Enclosing, the present study provides original insight into the molecular and genetic responses of L. reuteri PL503 to the toxic effects of MDA, one of the most common lipid peroxidation products. This bacterium is able to detoxify MDA and hence, exert a potential health benefit in the gastrointestinal tract. Contributing to identify some of the underlying genetic and biochemical mechanisms facilitates the development targeted prophylactic and treatment strategies involving these and other probiotic bacteria.

Summary
This study aimed to provide insight into the molecular and genetic mechanisms implicated in the responses of Lactobacillus reuteri against the oxidative stress induced by malondialdehyde (MDA) by analysing protein oxidation and assessing the uspA and the dhaT genes. Four experimental groups were evaluated depending on the concentration of MDA added in Man, Rogosa and Sharpe (MRS) broth: Control (L. reuteri), 5 µM (L. reuteri + 5 µM MDA), 25 µM (L. reuteri + 25 µM MDA) and 100 µM (L. reuteri + 100 µM MDA). Three replicates were incubated at 37 °C for 24 h in microaerophilic conditions and sampled at 12, 16, 20 and 24 h. The upregulation of the uspA gene by L. reuteri indicates the recognition of MDA as a potential DNA‐damaging agent. The dhaT gene, encoding a NADH‐dependent‐oxidoreductase, was also upregulated at the highest MDA concentrations. This gene was proposed to play a role in the antioxidant response of L. reuteri. The incubation of L. reuteri with MDA increased the production of ROS and caused thiol depletion and protein carbonylation. L. reuteri is proposed to detoxify pro‐oxidative species while the underlying mechanism requires further elucidation.
Introduction
Oxidative stress is a redox deregulation typically caused by an imbalance between pro‐oxidants and the antioxidant defences, which is manifested as damage to molecules of biological significance by radical and non‐radical species (Finkel and Holbrook, 2000). The biological consequences of the oxidative damage to lipids, proteins and the DNA, involve impaired physiological processes and the onset of assorted health disorders. Profuse literature supports the implication of persistent oxidative stress on severe health disorders such as diabetes (Asmat et al., 2016), neurodegenerative and cardiovascular diseases (Lin and Beal, 2006), and several types of cancer (Valko et al, 2005; Reuter et al, 2010). The colon has been identified to be particularly sensitive to oxidative stress (Sanders et al., 2014) and such condition is involved in the onset of a number of pathological conditions at this location, including ulcerative colitis, Crohn’s and inflammatory bowel diseases, and colorectal cancer (Gackowski et al., 2002; Zhu and Li, 2012). The intestinal mucosa is permanently exposed to the pro‐oxidant action of dietary lipid and protein oxidation products (~ luminal oxidative stress) which may contribute to impair the redox status of the bowel (Estévez and Luna, 2017). Malondialdehyde (MDA), a remarkable lipid oxidation product in muscle foods and oxidized oils, is able to bind to DNA leading to the formation of etheno‐modified DNA bases, with these adducts being found in organs with diseases related to enduring inflammatory conditions that may eventually cause malignancies (Bartsch and Nair, 2005). Owing to the pathological effects of luminal oxidative stress, dietary antioxidants have been proposed to inhibit and/or alleviate the symptoms of some of these health disorders (Spyropoulos et al., 2011). On this line, supplementation with probiotic bacteria has been proposed as a feasible strategy to counteract the oxidative stress in the gastrointestinal tract (GIT) in humans (Spyropoulos et al., 2011).
Probiotics are defined as ‘live microbes which, when administered in adequate amounts, confer a health benefit to the host’ (FAO, 2002). As a natural colonizer of the GIT tract, Lactobacillus reuteri has been widely used as a dietetic supplement to promote gut health in humans (Hjern et al., 2020; Wang et al., 2020). Oral administration of L. reuteri decreases the onset and severity of inflammatory and infectious disorders in the GIT and contributes to a balanced colonic microbiota (Wang et al., 2020). In relation to the underlying mechanisms of its probiotic effects, L. reuteri has been found to diminish colonic oxidative stress by reducing the formation and accumulation of oxidation products in the lumen (Amaretti et al, 2013). The positive effects of L. reuteri on the oxidative and health status of human colon are documented (Petrella, 2016) and include protection against a number of pathological conditions such as colorectal cancer (Bistas et al, 2020), inflammatory bowel diseases (Wang et al, 2020) and cardiovascular disorders (Kazemian et al, 2020). Yet, the precise biochemical and genetic responses of L. reuteri against pro‐oxidative conditions caused by luminal oxidation products (such as MDA) are not fully understood.
Pathogenesis of MDA not only involves the formation of DNA adducts, this lipid peroxidation product is known to bind to proteins and induce a number of oxidative modifications (Esterbauer et al, 1993). From a medical standpoint, the accumulation of oxidized proteins is a pathological hallmark of ageing and chronic diseases (Estévez and Luna, 2017) and protein oxidation is known to play central role in the pathogenesis of gut disorders driven by oxidative stress (Gackowski et al, 2002; Zhu and Li, 2012). Though much less studied than in humans, the occurrence of protein oxidation in bacteria has also been linked to impaired growth and senescence (Ezraty et al., 2017). On this line, the modifications induced by MDA in bacterial proteins and their biological consequences, are unknown. Furthermore, the precise biological mechanisms activated by L. reuteri in response to an MDA‐induced oxidative stress are poorly documented. In this regard, the study of the regulation of particular stress‐related genes seems crucial to comprehend the influence of external sources of oxidative stress on particular metabolic pathways and biological functions. Previous reports have documented that particular genes such as uspA and the dhaT are activated in L. reuteri in response to an oxidative threat caused by reactive oxygen species (ROS; Arcanjo et al, 2019). The universal stress protein A (uspA) superfamily is an ancient and conserved group of proteins found in assorted microorganisms, insects and plants (Kvint et al., 2003). The precise roles of Usp proteins in biological systems remain unclear; yet, they seem to be involved in the defence against DNA‐damaging agents (Kvint et al, 2003). The dhaT gene expression leads to the synthesis of a 1,3 propanediol oxidoreductase (1,3‐PDO), which was proposed by Arcanjo et al. (2019) to be involved in the protection of L. reuteri against oxidative stress. Arcanjo et al. (2019) also reported that protein carbonylation could play a relevant role as indicator of oxidative damage in bacteria and as a potential signalling mechanism. Yet, the connection between oxidative stress, gene expression and protein oxidation in probiotic bacteria is poorly understood.
This study aimed to provide insight into the molecular and genetic mechanisms involved in the responses of L. reuteri against MDA‐induced oxidative stress by analysing protein oxidation and assessing the uspA and the dhaT genes.
Results
Regulation of uspA and dhaT genes by L. reuteri in response to MDA
Fig. 1A shows the relative expression (2‐ΔΔC T) of the L. reuteri PL503 uspA gene during the incubation assay in the presence of different concentrations of MDA. The exposure to MDA caused an upregulation of the uspA gene, as shown in Fig. 1A. This event had a dose‐effect (12 h) which affected not only the intensity of the gene overexpression but also the time‐frame of the occurrence of such biological response. The lowest concentration (5 µM) apparently led to a fast and limited response already at 12 h. The relative expression of the uspA gene in L. reuteri PL503 incubated with 25 and 100 µM of MDA showed a similar response 4 h later, while such expression had a peak after 20 h of incubation being significantly more intense with 25 and 100 µM of MDA than in the absence or 5 µM of MDA.
