Summary
Antibiotic resistance in Helicobacter pylori has been growing worldwide with current treatment regimens. Development of new compounds for treatment of H. pylori infections is urgently required to achieve a successful eradication therapy in the future. Armeniaspirols, a novel class of natural products isolated from Streptomyces armeniacus, have been previously identified as antibacterial agents against Gram‐positive pathogens. In this study, we found that armeniaspirol A (ARM1) exhibited potent antibacterial activity against H. pylori, including multidrug‐resistant strains, with MIC range values of 4–16 μg ml‐1. The underlying mechanism of action of ARM1 against H. pylori involved the disruption of bacterial cell membranes. Also, ARM1 inhibited biofilm formation, eliminated preformed biofilms and killed biofilm‐encased H. pylori in a dose‐dependent manner. In a mouse model of multidrug‐resistant H. pylori infection, dual therapy with ARM1 and omeprazole showed efficient in vivo killing efficacy comparable to the standard triple therapy, and induced negligible toxicity against normal tissues. Moreover, at acidic pH 2.5, ARM1 exhibited a much more potent anti‐H. pylori activity than metronidazole. Thus, these findings demonstrated that ARM1 is a novel potent anti‐H. pylori agent, which can be developed as a promising drug lead for treatment of H. pylori infections.
ARM1, a natural product isolated from S . armeniacus, showed potent antimicrobial activity against H . pylori both in vivo and in vitro. It is a possible drug candidate not only inhibiting planktonic growth and biofilm formation but also disrupting the mature biofilm by permeabilizing bacterial membranes.ARM1 might provide a promising drug lead for the development of anti‐H . pylori therapy.
Introduction
Helicobacter pylori is a ubiquitous Gram‐negative, spiral and microaerophilic bacterium that selectively colonizes in human gastric mucosa, and affects more than 50% of the world’s population (Warren and Marshall, 1983; Peek and Blaser, 2002). It is closely associated with the incidence of chronic gastritis, peptic ulcers, gastric carcinoma and mucosa‐associated lymphoid tissue (MALT) lymphomas (Amieva and Peek, 2016; Zhang et al., 2016). In most cases, H. pylori infection is acquired in early childhood and adolescence, and persists lifelong if left untreated (Mamishi et al., 2016). The eradication therapy of H. pylori infection has been recommended in all H pylori‐positive patients with peptic ulcer disease by all of the consensus conferences around the world. It has also been proposed to reduce the risk of gastric cancer (Sugano et al., 2015; Choi et al., 2020). First‐line treatment of H. pylori infection consists of triple and quadruple therapies. The standard triple treatment consists of a proton pump inhibitor (PPI) and two broad‐spectrum antimicrobials (amoxicillin, clarithromycin, levofloxacin and metronidazole), while quadruple therapy contains one additional drug bismuth (Chey et al., 2017; Georgopoulos and Papastergiou, 2020). However, worldwide increasing resistance to key antibiotics is currently the primary concern in the field of H. pylori infection, leading to a high incidence of failures in H. pylori eradication (Fabrega et al., 2009; Papastergiou et al., 2014). Results of a meta‐analysis study in 2018 showed that the primary and secondary resistance rates to clarithromycin, metronidazole and levofloxacin have now reached alarming levels (≥ 15%) in almost all World Health Organization (WHO) regions (Savoldi et al., 2018). Due to the rapid development of antibiotic resistance, the failure rate of triple therapy has increased to more than 20% in many parts of the world (Thung et al., 2016). Therefore, there is a strong need to develop novel antibacterial strategies against H. pylori.
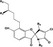
The development of drugs derived from natural microbial sources is receiving considerable attention particularly because of their potential to inhibit bacterial growth (Moloney, 2016). Halogenated pyrroles isolated from Streptomyces sp. have been shown to possess potent antimicrobial activities against a series of Gram‐positive bacteria and fungi (Trew et al., 2000; Hughes et al., 2008). Armeniaspirols, with a unique chlorinated spiro[4.4]non‐8‐ene scaffold (Table 1), are a new class of natural antibacterial products isolated from Streptomyces armeniacus (Couturier et al., 2012). In our recent work, we identified four enzymes involved in the biosynthesis of armeniaspirol A‐C (1 − 3) isolated from S. armeniacus DSM 43125 and manipulated their activity to generate nine new analogues, 7–15 (Table 1) (Qiao et al., 2019). Particularly, armeniaspirols 1–6 displayed potent activity against a range of multidrug‐resistant Gram‐positive bacterial pathogens, including Staphylococcus aureus, Enterococcus faecium and Bacillus subtilis (Couturier et al., 2012; Dufour et al., 2012; Qiao et al., 2019). However, no inhibitory activity against Gram‐negative bacteria, including Acinetobacter baumannii, Pseudomonas aeruginosa, Klebsiella pneumonia, Salmonella typhimurium, Shigella dysenteriae and Escherichia coli, was observed for all armeniaspirols 1–15 (Qiao et al., 2019).
Table 1.
MICs of ARM1 and its derivatives against H. pylori standard strain G27 and multidrug‐resistant strain Hp159.
| Parent structure | Compound | R1 | R2 | R3 | R4 | MIC | |
|---|---|---|---|---|---|---|---|
| (μg ml‐1) | |||||||
| G27 | Hp159 | ||||||
|
ARM1 | H | CH3 | CH3 | Cl | 8 | 8 |
| 2 | CH3 | H | CH3 | Cl | 8 | 4 | |
| 3 | CH3 | CH3 | CH3 | Cl | 8 | 4 | |
| 7 | H | CH3 | H | Cl | 32 | 16 | |
| 8 | CH3 | H | H | Cl | 32 | 16 | |
| 9 | CH3 | CH3 | H | Cl | 16 | 16 | |
| 10 | H | CH3 | H | H | 8 | 8 | |
| 11 | CH3 | H | H | H | 16 | 8 | |
| 12 | CH3 | CH3 | H | H | 16 | 16 | |
| 13 | H | CH3 | CH3 | H | 4 | 8 | |
| 14 | CH3 | H | CH3 | H | 4 | 8 | |
| 15 | CH3 | CH3 | CH3 | H | 4 | 4 | |
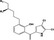
|
4 | H | CH3 | – | – | 8 | 4 |
| 5 | CH3 | H | – | – | 8 | 8 | |
| 6 | CH3 | CH3 | – | – | 8 | 16 | |
| MTZ | 2 (S) | 16 (R) | |||||
| CLR | 0.004 (S) | 2 (R) | |||||
| LVX | 0.5 (S) | 8 (R) | |||||
| AMX | 0.125 (S) | 0.125 (S) | |||||
Breakpoints are according to EUCAST (European Committee on Antimicrobial Susceptibility Testing) guidelines. S, drug sensitive; R, drug resistant; MTZ, metronidazole; CLR, clarithromycin; LVX, levofloxacin; AMX, amoxicillin.
Based on this, the aim of the present study was to investigate whether armeniaspirols have anti‐H. pylori activities. Herein we report that armeniaspirol A (ARM1) possesses a potent in vitro antibacterial activity against various standard and drug‐resistant strains of H. pylori. ARM1 also showed better antibiofilm activity with killing biofilm‐encased H. pylori cells than metronidazole. Furthermore, in vivo efficacy studies showed that oral administration of ARM1 plus omeprazole could significantly reduce H. pylori burden, with comparable efficacy to the triple‐therapy approach. These findings indicate that ARM1 can be used as a potential antimicrobial agent for H. pylori clearance and treatment.
Results
ARM1 possesses potent antibacterial effect on drug‐sensitive and ‐resistant H. pylori strains in vitro
The MICs of fifteen armeniaspirol derivatives against H. pylori standard strain G27 and multidrug‐resistant strain Hp159 were 4–32 μg ml‐1 (Table 1), suggesting that these compounds have potential anti‐H. pylori abilities in vitro. Considering the highly available fermentation yield of ARM1 (approximately 1 mg l‐1) compared with other compounds (approximately 0.2–0.5 mg l‐1), we focus on it for further assessing the antibacterial activity against H. pylori clinical isolates. ARM1 showed similar antibacterial activity for all 13 clinical strains including antibiotic‐susceptible and resistant isolates, with MIC values of 4–16 μg ml‐1 (Table 2).
