Abstract
Super-short rotavirus strains that have a rearranged gene segment 11 are rarely found in humans, and only five isolates, all from Southeast Asia, have been described in the literature. We report the first isolation in Japan from an infant with severe diarrhea of a rotavirus possessing a super-short RNA pattern. This strain, designated AU19, had a G1 VP7 and is also the first isolate in Japan that possesses a P2[6] VP4. Furthermore, the P2[6] VP4 carried by AU19 was divergent in the hypervariable region of the amino acid sequence from the P2A[6] VP4s carried by asymptomatic neonatal strains or from the P2B[6] VP4 carried by porcine rotavirus strain Gottfried. Thus, AU19 is likely to represent a new VP4 subtype, which we propose to call P2C. Given the recent emergence of the P2[6] VP4s in India, Brazil, and the United States and the role of VP4 in protective immunity, further scrutiny is justified to see whether the emergence of the previously underrepresented P2[6] VP4 serotype is related to this new P2 subtype.
Group A rotavirus, belonging to the genus Rotavirus within the family Reoviridae, is the single most important etiological agent of acute gastroenteritis in infants and young children worldwide (15). The rotavirus genome consists of 11 segments of double-stranded RNA which are separated upon polyacrylamide gel electrophoresis. The RNA migration pattern, termed electropherotype, is unique to each isolate and has been used extensively in the study of rotavirus molecular epidemiology (12). Among a myriad of electropherotypes, long, short, and super-short RNA patterns are identified based on the relative migration rates of gene segments 10 and 11 (15). Both short and super-short rotaviruses have a rearranged gene segment 11 (4, 24). However, super-short human rotaviruses are very rare, and only five strains isolated in Indonesia and Thailand were described previously in the literature (1, 25, 41). Here, we report the first isolation in Japan from an infant with severe diarrhea of a human rotavirus possessing a super-short RNA pattern. This strain, designated AU19, shared neither G nor P serotype with the previously identified super-short human rotaviruses but was found to possess serotype G1P2[6], making AU19 further unique in that it is the first P2[6] isolate in Japan.
Cell culture adaptation of rotaviruses was performed essentially as described previously (17).
G and P serotypes were initially predicted by the typing method based on reverse transcription-PCR according to the published methods (10, 11). The molecularly predicted serotypes were then confirmed by plaque reduction neutralization assays with monoclonal antibodies. Monoclonal antibodies to identify G1 and P2 serotypes were 5E8 (30) and HS11 (31), respectively. Hyperimmune sera were also used to determine the G serotype and to examine the serologic relatedness via VP4 with the reference strains possessing the P2 VP4. Plaque reduction neutralization assays were performed as previously described (29). Briefly, approximately 40 PFU of the virus, which had been activated for 1.5 h with 10 μg of trypsin (type IX; Sigma Chemical Company, St. Louis, Mo.) per ml, were incubated for 1 h at 37°C with serially diluted monoclonal antibody or hyperimmune serum. Each mixture was inoculated into the monolayer cells in six-well plastic dishes and overlaid with agarose containing 0.5 μg of trypsin per ml. When plaques were developed, the number of plaques was counted after neutral red staining. Neutralization titer was expressed as the highest dilution of serum neutralizing 60% or more of the input virus. The hyperimmune antisera to AU19 were made in guinea pigs by three monthly injections of the purified AU19 virions emulsified with complete Freund’s adjuvant (for priming) or incomplete Freund’s adjuvant (for booster injections).
Virus particles were purified from virus-infected cell cultures by pelleting at 36,000 rpm for 3 h in a Beckman type 45Ti rotor followed by sedimentation through 30% (wt/vol) sucrose at 38,000 rpm for 3 h in a Beckman type SW41Ti rotor.
