 2012).
Another valid strategy to reduce unwanted IS insertions into plasmids could be to identify the motifs or combinations of genetic components often hit by ISes. These could be prone to insertions by providing an integration target site, by posing a burden to the host cell that is most simply relieved by an insertion event or by both of the above mechanisms. Studies like ours may provide clues to plasmid engineers in the future on which motifs or combinations thereof to avoid including in their constructs. In our opinion, however, the relatively small number of hits coming from this single study is not sufficient to draw conclusions, or should be used cautiously for this purpose. For example, nearly 100% of plasmids with a pGWB14 vector backbone carry an IS1, seemingly pinpointing a motif with a deterministic IS‐acquiring effect. However, these plasmids all originate from the same deposit, possibly indicating that an insertion event that happened in the early stages of vector construction yielded a series of plasmid derivatives carrying the same insertion.
As a supplementary output, our BLAST results can also be used to infer the transposition activities of various bacterial mobile genetic elements. We used the ISes found in various strains of E. coli as queries, for this is the most commonly used host for plasmid cloning (Lodish et al., 2000). We observed high activities for IS1A and IS10R, intermediate activities for IS5 and low for Tn1000, IS1F and IS2. At an earlier stage of our analysis, we also found a case of IS4 inserted into a deposited plasmid, but this was eventually replaced by the donors. To infer how much the observable activities depend on the experimental setup, we compared the detected IS frequencies with the composition of IS transposition events reported by other investigators in E. coli. Our results showed a strong correlation (R = 0.88, P < 0.0002) with the data of Sousa et al. (2013), a mutation accumulation study that enumerated chromosomal transposition events detectable by sequencing 50 lines of E. coli, each cultured for 1610 generations (Fig. S4). In another analysis focusing on a single target plasmid, Rugbjerg et al. (2018) reported the fraction of mutants of plasmid pMVA1 attributable to each IS type (Fig. S8 of Rugbjerg et al., 2018), which also shows a good correlation with our observations (R = 0.69, P = 0.012) (Fig. S5). However, in a mutation accumulation experiment involving 520 lines and totalling to 2.2 million generations, the observed composition of chromosomal IS transposition events did not show a significant correlation with our results (R = 0.48; P = 0.11) (Lee et al., 2016) (Fig. S6). The well‐known adaptation experiment involving the liquid culturing of four E. coli strains for 50,000 generations each also produced IS transposition events that markedly differed in composition from the relative IS frequencies detected in the Addgene plasmids (R = 0.09; P = 0.79) (Consuegra et al., 2021). In the latter analysis, IS150 transposition caused the greatest number of new insertions, exceeding those of IS1 more than threefold (Fig. S7). Overall, the relatively high activity of IS1 is the only common feature of the five mentioned analyses, including ours. Therefore, it seems likely that the composition of IS transposition events is highly dependent on the experimental setup used for its analysis. Some of the major differences among the listed experiments are the presence or lack of selection (in directed evolution and mutation accumulation experiments, respectively), the analysis of plasmids or the entire chromosome as potential targets and the repertoire of IS elements present in the starting strain. A marked example for the latter factor is the lack of IS5 and IS10 elements in E. coli REL606 (Consuegra et al., 2021) and the lack of IS10 in E. coli PFM2 (Lee et al., 2016), which were prominent contributors to the mutational spectrum in the other three studies.
One last question to be addressed by our investigation is whether the frequency of ISes in the distributed plasmids is relevant to the operators and users of these repositories. Although the frequency is small (1.12% of plasmids carry at least one IS), the large size of the repositories yields a relatively high number (> 500) of plasmids. To date, no deleterious effect of unexpected IS elements has been reported, the theoretical possibility, however, cannot be excluded. Even without a complete inactivation, a smaller change in plasmid function could impair laboratory‐to‐laboratory reproducibility and act against the much promoted process of standardization in molecular and synthetic biology (Endy, 2005). The overall transposition rate of IS elements has been measured to be ≈ 10‐4 transpositions/genome/generation (Sousa et al., 2013; Lee et al., 2016). This value is comparable to the general mutation rate of bacterial cultures corresponding to population sizes used in molecular biology experiments (Krasovec et al., 2017), suggesting that IS transposition will likely contribute to the mutational repertoire of a cell. Therefore, one can anticipate a significant improvement in the genetic stability of host bacteria by the removal of IS elements. In addition, both transposons (Hamamoto et al., 2020; Hooton et al., 2021) and IS elements (Feher et al., 2012) have been described to transpose from plasmids to the chromosome. In a specific case, the complete interspecies horizontal gene transfer could be attributable to a mobile element transposing in and out of a conjugative plasmid (Hall et al., 2017). The chromosomal acquisition of mobile elements originating from plasmids transformed into the host cell of the end user is therefore a realistic scenario, but can nevertheless be avoided by the use of plasmids derived from IS‐free hosts.