Fig. 1.
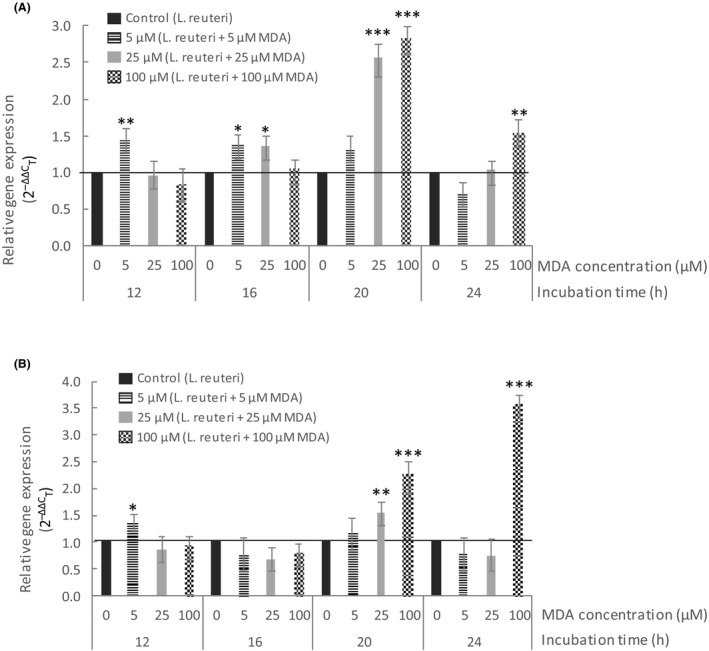
Relative expression (2‐ΔΔC T) of the uspA (A) and dhaT (B) genes in Lactobacillus reuteri PL503 as affected by increasing concentrations of malondialdehyde (MDA) for up to 24 h. Means for each experimental group are calculated from analyses applied to three biological replicates and all analyses were technically duplicated. Black line at 2‐ΔΔC T = 1 denotes standardized expression rate for CONTROL group at each sampling time (calibrator). 2‐ΔΔC T < 1 denotes suppression of gene expression; 2‐ΔΔC T > 1 denotes activation of gene expression. Asterisks on top of bars denote significant diferences in paired Student’s t‐tests performed to compare each MDA concentration with CONTROL [MDA = 0]: *P ≤ 0.05; **P ≤ 0.01; and ***P ≤ 0.001.
Overall, no significant changes in the relative expression of the dhaT gene were observed during the first two samplings. Yet, a dose‐dependent effect of the incubation with MDA on the dhaT gene expression was found at 20 h of incubation (Fig. 1B). In particular, the relative transcription of the dhaT gene significantly increased in the presence of 25 and 100 µM of MDA at 20 h. At the final sampling, the relative expression of the dhaT gene significantly increased in the bacterium challenged with the highest concentration of MDA.
Induction of oxidative stress in L. reuteri by MDA
To investigate the ability of MDA to induce oxidative stress in L. reuteri PL503, ROS generation was assessed in bacteria by flow cytometry. Figure 2 shows how MDA exposure increased the percentage of bacteria suffering from oxidative stress in a dose‐dependent manner. To assess the oxidative damage caused by MDA incubation in L. reuteri PL503, both lipid and protein oxidation markers were quantified.
Fig. 2.

Production of reactive oxygen species (ROS) in Lactobacillus reuteri PL503 grown in MRS broth with increasing concentrations of malondialdehyde (MDA) for 24 h. A: DNA containing bacterium (debris and nonbacterium events are removed); B: Gate for bacterium; C‐F: X‐axis bacterium (DNA +) Y‐axis ROS; C: CONTROL; D: L. reuteri PL503 incubated with 5 µM MDA; E: L. reuteri PL503 incubated with 25 µM MDA; F: L. reuteri PL503 incubated with 100 µM MDA. Analyses were applied to three biological replicates and all analyses were technically duplicated.
The initial concentration of TBARS found in the cultures accurately reflects the three levels of MDA used for challenging L. reuteri PL503 (Fig. 3A). The basal TBARS concentration in CONTROL cultures (~ 1 mg l−1) could be the result of the occurrence of lipid peroxidation in the bacterium under physiological conditions and did not change significantly during the assay (P > 0.05). In the cultures challenged with MDA, the initial concentration significantly decreased (P < 0.05) in the range of 22–26% with this depletion being a likely reflection of the reaction of MDA with other biomolecules.
Fig. 3.

Concentration of TBARS (A); allysine (B) and Schiff bases (C) (means ± standard deviation) in Lactobacillus reuteri PL503 grown in MRS broth with increasing concentrations of malondialdehyde (MDA) during an incubation period for up to 24 h. Means for each experimental group are calculated from analyses applied to three biological replicates and all analyses were technically duplicated. Different letters at the same sampling time denote significant differences between means within the same sampling point in ANOVA (P ≤ 0.05).
The evolution of the concentration of allysine, a recognized marker of protein oxidation, was assessed in L. reuteri PL503 during the incubation period (37 °C/24 h) and results are shown in Fig. 3B. The evolution of allysine in CONTROL group of L. reuteri PL503 shows a significant increase (P < 0.05) from 0.7 to 1.65 nmol allysine per mg protein. As compared to CONTROL, the exposure to MDA caused a significant increase (P < 0.001) in the concentration of allysine in proteins from L. reuteri PL503 and such increase followed a dose‐dependent fashion for 16 h, suggesting a direct implication of MDA in the carbonylation of proteins from L. reuteri PL503. After that sampling, the behaviour varied between experimental groups. In L. reuteri PL503 grown in the presence of 5 and 25 µM of MDA, the increase of allysine was sustained during the complete assay reaching the highest concentration at 24 h (3.7 and 5.2 nmol allysine/mg protein respectively). When exposed to the highest MDA concentration (100 µM) the concentration of allysine peaked at 16 h after which a decrease was observed so that basal levels (< 1 nmol allysine mg−1 protein) were found at the end of the incubation period. In the present study, the formation of Schiff bases (Fig. 3C) was dependent on the presence of MDA. Yet, it is worth noting that no clear dose‐dependent effect was observed and that the evolution of its concentration during the assay was erratic.
To verify whether MDA is able or not to induce allysine formation via an oxidative deamination mechanism similar to that exerted by other dicarbonyls (i.e. glyoxal), MDA (0.25 µM) was incubated with assorted proteins (5 mg ml−1) at 37 °C (HSA, HH) and 80 °C (LAC) for 24 h. As compared to CONTROL groups (protein suspensions with no added MDA), protein suspensions incubated with MDA had significantly lower concentrations of allysine (Fig. 4A). Conversely, MDA caused a significant increase in the concentration of Schiff bases (Fig. 4B).
Fig. 4.

Concentration of allysine (A) and Schiff bases (B) (means ± standard deviation) in human serum albumin (HSA), human haemoglobin (HH) and β‐lactoglobulin (LAC) (5 mg ml−1) after incubation with MDA (0.25 µM) at 37 °C (HSA, HH) and 80 °C (LAC) for 24 h. Means for each experimental group are calculated from analyses applied to three biological replicates and all analyses were technically duplicated. Different letters denote significant differences between means within the same sampling point in ANOVA (P ≤ 0.05).