Table 2.
Antibacterial activity of ARM1 against H. pylori clinical isolates.
| Strains | MIC (μg ml‐1) | ||
|---|---|---|---|
| MTZ | CLR | ARM1 | |
| JIGC00347 | 8 (S) | 2 (R) | 4 |
| JIGC00366 | 16 (R) | 0.06 (S) | 8 |
| JIGC00365 | 4 (S) | 0.032 (S) | 8 |
| YRES723 | 16 (R) | < 0.016 (S) | 8 |
| BYES720 | 16 (R) | 2 (R) | 16 |
| YRES811 | 2 (S) | < 0.016(S) | 8 |
| BYES771D | 16 (R) | 0.5 (S) | 16 |
| YRES00962 | 2 (S) | < 0.016 (S) | 8 |
| YRES731 | 2 (S) | 1 (R) | 8 |
| YRES665 | 16 (R) | 2 (R) | 8 |
| JRES00015 | 16 (R) | 4 (R) | 8 |
| YRES1226D | 32 (R) | 1 (R) | 16 |
| YR9009D | 1 (S) | 1 (R) | 8 |
| BHKS159 | 16 (R) | 2 (R) | 8 |
CLR, clarithromycin; MTZ, metronidazole; R, drug resistant; S, drug sensitive.
Breakpoints are according to EUCAST (European Committee on Antimicrobial Susceptibility Testing) guidelines.
Time‐dependent killing experiments were then performed with H. pylori G27 exposed to ARM1 to determine its effectiveness as a bactericidal agent. For quality control and comparative analyses, the antibiotic MTZ was also tested. Both compounds exhibited concentration‐dependent and time‐dependent killing kinetics (Fig. 1). With ARM1 at 1 × MIC, a 1‐log decrease in cell count was observed at 8 h, followed by some increase in growth till 48 h, indicating no effect at this dose. At 2 × MIC and 4 × MIC, ARM1 treatment was found to cause 1‐ and 2‐log reduction in viable counts after 4 h, and no recoverable colonies were present after 12 h of exposure at 4 × MIC. For the highest dose (8 × MIC), a sharply significant 5‐log decrease within 8 h and no recurrence in viability until 48h were observed (Fig. 1A). MTZ exhibited an almost identical kinetics profile, although the total killing was delayed compared to ARM1 (Fig. 1B). In addition, we used a clinical isolate, Hp159 strain, to confirm that ARM1 was highly effective in a concentration‐dependent and time‐dependent manner (Fig. S1). Taken together, ARM1 exhibited good bactericidal activity (>3‐log decrease in viable cell count) after 24 h and is more effective than the antibiotic MTZ as far as killing efficacy is concerned.
Fig. 1.
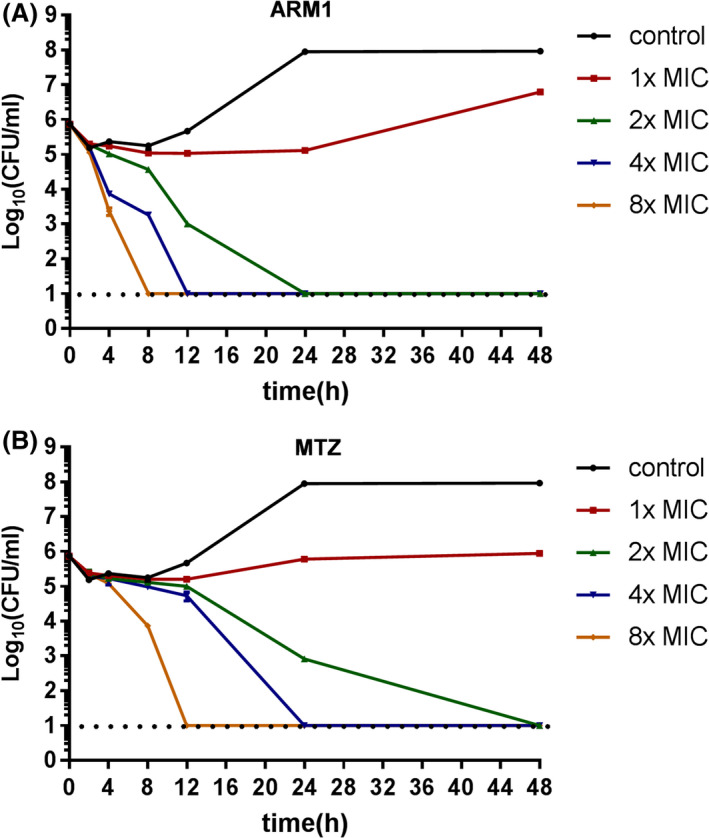
Kill kinetics of H. pylori G27 by ARM1. Time‐ and dose‐dependent killing of the bacteria by ARM1 (A) and MTZ (B) were evaluated. The MIC of ARM1 and MTZ was considered as 8 and 1 μg ml‐1 respectively. If no colonies were present, calculations were made using the limit of detection (101 CFU ml‐1).
The development of resistance is a great challenge that influences the efficacy of eradication regimens. We then evaluated the development of resistance through serial passaging in the presence of subinhibitory concentrations of ARM1. Development of resistance was not observed in H. pylori during continuous serial passaging with ARM1 over 27 d (Fig. S2). In contrast, H. pylori rapidly developed resistance to MTZ after the first three passages, resulting in a 64‐fold increased MIC over 27 days (Fig. S2). These results demonstrate that ARM1 has a very low tendency to induce drug resistance in H. pylori.
ARM1 rapidly permeabilizes H. pylori membranes and causes cell lysis
The bactericidal mechanism of ARM1 was explored by the bacterial morphology observation as well as vesicle leakage assessment. H. pylori G27 cells were exposed to 8, 32 and 64 μg ml‐1 of ARM1 for 2 and 4 h, respectively, and the membrane morphology was examined by Transmission Electron Microscopy (TEM). As shown in Fig. 2A, treatment with 8 μg ml of ARM1 induced the detachment of outer membranes and bleb‐like structures in the cytoplasm of H. pylori cells. Moreover, the swelling outer membrane resulted in extensive deformity of the organism and even whole cell lysis when exposed to 32 and 64 μg ml‐1 of ARM1. Note that the organism changed its appearance from spiral shape to irregular‐shaped form accompanied with leakage of cytoplasmic contents after exposure to 32 μg ml‐1 of ARM1 (Fig. 2A). As a control experiment, DMSO‐treated bacteria remained decent cell wall integrity and dense cytoplasm. In addition, the observed membrane destructive features of ARM1 showed similar efficacy to those of polymyxin, suggesting a disruption effect of ARM1 on H. pylori membranes.
Fig. 2.
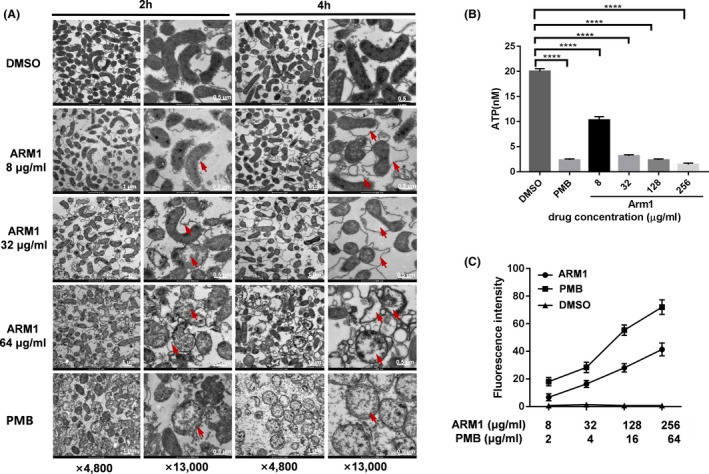
ARM1 permeabilizes H. pylori cell membranes.