Genomic RNA was extracted with phenol-chloroform from purified virions and reverse transcribed with a pair of primers, HumCom5 (5′-CTCTCGATGGTCCATATCAACC-3′) and HumCom3 (5′-TCCTTGTATTCTGAATTGGTGG-3′) to obtain the cDNA containing the hypervariable region of VP4 (amino acids [aa] 71 to 203) (11). The cDNA was amplified by PCR with the same primer pair, cloned into pCR2.1 vector (Invitrogen, San Diego, Calif.), and sequenced on an ABI PRISM 310 automated DNA sequencer (The Perkin-Elmer Corporation, Foster City, Calif.). Three independent cDNA clones were sequenced mostly on both strands. Sequence analysis was performed with the aid of the GeneWorks version 2.5 software package (IntelliGenetics, Campbell, Calif.). A phylogenetic tree based on the neighbor-joining method of Saitou and Nei (34) was drawn with the N-J plot program in the Clustal W package (39).
The reference rotavirus strains used in this study were Wa (G1, P1A[8]) (42), KUN (G2, P1B[4]) (17), AU64 (G1, P1B[4]) (27), M37 (G1, P2A[6]) (14), AU007 (G1, P1A[8]) (28), 1076 (G2, P2A[6]) (14), ST3 (G4, P2A[6]) (14), VA70 (G4, P1A[8]) (7), 69M (G8, P4[10]) (25), and SA11 (G3, P5B[2]) (22).
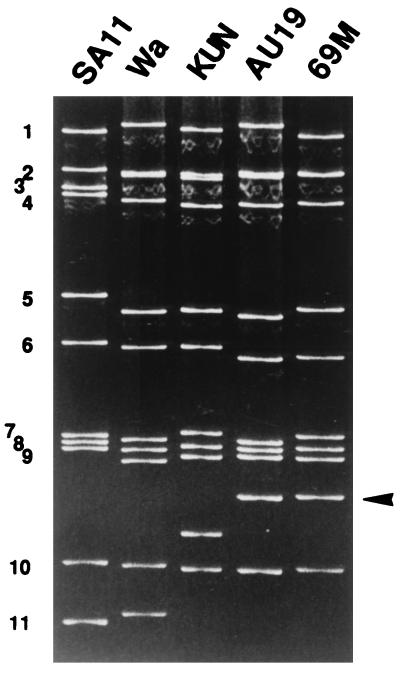
Stool specimen 97A019 was obtained from a 13-month-old girl who was admitted to the Akita University Hospital in September 1997 because of severe diarrhea and dehydration. Since this specimen was shown by a latex agglutination assay to contain rotavirus, the genomic RNA was extracted and the electropherotype was determined by polyacrylamide gel electrophoresis. This routine analysis disclosed the presence of a rotavirus strain with a super-short pattern. This strain, designated AU19, was culture adapted and triply plaque purified before serologic and molecular characterization. The electropherotype of this triply plaque-purified AU19 (Fig. 1) was identical with that of the RNA extracted from stool specimen 97A019 (data not shown). We therefore excluded the possibility of genome rearrangement during the culture adaptation.
FIG. 1.
The electropherotype of AU19 in comparison with reference laboratory strains. SA11 and Wa possess long RNA patterns, KUN possesses a short RNA pattern, and 69M is the prototype super-short strain. Note the lowest migration rates of gene segment 10 of AU19 as well as 69M (indicated by an arrowhead).
The G and P serotypes of AU19 were predicted by the reverse transcription-PCR performed according to the method of Gouvea et al. (10) and Gunasena et al. (11), respectively. Both G and P typing assays yielded unambiguous results indicating that AU19 belonged to G1 and P2[6] (data not shown).