In conclusion, we have seen that spontaneous IS transposition into plasmids during construction in the depositor laboratories and storage at plasmid repositories has a measurable frequency, reflecting the activities of E. coli‐derived transposable elements. While our findings indicate that IS element contamination in plasmids is not widespread (Fig. 6), use of IS‐free hosts for both plasmid construction and propagation could be a viable solution to avoid this type of mutagenesis, and thereby delay emergence of plasmid mutants.
Experimental procedures
Molecular biology methods
Plasmid transformation, growth of microbial cultures, plasmid preparation as well as restriction digestion and agarose gel electrophoresis of DNA was carried out according to established protocols (Sambrook et al., 1987). Antibiotics were used in the following concentrations: chloramphenicol (Cm): 25 μg ml‐1, ampicillin (Ap): 100 μg ml‐1 and kanamycin (Km): 25 μg ml‐1. Chemicals were obtained from Sigma‐Aldrich (St. Louis, MO, USA), unless otherwise specified. Restriction enzymes were provided by Thermo Scientific (Waltham, MA, USA).
ABRC stock donation and quality control procedure
Plasmid donations as two identical −80°C stocks are requested from donors. One copy is stored as the ‘original’ and the second as the ‘distribution’ copy of a plasmid. Two or three single colonies derived from each stock are analysed as part of ABRC quality control (QC). The analysis may include a restriction digest, PCR or sequencing depending on the type of QC. If it is necessary to generate a new distribution stock of a plasmid, for example, following a complaint in which a problem was identified in the existing distribution stock, it is prepared from a single colony derived from the original glycerol stock received from the donor and is analysed as part of ABRC QC.
Addgene stock donation and quality control procedure
Plasmid donations are accepted in the form of a small‐scale plasmid preparation. These are sequenced by next‐generation sequencing, and the obtained sequence is aligned with the theoretical sequence provided by the donor. Upon major discrepancies (e.g. unexpected insertions of transposable elements) or minor discrepancies at critical loci, the donors are asked to replace the donation with a correct version. Ultimately, the plasmid is transformed into a suitable E. coli host, and a single colony is used to grow a culture that is stored as a glycerol stock at −80°C. For each event of distribution, a stab culture is generated from the glycerol stock using LB agar containing the appropriate antibiotic.
PCR analysis of plasmids
To localize IS5 insertions in plasmid 44, ~ 1 ng samples of plasmid were amplified in a series of PCR reactions that combinatorially applied IS5‐specific primers (IS5ki1 or IS5ki2) paired with vector‐specific primers (pf183, pf2031, pf4045, pf6000 or pf8519) (primers listed in Table S1). If a PCR product was obtained, it was Sanger sequenced using the IS5‐specific primer to identify the exact point of insertion. The same approach was used to localize IS5 insertions in plasmid 27, but the vector‐specific primers were pm78, pm2063, pm4007, pm6020, pm8017, pm10057 and pm12019. The screening of pCambia, pBIN20 and pFGC5941‐derivative plasmids for the presence of IS insertions in between the replication origin and the Km resistance gene (shown in Figs 2, 3 and 4A) was carried out by PCR amplification using primers IS5‐flank‐ColE1‐F1 and IS5‐flank‐KanR‐R1 (Table S1). For all PCR reactions, the annealing temperature was 57°C and the elongation time was 90 s. Taq polymerase and dNTP mix were obtained from Thermo Scientific.
Bioinformatic methods
Variant analysis
Plasmid 27 was propagated in MDS42 for a total of 70 generations using a serial transfer culture. Upon each transfer, the culture was diluted 1000‐fold, warranting a 1000‐fold expansion in the subsequent growth phase, which corresponds to approx. 10 generations of growth (=log21000). Plasmid samples were extracted and purified following the 10th and 70th generations with the GeneJet Plasmid Purification kit (Thermo Scientific). Barcoded whole‐genome sequence libraries were generated for the gen10 and gen70 samples with the NexteraTm DNA Flex Library Prep kit (Illumina, San Diego, CA, USA) and sequenced on a paired end 151 bp Illumina MiSeq run. Sequence quality was initially assessed based on sequencing metrics from Illumina BaseSpace.