A. Morphology changes of H. pylori cells following exposure to DMSO, ARM1 or polymyxin B (PMB) observed by TEM. Membrane permeabilized agent PMB (10 µg ml‐1) served as a positive control which showed characteristically cell deformation and lysis with cytoplasmic contents leakage. Arrows indicate cell lysis, bleb formation, cell wall damage or separation between the cell wall and inner membrane. The data are representative images from two independent experiments.
B. Determination of intracellular ATP levels of H. pylori cells after exposure to various concentrations of ARM1. Statistical differences between the control (DMSO) and AMR1 treatment groups were analysed by one‐way ANOVA followed by Dunnett's t‐test (****P < 0.0001).
C. Uptake of N‐phenylnapthylamine (NPN) by H. pylori G27 cells after 30 min treatment of DMSO (negative control), PMB (positive control) and various concentrations of ARM1. Data represent medians ± SD of the results from three independent experiments.
The permeabilization of bacterial membrane induced by ARM1 was further demonstrated by ATP level and NPN uptake assays. As illustrated in Fig. 2B, after 2 h exposure of ARM1, the intracellular ATP levels in the H. pylori G27 were found to drop significantly in a concentration‐dependent manner. The treatment with 32 μg ml‐1 of ARM1 (4 × MIC) exhibited an almost comparable ATP leaking effect to the antibiotic polymyxin B (PMB), indicating that ARM1 could affect inner membrane integrity and/or intracellular ATP synthesis. We also used the hydrophobic fluorescence probe N‐phenyl‐1‐naphthylamine (NPN) to examine the effect of ARM1 on the outer membrane of H. pylori in response to ARM1 treatment. H. pylori G27 treated with increasing concentrations of ARM1 showed a significant increase in the fluorescence signal of NPN, similar to that treated with polymyxin B, which disrupts the outer membrane permeability barrier (Fig. 2C), indicating that ARM1 permeabilized the outer membranes of H. pylori. Collectively, these data suggested that the mechanism of ARM1 action against H. pylori involved the disruption of bacterial cell membranes.
ARM1 has antibiofilm activity and kills biofilm‐encased H. pylori
Biofilm formation has been suggested to play an important role in H. pylori colonization and resistance to antibacterial therapy (Carron et al., 2006; Yonezawa et al., 2015; Hathroubi et al., 2018). Thus, eradicating biofilms of H. pylori could become an effective strategy to improve the eradication rate. We first used the crystal violet assay to evaluate the ability of ARM1 to prevent biofilm formation. ARM1 and MTZ had similar biofilm inhibition pattern against H. pylori. Both even inhibited biofilm formation of H. pylori G27 at low concentrations (0.5 × MIC), and the biofilm formation could be reduced up to 54% and 49% at 1 × MIC respectively (Fig. 3A). Furthermore, preformed biofilms of H. pylori G27 were also eliminated by ARM1 in a dose‐dependent manner. ARM1 at 4 × MIC disrupted the preformed biofilm of H. pylori by 66% (Fig. 3B).
Fig. 3.
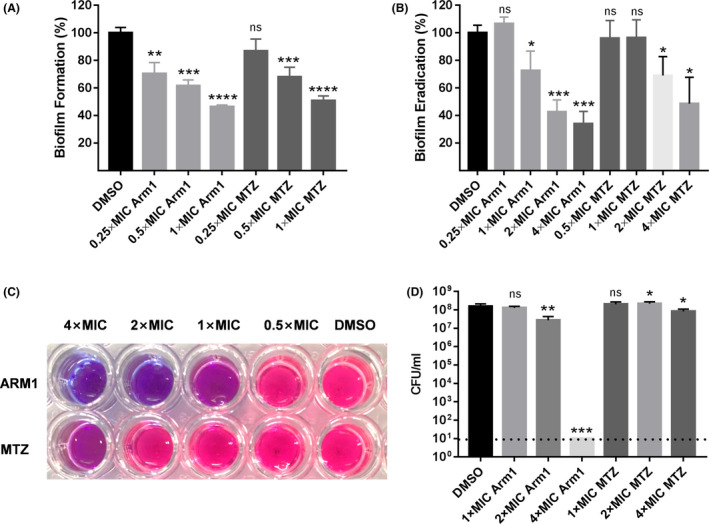
Antibiofilm activity of ARM1. The ability of ARM1 to inhibit biofilm formation (A), and to eliminate preformed biofilms (B) of H. pylori G27 was tested using crystal violet assays. The impact on the cell viability of H. pylori G27 within biofilms by ARM1 was assessed by Alamar Blue assay (C) and viable colony count methods (D). MTZ and DMSO served as positive and negative controls respectively. Data are represented as averages ± SD of three independent experiments analysed by Student's t‐test. If no colonies were present, calculations were made using the limit of detection. *P < 0.05; **P < 0.01; ***P < 0.001. ‘ns’ represents no significant difference (P > 0.05).
The Alamar Blue assays were then performed to examine the effect of ARM1 on the cell viability of preformed biofilms. Compared with MTZ, ARM1 at 16 mg l‐1 (2 × MIC) killed almost all H. pylori G27 cells within the biofilms (Fig. 3C). The complete eradication was further confirmed by performing viable cell counts. As shown in Fig. 3D, treatment with ARM1 at 4 × MIC caused a > 7‐log reduction in bacterial counts, which was much more effective than MTZ. Taken together, these data suggested that ARM1 was able to inhibit biofilm formation and to kill biofilm‐encased H. pylori.
ARM1 kills H. pylori in vivo
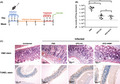
The in vivo antibacterial activity of the ARM1 was evaluated in the mouse infection model (Fig. 4A). The mouse was infected by H. pylori BHKS159 (Huang et al., 2019), a mouse‐adapted multidrug‐resistant strain, by oral gavage every other day for four times. At 3 weeks after inoculation, the mice were randomly divided into three groups and treated with phosphate‐buffered saline (PBS) (control), triple therapy (omeprazole plus amoxicillin and clarithromycin [OPZ + AC]) and dual therapy (omeprazole plus ARM1 [OPZ + ARM1]). In this study, therapeutic efficacy was evaluated by enumerating and comparing H. pylori counts in mouse stomach. Remarkably, after 5 day drug treatment, administration of OPZ + ARM1 (28.5 mg kg‐1) significantly impacted stomach bacteria load (Fig. 4B); the corresponding median colony‐forming units (cfu) dropped significantly from 2.5 × 105 to 1.1 × 103 CFU g‐1, representing 99.56% inhibition of stomach colonization compared with the PBS group. Likewise, triple‐therapy treatment also decreased the bacteria load to 1.3 × 103 CFU g‐1, showing similar bacteria‐killing ability with the dual therapy with OPZ + ARM1 (Fig. 4B). Additionally, histopathological examination of fixed stomach sections revealed that OPZ + ARM1 treatment alleviated stomach inflammation with reduced infiltration of inflammatory cells into the stomach epithelial cells (Fig. 4C). Moreover, OPZ + ARM1 administration did not show apparent gastric epithelial apoptosis compared to the PBS and OAC‐treated groups analysed by the TUNEL assay (Fig. 4C), indicating that ARM1 did not induce obvious apoptosis in vivo. Taken collectively, these results suggested a high efficacy of ARM1 in clearance of drug‐resistant H. pylori in vivo.
Fig. 4.
