Abstract
While COVID-19 infection during pregnancy is common, fetal transmission is rare, suggesting that intrauterine mechanisms form an effective blockade against SARS-CoV-2. Key among these is the decidual immune environment of the placenta. We hypothesize that decidual leukocytes are altered by maternal SARS-CoV-2 infection in pregnancy and that this decidual immune response is shaped by the timing of infection during gestation. To address this hypothesis, we collected decidua basalis tissues at delivery from women with symptomatic COVID-19 during second (2nd Tri COVID, n = 8) or third trimester (3rd Tri COVID, n = 8) and SARS-CoV-2-negative controls (Control, n = 8). Decidual natural killer (NK) cells, macrophages and T cells were evaluated using quantitative microscopy, and pro- and anti-inflammatory cytokine mRNA expression was evaluated using quantitative reverse transcriptase PCR (qRT-PCR). When compared with the Control group, decidual tissues from 3rd Tri COVID exhibited significantly increased macrophages, NK cells and T cells, whereas 2nd Tri COVID only had significantly increased T cells. In evaluating decidual cytokine expression, we noted that IL-6, IL-8, IL-10 and TNF-α were significantly correlated with macrophage cell abundance. However, in 2nd Tri COVID tissues, there was significant downregulation of IL-6, IL-8, IL-10, and TNF-α. Taken together, these results suggest innate and adaptive immune responses are present at the maternal-fetal interface in maternal SARS-CoV-2 infections late in pregnancy, and that infections earlier in pregnancy show evidence of a resolving immune response. Further studies are warranted to characterize the full scope of intrauterine immune responses in pregnancies affected by maternal COVID-19.
Keywords: COVID-19 in pregnancy, Decidual leukocytes, Cytokines, SARS-CoV-2, Vertical transmission
1. Introduction
Healthy pregnancies have been challenged by the pandemic of COVID-19, the illness caused by SARS-CoV-2. Disease severity correlates with expression of proinflammatory cytokines, i.e. ‘cytokine storm’ (Song et al., 2020). Pregnant women develop a spectrum of COVID-19 symptoms ranging from asymptomatic carriage to critical illness (Allotey et al., 2020, Dubey et al., 2020, Zambrano et al., 2020) and are at increased risk for preterm birth, preeclampsia, stillbirth, and low birth weight (Wei et al., 2021).
Nonetheless, vertical transmission of SARS-CoV-2 from human mother to fetus during pregnancy is rare (Facchetti et al., 2020, Juan et al., 2020, Raschetti et al., 2020, Al-Matary et al., 2021, Kotlyar et al., 2021, Tolu et al., 2021) with positive neonatal rates ranging between 0.5% and 2.5%. Interestingly, SARS-CoV-2 has been detected in placental tissues and can invade human placental trophoblast cells (Sharps et al., 2020, Taglauer et al., 2020, Kotlyar et al., 2021, Lu-Culligan et al., 2021, Ouyang et al., 2021). However, given the rarity of fetal transmission, additional factors within the maternal-fetal interface are likely preventing the transplacental passage of SARS-CoV-2. Of central importance are intrauterine immune responses, governed by maternal decidual leukocytes adjacent to the villous placenta. Among these, decidual T cells, NK cells, and macrophages are known to respond to viral infection at the maternal-fetal interface, either through cellular or cytokine-mediated responses (Crespo et al., 2016, Yockey and Iwasaki, 2018, Jabrane-Ferrat, 2019, Crespo et al., 2020, Parker et al., 2020, Granja et al., 2021).
Previous studies have identified inflammatory responses (Lu-Culligan et al., 2021) and sexually dimorphic alteration in interferon response genes in the human placenta in villous placental tissues from pregnant individuals with COVID-19 in the third trimester (Bordt et al., 2021). However, leukocyte and cytokine profiles of the decidual-specific immune response to maternal SARS-CoV-2 require more specific characterization, particularly in infections earlier in pregnancy.
We hypothesized that maternal immune cells in the decidua respond to COVID-19 via leukocyte and cytokine alteration which may be impacted by the timing of maternal infection in pregnancy. We thus evaluated decidual leukocyte and decidual tissue cytokine profiles from women with symptomatic COVID-19 infections in their 2nd or 3rd trimesters of pregnancy as compared to controls to characterize the trajectory of the decidual immune responses against SARS-CoV-2 at the maternal-fetal interface.
2. Materials and methods
2.1. Study enrollment
2.1.1. Setting
Boston Medical Center (BMC). All patient enrollment and tissue collection was approved by the Boston University Medical Campus Institutional Review Board.
2.1.2. Retrospective cohort (April-May 2020)
Collection from placentas designated for pathology from women who tested positive via SARS-CoV-2 nasopharyngeal PCR testing at delivery or from contemporary controls who tested negative. Samples were obtained without informed consent, and the IRB waiver of informed consent stipulated collection of limited demographic data.
2.1.3. Prospective cohort study (July 2020-April 2021)
Inclusion criteria included age minimum of 18 years, singleton pregnancy, and English/Spanish speaking. COVID-19 group: mothers with a positive nasopharyngeal PCR test for SARS-CoV-2 infection at any point during pregnancy. Of note SARS-CoV-2 genomes were not sequenced in these patients, but data collection occurred prior to emergence of SARS-CoV-2 Delta and Omicron variants (Anon, 2022). Control group: mothers with no documentation of SARS-CoV-2 infection at any point during pregnancy.
2.1.4. Data collection
Demographic and clinical variables were obtained from the electronic medical record (EMR) and recorded in a secure, de-identified RedCap Database (www.project-redcap.org).
2.2. Sample collection
2.2.1. Retrospective cohort
Tissues were placed in 4% buffered formalin at time of delivery and evaluated by board-certified pathologists. Full thickness placental biopsies were dissected from formalin fixed placental tissues, soaked in 18% sucrose, embedded in optimal cutting temperature (OCT) compound (Fisher) and frozen at − 80 °C.
2.2.2. Prospective cohort
Fresh placental tissue was collected within 6 h after delivery. Decidual tissue biopsies were obtained according to established methods (Xu et al., 2015), flash-frozen and stored at − 80 °C. Placental full-thickness biopsies were dissected and fixed with formaldehyde/sucrose for 72 h, embedded in OCT (Fisher) and frozen at − 80 °C.