The concentration of free thiols in L. reuteri PL503 during the incubation assay (37 °C/24 h) is shown in Fig. 5. No significant changes in the thiol concentration were observed in CONTROL samples during the first 12 h. During the second half of the assay, a quantitatively small but significant increase of thiols was detected. The incubation of L. reuteri PL503 in the presence of MDA caused a significant dose‐dependent increase of free thiols during the first 16 h of assay. In the following sampling points, the evolution of thiols remained stable in bacteria treated with 100 µM of MDA and significantly declined in bacteria exposed to 5 and 25 µM of MDA.
Fig. 5.

Concentration of free thiols (means ± standard deviation) in Lactobacillus reuteri PL503 grown in MRS broth with increasing concentrations of malondialdehyde (MDA) during an incubation period for up to 24 h. Means for each experimental group are calculated from analyses applied to three biological replicates and all analyses were technically duplicated. Different letters at the same sampling time denote significant differences between means within the same sampling point in ANOVA (P ≤ 0.05).
Discussion
Regulation of uspA and dhaT genes by L. reuteri in response to MDA
The increasing applied doses of MDA (5 µM, 25 µM, 100 µM) did not compromise the survival of L. reuteri, as the counts remained stable during the entire experimental assay (37 °C/24 h). This finding reflects the ability of L. reuteri to activate mechanisms to neutralize the potential harmful effects of the sublethal concentrations of this lipid peroxidation product. In the present study, these mechanisms were firstly assessed by the analysis of the relative transcription of stress‐related genes.
uspA gene
Considering the role of the uspA gene in the defence against DNA‐damaging agents (Kvint et al., 2003), the upregulation of such gene as a response to the challenge of MDA was expected. MDA is known to form adducts with assorted biomolecules, including the DNA (Esterbauer et al., 1993). Marnett (1999) reported that MDA reacts with DNA to form adducts to deoxyguanosine and deoxyadenosine. However, the major adduct to DNA is a pyrimidopurinone called M1G, which has been also identified in human liver, white blood cells, pancreas and breast tissues, and is considered a significant contributor to cancer linked to dietary factors (Niedernhofer et al., 2003). It is hence, relevant to find out whether colonic microbiota is able to detoxify and/or counteract the noxious effects of MDA. In bacteria, M1G has been also found to lead to mutagenesis, which is repaired by the nucleotide excision repair pathway (Marnett, 1999). The threat of such mutagenesis could have caused an upregulation of the uspA gene, as shown in Fig. 1A. These results are, however, divergent to those reported by Oberg et al. (2015) who observed a significant inhibition of the uspA gene expression in Bifidobacterium longum exposed to a hydroxyl radical generating system. Conversely, an ATP‐dependent metallo‐protease was found to be upregulated to likely protect membrane proteins against radical‐mediated oxidative damage. The specificity of the genetic responses of a given bacterium challenged with different pro‐oxidant threats seems plausible since the damage caused by MDA via adduct formation to biomolecules (including proteins and DNA) is fairly different from the severe oxidative damage caused by ROS.
dhaT gene
While the uspA gene encodes an assorted collection of proteins, the expression of the dhaT gene leads to the synthesis of a single protein with definite function, the 1,3 propanediol oxidoreductase (1,3‐PDO; Schaefer et al, 2010). This enzyme plays a relevant role in stress situations involving energetic demand since 1,3‐PDO facilitates the main carbohydrate fermentation pathway (6‐phosphogluconate/phosphoketolase; 6‐PG/PK) through the production of NAD+ (required for glucose fermentation) from NADH in the conversion of 3‐hydroxypropionaldehyde (3‐HPA) (its substrate) into 1,3 propanediol (1,3‐PD) under anaerobic conditions. Additionally, 3‐HPA, also known as reuterin, is known to be profusely excreted to the surrounding environment under stress situations, imparting strong antimicrobial properties (Schaefer et al, 2010).
The underlying mechanisms by which L. reuteri may try to protect against MDA‐induced biological damage through the activation of the 3‐HPA pathway should be a matter of thoroughly analysis. The 3‐HPA pathway, as previously reported, requires the presence of glycerol, commonly added as growth promoter in Lactobacillus cultures (Talarico et al, 1988). Since L. reuteri from the present experiment had no access to glycerol, and the production of 3‐HPA, preferential target of 1,3‐PDO, may not be present in the media, the purpose of the upregulation of the gene encoding the 1,3‐PDO remains indefinite. It is clear, however, that such enzyme may be implicated in a protection mechanism against MDA‐induced oxidative stress that reasonably involves the NADH/NAD+ redox pair and that 3‐HPA may not be the unique substrate for 1,3‐PDO. In a previous study (Arcanjo et al., 2019), in which L. reuteri was challenged with H2O2 (0.5 mM) in the absence of glycerol, a similar effect on the expression of the dhaT gene was observed. The authors hypothesized whether the NAD+‐dependent activity of the 1,3‐PDO may be able to detoxify H2O2 in the presence of NADH in accordance to the pathway proposed in Fig. 6. Interestingly, Lactobacillus spp. have been also found to be able to generate H2O2 and other ROS via implication of NAD(P)H oxidoreductases (Hertzberger et al, 2014). The incubation of L. reuteri PL503 in the presence of MDA led to increase the production ROS as shown in Fig. 2. The analysis of the bacterium with flow cytometry showed that increasing concentrations of MDA led to accumulative collection of cells with significant generation of ROS. These results originally prove that MDA induces the formation of diverse radical species in L. reuteri PL503 plausibly via production of H2O2, the most common source of hydroxyl radical in biological systems (Davies, 2005). The production of H2O2 by Lactobacillus spp., already reported in literature, seems to be promoted by the presence of electron acceptors such as O2 and fructose (Mane, 2016). MDA, as a potent electron acceptor, may also have such effect on L. reuteri PL503 supporting the connection between the lipid peroxidation product, the impairment of the redox status of the cell, the production of H2O2 and the upregulation of the gene encoding a NADH‐dependent oxidoreductase (Fig. 6). However, the underlying mechanism by which MDA would promote ROS formation in L. reuteri requires further elucidation. Although Lactobacillus spp. can produce antimicrobial peptides, bacteriocins and several organic compounds, releasing H2O2 seems to be central for antimicrobial and restorative processes (Singh et al, 2018). This has been also highlighted as a relevant probiotic mechanism as ROS production by Lactobacillus spp. has been shown to promote epithelial restitution during colitis and alleviate inflammation in human mucosa (Singh et al, 2018). Finally, physiological production of radical species induced by pro‐oxidative compounds such as MDA can contribute cells to minimize oxidative stress. This mechanism may involve radical species acting as signalling molecules or inducing subtle modifications in proteins that may, in turn, act as signalling molecules that could eventually enhance endogenous antioxidant mechanisms in the bacterium and in the host (Martín and Suarez, 2010). While the H2O2 production in Lactobacillus spp. is well documented, the mechanisms are not definite though it may involve the conjunction of NADPH and oxidoreductase enzymes (proposed mechanism in Fig. 6). The overexpression of the dhaT gene, alleged to protect against H2O2‐induced oxidative stress, may respond to the necessity of the bacterium to counteract the potential damage that such pro‐oxidant species may exert in its own biomolecules.
Fig. 6.

General scheme of the proposed mechanisms whereby malondialdehyde (MDA) damages biomolecules in Lactobacillus reuteri PL503 (red lines) and mechanism by which the bacterium may protect against MDA‐induced oxidative stress (green lines).