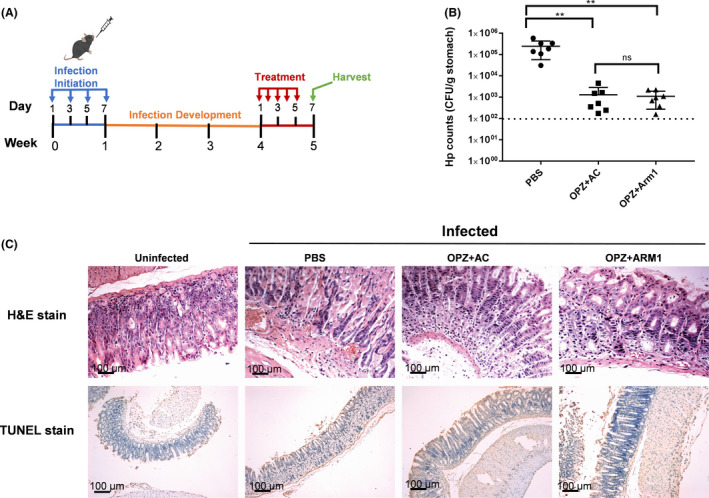
In vivo effect of ARM1 on H. pylori infection in C57BL/6J mice.
A. The mice infection and treatment schedule utilized to investigate in vivo efficacy of ARM1 against H. pylori BHKS159.
B. Quantification of bacterial burden in the stomach of H. pylori‐infected mice, euthanatized 48 h after the last treatment with PBS, triple therapy (OPZ + AC), omeprazole and ARM1 (OPZ + ARM1) respectively. Error bars represent the SD derived from 7 infected mice per group. If no colonies were present, calculations were made using the limit of detection (102 CFU g‐1). **P < 0.01. ‘ns’ represents no significant difference (P > 0.05).
C. Histological staining analysis of mice after the indicated treatments. Representative images of H&E or TUNEL stained stomach from infected mice receiving different treatments in vivo. TUNEL stain showed that OPZ + ARM1 treatment had the same safety level as control (n ≥ 5, scale bar = 100 μm).
ARM1 performed potent antibacterial effect at acidic condition (pH = 2.5)
H. pylori colonizes the acidic gastric environment and survives brief exposure to pHs of < 4. We then re‐evaluated the anti‐H. pylori activity of ARM1 in an acid survival analysis to closely mimic gastric conditions. The experiment was carried out at pH 2.5 in the presence of 10 mM Urea. When the bacteria were treated with ARM1 and MTZ at a lower concentration (0.5 μg ml‐1), both had no significant effect on the survival rates compared with the DMSO group (Fig. 5). However, notable bacterial killing was achieved by ARM1 at 2 and 8 μg ml‐1. Especially, when treated with 8 μg ml‐1, the survival rate of the ARM1‐treated group was 0.0013%, 3 orders of magnitude lower than that of MTZ‐treated group (Fig. 5). Thus, ARM1 exhibited much more potent anti‐H. pylori activity under extremely acidic conditions than MTZ.
Fig. 5.
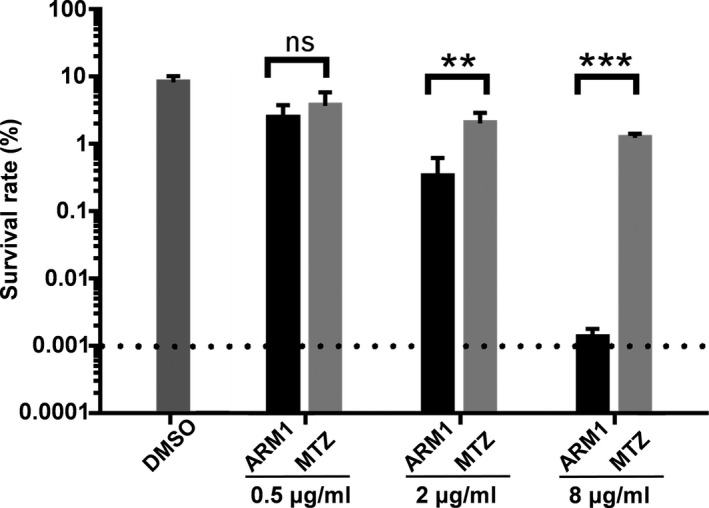
The survival rate of H. pylori G27 after incubation with ARM1 or MTZ for 30 min at pH 2.5. ARM1 or MTZ at various concentrations was incubated with H. pylori G27 in BHI broth (pH2.5) in the presence of fresh urea (10 mM) and 10% FBS. The bacterial count was determined by counting colony‐forming units (cfu) of live bacteria with agar plating. The data are represented as average ± SD of three independent experiments and analysed by Student's t‐test. **P < 0.01, ***P < 0.001. ‘ns’ represents no significant difference (P > 0.05).
Safety evaluation of ARM1
At last, the toxicity of ARM1 was investigated. ARM1 exhibited no significant cytotoxicity to human normal hepatocyte LO2 cells (IC50 = 147.5 μg ml‐1) with 96.5% survival rate when treated with 100 μg ml‐1 of ARM1 (Fig. S3). Moreover, the low toxicity of ARM1 was further validated by treatment with 10 times the effective dosage in vivo. Within 5 day overdose treatment of ARM1, no serious adverse effects such as diarrhoea and loss of appetite or body weight were observed (Fig. S4A). Also, there were no obvious differences in the histopathologic examination of mouse major organs including liver, kidney, spleen and stomach treated with ARM1 and PBS (Fig. S4B), which supported that ARM1 had low toxicity in vivo. In addition, the drug‐like properties of ARM1 were predicted by SwissADME in silico. ARM1 showed excellent drug‐like properties, including high oral absorption, high bioavailability and moderate toxicity (Table S1). Taken together, ARM1 could be developed as a potential anti‐H. pylori therapeutic agent with minimal toxicity profiles.
Discussion
In recent years, there has become more challenging to the clinical treatment of H. pylori due to the prevalence of antibiotic resistance (Hu et al., 2016; Zanotti and Cendron, 2019; Pichon et al., 2020). Hence, the discovery and development of new anti‐H. pylori agents without cross‐resistance to current drugs are highly impending and important. In this study, we demonstrated that ARM1, a natural product isolated from S. armeniacus, targets H. pylori in vivo and in vitro as a bactericidal agent. A comparable killing efficacy of dual therapy (ARM1 plus PPI) was evaluated in reducing H. pylori bacterial loads in the stomach of drug‐resistant H. pylori‐infected mice, compared with the current worldwide standard treatment, triple therapy. In addition, an additive effect between ARM1 and some conventional antibiotic against H. pylori was found (Table S2), indicated that ARM1 could be developed as a good drug synergist candidate in the triple‐therapy regimen used in clinical practice for future.
H. pylori produces a well‐structured biofilm, which can impact the efficacy of antibiotic treatment and clearance by the host immune systems (Cammarota et al., 2012; Hathroubi et al., 2018). Thus, targeting H. pylori biofilm infections can provide an effective strategy to overcome H. pylori eradication treatment failures. In the present study, ARM1 alone not only prevented H. pylori biofilm formation but also eliminated the embedded bacteria in the biofilm. Moreover, the killing efficacy of ARM1 was even superior to that of the clinical drug MTZ. Such compound with antibiofilm activity might provide advantages in resolving multidrug‐resistant H. pylori infections.
As demonstrated in our study, ARM1 permeabilized and disrupted the outer and inner membranes of H. pylori, resulting in a rapid killing effect. The highly destructive action for bacterial killing may explain the low tendency to develop resistance against H. pylori (Xu et al., 2017; Huang et al., 2019). In addition, the biofilm formation of H. pylori has been suggested to be highly associated with outer membrane proteins (Yonezawa et al., 2017). Therefore, the membrane structural disruption caused by ARM1 treatment could also probably destabilize membrane proteins and thus induce biofilm collapse.