2.3. Immune cell analysis
2.3.1. Immunohistochemistry
Ten micron thick placental tissue sections were washed with PBS-T, permeabilized, and incubated with Histostain Plus Broad Spectrum Blocking Solution (#859043, Novex by LifeTechnologies). Primary antibodies were diluted 1:100 in PBS-T as follows: single labeling: CD14 (mouse anti-human, 14–149–82, Invitrogen), CD56 (mouse anti-human, 14–0567–82, Invitrogen); double labeling: SARS CoV-2 spike glycoprotein (rabbit anti-human, ab272504 Abcam), CD3 (mouse anti-human, 14–0038–82, Invitrogen). Fluorescently labeled secondary antibodies were diluted 1:500 in PBS-T as follows: single labeling: Alexa 594 (anti-mouse, ab150108 Abcam); double labeling: AlexaFluor 594 (anti-mouse, ab150108 Abcam), AlexaFluor 647 (anti-rabbit, A21244, LifeTechnologies). Control slides were incubated with secondary antibodies alone. Washed slides were cover slipped with Prolong Gold with DAPI (ThermoFisher). All cohort slides were stained in bulk and imaged within 24–48 h of staining.
2.3.2. Microscopy
Images were acquired on a Nikon deconvolution wide-field epifluorescence microscope using NIS-Elements Software (Nikon). Decidua basalis images were obtained by automated acquisition at 200x of 4 tiled images from 5 randomized areas per slide. Exposure times were standardized for each target to ensure accurate comparative quantification.
2.3.3. Quantitative image analysis
Image area and integrated density were measured via ImageJ software (imagej.net) for each immunofluorescent 200x image (n = 5/slide) along with mean fluorescence values from 5 randomly selected background readings per image cohort to calculate a corrected total cell fluorescence (CTCF) per published protocols (Kenan et al., 2020, Taglauer et al., 2020, Benarroch et al., 2021). An average CTCF was calculated from secondary only negative control slides (n = 3 per staining assay) and used to calculate a final Fluorescence Ratio: target antigen CTCF/secondary only control CTCF. All ImageJ analysis and calculations were performed on blinded samples.
2.4. Gene expression analysis
2.4.1. RNA isolation and cDNA synthesis
RNA was isolated from frozen placental decidual biopsies (50–75 μg) using the RNAqueous™ kit (#AM1912, Invitrogen). RNA quantity and purity was assessed using a Nanodrop™ spectrophotometer (ThermoFisher). cDNA was generated from equal amounts of RNA (800 ng) using a RevertAid First Strand cDNA synthesis kit (ThermoFisher).
2.4.2. qRT-PCR
PrimePCR assays for IGF-BP1, HLA-A, IL-1β, IL-6, IL-8, IL-10, IFN-γ, TNF-α, and GAPDH (for normalization; #10025636, BIO-RAD) and iTaq™ Universal SYBR Green Supermix (#1725120, BIO-RAD) utilizing a BIO-RAD thermocycler per manufacturer instructions. Assays were run in technical triplicate and no-reverse transcriptase controls were included to ensure no DNA contamination. Fold changes were calculated for COVID-19 groups relative to the control group using the standardized ΔΔCT method (Livak and Schmittgen, 2001).
2.5. Statistical analysis
2.5.1. Clinical and demographic data
Categorical variables were reported as percentages [n (%)] and continuous variables were reported as either mean with standard deviation [mean(sd)] or median with interquartile range [median (IQR)], where appropriate. For Table 1, P-values for continuous variables were generated using t-test (indicated by ( * ), ANOVA test (indicated by ** ) or Kruskal Wallis rank test (indicated by *** ). P-values for categorical variables were generated using the Fisher exact tests. Differences were considered significant at α = 0.05. For Supplemental Table 1, P-values were generated using t-tests. Analyses were conducted using R version 3.6.1 (www.r-project.org).
Table 1.
Demographic and clinical data from prospective COVID-19 cohort.
| Demographic | 2nd Trimester COVID (N = 8) | 3rd Trimester COVID (N = 8) | Control (N = 8) | P-Value |
|---|---|---|---|---|
| Gestational age at infection (weeks) – Mean (SD) | 18.7 (4.1) | 30.0 (3.1) | N/A | 2.4e-05 * |
| Duration between infection and delivery (weeks) – Mean (SD) | 21.2 (4.4) | 10.0 (4.2) | N/A | 2.4e-04 * |
| Maternal COVID severity – N (%) | N/A | |||
| Asymptomatic | 0 | 0 | ||
| Mild/moderate symptoms | 8 | 7 | ||
| Hospitalized | 0 | 1 (12.5%) | ||
| ICU | 0 | 0 | ||
| Maternal age (years) – Mean (SD) | 30.0 (5.0) | 30.9 (5.1) | 30.4 (6.6) | 0.95 ** |
| Gravida (number) – Mean (SD) | 3.1 (2.0) | 1.9 (0.8) | 2.7 (1.5) | 0.25 ** |
| Para (number) – Mean (SD) | 1.5 (1.4) | 0.6 (0.7) | 0.6 (0.7) | 0.16 ** |
| Maternal race – N (%) | ||||
| Black | 1 (12.5%) | 1 (12.5%) | 0 | 0.90 |
| White | 3 (37.5%) | 2 (25%) | 2 (25%) | |
| Other | 4 (50%) | 5 (62.5%) | 6 (75%) | |
| Maternal ethnicity – N (%) | 5 (62.5%) | 6 (75%) | 6 (75%) | 1 |
| Hispanic/Latino | ||||
| Maternal chronic health conditionsa – N (%) | 3 (37.5%) | 4 (50%) | 5 (62.5%) | 0.87 |
| Pregnancy complicationsb – N (%) | 7 (87.5%) | 8 (100%) | 5 (62.5%) | 0.27 |
| Delivery by c-section – N (%) | 2 (25%) | 3 (37.5%) | 0 | 0.30 |
| Maternal WBC count at delivery – (cells/mL x 1000) – Median (IQR) | 8.7 [6.2–9.2] | 11.2 [9.4–14.2] | 8.2 [6.2–11.93] | 0.40 *** |
| Gestational age at birth (weeks) – Mean (SD) | 39.9 (1.1) | 40.0 (1.9) | 38.6 (0.9) | 0.09 ** |
| Infant sex – N (%) | ||||
| Male | 5 (62.5%) | 2 (25%) | 3 (37.5%) | 1 |
| Female | 3 (37.5%) | 6 (75%) | 5 (62.5%) | |
| Birth weight (grams) – Mean (SD) | 3453 (327) | 3446 (411) | 3353 (459) | 0.86 ** |
| 5-minute APGAR – median (IQR) | 9 [9–9] | 9 [9–9] | 9 [8.75–9.00] | 0.12 * ** |
| Required NICU admission – N (%) | 1 (12.5%) | 0 | 1 (12.5%) | 1 |
| Infant COVID-19 nasal swab positive results | 0 | 0 | 0 | N/A |
P-values for continuous variables were generated using t-test (indicated by (*), ANOVA test (indicated by ** ), or Kruskal Wallis rank test (indicated by *** ). P-values for categorical variables were generated using the Fisher exact tests.