An initial round of adaptor trimming was included as part of the MiSeq run, and a subsequent round of trimming was performed with Trimmomatic (Bolger et al., 2014) to remove any remaining adaptor sequences and low‐quality bases. Trimmomatic was run in paired‐end mode with options LEADING:3, TRAILING:3, SLIDINGWINDOW:4:15 and MINLEN:50. Adaptor‐ and quality‐trimmed sequence data were then evaluated with FastQC (Andrews, 2010).
A reference ‘genome’ was obtained by combining the genome for plasmid 27 (obtained from the depositor’s website and converted to FASTA format), the E. coli MDS42 genome (GenBank accession GCA_000350185.1) and the 18 Insertion Sequences of interest, which are available in Table 2 and File S1. Alignments of adaptor‐ and quality‐trimmed paired‐end reads from each of the two time points were performed with HiSat2 v2.1.0 (Kim et al., 2019). Duplicate reads were marked in the sam files with the markDuplicates function in Picard (Broad‐Institute, 2019) after sorting in SAMtools (Li et al., 2009), and variants with respect to the reference plasmid 27 sequence were identified using the HaplotypeCaller function from the GATK v4.1.2.0 (McKenna et al., 2010). Variants were called for each of the two time points assuming the reads came from a diploid sample. This allows for detection of a potential mixture of haplotypes within each plasmid population, which is expected after recent mutations arise. Aside from the designation of ploidy, runs of the GATK were performed using default values, and were based on duplicate‐marked alignment files. We called variants separately with the BCFtools (Danecek et al., 2021) workflow (BCFtools‐1.11 functions mpileup and call) to evaluate the sensitivity of our inferences to the variant calling approach. Results were broadly consistent between the two analyses, and only results from the GATK workflow are presented. Variants associated with the plasmid were then filtered from the vcf outfiles for analysis.
In order to evaluate whether any IS elements had been acquired in either of the plasmid samples (gen10 or gen70), the alignment outputs were filtered for uniquely mapping reads and assessed for hits to each of the 18 IS elements. The identities of the multiply mapped reads were also determined. This latter set of reads was expected to contain hits to IS1, as an IS1 element is included in both the plasmid 27 genome and the set of targeted IS elements.
All scripts used for performing the analyses described above are available at https://github.com/mikesovic/Brkljacic_etal. Raw sequence data are available from NCBI's sequence read archive (SRA) under BioSample accessions SAMN17496650 and SAMN17496651 associated with BioProject PRJNA694110.
BLAST analyses
NCBI BLAST+ (version 2.10.0) (Camacho et al., 2009) command line applications were used to identify plasmid sequences containing IS sequences. First, we used the ‘makeblastdb’ command to create a custom BLAST database containing 47 877 full, circular plasmid sequences from Addgene’s collection. Next, the set of 18 IS sequences, provided in File S1, were used as query sequences for a standard BLASTN search using default parameters. Each IS sequence was individually aligned against the custom database of plasmid sequences. The output from the BLAST search included the IS name, query alignment length, query start and end positions, length of the database element (plasmid sequence), number of identical nucleotide matches and per cent identity across the alignment.
The initial set of Blast hits was filtered in R v3.6.1 (R‐Core Team, 2019). First, the IS subclasses (i.e. IS1A, IS1b, IS1D, etc.) for the hits were binned into their respective major IS class (i.e. IS1) and unique IS/plasmid combinations were identified. For any IS/plasmid combination that had multiple valid hits, the set of hits was ordered by the alignment length as a proportion of IS element length (any values > 100%, corresponding to alignments containing indels, were rounded down to 100%, as they represent full length alignments). Per cent identity for the alignments was used as a secondary sorting factor to break ties, and the top hit (longest/best alignment) for each IS/plasmid combination was retained. In order to help ensure that hits represented functional IS elements, the data were subsequently filtered to include just the hits in which the alignment covered at least 95% of the IS sequence length.
Conflict of interest
Jason Niehaus was employed by Addgene. The other authors declare no conflict of interest.
Supporting information
Table S1. Primers used in this study.
Table S2. Variants in the 70th generation detected in the diploid analysis of plasmid 27 relative to the reference sequence (n=18).
Fig. S1. Testing the homogeneity of plasmids acquired from repositories. A: Plasmid preparations of cultures grown from ten colonies, obtained by plating the stab culture of the strain carrying plasmid 44, acquired from ABRC. Restriction digestion was carried out with HindIII and EcoRI enzymes. Expected bands: 7823 bp, 1946 bp, 1637 bp. Extra bands are marked by white arrowheads in lanes 7 and 10. B: Plasmid preparations of cultures grown from ten colonies, obtained by plating the stab culture of the strain carrying plasmid 27, acquired from ABRC. Restriction digestion was carried out with NheI and EcoRI enzymes. Expected bands: 9491 bp, 3050 bp, 1862 bp. Unexpected bands are marked by white arrowheads in lane 1. M: GeneRuler 1 kbp DNA Ladder (Thermo Scientific).