To confirm the molecular prediction, standard plaque reduction neutralization assays were performed with both serotype-specific monoclonal antibodies and hyperimmune sera raised against AU19 as well as reference strains. Serotype G1-specific monoclonal antibody 5E8 neutralized AU19 at a titer of 21,914. The definition for including a given strain in an established G serotype is a reciprocal 20-fold or smaller difference in neutralization titer when the strain is tested against reference strains representing the established serotype (15). Thus, AU19 was tested by reciprocal neutralization assays against two G1 strains routinely used in our laboratory, i.e., AU64 (G1, P1B[4]) and AU007 (G1, P1A[8]). As shown in Table 1, anti-AU19 hyperimmune serum neutralized AU64 and AU007 at a titer 16-fold lower than the homologous neutralization titer, while anti-AU64 hyperimmune serum neutralized AU19 at the same titer as the homologous titer and anti-AU007 hyperimmune serum neutralized AU19 at a titer fourfold lower than the homologous titer. Thus, two-way cross-neutralization was observed between AU19 and two known G1 strains, and AU19 was established to belong to G1.
TABLE 1.
Serologic characterization of AU19 by the plaque reduction neutralization test
| Virus | Serotype | Reciprocal of neutralization titer of antiserum to:
|
||
|---|---|---|---|---|
| AU19 | AU64 | AU007 | ||
| AU19 | 102,400 | 25,600 | 25,600 | |
| AU64 | G1, P1B[4] | 6,400 | 25,600 | NTa |
| AU007 | G1, P1A[8] | 6,400 | NT | 102,400 |
NT, not tested.
As to the P serotype of AU19, serotype P2-specific monoclonal antibody HS11 neutralized AU19 at a titer of 1,290. Although the serologic definition of P type equivalent to that of G serotype is not established, it is known that a shared P serotype specificity sometimes results in one-way or, less frequently, two-way neutralization between strains possessing different G serotypes (13, 23). To examine whether there is any neutralization by way of VP4, we tested four strains possessing various combinations of G and P serotypes against anti-AU19 hyperimmune serum (Table 2). Anti-AU19 neutralized M37 at a titer 16-fold lower than the homologous neutralization titer, but this neutralization was considered to be mediated by VP7, because anti-AU19 failed to neutralize either 1076 or ST3 within the 20-fold difference from the homologous titer. Both 1076 and ST3 belong to P2, but they have G serotypes different from that of AU19. Given that anti-AU19 did not at all neutralize VA70, whose G and P serotypes were different from those of AU19, we could not exclude the possibility that the very weak neutralization (1/64 of the homologous neutralization) of anti-AU19 against 1076 and ST3 was mediated by VP4.
TABLE 2.
Neutralization activity of the anti-AU19 hyperimmune serum against the reference strains
| Strain | G type | P type | Reciprocal of neutralization titer |
|---|---|---|---|
| AU19 | G1 | 102,400 | |
| M37 | G1 | P2A[6] | 6,400 |
| 1076 | G2 | P2A[6] | 1,600 |
| ST3 | G4 | P2A[6] | 1,600 |
| VA70 | G4 | P1A[8] | <800 |
The absence of significant neutralization between AU19 and the reference P2 strains may reflect only the weaker immunogenicity of VP4. Rotavirus hyperimmune sera usually contain more antibodies directed at VP7 than at VP4 (5). An alternative possibility is that the VP4 sequence of AU19 may be slightly divergent from those of the reference P2 strains. To test the latter hypothesis, we sequenced the VP4 of AU19 and compared the deduced amino acid sequence of the hypervariable region (spanning aa residues 71 to 203) with those of previously sequenced P2[6] strains. As shown in Table 3, AU19 had amino acid identities of 73 to 80% with other P2[6] strains, whereas amino acid identities between M37 and other P2[6] strains except AU19 and porcine rotavirus strain Gottfried were much higher (95 to 98%). Porcine rotavirus Gottfried was shown by serology to be slightly different from the neonatal P2A strains, and hence it was assigned to P2B. Upon entire VP4 sequence comparison, Gottfried was 87 to 88% identical with the neonatal strains and classified in the P[6] genotype (9). However, Gottfried was only 69 to 70% identical with the P2A strains at this hypervariable region (Table 3).
TABLE 3.