Chronic health conditions included autoimmune disease, diabetes, hepatitis B, hepatitis C, HSV, HIV, hypertension, obesity, thyroid disease, substance use disorder, or other
Pregnancy complications included chorioamnionitis, gestational diabetes, hypertensive disorder of pregnancy, fetal growth restriction, placenta previa, preterm labor, unexplained vaginal bleeding, or other
2.5.2. Laboratory data
P-values for quantitative immunofluorescence were generated using ANOVA with Tukey’s multiple comparison’s test. P-values for fold-changes by qRT-PCR results were generated using the one-sample Wilcoxon Signed Rank Test compared to a theoretical median of 1.0. Pearson test was used to generate P-values for correlations between cytokine fold-changes and immune cell abundance. Differences were considered significant at α = 0.05. All laboratory statistical analysis was performed using Prism 9 software (GraphPad).
3. Results
3.1. Decidual macrophages and NK cells are increased in 3rd Trimester COVID 19 infections
We first assessed decidual leukocytes in the retrospective cohort. There were no significant differences in gestational age at birth, birth weight, or maternal age between the COVID-19 infected (n = 8) and control (n = 5) groups (Supplemental Table 1). Of note, multiple infants in this cohort tested positive for SARS-CoV-2 by nasopharyngeal PCR after birth (Supplemental Table 1). In comparison to controls, decidua basalis samples from women with COVID-19 had significantly increased macrophage (Supplemental Fig. 1A and B) and NK cell populations (Supplemental Fig. 1C and D). There were no differences between female and male infants (data not shown).
3.2. The abundance of decidual macrophages, NK cells, and T cells in the differs based on gestational timing of COVID-19 infection
Decidual leukocyte abundance was examined further in our prospective cohort, with 16 total in the COVID-19 and 8 in the control group. All participants with COVID-19 were symptomatic in this study, with one requiring hospitalization. There were no significant differences in maternal or infant demographics (Table 1). None of the infants in the COVID-19 groups tested positive for SARS-CoV-2 by nasopharyngeal PCR, but testing was only performed in mothers with a symptomatic infection at time of delivery. Further, there were no significant differences in placental pathology reports observed in the COVID-19 compared to control groups (Supplemental Table 2).
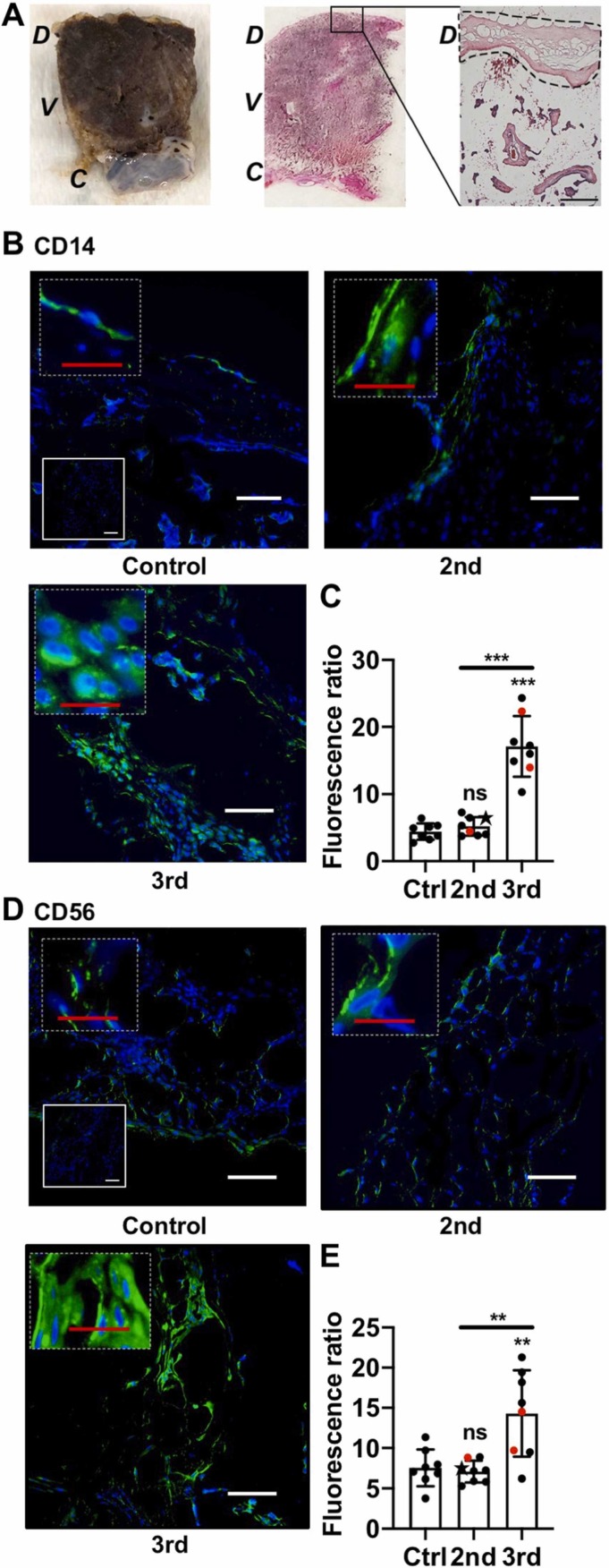
We focused our analysis on the decidua basalis ( Fig. 1A). Evaluation of decidual CD14 staining showed that decidual macrophages were significantly increased in the 3rd Tri COVID group (Fig. 1B & C), confirming our retrospective study results. In contrast, there was no difference in macrophage cell staining between Control and 2nd Tri COVID groups (Fig. 1B & C). Analysis of CD56 staining demonstrated a significant increase in decidual NK cells in the 3rd Tri COVID group, whereas there were no differences in NK cell abundance between Control and 2nd Tri COVID groups (Fig. 1D & E).