Fig. S2. Testing the homogeneity of plasmids acquired from repositories. A: Plasmid preparations of cultures grown from ten colonies, obtained by plating the stab culture of the strain carrying plasmid 45, acquired from Addgene. PvuII and SpeI enzymes were used for the restriction digestion. Expected bands: 5907 bp, 2747 bp. Green arrowheads mark the positions where bands are expected. Unexpected bands are visible in all lanes. B: Plasmid preparations of cultures grown from ten colonies, obtained by plating the stab culture of the strain carrying plasmid 46, acquired from Addgene. EcoRI and EcoRV enzymes were used for the restriction digestion. Expected bands: 5447 bp, 1399 bp. Only expected bands are visible in all lanes. M: GeneRuler 1 kbp DNA Ladder (Thermo Scientific).
Fig. S3. Stability of the restriction pattern of plasmids propagated in E. coli BLK16. The numbers on top of each gel photo represent the number of generations the culture had gone through at time of sampling. (A) plasmid 44, digested with HinDIII and EcoRI. Expected bands: 7823 bp, 1946 bp, 1637 bp. Only expected bands are visible in all lanes. (B) plasmid 27, digested with EcoRI + NheI. Expected bands: 9491 bp, 3050 bp, 1862 bp. Only expected bands are visible in all lanes. M: GeneRuler 1 kbp DNA Ladder (Thermo Scientific).
Fig. S4. The correlation between the IS transposition activities measured by Sousa et al. (2013) and the number of imperfect matches of the respective IS elements found in Addgene sequencing data. Numbers for IS1A and IS1F have been combined as IS1. R=0.88, p=.00016.
Fig. S5. The correlation between the IS transposition activities measured by Rugbjerg et al. (2018) by analysis of mutant pMVA1 plasmids and the number of imperfect matches of the respective IS elements found in Addgene sequencing data. Data for IS1A and IS1F have been combined as IS1. R=0.69, p=.012.
Appendix S1. Introducing the IS elements of Escherichia coli.
Table S3. The identified copy numbers of IS elements identified in this study in various E. coli strains.
Appendix S2. Analyzing the mutations of plasmid 27.
Fig. S6. The region of plasmid 27 encoding the N‐terminal of the KmR gene.
Appendix S3. Features of the IS‐containing plasmid set.
Fig. S7. Comparing the copy number composition of the plasmid sets. The fraction of high, low or unknown copy‐number plasmids are shown for the entire Addgene collection (blue) or the IS‐containing subset (orange). Comparisons were carried out either considering all IS‐containing plasmids (A) or omitting those with a pGWB14 backbone (B).
Fig. S8. Comparing the size composition of the plasmid sets. The fraction of plasmids falling into the size ranges indicated on the X‐axis are shown for the entire Addgene collection (blue) or the IS‐containing subset (orange). Comparisons were carried out either considering all IS‐containing plasmids (A) or omitting those with a pGWB14 backbone (B).
Fig. S9. Comparing the antibiotic resistance composition of the plasmid sets. The fraction of plasmids carrying the indicated resistance gene or genes are shown for the entire Addgene collection (blue) or the IS‐containing subset (orange). Comparisons were carried out either considering all IS‐containing plasmids (A) or omitting those with a pGWB14 backbone (B).
Acknowledgements
We thank György Pósfai and Emma Knee for helpful discussions. We thank Akasia Collins for PCR analysis presented in Fig. 2. T.F. was supported by the National Research, Development, and Innovation Office of Hungary (NKFIH) Grant No. K119298 and the GINOP‐2.3.2‐15‐2016‐00001. The ABRC (D.E.S.) was supported by the National Science Foundation (NSF) grants DBI‐1756439 and DBI‐1561210.
Microbial Biotechnology (2022) 15(2), 455–468
Funding Information
T.F. was supported by the National Research, Development, and Innovation Office of Hungary (NKFIH) Grant No. K119298 and the GINOP‐2.3.2‐15‐2016‐00001. The ABRC (D.E.S.) was supported by the National Science Foundation (NSF) grants DBI‐1756439 and DBI‐1561210.
Contributor Information
Tamás Fehér, Email: fehert@brc.hu.
David E. Somers, Email: somers.24@osu.edu.