Based on previous studies, armeniaspirols demonstrated no antimicrobial activity against many Gram‐negative bacteria (Couturier et al., 2012; Qiao et al., 2019). Thus, besides the cell membrane disrupting mode, we speculate there may also have alternative mechanisms for the bactericidal activity of armeniaspirole against H. pylori. Therefore, ARM1 likely targets a specific H. pylori protein absent in other Gram‐negative bacteria. It should be also mentioned that under acidic condition (pH = 2.5), ARM1 showed much more potent anti‐H. pylori activity than MTZ in the presence of urea (Fig. 5). This stronger antibacterial activity of ARM1 should not be attributed to changes in chemical structure under acidic condition, since the compound remains stable when exposed to pH2.5 (data not shown). H. pylori can utilize a complex and robust acid resistance system centred around the nickel enzyme urease, to survive the harsh acidic conditions of the stomach (Scott et al., 2002; Krulwich et al., 2011). It is highly possible that ARM1 interfere with this survival strategy in the acidic conditions, thus reducing survival rate of H. pylori. In view of the fact that the complex acid resistance system involves a variety of proteins, even unknown, ARM1 might act by regulating the urease expression and urease maturation or inhibiting its activity via binding to distinct proteins. Taken together, the robust anti‐H. pylori activity conferred by ARM1 can be explained by the unique capability of ARM1 to disrupt membrane integrity and to suppress the acid resistance of H. pylori.
In summary, we report that ARM1, a natural product isolated from S. armeniacus, showed potent antimicrobial activity against H. pylori both in vivo and in vitro. It is a possible drug candidate not only inhibiting planktonic growth and biofilm formation but also disrupting the mature biofilm by permeabilizing bacterial membranes. Moreover, the early toxicity and safety evaluations also show that ARM1 exhibits a relative safety index. ARM1 might provide a promising drug lead for the development of anti‐H. pylori therapy.
Experimental procedures
Chemicals
The cultivation of S. armeniacus strains and the purification of the compounds from the culture were carried out as previously described (Qiao et al., 2019). The S. armeniacus strains (the wild‐type strain DSM 43125 and mutant strains) were cultured in the liquid ISP‐2 medium (0.4% yeast extract, 1% malt extract, 0.4% glucose, pH 7.0) containing 2 g l‐1 CaCO3 at 30°C and 200 rpm for 3 days. Then, 10% (v/v) of the culture was transferred in 50 ml of fresh ISP‐2 medium and fermented at 30°C and 200 rpm for 6 days. The culture was separated from the biomass fraction and the supernatant fraction by centrifugation. The supernatant fraction was extracted with an equal volume of ethyl acetate twice. The biomass fraction was extracted with methanol, disrupted by sonication and centrifuged to remove the cell debris. The retained supernatant was concentrated on a rotovap, resuspended in water and extracted with an equal volume of ethyl acetate twice. The combined organic extracts were dried over anhydrous Na2SO4, filtered and concentrated on a rotovap. The residue was resuspended in methanol and filtered with a microporous membrane (0.22 μm, nylon). The compounds were purified first by using HPLC (Beijing QingBoHua Technologies, Beijing, China) with a Daisogel C18 reversed‐phase HPLC column (250 × 20 mm, 10 μm). The HPLC conditions (mobile phase A: water; mobile phase B: methanol; UV detection λ:300 nm) were as follows: 10−90% B for 35 min, 90% B for 15 min, 90−10% B for 5 min and 10% B for 10 min at a flow rate of 10 ml min‐1. The obtained compounds were concentrated on a rotovap and further purified by using HPLC (Shimadzu, SPD‐M20A/LC‐20AT, Kyoto, Japan) with a Thermo Scientific C18 reversed‐phase HPLC column (250 × 10 mm, 8 μm). The HPLC conditions (mobile phase A: water; mobile phase B: acetonitrile; UV detection λ: 300 nm) were as follows: 70% B for 30 min. The purified compounds were concentrated on a rotovap and dried on a freeze‐dryer. 70 mg ARM1 (C18H19Cl2NO4; UV/vis λmax 208, 232, 299; HR‐ESI‐MS m/z 382.06818 ([M − H] −, calcd m/z 382.06129) was purified from the culture of the wild‐type strain DSM 43125 approximately 71 L. The following strains were cultured about 2–4 l respectively to purify the compounds 2‐15 (approximately 1 mg each compound) used in this study : the wild‐type strain DSM 43125 for 2‐3, the Δarm9Δarm15 mutant strain QY13 for 4‐6, the Δarm16 mutant strain QY6 for 7‐9 and the Δarm9 mutant strain QY4 for 10‐15.
Bacterial strains and culture media
H. pylori strains G27 (Baldwin et al., 2007), Hp159 (Huang et al., 2019) and 13 clinical isolates were used in this study and were routinely cultured either in brain heart infusion (BHI) broth (Becton, Dickinson, Sparks, MD, USA) medium containing 10% fetal calf serum (FCS) or in Columbia blood agar (Oxoid, Basingstoke, UK) plates containing 5% FCS, supplemented with Dent selective supplement (Oxoid). All of the plates or media were incubated at 37°C for 48 to 72 h under microaerophilic conditions (10% CO2, 85% N2, and 5% O2 and 90% relative humidity) using a double‐gas CO2 incubator (Binder, model CB160, Tuttlingen, Germany). A total of 13 clinical strains of H. pylori were isolated from biopsy samples from patients with gastric cancer or gastric ulcer, obtained from the First Affiliated Hospital of Nanchang University and the First Affiliated Hospital of Nanjing Medical University, China, using standard protocols. The strains were identified based on colony appearance, Gram staining and positive reactions in the rapid urease test.
Drug susceptibility test
Antibacterial activities were evaluated by minimum inhibitory concentration (MIC) determination. Briefly, twofold serial dilutions of compounds were prepared in a 96‐well microtitre plate containing 100 μl of BHI broth containing 10% FCS. An overnight H. pylori liquid culture was diluted in BHI broth and was inoculated into each well with a final concentration of 5 × 105 CFU ml‐1. The plates were incubated for 2 days in a microaerophilic atmosphere at 37°C. After incubation, the plates were examined visually, and the MIC was determined to be the lowest concentration with no turbidity in well. For quality control and comparative analysis, the antibiotic metronidazole was also tested with each batch of ARM1. All MIC assays were performed in at least triplicate.
Checkerboard microdilution assay
The standard checkerboard microdilution assay was used for evaluation of antimicrobial interactions between ARM1 and each antibiotic (AMX, CTR, LVX or MTZ) against H. pylori strains (Tanaka et al., 2002). The test compounds represented by each antibiotic were serially diluted on the x‐axis, ranging from 1 to 1/64 × MIC, with increasing concentrations of ARM1 ranging from 1/32 to 1 × MIC on the y‐axis. For evaluation of synergistic effect of ARM1 (A) with the antibiotic tested (B), the fractional inhibitory concentrations (FICs) were calculated as follows: ∑FIC = FICA+ FICB where FICA = MICA (in the presence of B)/MICA (alone), and FICB = MICB (in the presence of A)/MICB (alone). The FIC indices were interpreted as follows: ≤0.5, synergy; 0.5–1, additive; 1–4.0, indifference; >4.0, antagonism. All MIC determinations were performed in triplicate for each strain.
Time‐kill kinetics assay
10 ml of BHI containing 10% FCS in the presence of ARM1 (at 1×, 2×, 4×, or 8 × MIC) or an equivalent amount of DMSO (which served as a vehicle control) was inoculated with H. pylori strain G27 obtained from an overnight culture to yield an initial cell density of ~ 106 CFU ml‐1. The culture was shaken at 150 rpm at 37°C inside the double‐gas CO2 incubator as stated before. At different time points (0, 2, 4, 8, 12, 24 and 48 h), aliquots (100 μl) of culture were taken out to monitor the growth by direct microscopic examination, and the viable cells were counted by inoculating 10‐fold serially diluted suspensions onto Columbia blood agar plates. Colonies were counted after 4 days of incubation, and the bactericidal activities were assessed in terms of decrease in cell count (CFU ml‐1). The rate and extent of killing were determined by plotting viable counts (log10 CFU ml‐1) against time (h).