Fig. 1.
Innate immune cell decidual infiltrates following COVID-19 in pregnancy. (A) Representative images demonstrating focused areas of decidual tissue for fluorescent microscopy analysis. D: Decidua, V: Villous tissue, C: chorionic plate. Black square denotes area of enlarged H&E image. Dotted outline denotes representative area of focused decidual tissue survey. Scale bar: 200 µm. (B, D) Representative images (200x) of decidual areas stained for (B) CD14 (green) or (D) CD56 (green) immunofluorescence. White scale bar = 50 µm. Dashed insets are higher magnification, with red scale bar = 12.5 µm. Solid insets: secondary-only controls. (C, E) Graphical analysis of comparative fluorescence quantitation of (C) CD14 or (E) CD56. Red symbols indicate placentas with positive staining for SARS-CoV-2 Spike protein. Star symbol indicates placenta from participant who required hospitalization for COVID-19. 2nd = 2nd Tri COVID (n = 8); 3rd = 3rd Tri COVID (n = 8); control/ctrl = negative for COVID (n = 8). ** p < 0.01; ** p < 0.001. Scale bar: 50 µm; insets: secondary-only controls.(For interpretation of the references to colour in this figure, the reader is referred to the web version of this article.)
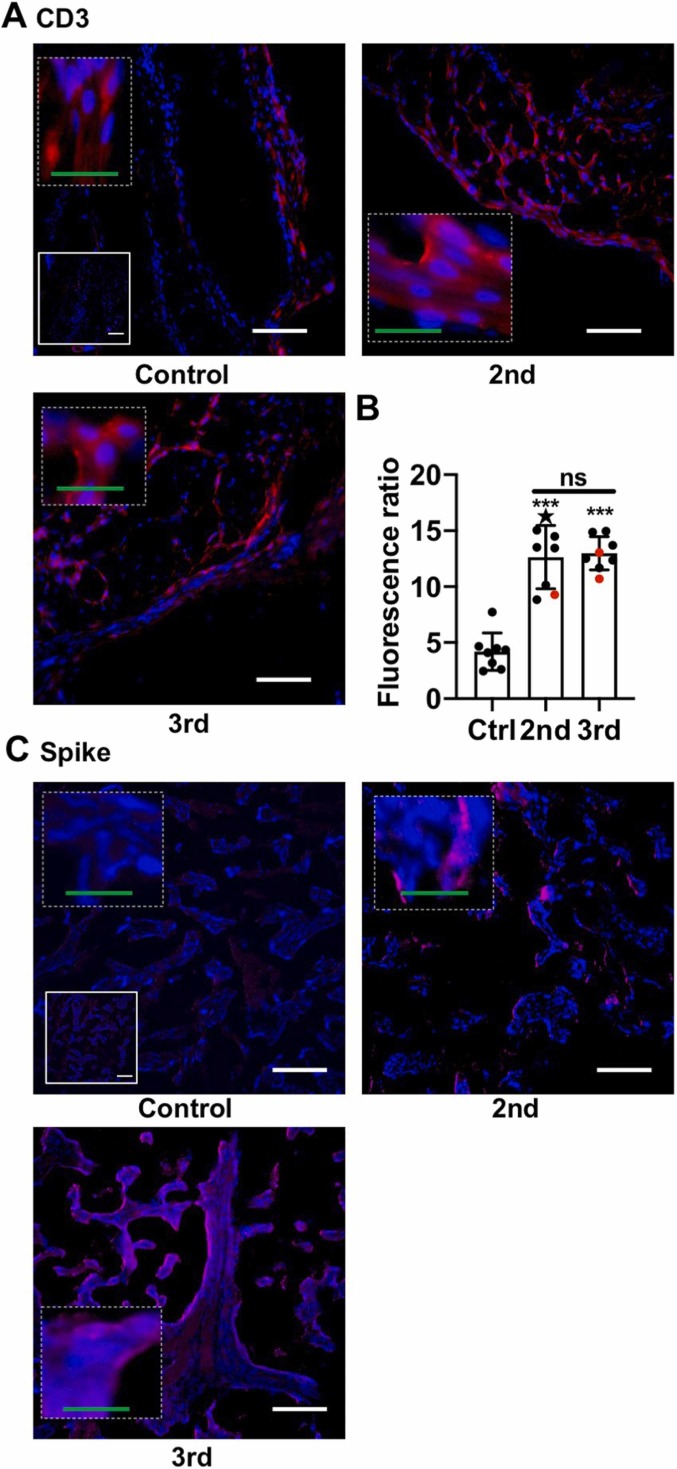
Similar to macrophage and NK cells, CD3+ T cell staining was significantly increased in the 3rd Tri COVID group ( Fig. 2A & B). However, in contrast to macrophage and NK cell staining, the T cell infiltrate was also increased in 2nd Tri COVID as compared to Control group (Fig. 2A & B). There was a small, but significant, difference in CD14 staining comparing male and female infants in Control but not COVID-19 groups (Supplemental Fig. 2); no gender-specific differences were found in NK cells and T cells (Data not shown). As multiple pregnancies have been shown to impact decidual immune infiltrates in humans (Gamliel et al., 2018), we performed a correlation analysis between immune cell infiltrates and number of prior pregnancies and deliveries and found no significant correlations (Data not shown). Finally, we assessed whether there was evidence of SARS-CoV-2 infection in the placenta at time of delivery by performing immunofluorescence for the Spike protein. Two placentas from the 3rd Tri COVID and one from the 2nd Tri COVID group stained positive for Spike protein (Fig. 2C).
Fig. 2.