Comparison of amino acid sequence identities between P2[6] strains
| Strain | P type | G type | % Amino acid identity with:
|
||
|---|---|---|---|---|---|
| AU19 | M37 | Gottfried | |||
| AU19 | P2[6] | G1 | 100 | 78 | 73 |
| M37 | P2A[6] | G1 | 78 | 100 | 70 |
| VE7156 | P2[6] | G1 | 78 | 98 | 70 |
| 1076 | P2A[6] | G2 | 78 | 95 | 70 |
| SC2 | P2[6] | G2 | 77 | 95 | 69 |
| McN | P2A[6] | G3 | 79 | 95 | 70 |
| RV3 | P2[6] | G3 | 80 | 96 | 70 |
| MtA5 | P2A[6] | G3 | 80 | 96 | 70 |
| MtB2 | P2A[6] | G4 | 80 | 96 | 70 |
| ST3 | P2A[6] | G4 | 77 | 96 | 70 |
| Gottfried | P2B[6] | G4 | 73 | 70 | 100 |
When the AU19 sequence was aligned with the other P2 sequences, 12 unique substitutions were identified at the hypervariable region (data not shown). However, none of these unique substitutions coincided with the neutralization epitopes identified previously by the neutralization escape mutants (21).
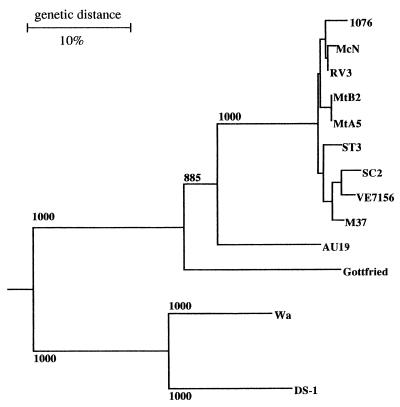
The relationships between AU19 and the P2A strains as well as the Gottfried strain were confirmed by phylogenetic analysis. In the phylogenetic tree drawn based on the amino acid sequences of the hypervariable region (Fig. 2), AU19 was placed on the lineage clearly distinct from either the P2A strains or porcine strain Gottfried (P2B). This tree also indicates that all P2[6] strains including AU19 and Gottfried belong to a single cluster with a high bootstrap probability that is clearly distinct from other VP4 genotypes such as P[8] (Wa) and P[4] (DS-1).
FIG. 2.
A phylogenetic tree for the hypervariable region of VP4 constructed by the neighbor-joining method. The horizontal distance between two strains is proportional to the genetic distance between these two strains. The distance is expressed as the number of substitutions per 100 sites (percent divergence). Bootstrap probabilities equal to or greater than 850 (of 1,000 trials) are indicated.
The major characteristics of AU19, a human rotavirus strain possessing a super-short RNA pattern isolated for the first time outside Southeast Asia, are that it possesses a variant of genotype P[6] VP4 and that it probably represents a new P2 subtype.
Rotavirus serotypes are defined by the antigenic specificity of two outer capsid proteins, i.e., VP4 and VP7 (15). While the numbering of the G serotype agrees with the corresponding VP7 genotype, the VP4 genotype is designated by including the number in square brackets after the P term (5). Human rotaviruses carrying P2[6] VP4 were initially isolated from asymptomatic neonates (6, 8, 14). Subsequent studies have shown, however, that symptomatic children also shed rotaviruses possessing the P2[6] VP4 (2, 18, 26, 32, 33, 36–38). Within these P2[6] strains, irrespective of whether they were isolated from symptomatic children or from asymptomatic neonates, the amino acid sequences are highly conserved, with identities in the VP8* region (which contains the hypervariable region) between 92.7 and 99.7% (35). Similarly, the hypervariable region of both nine asymptomatic strains and five symptomatic strains from Australia obtained between 1974 and 1986 was shown to have only a few amino acid substitutions, and their sequences are highly homologous to that of the prototype strain RV3 (16).