The pathogenesis of particular chemical species depends on their ability to establish molecular interactions with biomolecules from the host and as a result, impair biological processes. MDA is known to exert harmful effects by adducting biomolecules such as DNA and proteins (Bartsch and Nair, 2005). While the adducts with DNA and the corresponding MDA‐induced mutations have been known for some time in bacteria (Draper et al., 1986), the impact of MDA on proteins of biological relevance is not so well understood. Protein oxidation is not only mediated by ROS, as oxidizing lipids and final lipid oxidation products have been also identified as potential initiators of oxidative reactions in proteins (Davies, 2005). The damage caused to proteins in pro‐oxidative environments has been emphasized as one of the most salient causes of ageing and disease in humans (Davies, 2005; Estévez and Luna, 2017). Protein oxidation also has devastating effects on the structure and functionality of bacteria which may even lead to bacterial senescence and cell death (Ezraty et al., 2017). Interestingly, the oxidative damage to proteins plays a role in the bacterial response to oxidative stress (Ezraty et al., 2017) as proteins are activated by oxidative means to trigger specific antioxidant mechanisms. In order to provide further insight into the effects of MDA on L. reuteri, protein oxidation was assessed through the quantification of a specific protein carbonyl (allysine) and the formation of Schiff bases in bacterial proteins. Free thiols, as relevant redox‐active moieties in proteins, were also quantified.
Protein oxidation
Allysine is the most abundant protein carbonyl in biological systems and typically used as indicator of the oxidative damage to proteins (Estévez, 2011). In tissues from mammals, a concentration of 1 nmol carbonyls per mg protein has been reported as physiological while significant increases are commonly reflecting oxidative stress conditions (Akagawa et al., 2002). This is, to our knowledge, the first time that such specific protein oxidation product is quantified in cultured bacteria as marker of oxidative stress. Measuring protein carbonylation using the routine spectrophotometric DNPH method, Ballesteros et al. (2001) proposed this expression of the oxidative damage to proteins as a reflection of bacterial senescence as oxidized proteins accumulate in non‐proliferating bacteria. This is consistent with the evolution of allysine in CONTROL group of L. reuteri PL503. The present results show that allysine, formed in bacteria, as in eukaryotes, may be used as a reliable indicator of protein oxidation. Allysine is formed in proteins as an outcome of oxidative deamination of the ɛ‐amino group in lysine residues and that oxidation pathway can be initiated by i) radical species (i.e. hydroxyl radical) (Utrera and Estevez, 2013) or by dicarbonyls from the Maillard reaction (i.e. glyoxal/methylglyoxal) (Akagawa et al., 2002). MDA was found to promote the formation of allysine in L. reuteri PL503 and we performed additional analyses to find out the underlying mechanism of such oxidative damage. It is known that MDA reacts with ɛ‐amino group in lysine residues but the formation of allysine as an outcome of such reaction is not described in the literature. The additional assay carried out with human and bovine proteins (Fig. 4A,B) confirmed that MDA is not able to induce the oxidative deamination of alkaline amino acids via the Maillard mechanism previously reported. Instead, the reaction of MDA with such residues is known to yield Schiff bases and stable protein crosslinks (Requena et al., 1997; Estévez et al., 2019). While the formation of such fluorescent structures was the most likely fate of MDA residues in isolated human and bovine proteins, the evolution of Schiff bases in bacteria was irregular and considerably low as compared to other studies carried out in animal proteins (Utrera and Estevez, 2013). It is then reasonable to hypothesize that the significant decrease in MDA, previously stated (Fig. 3A), may respond to reactions with other biomolecules, including DNA, which would explain the fast and dose‐dependent overexpression of the uspA gene, involved in the defence against DNA‐damaging agents. These results suggest that MDA promoted allysine formation in L. reuteri PL503 through mechanisms that likely involve a ROS‐mediated pathway. This hypothesis is supported by the increase of ROS detected by flow cytometry in the bacterium treated with MDA (Fig. 2). This mechanism may involve the previous formation of H2O2 and its subsequent decomposition through the Fenton reaction into hydroxyl radicals (Proposed mechanism depicted in Fig. 6). The radical‐mediated oxidative deamination of lysyl residues is, so far, the most plausible mechanism behind the formation of allysine in L. reuteri PL503 incubated with MDA.
The biological significance of protein carbonylation should be another issue of discussion. As an irreversible modification in proteins in a pro‐oxidative environment, protein carbonylation is typically regarded as a reaction of negative biological consequences. Carbonylated proteins can be dysfunctional and may be tagged to removal as their accumulation causes impaired homeostasis that leads to chronic dysfunction and apoptosis (Shacter, 2000). On the other hand, carbonylated proteins may also act as signalling molecules, which may trigger specific pathways, aimed to preserve homeostasis control senescence (Shacter, 2000). Both circumstances may be applied to the present experiment. The initial increase in protein carbonyls in the MDA‐treated bacterium (16 h) was followed by a notable decrease of allysine in L. reuteri PL503 exposed to the highest MDA concentration. In these samples, at concentrations around 3 nmol allysine mg−1 protein, such bacterium may have activated the dhaT gene, clearly noticeable in the following sampling times. The NADH‐dependent oxidoreductase decoded by this gene may have contributed to detoxify pro‐oxidant species such as H2O2 and hence, inhibiting the enduring carbonylation observed in L. reuteri treated with lower doses of MDA (Fig. 6). The allysine decline by the end of the assay in L. reuteri PL503 treated with 100 µM MDA can only respond to the removal of carbonylated proteins through a mechanism (not identified in the present study) that could have been likely activated along with the dhaT pathway. It is worth noting that these mechanisms were not present in the bacterium incubated with 5 and 25 µM as allysine concentrations higher than 3 nmol allysine mg−1 protein were only reached at the end of the assay and regrettably the events that could have happened in hypothetical further sampling times are ignored. The hypothesis that the dhaT gene could have been activated by H2O2 and/or the effect of the former on protein carbonylation is supported by previous considerations made by Ezraty et al. (2017) and Arcanjo et al. (2019). The latter authors, in particular, observed how the accumulation of carbonyls in L. reuteri PL503, as a irreversible modification in oxidized proteins, triggered the upregulation of the dhaT gene in the presence of H2O2 and resveratrol.
Free thiols
Sulfur‐containing amino acids such as cysteine (Cys) and methionine (Met) are particularly sensitive to oxidation and thiol depletion is a typical feature in oxidized proteins. While the oxidation of thiols in the active site of enzymes may lead to dysfunction, irrelevant sulfur‐containing amino acids are known to act as endogenous antioxidants by offering a sacrificial loss to ROS and protecting other amino acids with relevant biological significance (Estévez et al, 2020). This double role of thiols was investigated in the present experiment. A paired balance between thiol oxidation and repair/de novo synthesis of proteins occurred during the first 12 h in CONTROL bacteria owing to the absence of oxidative stress. During the second half of the assay, the timely coincidence of thiol accretion with the increase of carbonylation in CONTROL samples may respond to a physiological strategy to keep a balanced redox status in a senescent cell. The pro‐oxidant changes induced by MDA in bacteria, including the formation of protein carbonyls, plausibly triggered the accretion of thiol groups by de novo synthesis of sulfur‐containing proteins/peptides and as a result, protect the bacterium against potential pro‐oxidant threats (Fig. 5). Thiols are typically regarded as elements of antioxidant protection in eukaryotes and also, in lactic acid bacteria (Schaefer et al., 2010; Xiao et al., 2011). Yet, the underlying molecular mechanisms behind the synthesis of thiol‐containing species remain indefinite and needs further elucidation.