Plasma membrane permeability assay
The intracellular ATP level was determined as an indicator of plasma membrane permeability. Briefly, an overnight liquid culture of H. pylori G27 was diluted to an OD600 of 0.3, and the bacterial cells were treated with polymyxin B (10 μg ml‐1) or ARM1 (8, 16, 128 and 256 μg ml‐1) for 30min. An equivalent amount of DMSO was used as a vehicle control. After treatment, each bacterial suspension was centrifuged, the supernatant was separated from the cell pellet, and the cell pellet was resuspended in 1 ml of PBS. 100 μl aliquot of BacTiter‐Glo reagent in the microbial cell viability assay kit (Promega, Madison, WI, USA) with 500 μg of lysostaphin was added to the resuspended cell pellet. The mixture was shaken for 2 min, and luminescence was determined by a Synergy HTX multimode microplate reader (BioTek Instruments, Winooski, VT, USA). The concentration of ATP was determined by comparison to the luminescence obtained from the addition of the BacTiter‐Glo reagent to an ATP standard curve.
Outer membrane permeability assay
NPN uptake assays were performed to determine the ability of ARM1 to permeabilize H. pylori outer membranes. Briefly, an overnight liquid culture of H. pylori G27 was harvested by centrifugation (4700 rpm, 10 min), washed twice in 5 mM 4‐(2‐hydroxyethyl)‐1‐piperazineethanesulfonic acid (HEPES) buffer (pH7.4) and then resuspended to an OD at 600 nm of 1 in 5 mM HEPES. Aliquots (100 μl) of the cell suspension were added to each well of 96‐well microtitre plates containing 50 μl of ARM1 at various concentrations (8, 32, 128 and 256 µg ml‐1), while the polymyxin B and DMSO served as positive and negative controls respectively. After addition of 50μl NPN (40 μM), measurements were implemented taken after 30 min incubation at 37°C. Fluorescence measurements were taken using a Synergy HTX multimode microplate reader with an excitation wavelength of 350 nm and an emission wavelength of 420 nm. All NPN assays were performed in triplicate.
The antibiofilm assay
Biofilm formation assays were carried out as described previously (Hathroubi, et al., 2018), with small modification. H. pylori G27 was grown overnight, diluted to an optical density at 600 nm (OD600) of 0.15 with fresh Brucella broth containing 10% FCS and then used to fill triplicate wells of a sterile 96‐well polystyrene microtitre plate. Various doses of ARM1 or metronidazole (MTZ, served as a positive control) were added to the wells above and an equivalent amount of DMSO was used as a vehicle control. Following 48 h of static incubation under microaerobic and 37°C, the culture medium was removed by aspiration, and the wells were washed twice with phosphate‐buffered saline (PBS). Plates were incubated for 10 min at 37°C for dry. The wells were then filled with 250 μl of crystal violet (0.1% wt/vol), and biofilm inhibition was measured using the crystal violet staining method (Hathroubi et al., 2018). To assess the ability of ARM1 to eradicate preformed biofilms, H. pylori G27 cells were cultured in microtitre plates for 2 days to allow for biofilm formation, and the medium was aspirated and then washed with PBS twice. Fresh Brucella broth containing 10% FCS and various doses of ARM1 or MTZ were added to the wells for another 24 h. The dispersal of mature biofilms was measured using the crystal violet staining method.
Quantifying effect of ARM1 on bacterial viability in biofilms
The bacterial viability within the biofilm was determined by Alamar Blue assay (Pettit et al., 2005), which is a colorimetric assay involving the cellular reduction of a blue phenoxazin dye, resazurin to the pink fluorescent compound resorufin. Biofilms of the H. pylori G27 cells were prepared and treated followed by 24 h under microaerobic conditions described in the previous section. First, 10% Alamar Blue solution was added to each well. Plates were then incubated for 2 h at room temperature in the dark, and images were taken by a camera. The resorufin fluorescence was measured (λexcitation, 530 nm; λemission, 590 nm) thereafter using a Synergy HTX multimode microplate reader. The fluorescence values of the samples were corrected by subtracting the average fluorescence value of Alamar Blue in uninoculated wells (blank). The percentage of surviving biofilm cells was calculated relative to the control treatment.
The effect of ARM1 on the cell viability of preformed biofilms was also investigated using colony count methods. Biofilms of the H. pylori G27 cells were prepared and treated followed by 24 h under microaerobic conditions described in the previous section. After incubation, additives were removed, and the biofilms were detached by scraping wells completely with a 200 μl pipette tip across the well. The scraped biofilms were then homogenized in solution by repeatedly pipetting the solution several times. Finally, the homogenized biofilm suspensions were serially diluted, spread out onto Columbia blood agar plates containing Dent supplement, and then incubated at 37°C under microaerobic conditions for 4 days. Viable bacterial colonies on plates were enumerated and expressed as CFU ml‐1.
Drug resistance study
H. pylori G27 was inoculated with fresh BHI containing 10% FCS and incubated at 37°C under continuous shaking. Cells were grown to log phase and dispensed into 96‐well microtitre plates with 100 μl fresh medium per well (adjusted to approximately 2 × 106 cells). Metronidazole or ARM1 was added at a concentration of 0.5 × MIC. After a 3 day incubation at 37°C under continuous shaking, growth was determined with a microplate reader at an optical density at 600 nm, and cells were used to inoculate the subsequent cultures. Alterations of MICs were recorded after every cycle during the continued exposure of 27 days with nine cycles of repetition. All measurements were performed with biological replicates.
Antibactericidal activity of ARM1 under acidic condition (pH 2.5)
H. pylori G27 was grown overnight, and diluted to an optical density at 600 nm (OD600) of 0.3 with 1 ml fresh BHI broth containing 10% FCS and 10mM Urea (adjusted to pH 2.5 with HCl). Various doses of ARM1 or metronidazole (MTZ, served as a positive control) were added to the medium above, and an equivalent amount of DMSO was used as a vehicle control. After 30 min of incubation in a microaerobic environment, the samples were harvested by centrifugation, washed once with PBS and resuspended with fresh BHI. The homogenate was serially diluted and spotted onto a Columbia blood agar plate supplemented with Dent selective supplement. The plates were then incubated at 37°C under microaerobic conditions for 4–5 days, and bacterial colonies were enumerated, and acid survival was expressed as a percentage of the number of viable cells after incubation at pH 2.5 relative to that before the challenge.
Cytotoxicity in LO2 cell line
The in vitro cytotoxicity investigation was carried out by CCK‐8 assays according to the manufacturer's instructions. Confluent human normal liver cells LO2 (American Type Culture Collection, ATCC) in good condition were cultured in 96‐well plates (5 × 103 cells per well) with RPMI 1640 medium containing 10% FBS and treated with ARM1 at various concentrations (16, 32, 64, 128 and 256 µg ml‐1) for 48 h. Then, the absorbance of each well at 490 nm was measured by a Synergy HTX multimode microplate reader. The inhibition rate and cell viability were normalized by the value of the DMSO‐treated cells. The IC 50 value was calculated using GraphPad Prism 6 software (San Diego, CA, USA).
Transmission electron microscopy
The effects of ARM1 on the cell structure of H. pylori G27 were examined with TEM. Sample processing was performed essentially according to a published procedure (Hathroubi et al., 2020). Briefly, an overnight culture of H. pylori G27 was treated with ARM1 (8, 32 and 64 μg ml‐1) and polymyxin B (10 µg ml‐1) served as positive control incubated for 2 h. PBS was used as a negative control. Samples were centrifuged at 1000 g for 10 min and bacterial pellets fixed by resuspending in 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4). Bacterial pellets were then embedded in 2% agarose and postfixed with 1% osmium tetroxide overnight at room temperature. After washing, samples were dehydrated multiple times in increasing concentrations of ethanol and embedded in Durcupan resin (Sigma‐Aldrich, St. Louis, MO, USA). Fifty‐five‐nanometer sections were examined by a JEM‐1200 transmission electron microscope (JEOL; Akishima, Tokyo, Japan).