Porcine rotavirus strain Gottfried, originally isolated from the intestinal contents of a suckling pig with diarrhea (3), was shown to have VP4 that was 87 to 88% homologous to the VP4s of the P2[6] asymptomatic neonatal strains (9). Based on one-way neutralization, the Gottfried strain was proposed to be a subtype of P2, i.e., P2B, and the P2 specificity shared by all asymptomatic neonatal strains was proposed to be P2A (20).
A comparison of the deduced VP4 amino acid sequences of a wide variety of rotaviruses shows a hypervariable region spanning aa 71 to 203 (5), with P serotype specificity mapping within this hypervariable region (aa 92 to 192) (19). Thus, a comparison of the deduced amino acid sequences of the hypervariable region of VP4 can provide information closely associated with the P type-specific neutralization. High amino acid homologies in this hypervariable region (94 to 100%) observed between P2A[6] strains indicate that they all belong to a single P2A subtype, although neutralization assays using hyperimmune sera raised against baculovirus-expressed VP4 proteins have not been performed thus far on any of the symptomatic P2[6] strains. The sequence of AU19 shows that it is almost equally distant from both P2A and P2B strains and that the distances were significantly greater than those commonly observed among individual members belonging to the same subtype (Table 3). On the other hand, AU19 is a member of the P2 strains because one monoclonal antibody that was shown to neutralize P2 strains (30) neutralized AU19. Taken together, these observations strongly suggest that AU19 belongs to neither P2A nor P2B but represents a previously unidentified P2 subtype, which we propose to call P2C.
The epidemiological significance of this single isolate is not clear. A recent nationwide surveillance in the United States showed that P[6] strains were identified in 6.9% of rotavirus-positive specimens from diarrheal children (33). The G serotype of these P[6] strains was reported to be either G1 (1.4%) or G9 (5.5%). Whether any of the G1P[6] strains had super-short RNA patterns was not reported. It is not known, either, whether any of the P[6] VP4s had a variant P2[6] similar to AU19. However, an increasing number of P2[6] strains from symptomatic children have been reported in India and Brazil. In India, 43% (27 of 63) of rotavirus-positive stool specimens contained P[6] strains (32), and 13% (18 of 139) were genotyped as P[6] in Brazil (40). Since VP4 plays a role in protective immunity, it warrants further scrutiny whether the apparent increase in the prevalence of the previously underrepresented P2[6] VP4 serotype is related to the emergence of this new P2 subtype.
Nucleotide sequence accession number.
The nucleotide sequence of the hypervariable region of AU19 VP4 has been deposited in the GenBank, EMBL, and DDBJ sequence databases and given accession no. AB017917.
Acknowledgments
We acknowledge the excellent technical assistance provided by Reietsu Ito and Yoko Nakamura.
This study was supported in part by a grant-in-aid for Scientific Research from the Ministry of Education, Science, Sport and Culture, Japan.