The depletion of thiols observed in the following sampling points may respond to two divergent circumstances depending on the concentration of MDA. In cultures with 100 µM MDA, the control of the oxidative threat through a timely genetic response (overexpression of the uspA and particularly the dhaT genes, among others) allowed lowering the amount of protein carbonyls to physiological levels and hence, the accretion of thiols may not be required anymore and was also lowered at basal levels. In the groups of L. reuteri PL503 treated with 5 and 25 µM, an insufficient genetic response did not avoid MDA‐induced ROS generation and the subsequent protein carbonylation, and hence, the drop in thiols may respond to the consumption of such moieties in the sacrificial loss aforementioned. In probiotic bacteria such as L. reuteri, this mechanism may contribute to their capability to counteract ROS and hence, defend themselves and the host against oxidative stress.
Enclosing, the present study provides original insight into the molecular and genetic responses of L. reuteri PL503 to the toxic effects of MDA, one of the most common lipid peroxidation products. This bacterium is able to detoxify MDA and hence, exert a potential health benefit in the gastrointestinal tract. Contributing to identify some of the underlying genetic and biochemical mechanisms facilitate the development targeted prophylactic and treatment strategies involving this and other probiotic bacteria. While the uspA and dhaT genes have been found to be strongly affected by MDA and likely play a relevant role in the response of the probiotic bacteria to oxidative stress, other genes and metabolic pathways may have been affected. Therefore, further genomic studies are required to unveil which other genes and metabolic routes may be involved in the responses of probiotic bacteria to the oxidative stress induced by MDA. The health benefits of dietary supplementation may be proven in further clinical studies.
Experimental procedures
Chemicals and raw material
All chemicals and reagents used in this study were of American Chemical Society (ACS) analytical grade and purchased from Sigma Chemicals (Sigma‐Aldrich, Germany), Scharlab S.L. (Spain), Pronadisa (Conda Laboratory, Spain), Applied Biosystems (USA), Epicentre (USA) and Acros Organics (Spain). L. reuteri PL503 isolated from pig faeces was previously identified by 16S rRNA gene sequencing (Ruiz‐Moyano et al., 2008).
Experimental setting
Stock cultures of L. reuteri PL503 were stored at −80 °C in Man, Rogosa and Sharpe (MRS) broth supplied with glycerol to a final concentration of 20% (v/v). Before experimental use, L. reuteri PL503 was subcultured twice under microaerophilic conditions at 37 °C for 24 h in MRS broth supplemented with 0.5% of diluted acetic acid (10%, v/v). Four experimental groups were considered depending on the concentration of MDA added: CONTROL (L. reuteri), 5 µM (L. reuteri + 5 µM MDA), 25 µM (L. reuteri + 25 µM MDA) and 100 µM (L. reuteri + 100 µM MDA). Three replicates were carried out for each treatment. Experimental tubes were inoculated with 100 µl of the last overnight culture of L. reuteri PL503 in MRS broth and incubated at 37 °C for up to 24 h in microaerophilic conditions. The bacterial counts ranged from 9.6 to 10 log cfu ml−1 during the entire assay and no significant changes were observed between experimental groups. Samples of the cultures were collected in four times (12, 16, 20 and 24 h) from the inoculation. For further protein analyses, culture medium was removed by washing with a phosphate‐buffered saline (PBS, pH 7.4) solution twice. For counting of viable cells, 100 µl of L. reuteri PL503 was inoculated on MRS agar at the same sampling time and conditions as the experimental tubes.
Gene expression studies
Extraction of RNA
After each incubation time, 1 ml samples of each treatment were frozen and stored at –80 °C. RNA was extracted using the MasterPure™ RNA purification kit (Epicentre), which includes DNase treatment, as described by the manufacturer. Pure RNA was eluted in 35 μl Tris (200 mM) EDTA (20 mM) buffer (pH 8) and kept at –80 °C until required. The RNA concentration (ng µl−1) and purity (A260/A280 ratio) were spectrophotometrically determined using a 1.5 μl aliquot on the Nanodrop 2000 (Thermo Scientific, Waltham, MA, USA).
cDNA synthesis
cDNA was synthesized using about 500 ng of total RNA following the instructions of the PrimeScript™ RT Reagent kit (Takara Bio Inc., Kusatsu, Japan). cDNA was stored at −20 °C until further use.
Real‐time PCR analysis of gene expression
The uspA and dhaT genes were selected for relative expression studies using real‐time PCR (qPCR). The 16S gene was used as reference gene. The qPCR assays were performed and monitored in a ViiA™ 7 Real‐Time system (Applied Biosystems, Foster City, CA, USA) using MicroAmp optical 96‐well reaction plates, sealed with optical adhesive covers (Applied Biosystems). qPCR results were analysed using the Software ViiA™ 7 RUO v1.2.4. (Applied Biosystems). The SYBR Green technology was used. The reaction mixture (final volume, 12.5 µl), contained 2.5 µl of cDNA, 6.25 µl of SYBR® Premix Ex Taq™ (Takara Bio Inc.), 0.625 µl of ROX™ Reference Dye (Takara Bio Inc.) and 300 mM of each primer pair. Some primers were specifically develop for this study such as uspALr‐F1 (CTTGGGTAGCGTTCACCATT) and uspALr‐R1 (TGAAAAAGCGGTTGACACTG) for the uspA gene (annealing temperature: 60 °C) and Lr16S_F (CCGCTTAAACTCTGTTGTTG) and Lr16S_R (CGTGACTTTCTGGTTGGATA) for the 16S gene (annealing temperature: 55 °C). The primers LS67 (TGACTGGATCCTAATTTGGTCCTGGTGTTATTGC) and LS68 (TGACTGAATTCTTCCGGATCTTAGGGTTAGG) were designed in accordance to Schaefer et al. [50] for the dhaT gene (annealing temperature: 60 °C).
The qPCR programme consisted of initial denaturation step at 95 °C for 10 min at 95 °C; 40 cycles at a denaturation temperature of 95 °C for 15 s and annealing/extension temperatures of 55 °C and 60 °C for the 16S and target genes, respectively, during 30 s. After the final qPCR cycle, a melting curve was included by heating the product from 60 to 99 °C and continuous measurement of the fluorescence was performed to verify the qPCR products. All samples were analysed in triplicate, including control sample consisting of adding sterile ultrapure water instead of cDNA. The expression ratio was calculated using the 2−ΔΔC T method reported by Livak and Schmittgen (2001). The calibrator sample corresponded to the value of the expression of the experimental group CONTROL at each sampling time.
Study of in vitro reactivity of MDA with proteins
In order to evaluate the ability of MDA to induce carbonylation in proteins, three proteins, namely, human serum albumin (HSA), human haemoglobin (HH) and bovine β‐lactoglobulin (LAC) (5 mg ml−1, final concentration) were dissolved in 100 mM phosphate buffer pH 6.5 and incubated separately with MDA (0.25 µM) at 37 °C (HSA, HH) and 80 °C (LAC in an oven at constant stirring for 24 h. Proteins were selected based on their previously reported susceptibility to oxidative damage (Luna and Estévez, 2018, 2019). MDA concentration was set at preliminary tests aimed to find sublethal concentrations within the range found in colonic digests. Samples were taken at 24 h for the quantification of allysine and Schiff bases. The preparation of six experimental units corresponding to the reaction units (HSA‐MDA, HH‐MDA and LAC‐MDA) and the corresponding controls (proteins without MDA) were replicated three times in corresponding independent assay and all analyses were repeated three times in each same experimental unit (9 measurements per analysis and per treatment to calculate means and standard deviations).