T cells and SARS-CoV-2 Spike protein following COVID-19 in pregnancy. Imaging of placental decidua for SARS-CoV-2 Spike protein. (A) Representative images (200x) of decidual areas stained for CD3 (red) immunofluorescence. (B) Graphical analysis of comparative fluorescence quantitation of CD3. Red symbols indicate placentas with positive staining for SARS-CoV-2 Spike protein. Star symbol indicates placenta from participant who required hospitalization for COVID-19. 2nd = 2nd Tri COVID (n = 8); 3rd = 3rd Tri COVID (n = 8); control/ctrl = negative for COVID (n = 8). ** p < 0.01; ** p < 0.001. (C) Representative immunofluorescence images (200x) of the three placental decidua tissues that stained positive for SARS-CoV-2 Spike protein (red). For A and C, white scale bar = 50 µm. Dashed insets are higher magnification, with green scale bar = 12.5 µm. Solid insets: secondary-only controls.(For interpretation of the references to colour in this figure, the reader is referred to the web version of this article.)
3.3. Decidual cytokine expression is positively correlated with macrophage and NK cell abundance
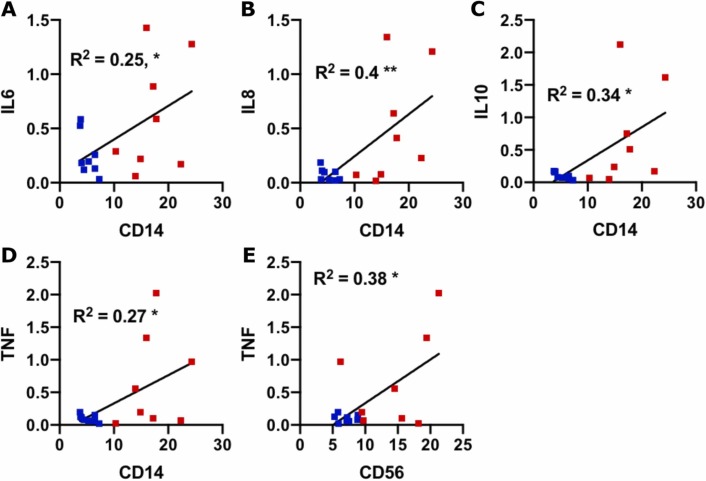
We next chose to evaluate a targeted panel of cytokines which had been identified in human SARS-CoV-2 immune responses in other organs (Leisman et al., 2020, Tang et al., 2020) and known to be present within the maternal-fetal interface (Mori et al., 2016). First, to evaluate decidual biopsy purity, tissues were assayed for the expression of decidual marker IGF-BP1 (Giudice et al., 1992, Han et al., 1996, Okada et al., 2018)and for the presence of HLA-A, which would be absent or low with significant trophoblast contamination (Apps et al., 2009). Expression of both markers were detected at high levels (Supplemental Fig. 3), suggesting a predominant component of decidual mRNA. For cytokine evaluation, we conducted a correlation analysis between cytokine expression and decidual leukocyte abundance in the COVID groups. We found that there was a significant positive correlation between IL-6, IL-8, IL-10, and TNF-α expression and macrophage abundance, and a significant positive correlation between TNF-α and NK cell abundance ( Fig. 3). No significant correlations were found among cytokine expression and T cell abundance (Supplemental Fig. 4).
Fig. 3.
Correlation between cytokine abundance and immune cells. (A–E) Scatter plots of the indicated cytokines (Y axis; fold change relative to control) and immune cell markers (X axis; fluorescence ratio) for each placental sample (COVID-19 +; n = 16). Blue symbols = 2nd Tri COVID; red symbols = 3rd Tri COVID. * , p < 0.05; ** , p < 0.01.
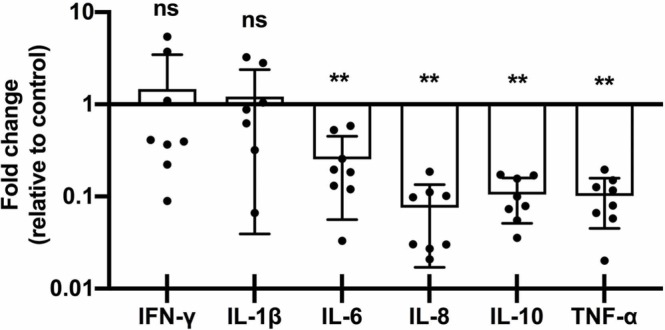
3.4. IL-6, IL-8, IL-10 and TNF-α are downregulated in decidual tissues from 2nd trimester COVID-19 infections
When evaluating total cytokine changes, the 2nd-Tri COVID group showed a significant decrease in IL-6, IL-8, IL-10, and TNF-α and no change in abundance for IL-1β or IFN-γ ( Fig. 4). Further, in the 3rd-Tri COVID tissues, there were no differences in expression of IL-1β, IL-6, IL-10, TNF-α, or IFN-γ, and a significant downregulation of IL-8 (Supplemental Figure 5). Finally, there were no differences in cytokine levels based on the gender of the infant (data not shown).
Fig. 4.
Placental cytokine mRNA downregulation in 2nd Tri COVID group. Fold-changes in cytokine abundance for IFN-γ, IL-1β, IL-6, IL-8, IL-10, and TNF-α by qRT-PCR of placental decidua from women with 2nd trimester COVID infection (2nd, n = 8) calculated as fold-change compared to control (n = 8). ns = not significant; ** p < 0.01.
4. Discussion
In the current study, we identified an association between COVID-19 in pregnancy and an increased immune cell abundance in the decidua basalis, with the specific cell types dependent on gestational timing of SARS-CoV-2 infection. Further, we identified that levels of multiple cytokine transcripts correlated with macrophage abundance, driven by 3rd trimester COVID-19 infections. In contrast, there was an overall trend of downregulation in the pro and anti-inflammatory cytokine production in pregnancies with 2nd trimester maternal COVID-19 disease. These results provide new insight into SARS-CoV-2 immune responses at the maternal-fetal interface and highlight the importance of evaluating tissues from various gestational stages of maternal COVID-19 infections.
Our immune cell analysis is consistent with our previously reported findings in this same cohort identifying gross pathology of intervillous and subchorionic fibrosis (Taglauer et al., 2020). Importantly, this study demonstrates an association between COVID-19 infection and increased immune cells that is dependent on the gestational age at which infection occurred. The inclusion of a second-trimester infection group is particularly valuable, as there is very limited knowledge of second-trimester SARS-CoV-2 infection and long-term immune consequences at the maternal-fetal interface.