References
- Amster, O. , and Zamir, A. (1986) Sequence rearrangements may alter the in vivo superhelicity of recombinant plasmids. FEBS Lett 197: 93–98. [DOI] [PubMed] [Google Scholar]
- Andrews, S. (2010) FastQC: A Quality Control Tool for High Throughput Sequence Data. Cambridge, UK: Babraham Bioinformatics, Babraham Institute. [Google Scholar]
- Blumenthal, R.M. , Gregory, S.A. , and Cooperider, J.S. (1985) Cloning of a restriction‐modification system from Proteus vulgaris and its use in analyzing a methylase‐sensitive phenotype in Escherichia coli . J Bacteriol 164: 501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger, A.M. , Lohse, M. , and Usadel, B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broad‐Institute . (2019) Picard Toolkit. GitHub Repository. URL http://broadinstitute.github.io/picard/.
- Camacho, C. , Coulouris, G. , Avagyan, V. , Ma, N. , Papadopoulos, J. , Bealer, K. , and Madden, T.L. (2009) BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, P.E. Jr , Horzempa, J. , O'Dee, D.M. , Robinson, C.M. , Neophytou, P. , Labrinidis, A. , and Nau, G.J. (2009) Global transcriptional response to spermine, a component of the intramacrophage environment, reveals regulation of Francisella gene expression through insertion sequence elements. J Bacteriol 191: 6855–6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.H. , and Yeh, H.T. (1997) The seventh copy of IS1 in Escherichia coli W3110 belongs to the IS1 A (IS1E) type which is the only IS1 type that transposes from chromosome to plasmids. Proc Natl Sci Counc Repub China B 21: 100–105. [PubMed] [Google Scholar]
- Choi, J.W. , Yim, S.S. , Kim, M.J. , and Jeong, K.J. (2015) Enhanced production of recombinant proteins with Corynebacterium glutamicum by deletion of insertion sequences (IS elements). Microb Cell Fact 14: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consuegra, J. , Gaffe, J. , Lenski, R.E. , Hindre, T. , Barrick, J.E. , Tenaillon, O. , and Schneider, D. (2021) Insertion‐sequence‐mediated mutations both promote and constrain evolvability during a long‐term experiment with bacteria. Nat Commun 12: 980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorgo, B. , Feher, T. , Timar, E. , Blattner, F.R. , and Posfai, G. (2012) Low‐mutation‐rate, reduced‐genome Escherichia coli: an improved host for faithful maintenance of engineered genetic constructs. Microb Cell Fact 11: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek, P. , Bonfield, J. K. , Liddle, J. , Marshall, J. , Ohan, V. , Pollard, M. O. , et al. (2021) Twelve years of SAMtools and BCFtools. GigaScience 10: giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevinek, P. , Baldwin, A. , Lindenburg, L. , Joshi, L.T. , Marchbank, A. , Vosahlikova, S. , et al. (2010) Oxidative stress of Burkholderia cenocepacia induces insertion sequence‐mediated genomic rearrangements that interfere with macrorestriction‐based genotyping. J Clin Microbiol 48: 34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenbaum, Z. , and Livneh, Z. (1998) UV light induces IS10 transposition in Escherichia coli . Genetics 149: 1173–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endy, D. (2005) Foundations for engineering biology. Nature 438: 449–453. [DOI] [PubMed] [Google Scholar]
- Fan, C. , Wu, Y.‐H. , Decker, C.M. , Rohani, R. , Gesell Salazar, M. , Ye, H. , et al. (2019) Defensive function of transposable elements in bacteria. ACS Synth Biol 8: 2141–2151. [DOI] [PubMed] [Google Scholar]
- Feher, T. , Cseh, B. , Umenhoffer, K. , Karcagi, I. , and Posfai, G. (2006) Characterization of cycA mutants of Escherichia coli. an assay for measuring in vivo mutation rates. Mutat Res 595: 184–190. [DOI] [PubMed] [Google Scholar]
- Feher, T. , Karcagi, I. , Blattner, F.R. , and Posfai, G. (2012) Bacteriophage recombineering in the lytic state using the lambda red recombinases. Microb Biotechnol 5: 466–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, C. , Larhammar, D. , Servenius, B. , Rask, L. , and Peterson, P.A. (1986) Spontaneous insertions into cosmid vector ‐ a warning. Gene 42: 215–219. [DOI] [PubMed] [Google Scholar]
- Geng, P. , Leonard, S.P. , Mishler, D.M. , and Barrick, J.E. (2019) Synthetic genome defenses against selfish DNA Elements Stabilize Engineered Bacteria against Evolutionary Failure. ACS Synth Biol 8: 521–531. [DOI] [PubMed] [Google Scholar]
- Hall, B.G. (1998) Activation of the bgl operon by adaptive mutation. Mol Biol Evol 15: 1–5. [DOI] [PubMed] [Google Scholar]
- Hall, B.G. (1999) Spectra of spontaneous growth‐dependent and adaptive mutations at ebgR . J Bacteriol 181: 1149–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, J.P.J. , Williams, D. , Paterson, S. , Harrison, E. , and Brockhurst, M.A. (2017) Positive selection inhibits gene mobilisation and transfer in soil bacterial communities. Nat Ecol Evol 1: 1348–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday, J.A. , and Glickman, B.W. (1991) Mechanisms of spontaneous mutation in DNA repair‐proficient Escherichia coli . Mutat Res 250: 55–71. [DOI] [PubMed] [Google Scholar]