REFERENCES
- 1.Albert M J, Unicomb L E, Bishop R F. Cultivation and characterization of human rotaviruses with “supershort” RNA patterns. J Clin Microbiol. 1987;25:183–185. doi: 10.1128/jcm.25.1.183-185.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bern C, Unicomb L, Gentsch J R, Banul N, Yunus M, Sack R B, Glass R I. Rotavirus diarrhea in Bangladeshi children: correlation of disease severity with serotypes. J Clin Microbiol. 1992;30:3234–3238. doi: 10.1128/jcm.30.12.3234-3238.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohl E H, Theil K W, Saif L J. Isolation and serotyping of porcine rotaviruses and antigenic comparison with other rotaviruses. J Clin Microbiol. 1984;19:105–111. doi: 10.1128/jcm.19.2.105-111.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dyall-Smith M L, Holmes I H. Gene-coding assignments of rotavirus double-stranded RNA segments 10 and 11. J Virol. 1981;38:1099–1103. doi: 10.1128/jvi.38.3.1099-1103.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Estes M K. Rotaviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1625–1655. [Google Scholar]
- 6.Flores J, Midthun K, Hoshino Y, Green K, Gorziglia M, Kapikian A Z, Chanock R M. Conservation of the fourth gene among rotaviruses recovered from asymptomatic newborn infants and its possible role in attenuation. J Virol. 1986;60:972–979. doi: 10.1128/jvi.60.3.972-979.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerna G, Passarani N, Battaglia M, Percivalle E. Rapid serotyping of human rotavirus strains by solid-phase immune electron microscopy. J Clin Microbiol. 1984;19:273–278. doi: 10.1128/jcm.19.2.273-278.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorziglia M, Green K, Nishikawa K, Taniguchi K, Jones R, Kapikian A Z, Chanock R M. Sequence of the fourth gene of human rotaviruses recovered from asymptomatic or symptomatic infections. J Virol. 1988;62:2978–2984. doi: 10.1128/jvi.62.8.2978-2984.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorziglia M, Nishikawa K, Hoshino Y, Taniguchi K. Similarity of the outer capsid protein VP4 of the Gottfried strain of porcine rotavirus to that of asymptomatic human rotavirus strains. J Virol. 1990;64:414–418. doi: 10.1128/jvi.64.1.414-418.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gouvea V, Glass R I, Woods P, Taniguchi K, Clark H F, Forrester B, Fang Z Y. Polymerase chain reaction amplification and typing of rotavirus nucleic acids from stool specimens. J Clin Microbiol. 1990;28:276–282. doi: 10.1128/jcm.28.2.276-282.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gunasena S, Nakagomi O, Isegawa Y, Kaga E, Nakagomi T, Steele A D, Flores J, Ueda S. Relative frequency of VP4 gene alleles among human rotaviruses recovered over a 10-year period (1982–1991) from Japanese children with diarrhea. J Clin Microbiol. 1993;31:2195–2197. doi: 10.1128/jcm.31.8.2195-2197.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes I H. Development of rotavirus molecular epidemiology: electropherotyping. Arch Virol. 1996;12(Suppl.):87–91. doi: 10.1007/978-3-7091-6553-9_10. [DOI] [PubMed] [Google Scholar]
- 13.Hoshino Y, Sereno M M, Midthun K, Flores J, Kapikian A Z, Chanock R M. Independent segregation of two antigenic specificities (VP3 and VP7) involved in neutralization of rotavirus infectivity. Proc Natl Acad Sci USA. 1985;82:8701–8704. doi: 10.1073/pnas.82.24.8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoshino Y, Wyatt R G, Flores J, Midthun K, Kapikian A Z. Serotypic characterization of rotaviruses derived from asymptomatic human neonatal infection. J Clin Microbiol. 1985;21:425–430. doi: 10.1128/jcm.21.3.425-430.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapikian A Z, Chanock R M. Rotaviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1657–1708. [Google Scholar]
- 16.Kirkwood D D, Coulson B S, Bishop R F. G3P2 rotaviruses causing diarrhoeal disease in neonates differ in VP4, VP7 and NSP4 sequence from G3P2 strains causing asymptomatic neonatal infection. Arch Virol. 1996;141:1661–1676. doi: 10.1007/BF01718290. [DOI] [PubMed] [Google Scholar]