The lack of differences macrophage and NK cell numbers compared to Control samples in 2nd Tri COVID group suggests a waning or active downregulation of the immune response over time but the finding that increased T cell infiltrates were present 3rd Tri and 2nd Tri COVID groups ranging weeks to months following infection is additionally intriguing. Additional cellular phenotyping and further characterization of the decidual immune response mechanisms over time is warranted to further evaluate these findings.
In cytokine analysis, we found no significant differences with the exception of IL-8 downregulation. While these results differ from an early report of global inflammatory signaling in placental tissues from 3rd trimester COVID-19 infections (Lu-Culligan et al., 2021), our findings are similar to more recently published data showing no changes in human placental expression of TNF-α, IL-1β, or IL-6 associated with COVID-19 and upregulation of IFN-γ only in male fetuses (Bordt et al., 2021). While our analysis may have missed the early window of inflammatory cytokine regulation, decidual immune responses to COVID-19 may not involve upregulation of these pro-inflammatory cytokines for fetal benefit given known pathologies associated with increased IFN-γ, IL-1β, IL-6, and TNF-α in mouse models (Yockey and Iwasaki, 2018).
The positive correlation between macrophage abundance and IL-6, IL-8, IL-10, and TNF-α expression may be due to production of these cytokines by maternal macrophages through established pathogen response pathways (Hoo et al., 2020). TNF-α was also positively correlated with NK cell abundance consistent with reports of human decidual NK cells responding to virally-infected decidual stromal cells by producing TNF-α (Crespo et al., 2016). Of note, despite these correlations, cytokines were generally unchanged or downregulated in the COVID-19 groups. Other drivers of cytokine expression, such as parturition, may have masked the impact of COVID-19 on cytokine expression.
Interestingly, proinflammatory cytokines were downregulated in the placenta following second-trimester COVID-19. This may be because the tissues were in the resolution phase post- infection. Alternatively the prominent T cell presence observed in the second trimester decidua could be transitioning to an “exhaustion phenotype” with decreased cytokine production. Future studies should interrogate for T cells markers of exhaustion in these samples, including PD-1, CTLA-4, and LAG3, characterizing human decidual CD8 + T Cells that express low cytolytic molecules but are capable of responding to infection (Taglauer et al., 2008, Van Der Zwan et al., 2018).
The current study has several limitations. First, our findings are from a relatively small sample size from a single clinical site. Only basic clinical demographic data was available from the retrospective cohort, and it is possible that the women had severe COVID-19 disease, limiting the generalizability of these results. Additionally, multiple infants tested positive for COVID-19 in the retrospective cohort, at rates higher than generally reported in the literature. These positive tests may represent postnatal transmission as isolation protocols which were still being developed during this time or the result of false positives as the sensitivity and specificity of the qRT-PCR testing was rapidly evolving during this early phase of the pandemic. In our prospective cohort, most women were asymptomatic at time of delivery and SARS-CoV-2 testing was accordingly not performed on their infants. The current study also analyzed only single cell surface leukocyte phenotype markers; flow cytometry analysis was not possible due to institutional biosafety restrictions at the time of this study. More detailed characterization of the decidual leukocyte populations in future studies will be imperative to understand their function. Finally, while we demonstrated high expression of decidua-specific mRNA markers in tissues for cytokine analysis, some villous placental contamination may have been present the mRNA analysis.
Overall, these results demonstrate that immune cell infiltrates in the placenta following COVID-19 are driven by timing of infection during gestation. While additional studies are required to more fully evaluate the intrauterine immune millieu in pregnancies affected by maternal COVID-19, these data provide early evidence supporting the role of decidual leukocytes in the physiologic response against SARS-CoV-2 at the maternal-fetal interface.
Funding
Boston University Clinical & Translational Science Institute1UL1TR001430 (E.M.W)., Lovejoy Resident Research Grant (L.J.J)., American Academy of Pediatrics Resident Research Award (L.J.J).
Acknowledgements
We thank staff of the Boston Medical Center Labor and Delivery Unit, Department of Pathology, Neonatal ICU, and Newborn Nursery for their help in obtaining clinical samples.
Declaration of interest
None.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jri.2022.103501.
Appendix A. Supplementary material
Supplementary material.Supplemental Figure 1: Placental maternal immune cell infiltrates from retrospective COVID-19 cohort. (A, C) Representative images (200x) of decidual areas stained for (A) CD14 (red) or (C) CD56 (red) immunofluorescence. (B, D) Graphical analysis of comparative fluorescence quantitation of (B) CD14 and (D) CD56. Red circles indicate dyads with an infant who tested positive for SARS-CoV-2 by nasal PCR swab at 24 or 48 h of life. SARS-CoV-2 positive (COVID, n = 8) or negative (control, n = 5). * p < 0.05; ** p < 0.01. Scale bar: 50 µm; insets: secondary-only controls.
.
Supplementary material.Supplemental Figure 2: Infant gender sub-analysis of CD14 fluorescence staining in prospective cohort. Data from Figure 2 were divided by infant gender assigned at birth. p < 0.05 by t-test.
.
Supplementary material.Supplemental Figure 3: Evaluation of decidual tissue purity for qRT-PCR analysis. (A–B) qPCR Ct values of (A) IGF-BP1 or (B) HLA-A expression in decidual biopsies from Control (SARS-CoV-2 negative), 2nd trimester COVID infection (2nd) or 3rd trimester COVID infection (3rd). Data are presented as mean ± SD. Differences across groups were assessed by ANOVA and there were no significant differences.
.
Supplementary material.Supplemental Figure 4: Non-significant correlations between cytokine abundance and immune cells. (A-G) Scatter plots of the indicated cytokines (Y axis; fold change relative to control) and immune cell markers (X axis; fluorescence ratio) for each placental sample (COVID-19 +; n = 16). Blue symbols = 2nd trimester; red symbols = 3rd trimester.
.
Supplementary material.Supplemental Figure 5: Placental cytokine expression in 3rd Tri COVID group. Fold-changes in cytokine abundance for IFN-γ, IL-1β, IL-6, IL-8, IL-10, and TNF-α by qRT-PCR of placental decidua from women with 3nd trimester COVID infection (n = 8) calculated as fold-change compared to control (n = 8). ns = not significant; * p < 0.05.
.
Supplementary material.Supplemental Table 1: Demographic data from retrospective COVID-19 cohort.