Analytical procedures
ROS generation by flow cytometry analyses
Flow cytometry detection of ROS in L. reuteri PL503 was performed as determined using previous published protocols (Ortega‐Ferrusola et al., 2017; Peña et al., 2018). In brief, L. reuteri PL503 (1 × 106) was extended in 1 ml of PBS and stained with CellRox Deep Red (ThermoFisher, Waltham, MA, USA; 5 μM; excitation and emission wavelengths, 644 and 645 nm respectively) for detection of the bacterium producing ROS, and Hoechst 33342 (0.5 μM) (excitation and emission wavelengths, 345 nm and 488 nm respectively; Sigma, Steinheim, Germany) to identify the bacterium and remove debris from the analysis. After thorough mixing, the cell suspension was incubated at room temperature in the dark for 25 min, washed in PBS and the samples were immediately run on the flow cytometer. Flow cytometry analyses were conducted using a Cytoflex® flow cytometer (Beckman Coulter, Brea, CA, USA) equipped with violet, blue and red lasers. The instrument was calibrated daily using specific calibration beads provided by the manufacturer. A compensation overlap was performed before each experiment, however, due to emission and excitation characteristics of the combination of probes used, spectral overlap was negligible. Files were exported as FCS files and analysed using FlowjoV 10.5.3 Software for Mac OS (Ashland, OR, USA). Unstained, single‐stained, and Fluorescence Minus One (FMO) controls were used to determine compensations and positive and negative events, as well as to set regions of interest.
Synthesis of allysine standard compound
N‐Acetyl‐L‐AAS (allysine) was synthesized from Nα‐acetyl‐L‐lysine using lysyl oxidase activity from egg shell membrane following the procedure described by Akagawa et al. (2002). Briefly, 10 mM Nα‐acetyl‐L‐lysine was incubated under constant stirring with 5 g egg shell membrane in 50 ml of 20 mM sodium phosphate buffer, pH 9.0 at 37 °C for 24 h. The egg shell membrane was then removed by centrifugation and the pH of the solution adjusted to 6.0 using 1 M HCl. The resulting aldehydes were reductively aminated with 3 mmol ABA (4‐ aminobenzoic acid) in the presence of 4.5 mmol sodium cyanoborohydride (NaBH3CN) at 37 °C for 2 h under stirring. Then, ABA derivatives were hydrolysed by 50 ml of 12 M HCl at 110 °C for 10 h. The hydrolysates were evaporated at 40 °C in vacuo to dryness. The resulting allysine‐ABA was purified by using silica gel column chromatography and ethyl acetate/acetic acid/water (20:2:1, v/v/v) as elution solvent. The purity of the resulting solution and authenticity of the standard compounds obtained following the aforementioned procedures were checked by using MS and 1H NMR (Estévez et al., 2009).
Quantification of allysine
Five hundred microlitres of culture were dispensed in 2 ml microtubes and treated with cold (4 °C) 10% Trichloroacetic acid (TCA) solution. Each microtube was vortexed and then subjected to centrifugation at 600 g for 5 min at 4 °C. The supernatants were removed and the pellets were incubated with the following freshly prepared solutions: 0.5 ml 250 mM 2‐(N‐morpholino) ethanesulfonic acid (MES) buffer pH 6.0 containing 1 mM diethylenetriaminepentaacetic acid (DTPA), 0.5 ml 50 mM ABA in 250 mM MES buffer pH 6.0 and 0.25 ml 100 mM NaBH3CN in 250 mM MES buffer pH 6.0. The tubes were vortexed and then incubated in water bath at 37 °C for 90 min. The samples were stirred every 15 min. After derivatization, samples were treated with a cold (4 °C) 50% TCA solution and centrifuged at 1200 g for 10 min. The pellets were then washed twice with 10% TCA and diethyl ether‐ethanol (1:1). Finally, the pellet was treated with 6N HCl and kept in an oven at 110 °C for 18 h until completion of hydrolysis. The hydrolysates were dried in vacuo in a centrifugal evaporator. The generated residue was reconstituted with 200 µl of milliQ water and then filtered through hydrophilic polypropylene GH Polypro (GHP) syringe filters (0.45 μm pore size, Pall Corporation, USA) for HPLC analysis.
A Shimadzu ‘Prominence’ HPLC apparatus (Shimadzu Corporation, Japan), equipped with a quaternary solvent delivery system (LC‐20AD), a DGU‐20AS online degasser, a SIL‐20A auto‐sampler, a RF‐10A XL fluorescence detector, and a CBM‐20A system controller, was used. An aliquot (1 μl) from the reconstituted protein hydrolysates was injected and analysed in the above‐mentioned HPLC equipment. AAS‐ ABA was eluted in a Cosmosil 5C18‐AR‐II RP‐HPLC column (5 µm, 150 × 4.6 mm) equipped with a guard column (10 × 4.6 mm) packed with the same material. The flow rate was kept at 1 ml min−1 and the temperature of the column was maintained constant at 30 °C. The eluate was monitored with excitation and emission wavelengths set at 283 and 350 nm respectively. Standards (0.1 μl) were run and analysed under the same conditions. Identification of both derivatized semialdehydes in the FLD chromatograms was carried out by comparing their retention times with those from the standard compounds. The peak corresponding to allysine‐ABA was manually integrated from FLD chromatograms and the resulting areas plotted against an ABA standard curve with known concentrations that ranged from 0.1 to 0.5 mMn (Utrera et al., 2011). Results were expressed as nmol of allysine per mg of protein.
Analysis of Schiff bases
The formation of Schiff bases (SB) was assessed using a LS‐55 PerkinElmer fluorescence spectrometer (PerkinElmer, Waltham, MA, USA). Prior to the analysis, reaction mixtures were diluted (1:20) with 8 M urea in 100 mM sodium phosphate buffer, pH 7. SB was excited at 350 nm and the emitted fluorescence recorded at 450 nm. The excitation and emission slit widths were set at 10 nm and the speed of data collection while scanning was of 500 nm per min. The height of the peaks corresponding to SB spectra was recorded. After taking into consideration the applied dilutions, the results were expressed as fluorescence units.
Analysis of protein thiols
To avoid possible contamination with thiols from the medium, 250 µl of each L. reuteri PL503 culture were washed twice with PBS and with ethanol:ethyl acetate (1:1). The pellet was resuspended in 250 µl of guanidine hydrochloride and added to the cuvette in a final volume of 1250 µl of guanidine hydrochloride. Absorbance was measured at 324 nm, pre and post addition of 250 µl of 4 DPS (4,4′‐Dipyridyl disulfide) in 12 mM HCl. Results were expressed as µmol of free thiol groups per mg of protein.
Analysis of thiobarbituric‐reactive substances
Malondialdehyde and other thiobarbituric‐reactive substances (TBARS) was measured in all samples from 200 µl of each L. reuteri PL503 culture, adding 500 µl thiobarbituric acid (0.02 M) and 500 µl trichloroacetic acid (10%), incubating during 20 min at 90 °C. After cooling, a 5 min centrifugation at 600 g was made and the supernatant was measured at 532 nm. Results are expressed as mg TBARS per L of sample.