- Hamamoto, K. , Tokunaga, T. , Yagi, N. , and Hirai, I. (2020) Characterization of blaCTX‐M‐14 transposition from plasmid to chromosome in Escherichia coli experimental strain. Int J Med Microbiol 310: 151395. [DOI] [PubMed] [Google Scholar]
- Hooton, S.P.T. , Pritchard, A.C.W. , Asiani, K. , Gray‐Hammerton, C.J. , Stekel, D.J. , Crossman, L.C. , et al. (2021) Laboratory stock variants of the archetype silver resistance plasmid pMG101 demonstrate plasmid fusion, loss of transmissibility, and transposition of Tn7/pco/sil Into the host chromosome. Front Microbiol 12: 723322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaleta, P. , O'Callaghan, J. , Fitzgerald, G.F. , Beresford, T.P. , and Ross, R.P. (2010) Crucial role for insertion sequence elements in Lactobacillus helveticus evolution as revealed by interstrain genomic comparison. Appl Environ Microbiol 76: 212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Paggi, J.M. , Park, C. , Bennett, C. , and Salzberg, S.L. (2019) Graph‐based genome alignment and genotyping with HISAT2 and HISAT‐genotype. Nat Biotechnol 37: 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krašovec, R. , Richards, H. , Gifford, D.R. , Hatcher, C. , Faulkner, K.J. , Belavkin, R.V. , et al. (2017) Spontaneous mutation rate is a plastic trait associated with population density across domains of life. PLoS Biol 15: e2002731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H. , Doak, T.G. , Popodi, E. , Foster, P.L. , and Tang, H. (2016) Insertion sequence‐caused large‐scale rearrangements in the genome of Escherichia coli . Nucleic Acids Res 44: 7109–7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Handsaker, B. , Wysoker, A. , Fennell, T. , Ruan, J. , Homer, N. , et al. (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S. , Haas, S. , Zemojtel, T. , Xiao, P. , Vingron, M. , and Li, R. (2011) Genome‐wide comparison of cyanobacterial transposable elements, potential genetic diversity indicators. Gene 473: 139–149. [DOI] [PubMed] [Google Scholar]
- Lodish, H. , Berk, A. , Zipursky, S.L. , Matsudaira, P. , Baltimore, D. , and Darnell, J. (2000) Molecular Cell Biology, 4th edn. New York: W. H. Freeman. [Google Scholar]
- Mahillon, J. , and Chandler, M. (1998) Insertion sequences. Microbiol Mol Biol Rev 62: 725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna, A. , Hanna, M. , Banks, E. , Sivachenko, A. , Cibulskis, K. , Kernytsky, A. , et al. (2010) The genome analysis toolkit: a mapreduce framework for analyzing next‐generation DNA sequencing data. Genome Res 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, J. , Reinert, H. , and Malke, H. (1989) Streptokinase mutations relieving Escherichia coli K‐12 (prlA4) of detriments caused by the wild‐type skc gene. J Bacteriol 171: 2202–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, K. , and Inouye, M. (1981) Inactivation of the Serratia marcescens gene for the lipoprotein in Escherichia coli by insertion sequences, IS1 and IS5; sequence analysis of junction points. Mol Gen Genet 183: 107–114. [DOI] [PubMed] [Google Scholar]
- Nyerges, A. , Balint, B. , Cseklye, J. , Nagy, I. , Pal, C. , and Feher, T. (2019) CRISPR‐interference‐based modulation of mobile genetic elements in bacteria. Synth Biol (Oxf) 4: ysz008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakeson, K.F. , Gil, R. , Clayton, A.L. , Dunn, D.M. , von Niederhausern, A.C. , Hamil, C. , et al. (2014) Genome degeneration and adaptation in a nascent stage of symbiosis. Genome Biol Evol 6: 76–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, M.K. , Lee, S.H. , Yang, K.S. , Jung, S.C. , Lee, J.H. , and Kim, S.C. (2014) Enhancing recombinant protein production with an Escherichia coli host strain lacking insertion sequences. Appl Microbiol Biotechnol 98: 6701–6713. [DOI] [PubMed] [Google Scholar]
- Pasternak, C. , Ton‐Hoang, B. , Coste, G. , Bailone, A. , Chandler, M. , and Sommer, S. (2010) Irradiation‐induced Deinococcus radiodurans genome fragmentation triggers transposition of a single resident insertion sequence. PLoS Genet 6: e1000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plague, G.R. , Dunbar, H.E. , Tran, P.L. , and Moran, N.A. (2008) Extensive proliferation of transposable elements in heritable bacterial symbionts. J Bacteriol 190: 777–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pósfai, G. , Plunkett, G. 3rd , Fehér, T. , Frisch, D. , Keil, G.M. , Umenhoffer, K. , et al. (2006) Emergent properties of reduced‐genome Escherichia coli . Science 312: 1044–1046. [DOI] [PubMed] [Google Scholar]
- R Core Team . (2019) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R‐project.org/.