.
Supplementary material.Supplemental Table 2: Pathology diagnoses from prospective COVID-19 cohort.
.
References
- Al-Matary A., Almatari F., Al-Matary M., AlDhaefi A., Alqahtani M.H.S., Alhulaimi E.A., et al. Clinical outcomes of maternal and neonate with covid-19 infection - multicenter study in saudi arabia. J. Infect. Public Health. 2021;14:702–708. doi: 10.1016/j.jiph.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allotey J., Stallings E., Bonet M., Yap M., Chatterjee S., Kew T., et al. Clinical manifestations, risk factors, and maternal and perinatal outcomes of coronavirus disease 2019 in pregnancy: living systematic review and meta-analysis. BMJ. 2020;370:m3320. doi: 10.1136/bmj.m3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anon, 2022. Gisaid tracking of variants. GISAID: 〈https://www.gisaid.org/hcov19-variants/〉.
- Apps R., Murphy S.P., Fernando R., Gardner L., Ahad T., Moffett A. Human leucocyte antigen (hla) expression of primary trophoblast cells and placental cell lines, determined using single antigen beads to characterize allotype specificities of anti-hla antibodies. Immunology. 2009;127:26–39. doi: 10.1111/j.1365-2567.2008.03019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benarroch Y., Juttukonda L., Sabharwal V., Boateng J., Khan A.R., Yarrington C., et al. Differential expression of rab5 and rab7 small gtpase proteins in placental tissues from pregnancies affected by maternal coronavirus disease 2019. Clin. Ther. 2021;43:308–318. doi: 10.1016/j.clinthera.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordt E.A., Shook L.L., Atyeo C., Pullen K.M., De Guzman R.M., Meinsohn M.C., et al. Maternal sars-cov-2 infection elicits sexually dimorphic placental immune responses. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abi7428. eabi7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo Â.C., Mulik S., Dotiwala F., Ansara J.A., Sen Santara S., Ingersoll K., et al. Decidual nk cells transfer granulysin to selectively kill bacteria in trophoblasts. Cell. 2020;182:1125–1139. doi: 10.1016/j.cell.2020.07.019. e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo Â.C., Strominger J.L., Tilburgs T. Expression of kir2ds1 by decidual natural killer cells increases their ability to control placental hcmv infection. Proc. Natl. Acad. Sci. USA. 2016;113:15072–15077. doi: 10.1073/pnas.1617927114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey P., Reddy S.Y., Manuel S., Dwivedi A.K. Maternal and neonatal characteristics and outcomes among covid-19 infected women: An updated systematic review and meta-analysis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2020;252:490–501. doi: 10.1016/j.ejogrb.2020.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchetti F., Bugatti M., Drera E., Tripodo C., Sartori E., Cancila V., et al. Sars-cov2 vertical transmission with adverse effects on the newborn revealed through integrated immunohistochemical, electron microscopy and molecular analyses of placenta. EBioMedicine. 2020;59 doi: 10.1016/j.ebiom.2020.102951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamliel M., Goldman-Wohl D., Isaacson B., Gur C., Stein N., Yamin R., et al. Trained memory of human uterine nk cells enhances their function in subsequent pregnancies. Immunity. 2018;48:951–962. doi: 10.1016/j.immuni.2018.03.030. e5. [DOI] [PubMed] [Google Scholar]
- Giudice LC, Dsupin BA, Irwin JC. Steroid and peptide regulation of insulin-like growth factor-binding proteins secreted by human endometrial stromal cells is dependent on stromal differentiation. J Clin Endocrinol Metab. 1992;75(5):1235–1241. doi: 10.1210/jcem.75.5.1385468. [DOI] [PubMed] [Google Scholar]
- Granja M.G., Oliveira A.C.R., de Figueiredo C.S., Gomes A.P., Ferreira E.C., Giestal-de-Araujo E., et al. Sars-cov-2 infection in pregnant women: Neuroimmune-endocrine changes at the maternal-fetal interface. Neuroimmunomodulation. 2021;28:1–21. doi: 10.1159/000515556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han VK, Bassett N, Challis JR. The expression of insulin-like growth factor (IGF) and IGF-binding protein (IGFBP) genes in the human placenta and membranes: evidence for IGF-IGFBP interactions at the feto-maternal interface. J Clin Endocrinol Metab. 1996;81(7):2680–2693. doi: 10.1210/jcem.81.7.8675597. [DOI] [PubMed] [Google Scholar]
- Hoo R., Nakimuli A., Vento-Tormo R. Innate immune mechanisms to protect against infection at the human decidual-placental interface. Front Immunol. 2020;11:2070. doi: 10.3389/fimmu.2020.02070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabrane-Ferrat N. Features of human decidual nk cells in healthy pregnancy and during viral infection. Front Immunol. 2019;10:1397. doi: 10.3389/fimmu.2019.01397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan J., Gil M.M., Rong Z., Zhang Y., Yang H., Poon L.C. Effect of coronavirus disease 2019 (covid-19) on maternal, perinatal and neonatal outcome: systematic review. Ultrasound Obstet. Gynecol. 2020;56:15–27. doi: 10.1002/uog.22088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenan S., Liang H., Goodman H.J., Jacobs A.J., Chan A., Grande D.A., et al. 5-aminolevulinic acid tumor paint and photodynamic therapy for myxofibrosarcoma: an in vitro study. J. Orthop. Surg. Res. 2020;15:94. doi: 10.1186/s13018-020-01606-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlyar A.M., Grechukhina O., Chen A., Popkhadze S., Grimshaw A., Tal O., et al. Vertical transmission of coronavirus disease 2019: a systematic review and meta-analysis. Am. J. Obstet. Gynecol. 2021;224:35–53. doi: 10.1016/j.ajog.2020.07.049. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisman D.E., Ronner L., Pinotti R., Taylor M.D., Sinha P., Calfee C.S., et al. Cytokine elevation in severe and critical covid-19: a rapid systematic review, meta-analysis, and comparison with other inflammatory syndromes. Lancet Respir. Med. 2020;8:1233–1244. doi: 10.1016/S2213-2600(20)30404-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu-Culligan A., Chavan A.R., Vijayakumar P., Irshaid L., Courchaine E.M., Milano K.M., et al. Maternal respiratory sars-cov-2 infection in pregnancy is associated with a robust inflammatory response at the maternal-fetal interface. Med (N. Y) 2021;2:591–610. doi: 10.1016/j.medj.2021.04.016. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M., Bogdan A., Balassa T., Csabai T., Szekeres-Bartho J. The decidua-the maternal bed embracing the embryo-maintains the pregnancy. Semin Immunopathol. 2016;38:635–649. doi: 10.1007/s00281-016-0574-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H, Tsuzuki T, Murata H. Decidualization of the human endometrium. Reprod Med Biol. 2018;17(3):220–227. doi: 10.1002/rmb2.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y., Bagalkot T., Fitzgerald W., Sadovsky E., Chu T., Martínez-Marchal A., et al. Term human placental trophoblasts express sars-cov-2 entry factors ace2, tmprss2, and furin. mSphere. 2021:66. doi: 10.1128/mSphere.00250-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker E.L., Silverstein R.B., Verma S., Mysorekar I.U. Viral-immune cell interactions at the maternal-fetal interface in human pregnancy. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.522047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschetti R., Vivanti A.J., Vauloup-Fellous C., Loi B., Benachi A., De Luca D. Synthesis and systematic review of reported neonatal sars-cov-2 infections. Nat. Commun. 2020;11:5164. doi: 10.1038/s41467-020-18982-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharps M.C., Hayes D.J.L., Lee S., Zou Z., Brady C.A., Almoghrabi Y., et al. A structured review of placental morphology and histopathological lesions associated with sars-cov-2 infection. Placenta. 2020;101:13–29. doi: 10.1016/j.placenta.2020.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song P., Li W., Xie J., Hou Y., You C. Cytokine storm induced by sars-cov-2. Clin. Chim. Acta. 2020;509:280–287. doi: 10.1016/j.cca.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taglauer E., Benarroch Y., Rop K., Barnett E., Sabharwal V., Yarrington C., et al. Consistent localization of sars-cov-2 spike glycoprotein and ace2 over tmprss2 predominance in placental villi of 15 covid-19 positive maternal-fetal dyads. Placenta. 2020;100:69–74. doi: 10.1016/j.placenta.2020.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taglauer E.S., Trikhacheva A.S., Slusser J.G., Petroff M.G. Expression and function of pdcd1 at the human maternal-fetal interface. Biol. Reprod. 2008;79:562–569. doi: 10.1095/biolreprod.107.066324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Liu J., Zhang D., Xu Z., Ji J., Wen C. Cytokine storm in covid-19: the current evidence and treatment strategies. Front. Immunol. 2020;11:1708. doi: 10.3389/fimmu.2020.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolu L.B., Ezeh A., Feyissa G.T. Vertical transmission of severe acute respiratory syndrome coronavirus 2: a scoping review. PLoS One. 2021;16 doi: 10.1371/journal.pone.0250196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Zwan A., Bi K., Norwitz E.R., Crespo Â.C., Claas F., Strominger J.L., et al. Mixed signature of activation and dysfunction allows human decidual cd8. Proc. Natl. Acad. Sci. USA. 2018;115:385–cd390. doi: 10.1073/pnas.1713957115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S.Q., Bilodeau-Bertrand M., Liu S., Auger N. The impact of covid-19 on pregnancy outcomes: a systematic review and meta-analysis. CMAJ. 2021;193:E540–E548. doi: 10.1503/cmaj.202604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Plazyo O., Romero R., Hassan S.S., Gomez-Lopez N. Isolation of leukocytes from the human maternal-fetal interface. J. Vis. Exp. 2015 doi: 10.3791/52863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yockey L.J., Iwasaki A. Interferons and proinflammatory cytokines in pregnancy and fetal development. Immunity. 2018;49:397–412. doi: 10.1016/j.immuni.2018.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrano L.D., Ellington S., Strid P., Galang R.R., Oduyebo T., Tong V.T., et al. Update: characteristics of symptomatic women of reproductive age with laboratory-confirmed sars-cov-2 infection by pregnancy status - united states, january 22-october 3, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:1641–1647. doi: 10.15585/mmwr.mm6944e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.Supplemental Figure 1: Placental maternal immune cell infiltrates from retrospective COVID-19 cohort. (A, C) Representative images (200x) of decidual areas stained for (A) CD14 (red) or (C) CD56 (red) immunofluorescence. (B, D) Graphical analysis of comparative fluorescence quantitation of (B) CD14 and (D) CD56. Red circles indicate dyads with an infant who tested positive for SARS-CoV-2 by nasal PCR swab at 24 or 48 h of life. SARS-CoV-2 positive (COVID, n = 8) or negative (control, n = 5). * p < 0.05; ** p < 0.01. Scale bar: 50 µm; insets: secondary-only controls.
Supplementary material.Supplemental Figure 2: Infant gender sub-analysis of CD14 fluorescence staining in prospective cohort. Data from Figure 2 were divided by infant gender assigned at birth. p < 0.05 by t-test.
Supplementary material.Supplemental Figure 3: Evaluation of decidual tissue purity for qRT-PCR analysis. (A–B) qPCR Ct values of (A) IGF-BP1 or (B) HLA-A expression in decidual biopsies from Control (SARS-CoV-2 negative), 2nd trimester COVID infection (2nd) or 3rd trimester COVID infection (3rd). Data are presented as mean ± SD. Differences across groups were assessed by ANOVA and there were no significant differences.
Supplementary material.Supplemental Figure 4: Non-significant correlations between cytokine abundance and immune cells. (A-G) Scatter plots of the indicated cytokines (Y axis; fold change relative to control) and immune cell markers (X axis; fluorescence ratio) for each placental sample (COVID-19 +; n = 16). Blue symbols = 2nd trimester; red symbols = 3rd trimester.
Supplementary material.Supplemental Figure 5: Placental cytokine expression in 3rd Tri COVID group. Fold-changes in cytokine abundance for IFN-γ, IL-1β, IL-6, IL-8, IL-10, and TNF-α by qRT-PCR of placental decidua from women with 3nd trimester COVID infection (n = 8) calculated as fold-change compared to control (n = 8). ns = not significant; * p < 0.05.
Supplementary material.Supplemental Table 1: Demographic data from retrospective COVID-19 cohort.
Supplementary material.Supplemental Table 2: Pathology diagnoses from prospective COVID-19 cohort.