Statistical analysis
Data from the analysis (n = 3) were collected and subjected to statistical analysis. The effect of different concentrations of MDA and incubation times on the chemical measurements, analyses of variance (ANOVA) was applied [spss v. 15.5, IBM (Endicott, NY, USA)]. The effect of MDA on the gene expression (ΔΔCT values) was analysed using paired Students’ t‐tests (spss v. 15.5). The statistical significance was set at P ≤ 0.05.
Author contributions
M.E. conceived the project and finalized the manuscript. M.E., M.J.A and A.R. designed the experiments and analysed the data. P.P. collected the samples, conducted the PCR and allysine analyses and drafted the manuscript. F.J.P. conducted the cell cytometry analysis. All authors contributed to interpreting the data, reviewed and approved the submission of the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Acknowledgements
Dr. Ruiz‐Moyano is acknowledged for the donation of L. reuteri PL503.
Microb. Biotechnol. (2022) 15(2), 668–682
Funding information
This study was funded by the Spanish Ministry of Economics and Competitiveness (SMEC) through the project AGL2017‐84586R.
References
- Akagawa, M. , Sasaki, T. , and Suyama, K. (2002) Oxidative deamination of lysine residue in plasma protein of diabetic rats: novel mechanism via the Maillard reaction. Eur J Biochem 269: 5451–5458. 10.1046/j.1432-1033.2002.03243.x [DOI] [PubMed] [Google Scholar]
- Amaretti, A. , Nunzio, M. , Pompei, A. , Raimondi, S. , Rossi, M. , and Bordoni, A. (2013) Antioxidant properties of potentially probiotic bacteria: in vitro and in vivo activities. Appl Microb cell Physiol 97: 809–817. 10.1007/s00253-012-4241-7 [DOI] [PubMed] [Google Scholar]
- Arcanjo, N.O. , Andrade, M.J. , Padilla, P. , Rodríguez, A. , Madruga, M.S. , and Estévez, M. (2019) Resveratrol protects Lactobacillus reuteri against H2O2 – induced oxidative stress and stimulates antioxidant defenses through upregulation of the dhaT gene. Free Radic Biol Med 135: 38–45. 10.1016/j.freeradbiomed.2019.02.023 [DOI] [PubMed] [Google Scholar]
- Asmat, U. , Abad, K. , and Ismail, K. (2016) Diabetes mellitus and oxidative stress—a concise review. Saudi Pharm J 24: 547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros, M. , Fredriksson, Å. , Henriksson, J. , and Nyström, T. (2001) Bacterial senescence: protein oxidation in non‐proliferating cells is dictated by the accuracy of the ribosomes. EMBO J 20: 5280–5289. 10.1093/emboj/20.18.5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch, H. , and Nair, J. (2005) Accumulation of lipid peroxidation‐derived DNA lesions: potential lead markers for chemoprevention of inflammation‐driven malignancies. Mutat Res 591: 34–44. 10.1016/j.mrfmmm.2005.04.013 [DOI] [PubMed] [Google Scholar]
- Bistas, K.G. , Bistas, E. , and Mogaka, E.N. (2020) Lactobacillus reuteri's role in the prevention of colorectal cancer: a review of literature. Univ Toronto Medl J 97: 29–36. [Google Scholar]
- Davies, M.J. (2005) The oxidative environment and protein damage. Biochim Biophys Acta 1703: 93–109. 10.1016/j.bbapap.2004.08.007 [DOI] [PubMed] [Google Scholar]
- Draper, H.H. , McGirr, L.G. , and Hadley, M. (1986) The metabolism of malondialdehyde. Lipids 21: 305–307 10.1007/BF02536418 [DOI] [PubMed] [Google Scholar]
- Esterbauer, H. , Wäg, G. , and Puhl, H. (1993) Lipid peroxidation and its role in atherosclerosis. Br Med Bull 49: 566–576. [DOI] [PubMed] [Google Scholar]
- Estévez, M. (2011) Protein carbonyls in meat systems: a review. Meat Sci 89: 259–279. 10.1016/j.meatsci.2011.04.025 [DOI] [PubMed] [Google Scholar]
- Estévez, M. , Geraert, P.‐A. , Liu, R. , Delgado, J. , Mercier, Y. , and Zhang, W. (2020) Sulphur amino acids, muscle redox status and meat quality: more than building blocks – Invited review. Meat Sci 163: 108087. [DOI] [PubMed] [Google Scholar]
- Estévez, M. , and Luna, C. (2017) Dietary protein oxidation: A silent threat to human health? Crit Rev Food Sci Nutr 57: 3781–3793. 10.1080/10408398.2016.1165182 [DOI] [PubMed] [Google Scholar]
- Estévez, M. , Ollilainen, V. , and Heinonen, M. (2009) Analysis of protein oxidation markers α‐Aminoadipic and γ‐Glutamic semialdehydes in food proteins using liquid chromatography (LC)‐Electrospray ionization (ESI)‐Multistage tandem mass spectrometry (MS). J Agric Food Chem 57: 3901–3910. 10.1021/jf804017p [DOI] [PubMed] [Google Scholar]
- Estévez, M. , Padilla, P. , Carvalho, L. , Martín, L. , Carrapiso, A. , and Delgado, J. (2019) Malondialdehyde interferes with the formation and detection of primary carbonyls in oxidized proteins. Redox Biol 26: 101277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezraty, B. , Gennaris, A. , Barras, F. , and Collet, J.‐F. (2017) Oxidative stress, protein damage and repair in bacteria. Nat Rev Microbiol 15: 385. [DOI] [PubMed] [Google Scholar]
- Finkel, T. , and Holbrook, N.J. (2000) Oxidants, oxidative stress and the biology of ageing. Nature 408: 239. [DOI] [PubMed] [Google Scholar]
- Food and Agricultural Organization of the United Nations and World Health Organization . (2002) Joint FAO/WHO working group report on drafting guidelines for the evaluation of probiotics in food.
- Gackowski, D. , Banaszkiewicz, Z. , Rozalski, R. , Jawien, A. , and Olinski, R. (2002) Persistent oxidative stress in colorectal carcinoma patients. Int J cancer 101: 395–397. 10.1002/ijc.10610 [DOI] [PubMed] [Google Scholar]
- Hertzberger, R. , Arents, J. , Dekker, H.L. , Pridmore, R.D. , Gysler, C. , Kleerebezem, M. , and de Mattos, M.J.T. (2014) H2O2 production in species of the Lactobacillus acidophilus group: a central role for a novel NADH‐dependent flavin reductase. Appl Environ Microbiol 80: 2229–2239. 10.1128/AEM.04272-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjern, A. , Lindblom, K. , Reuter, A. , and Silfverdal, S.‐A. (2020) A systematic review of prevention and treatment of infantile colic. Acta Paediatr Int J Paed 109: 1733–1744. [DOI] [PubMed] [Google Scholar]
- Kazemian, N. , Mahmoudi, M. , Halperin, F. , Wu, J.C. , and Pakpour, S. (2020) Gut microbiota and cardiovascular disease: opportunities and challenges. Microbiome 8: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvint, K. , Nachin, L. , Diez, A. , and Nyström, T. (2003) The bacterial universal stress protein: function and regulation. Curr Opin Microbiol 6: 140–145. 10.1016/S1369-5274(03)00025-0 [DOI] [PubMed] [Google Scholar]
- Lin, M.T. , and Beal, M.F. (2006) Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443: 787–795. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. , and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2‐ΔΔC T method. Methods 25: 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Luna, C. , and Estévez, M. (2018) Oxidative damage to food and human serum proteins: radical‐mediated oxidation vs. glyco‐oxidation. Food Chem 267: 111–118. 10.1016/j.foodchem.2017.06.154 [DOI] [PubMed] [Google Scholar]
- Luna, C. , and Estévez, M. (2019) Formation of allysine in β ‐lactoglobulin and myofibrillar proteins by glyoxal and methylglyoxal: impact on water‐holding capacity and in vitro digestibility. Food Chem 271: 87–93. 10.1016/j.foodchem.2018.07.167 [DOI] [PubMed] [Google Scholar]
- Mane, G.P. (2016) Effect of external electron acceptors on the growth of the L. reuteri DSM 17938. Master Thesis, Lund University, Sweden.