Anti‐H. pylori efficacy in vivo
Six‐week‐old specific‐pathogen‐free female C57BL/6 mice were used for this study. Each mouse was administered intragastrically with 0.4 ml of ~ 1×109 CFU ml‐1 H. pylori BHKS159 every 48 h, repeated four times (on days 1, 3, 5 and 7 respectively). At 3 weeks after inoculation, the mice were randomly assigned to three treatment groups (n = 7) to treat as follows: ARM1 plus omeprazole, triple therapy or PBS. Omeprazole (400 μmol kg‐1 day‐1) was fed through oral gavage 30 min before the administration of the assigned treatments. ARM1 (28.5 mg kg‐1) and triple‐therapy formulation (amoxicillin 28.5 mg kg‐1 and clarithromycin 14.3 mg kg‐1) were administered once daily for a consecutive 5 days by oral gavage. The control group received an equivalent volume of PBS. Forty‐eight hours after the last treatment, mice were euthanatized. The stomach, the liver, the spleen and the kidney were harvested from the abdominal cavity. The stomach was cut along the greater curvature, and the gastric content was removed and rinsed with BHI. The stomachs were cut into two longitudinal sections, used for assessment of bacterial colonization and histology/epithelial apoptosis, as described previously (Huang et al., 2019). Mouse body weight was monitored every day daily. For bacterial colonization, a gastric tissue section was suspended in 1 ml BHI and homogenized for H. pylori recovery. The homogenate was serially diluted and spotted onto a Columbia agar plate containing vancomycin (20 μg ml‐1), amphotericin (2 μg ml‐1) and bacitracin (30 μg ml‐1). The plates were then incubated at 37°C under microaerobic conditions for 4–5 days, and bacterial colonies were enumerated, and the outcome was expressed as cfu per gram of stomach.
In vivo toxicity study
Animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) of Nanjing Medical University (NJMU) and conducted in accordance with the international standards for animal welfare and institutional guidelines. Mice were obtained from the Animal Core Facility at NJMU and housed at the same place. ARM1 safety was evaluated in vivo by treatment of ARM1 overdose (285 mg kg‐1, 10‐fold effective dosage) in C57BL/6 female mice (n = 6) at 6–8 week of age. Mice were given a daily gavage of ARM1 or PBS for 5 consecutive days. Mice were sacrificed on day 7, and the stomach, the liver, the spleen and the kidney were harvested from the abdominal cavity for histological analysis. The longitudinal sections of each tissue were fixed in buffered paraffin, embedded in paraffin wax, sectioned at 5 μm and stained with haematoxylin and eosin (H&E) to analyse tissue inflammation. Mouse body weight was monitored during the experiment period by weighing the mice daily.
Statistical analysis
Statistical analyses were performed, and graphs were generated by using Graphpad prism 7.0 version (San Diego, CA, USA). For all experiments, statistical analyses were determined with Student's t‐test. A P value of < 0.05 was considered significance statistical difference.
Conflict of interest
All authors have no conflict of interest to declare.
Supporting information
Fig. S1. Kill kinetics of H. pylori Hp159 by ARM1. Time‐ and dose‐dependent killing of the bacteria by ARM1 (A) and MTZ (B) were evaluated. The MIC of ARM1 and MTZ was considered as 8 and 16 μg ml‐1, respectively. If no colonies were present, calculations were made using the limit of detection (101 CFU ml‐1).
Fig S2. Development of resistance to ARM1 in H. pylori G27. The fold change was the normalized ratio of the MIC obtained for a given subculture to the MIC obtained for first‐time exposure. ARM1 did not develop drug resistance over the 27 days of studies while metronidazole quickly acquired resistance on day 9. A representative of two independent experiments is shown.
Fig S3. Cell Cytotoxicity of ARM1 was assessed in human normal hepatocyte LO2 cells. After overnight culture, cells were incubated with ARM1 at predetermined concentrations for 48 h and were then treated with CCK‐8 reagent for assessment of cytotoxicity.
Fig S4. ARM1 had low toxicity in vivo. (A) After the mice were orally administrated with overdose (285 mg/kg, 10‐fold effective dosage) ARM1 once a day for 5 consecutive days, the mice body weights were monitored daily for 7 days. (B) At the seven days after the last treatment, the mice were sacrificed, and the stomach, the liver, the spleen and the kidney were removed from the abdominal cavity and examined by H&E stain.
Table S1. The predicted Drug‐like properties of ARM1.
Table S2. In vitro combinatory anti‐H. pylori effect of ARM1 with each antibiotic.
Acknowledgements
This work was supported by The National Key Research and Development Programs of China (No. 2018YFC0311003 to H. B.), The National Science Foundation of the Jiangsu Higher Education Institutions of China (No. 18KJA310002 to H. B.), the Jiangsu Specially Appointed Professor and Jiangsu Medical Specialist Programs of China (to H. B.) and Jiangsu Province ‘Innovative and Entrepreneurial Team’ Program (to H. B.). We thank Dr. Nina Salama, Dr. Karen Ottemann and Dr. Yong Xie for providing H. pylori strains.
Microb. Biotechnol. (2022) 15(2), 442–454
Contributor Information
Dongqing Zhu, Email: dzhu2011@whu.edu.cn.
Hongkai Bi, Email: hkbi@njmu.edu.cn.
References
- Amieva, M. , and Peek, R.M. Jr (2016) Pathobiology of Helicobacter pylori‐induced gastric cancer. Gastroenterology 150: 64–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin, D.N. , Shepherd, B. , Kraemer, P. , Hall, M.K. , Sycuro, L.K. , Pinto‐Santini, D.M. , and Salama, N.R. (2007) Identification of Helicobacter pylori genes that contribute to stomach colonization. Infect Immun 75: 1005–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarota, G. , Sanguinetti, M. , Gallo, A. , and Posteraro, B. (2012) Review article: biofilm formation by Helicobacter pylori as a target for eradication of resistant infection. Aliment Pharmacol Ther 36: 222–230. [DOI] [PubMed] [Google Scholar]
- Carron, M.A. , Tran, V.R. , Sugawa, C. , and Coticchia, J.M. (2006) Identification of Helicobacter pylori biofilms in human gastric mucosa. J Gastrointest Surg 10: 712–717. [DOI] [PubMed] [Google Scholar]
- Chey, W.D. , Leontiadis, G.I. , Howden, C.W. , and Moss, S.F. (2017) ACG clinical guideline: treatment of Helicobacter pylori infection. Am J Gastroenterol 112: 212–239. [DOI] [PubMed] [Google Scholar]
- Choi, I.J. , Kim, C.G. , Lee, J.Y. , Kim, Y.I. , Kook, M.C. , Park, B. , and Joo, J. (2020) Family history of gastric cancer and Helicobacter pylori treatment. N Engl J Med 382: 427–436. [DOI] [PubMed] [Google Scholar]
- Couturier, C. , Bauer, A. , Rey, A. , Schroif‐Dufour, C. , and Broenstrup, M. (2012) Armeniaspiroles, a new class of antibacterials: Antibacterial activities and total synthesis of 5‐chloro‐Armeniaspirole A. Bioorg Med Chem Lett 22: 6292–6296. [DOI] [PubMed] [Google Scholar]
- Dufour, C. , Wink, J. , Kurz, M. , Kogler, H. , Olivan, H. , Sable, S. , et al. (2012) Isolation and structural elucidation of armeniaspirols A‐C: potent antibiotics against gram‐positive pathogens. Chemistry 18: 16123–16128. [DOI] [PubMed] [Google Scholar]
- Fabrega, A. , Madurga, S. , Giralt, E. , and Vila, J. (2009) Mechanism of action of and resistance to quinolones. Microb Biotechnol 2: 40–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos, S. , and Papastergiou, V. (2020) An update on current and advancing pharmacotherapy options for the treatment of H. pylori infection. Expert Opin Pharmacother 12: 1–13. [DOI] [PubMed] [Google Scholar]