- Rawat, P. , Kumar, S. , Pental, D. , and Burma, P.K. (2009) Inactivation of a transgene due to transposition of insertion sequence (IS136) of Agrobacterium tumefaciens . J Biosci 34: 199–202. [DOI] [PubMed] [Google Scholar]
- Reynolds, A.E. , Felton, J. , and Wright, A. (1981) Insertion of DNA activates the cryptic bgl operon in E. coli K‐12. Nature 293: 625–629. [DOI] [PubMed] [Google Scholar]
- Rood, J.I. , Sneddon, M.K. , and Morrison, J.F. (1980) Instability in tyrR strains of plasmids carrying the tyrosine operon: isolation and characterization of plasmid derivatives with insertions or deletions. J Bacteriol 144: 552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugbjerg, P. , Myling‐Petersen, N. , Porse, A. , Sarup‐Lytzen, K. , and Sommer, M.O.A. (2018) Diverse genetic error modes constrain large‐scale bio‐based production. Nat Commun 9: 787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. , Fritch, E.F. , and Maniatis, T. (1987) Molecular Cloning. A Laboratory Manual. Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sawyer, S.A. , Dykhuizen, D.E. , DuBose, R.F. , Green, L. , Mutangadura‐Mhlanga, T. , Wolczyk, D.F. , and Hartl, D.L. (1987) Distribution and abundance of insertion sequences among natural isolates of Escherichia coli . Genetics 115: 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto, C.Y. , Menendez, M.C. , Perez, E. , Samper, S. , Gomez, A.B. , Garcia, M.J. , and Martin, C. (2004) IS6110 mediates increased transcription of the phoP virulence gene in a multidrug‐resistant clinical isolate responsible for tuberculosis outbreaks. J Clin Microbiol 42: 212–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa, A. , Bourgard, C. , Wahl, L.M. , and Gordo, I. (2013) Rates of transposition in Escherichia coli . Biol Lett 9: 20130838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez, G.A. , Renda, B.A. , Dasgupta, A. , and Barrick, J.E. (2017) Reduced mutation rate and increased transformability of transposon‐free Acinetobacter baylyi ADP1‐ISx. Appl Environ Microbiol 83: e01025‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon, M. , and Rocha, E.P. (2007) Causes of insertion sequences abundance in prokaryotic genomes. Mol Biol Evol 24: 969–981. [DOI] [PubMed] [Google Scholar]
- Umenhoffer, K. , Draskovits, G. , Nyerges, Á. , Karcagi, I. , Bogos, B. , Tímár, E. , et al. (2017) Genome‐wide abolishment of mobile genetic elements using genome shuffling and CRISPR/Cas‐assisted MAGE allows the efficient stabilization of a bacterial chassis. ACS Synth Biol 6: 1471–1483. [DOI] [PubMed] [Google Scholar]
- Umenhoffer, K. , Feher, T. , Baliko, G. , Ayaydin, F. , Posfai, J. , Blattner, F.R. , and Posfai, G. (2010) Reduced evolvability of Escherichia coli MDS42, an IS‐less cellular chassis for molecular and synthetic biology applications. Microb Cell Fact 9: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle‐Garcia, D. , Griffiths, L.M. , Dyer, M.A. , Bernstein, E. , and Recillas‐Targa, F. (2014) The ATRX cDNA is prone to bacterial IS10 element insertions that alter its structure. Springerplus 3: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, A. (2006) Periodic extinctions of transposable elements in bacterial lineages: evidence from intragenomic variation in multiple genomes. Mol Biol Evol 23: 723–733. [DOI] [PubMed] [Google Scholar]
- Watson, M.R. , Lin, Y.F. , Hollwey, E. , Dodds, R.E. , Meyer, P. , and McDowall, K.J. (2016) An improved binary vector and escherichia coli strain for agrobacterium tumefaciens‐mediated plant transformation. G3: Genes ‐ Genomes ‐ Genetics 6: 2195–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Kukita, C. , Humayun, M.Z. , and Saier, M.H. (2017) Environment‐directed activation of the Escherichia coli flhDC operon by transposons. Microbiology 163: 554–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primers used in this study.