- Marnett, L.J. (1999) Lipid peroxidation — DNA damage by malondialdehyde. Mutat Res 424: 83–95. [DOI] [PubMed] [Google Scholar]
- Martín, R. , and Suarez, J.E. (2010) Biosynthesis and degradation of H2O2 by vaginal lactobacilli. Appl Environ Microbiol 76: 400–405. 10.1128/AEM.01631-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer, L.J. , Daniels, J.S. , Rouzer, C.A. , Greene, R.E. , and Marnett, L.J. (2003) Malondialdehyde, a product of lipid peroxidation, is mutagenic in human cells. J Biol Chem 278: 31426–31433. 10.1074/jbc.M212549200 [DOI] [PubMed] [Google Scholar]
- Oberg, T.S. , Ward, R.E. , Steele, J.L. , and Broadbent, J.R. (2015) Transcriptome analysis of Bifidobacterium longum strains that show a differential response to hydrogen peroxide stress. J Biotechnol 212: 58–64. 10.1016/j.jbiotec.2015.06.405 [DOI] [PubMed] [Google Scholar]
- Ortega‐Ferrusola, C. , Anel‐López, L. , Martín‐Muñoz, P. , Ortíz‐Rodríguez, J.M. , Gil, M.C. , Alvarez, M. , et al. (2017) Computational flow cytometry reveals that cryopreservation induces spermptosis but subpopulations of spermatozoa may experience capacitation‐like changes. Reproduction 153: 293–304. 10.1530/REP-16-0539 [DOI] [PubMed] [Google Scholar]
- Peña, F.J. , Ortiz Rodriguez, J.M. , Gil, M.C. , and Ortega Ferrusola, C. (2018) Flow cytometry analysis of spermatozoa: Is it time for flow spermetry? Reprod Domest Anim 53: 37–45. 10.1111/rda.13261 [DOI] [PubMed] [Google Scholar]
- Petrella, C. (2016) Lactobacillus reuteri treatment and DSS colitis: new insight into the mechanism of protection. Acta Physiol 217: 274–275. 10.1111/apha.12719 [DOI] [PubMed] [Google Scholar]
- Requena, J.R. , Fu, M.X. , Ahmed, M.U. , Jenkins, A.J. , Lyons, T.J. , Baynes, J.W. , and Thorpe, S.R. (1997) Quantification of malondialdehyde and 4‐hydroxynonenal adducts to lysine residues in native and oxidized human low‐density lipoprotein. Biochem J 322: 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter, S. , Gupta, S.C. , Chaturvedi, M.M. , and Aggarwal, B.B. (2010) Oxidative stress, inflammation, and cancer: How are they linked? Free Radic Biol Med 49: 1603–1616. 10.1016/j.freeradbiomed.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz‐Moyano, S. , Martín, A. , Benito, M.J. , Pérez‐Nevado, F. , and Córdoba, M.D.G. (2008) Screening of lactic acid bacteria and bifidobacteria for potential probiotic use in Iberian dry fermented sausages. Meat Sci 80: 715–721. 10.1016/j.meatsci.2008.03.011 [DOI] [PubMed] [Google Scholar]
- Sanders, J.G. , Powell, S. , Kronauer, D.J.C. , Vasconcelos, H.L. , Frederickson, M.E. , and Pierce, N.E. (2014) Stability and phylogenetic correlation in gut microbiota: lessons from ants and apes. Mol Ecol 23: 1268–1283. 10.1111/mec.12611 [DOI] [PubMed] [Google Scholar]
- Schaefer, L. , Auchtung, T.A. , Hermans, K.E. , Whitehead, D. , Borhan, B. , and Britton, R.A. (2010) The antimicrobial compound reuterin (3‐hydroxypropionaldehyde) induces oxidative stress via interaction with thiol groups. Microbiology 156: 1589–1599. 10.1099/mic.0.035642-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shacter, E. (2000) Quantification and significance of protein oxidation in biological samples. Drug Metabol Rev 32: 307–326. [DOI] [PubMed] [Google Scholar]
- Singh, A.K. , Hertzberger, R.Y. , and Knaus, U.G. (2018) Redox biology hydrogen peroxide production by lactobacilli promotes epithelial restitution during colitis. Redox Biol 16: 11–20. 10.1016/j.redox.2018.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyropoulos, B. , Misiakos, E. , Fotiadis, C. , and Stoidis, C. (2011) Antioxidant properties of probiotics and their protective effects in the pathogenesis of radiation‐induced enteritis and colitis. Dig Dis Sci 56: 285–294. 10.1007/s10620-010-1307-1 [DOI] [PubMed] [Google Scholar]
- Talarico, T.L. , Casas, I.A. , Chung, T.C. , and Dobrogosz, W.J. (1988) Production and isolation of reuterin, a growth inhibitor produced by Lactobacillus reuteri . Antimicrob Agents Chemother 32: 1854–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utrera, M. , and Estevez, M. (2013) Impact of trolox, quercetin, genistein and gallic acid on the oxidative damage to myofibrillar proteins: the carbonylation pathway. Food Chem 141: 4000–4009. [DOI] [PubMed] [Google Scholar]
- Utrera, M. , Morcuende, D. , Rodríguez‐Carpena, J.G. , and Estévez, M. (2011) Fluorescent HPLC for the detection of specific protein oxidation carbonyls – α‐aminoadipic and γ‐glutamic semialdehydes‐ in meat systems. Meat Sci 89: 500–506. 10.1016/j.meatsci.2011.05.017 [DOI] [PubMed] [Google Scholar]
- Valko, M. , Morris, H. , and Cronin, M. (2005) Metals, toxicity and oxidative stress. Curr Med Chem 12: 1161–1208. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Zhou, C. , Huang, J. , Kuai, X. , and Shao, X. (2020) The potential therapeutic role of Lactobacillus reuteri for treatment of inflammatory bowel disease. Am J Transl Res 12: 1569–1583. [PMC free article] [PubMed] [Google Scholar]
- Xiao, M. , Xu, P. , Zhao, J. , Wang, Z. , Zuo, F. , Zhang, J. , et al. (2011) Oxidative stress‐related responses of Bifidobacterium longum subsp. longum BBMN68 at the proteomic level after exposure to oxygen. Microbiology 157: 1573–1588. 10.1099/mic.0.044297-0 [DOI] [PubMed] [Google Scholar]
- Zhu, H. , and Li, Y.R. (2012) Oxidative stress and redox signaling mechanisms of inflammatory bowel disease: updated experimental and clinical evidence. Exp Biol Med 237: 474–480. 10.1258/ebm.2011.011358 [DOI] [PubMed] [Google Scholar]