- Hathroubi, S. , Servetas, S.L. , Windham, I. , Merrell, D.S. , and Ottemann, K.M. (2018) Helicobacter pylori biofilm formation and its potential role in pathogenesis. Microbiol Mol Biol Rev 82: e00001–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hathroubi, S. , Zerebinski, J. , Clarke, A. , and Ottemann, K.M. (2020) Helicobacter pylori biofilm confers antibiotic tolerance in part via a protein‐dependent mechanism. Antibiotics (Basel) 9: 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y. , Zhang, M. , Lu, B. , and Dai, J. (2016) Helicobacter pylori and antibiotic resistance. A continuing and intractable problem. Helicobacter 21: 349–363. [DOI] [PubMed] [Google Scholar]
- Huang, Y. , Hang, X. , Jiang, X. , Zeng, L. , Jia, J. , Xie, Y. , et al. (2019) In vitro and in vivo activities of zinc linolenate, a selective antibacterial agent against Helicobacter pylori . Antimicrob Agents Chemother 63: e00004–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, C.C. , Prieto‐Davo, A. , Jensen, P.R. , and Fenical, W. (2008) The marinopyrroles, antibiotics of an unprecedented structure class from a marine Streptomyces sp. Org Lett 10: 629–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krulwich, T.A. , Sachs, G. , and Padan, E. (2011) Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol 9: 330–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamishi, S. , Eshaghi, H. , Mahmoudi, S. , Bahador, A. , Hosseinpour Sadeghi, R. , Najafi, M. , et al. (2016) Intrafamilial transmission of Helicobacter pylori: genotyping of faecal samples. Br J Biomed Sci 73: 38–43. [DOI] [PubMed] [Google Scholar]
- Moloney, M.G. (2016) Natural products as a source for novel antibiotics. Trends Pharmacol Sci 37: 689–701. [DOI] [PubMed] [Google Scholar]
- Papastergiou, V. , Georgopoulos, S.D. , and Karatapanis, S. (2014) Treatment of Helicobacter pylori infection: meeting the challenge of antimicrobial resistance. World J Gastroenterol 20: 9898–9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peek, R.M. Jr , and Blaser, M.J. (2002) Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer 2: 28–37. [DOI] [PubMed] [Google Scholar]
- Pettit, R.K. , Weber, C.A. , Kean, M.J. , Hoffmann, H. , Pettit, G.R. , Tan, R. , et al. (2005) Microplate Alamar blue assay for Staphylococcus epidermidis biofilm susceptibility testing. Antimicrob Agents Chemother 49: 2612–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon, M. , Broutin, L. , Touroult‐Jupin, P. , Cremniter, J. , Plouzeau, C. , Faure, J.P. , et al. (2020) First detection in Helicobacter suis of a mutation conferring resistance to clarithromycin in Helicobacter pylori: case report and review of the literature. Microb Drug Resist 26: 677–680. [DOI] [PubMed] [Google Scholar]
- Qiao, Y. , Yan, J. , Jia, J. , Xue, J. , Qu, X. , Hu, Y. , et al. (2019) Characterization of the biosynthetic gene cluster for the antibiotic armeniaspirols in Streptomyces armeniacus . J Nat Prod 82: 318–323. [DOI] [PubMed] [Google Scholar]
- Savoldi, A. , Carrara, E. , Graham, D.Y. , Conti, M. , and Tacconelli, E. (2018) Prevalence of antibiotic resistance in Helicobacter pylori: a systematic review and meta‐analysis in world health organization regions. Gastroenterology 155: 1372–1382 e1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, D.R. , Marcus, E.A. , Weeks, D.L. , and Sachs, G. (2002) Mechanisms of acid resistance due to the urease system of Helicobacter pylori . Gastroenterology 123: 187–195. [DOI] [PubMed] [Google Scholar]
- Sugano, K. , Tack, J. , Kuipers, E.J. , Graham, D.Y. , El‐Omar, E.M. , Miura, S. , et al. (2015) Kyoto global consensus report on Helicobacter pylori gastritis. Gut 64: 1353–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, M. , Isogai, E. , Isogai, H. , Hayashi, S. , Hirose, K. , Kimura, K. , et al. (2002) Synergic effect of quinolone antibacterial agents and proton pump inhibitors on Helicobacter pylori . J Antimicrob Chemother 49: 1039–1040. [DOI] [PubMed] [Google Scholar]
- Thung, I. , Aramin, H. , Vavinskaya, V. , Gupta, S. , Park, J.Y. , Crowe, S.E. , and Valasek, M.A. (2016) Review article: the global emergence of Helicobacter pylori antibiotic resistance. Aliment Pharmacol Ther 43: 514–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trew, S.J. , Wrigley, S.K. , Pairet, L. , Sohal, J. , Shanu‐Wilson, P. , Hayes, M.A. , et al. (2000) Novel streptopyrroles from Streptomyces rimosus with bacterial protein histidine kinase inhibitory and antimicrobial activities. J Antibiot 53: 1–11. [DOI] [PubMed] [Google Scholar]
- Warren, J.R. , and Marshall, B. (1983) Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet 1: 1273–1275. [PubMed] [Google Scholar]
- Xu, Y.F. , Lian, D.W. , Chen, Y.Q. , Cai, Y.F. , Zheng, Y.F. , Fan, P.L. , et al. (2017) In vitro and in vivo antibacterial activities of patchouli alcohol, a naturally occurring tricyclic sesquiterpene, against Helicobacter pylori infection. Antimicrob Agents Chemother 61: e00122–e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa, H. , Osaki, T. , and Kamiya, S. (2015) Biofilm formation by Helicobacter pylori and its involvement for antibiotic resistance. Biomed Res Int 2015: 914791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa, H. , Osaki, T. , Fukutomi, T. , Hanawa, T. , Kurata, S. , Zaman, C. , et al. (2017) Diversification of the AlpB outer membrane protein of Helicobacter pylori affects biofilm formation and cellular adhesion. J Bacteriol 199: e00729–e816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti, G. , and Cendron, L. (2019) Structural aspects of Helicobacter pylori antibiotic resistance. Adv Exp Med Biol 1149: 227–241. [DOI] [PubMed] [Google Scholar]
- Zhang, R.G. , Duan, G.C. , Fan, Q.T. , and Chen, S.Y. (2016) Role of Helicobacter pylori infection in pathogenesis of gastric carcinoma. World J Gastrointest Pathophysiol 7: 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Kill kinetics of H. pylori Hp159 by ARM1. Time‐ and dose‐dependent killing of the bacteria by ARM1 (A) and MTZ (B) were evaluated. The MIC of ARM1 and MTZ was considered as 8 and 16 μg ml‐1, respectively. If no colonies were present, calculations were made using the limit of detection (101 CFU ml‐1).
Fig S2. Development of resistance to ARM1 in H. pylori G27. The fold change was the normalized ratio of the MIC obtained for a given subculture to the MIC obtained for first‐time exposure. ARM1 did not develop drug resistance over the 27 days of studies while metronidazole quickly acquired resistance on day 9. A representative of two independent experiments is shown.
Fig S3. Cell Cytotoxicity of ARM1 was assessed in human normal hepatocyte LO2 cells. After overnight culture, cells were incubated with ARM1 at predetermined concentrations for 48 h and were then treated with CCK‐8 reagent for assessment of cytotoxicity.
Fig S4. ARM1 had low toxicity in vivo. (A) After the mice were orally administrated with overdose (285 mg/kg, 10‐fold effective dosage) ARM1 once a day for 5 consecutive days, the mice body weights were monitored daily for 7 days. (B) At the seven days after the last treatment, the mice were sacrificed, and the stomach, the liver, the spleen and the kidney were removed from the abdominal cavity and examined by H&E stain.
Table S1. The predicted Drug‐like properties of ARM1.
Table S2. In vitro combinatory anti‐H. pylori effect of ARM1 with each antibiotic.