Table S2. Variants in the 70th generation detected in the diploid analysis of plasmid 27 relative to the reference sequence (n=18).
Fig. S1. Testing the homogeneity of plasmids acquired from repositories. A: Plasmid preparations of cultures grown from ten colonies, obtained by plating the stab culture of the strain carrying plasmid 44, acquired from ABRC. Restriction digestion was carried out with HindIII and EcoRI enzymes. Expected bands: 7823 bp, 1946 bp, 1637 bp. Extra bands are marked by white arrowheads in lanes 7 and 10. B: Plasmid preparations of cultures grown from ten colonies, obtained by plating the stab culture of the strain carrying plasmid 27, acquired from ABRC. Restriction digestion was carried out with NheI and EcoRI enzymes. Expected bands: 9491 bp, 3050 bp, 1862 bp. Unexpected bands are marked by white arrowheads in lane 1. M: GeneRuler 1 kbp DNA Ladder (Thermo Scientific).
Fig. S2. Testing the homogeneity of plasmids acquired from repositories. A: Plasmid preparations of cultures grown from ten colonies, obtained by plating the stab culture of the strain carrying plasmid 45, acquired from Addgene. PvuII and SpeI enzymes were used for the restriction digestion. Expected bands: 5907 bp, 2747 bp. Green arrowheads mark the positions where bands are expected. Unexpected bands are visible in all lanes. B: Plasmid preparations of cultures grown from ten colonies, obtained by plating the stab culture of the strain carrying plasmid 46, acquired from Addgene. EcoRI and EcoRV enzymes were used for the restriction digestion. Expected bands: 5447 bp, 1399 bp. Only expected bands are visible in all lanes. M: GeneRuler 1 kbp DNA Ladder (Thermo Scientific).
Fig. S3. Stability of the restriction pattern of plasmids propagated in E. coli BLK16. The numbers on top of each gel photo represent the number of generations the culture had gone through at time of sampling. (A) plasmid 44, digested with HinDIII and EcoRI. Expected bands: 7823 bp, 1946 bp, 1637 bp. Only expected bands are visible in all lanes. (B) plasmid 27, digested with EcoRI + NheI. Expected bands: 9491 bp, 3050 bp, 1862 bp. Only expected bands are visible in all lanes. M: GeneRuler 1 kbp DNA Ladder (Thermo Scientific).
Fig. S4. The correlation between the IS transposition activities measured by Sousa et al. (2013) and the number of imperfect matches of the respective IS elements found in Addgene sequencing data. Numbers for IS1A and IS1F have been combined as IS1. R=0.88, p=.00016.
Fig. S5. The correlation between the IS transposition activities measured by Rugbjerg et al. (2018) by analysis of mutant pMVA1 plasmids and the number of imperfect matches of the respective IS elements found in Addgene sequencing data. Data for IS1A and IS1F have been combined as IS1. R=0.69, p=.012.
Appendix S1. Introducing the IS elements of Escherichia coli.
Table S3. The identified copy numbers of IS elements identified in this study in various E. coli strains.
Appendix S2. Analyzing the mutations of plasmid 27.
Fig. S6. The region of plasmid 27 encoding the N‐terminal of the KmR gene.
Appendix S3. Features of the IS‐containing plasmid set.
Fig. S7. Comparing the copy number composition of the plasmid sets. The fraction of high, low or unknown copy‐number plasmids are shown for the entire Addgene collection (blue) or the IS‐containing subset (orange). Comparisons were carried out either considering all IS‐containing plasmids (A) or omitting those with a pGWB14 backbone (B).
Fig. S8. Comparing the size composition of the plasmid sets. The fraction of plasmids falling into the size ranges indicated on the X‐axis are shown for the entire Addgene collection (blue) or the IS‐containing subset (orange). Comparisons were carried out either considering all IS‐containing plasmids (A) or omitting those with a pGWB14 backbone (B).
Fig. S9. Comparing the antibiotic resistance composition of the plasmid sets. The fraction of plasmids carrying the indicated resistance gene or genes are shown for the entire Addgene collection (blue) or the IS‐containing subset (orange). Comparisons were carried out either considering all IS‐containing plasmids (A) or omitting those with a pGWB14 backbone (B).