- 17.Kutsuzawa T, Konno T, Suzuki H, Kapikian A Z, Ebina T, Ishida N. Isolation of human rotavirus subgroups 1 and 2 in cell culture. J Clin Microbiol. 1982;16:727–730. doi: 10.1128/jcm.16.4.727-730.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larralde G, Flores J. Identification of gene 4 alleles among human rotaviruses by polymerase chain reaction-derived probes. Virology. 1990;179:469–473. doi: 10.1016/0042-6822(90)90317-k. [DOI] [PubMed] [Google Scholar]
- 19.Larralde G, Li B, Kapikian A Z, Gorziglia M. Serotype-specific epitope(s) present on the VP8 subunit of rotavirus VP4 protein. J Virol. 1991;65:3213–3218. doi: 10.1128/jvi.65.6.3213-3218.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li B, Gorziglia M. VP4 serotype of the Gottfried strain of porcine rotavirus. J Clin Microbiol. 1993;31:3075–3077. doi: 10.1128/jcm.31.11.3075-3077.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mackow E R, Shaw R D, Matsui S M, Vo P T, Dang M-N, Greenberg H B. The rhesus rotavirus gene encoding protein VP3: location of amino acids involved in homologous and heterologous rotavirus neutralization and identification of a putative fusion region. Proc Natl Acad Sci USA. 1988;85:645–649. doi: 10.1073/pnas.85.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malherbe H, Harwin R, Ulrich M. The cytopathic effects of vervet monkey viruses. S Afr Med J. 1963;37:407–411. [PubMed] [Google Scholar]
- 23.Matsuda Y, Isegawa Y, Woode G N, Zheng S, Kaga E, Nakagomi T, Ueda S, Nakagomi O. Two-way cross-neutralization mediated by a shared P (VP4) serotype between bovine rotavirus strains with distinct G (VP7) serotypes. J Clin Microbiol. 1993;31:354–358. doi: 10.1128/jcm.31.2.354-358.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsui S M, Mackow E R, Matsuno S, Paul P S, Greenberg H B. Sequence analysis of gene 11 equivalents from “short” and “super-short” strains of rotavirus. J Virol. 1990;64:120–124. doi: 10.1128/jvi.64.1.120-124.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuno S, Hasegawa A, Mukoyama A, Inouye S. A candidate for a new serotype of human rotavirus. J Virol. 1985;54:623–624. doi: 10.1128/jvi.54.2.623-624.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mphahlele M J, Steele A D. Relative frequency of human rotavirus VP4(P) genotypes recovered over a 10-year period from South African children with diarrhea. J Med Virol. 1995;47:1–5. doi: 10.1002/jmv.1890470102. [DOI] [PubMed] [Google Scholar]
- 27.Nakagomi O, Nakagomi T. Molecular evidence for naturally occurring single VP7 gene substitution reassortant between human rotaviruses belonging to two different genogroups. Arch Virol. 1991;119:67–81. doi: 10.1007/BF01314324. [DOI] [PubMed] [Google Scholar]
- 28.Nakagomi, T. Unpublished observation.
- 29.Nakagomi T, Nakagomi O, Streckert H-J. Bovine rotavirus strain 678 possesses VP4 of serotype P7[5] specificity. Arch Virol. 1997;142:1255–1262. doi: 10.1007/s007050050157. [DOI] [PubMed] [Google Scholar]
- 30.Padilla-Noriega L, Arias C F, Lopez S, Puerto F, Snodgrass D R, Taniguchi K, Greenberg H B. Diversity of rotavirus serotypes in Mexican infants with gastroenteritis. J Clin Microbiol. 1990;28:1114–1119. doi: 10.1128/jcm.28.6.1114-1119.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Padilla-Noriega L, Werner-Eckert R, Mackow E R, Gorziglia M, Larralde G, Taniguchi K, Greenberg H B. Serologic analysis of human rotavirus serotypes P1A and P2 by using monoclonal antibodies. J Clin Microbiol. 1993;31:622–628. doi: 10.1128/jcm.31.3.622-628.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramachandran M, Das B K, Vij A, Kumar R, Bhambal S S, Kesari N, Rawat H, Bahl L, Thakur S, Woods P A, Glass R I, Bhan M, Gentsch J R. Unusual diversity of human rotavirus G and P genotypes in India. J Clin Microbiol. 1996;34:436–439. doi: 10.1128/jcm.34.2.436-439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramachandran M, Gentsch J R, Parashar U D, Jin S, Woods P A, Holmes J L, Kirkwood C D, Bishop R F, Greenberg H B, Urasawa S, Gerna G, Coulson B S, Taniguchi K, Bresee J S, Glass R I The National Rotavirus Strain Surveillance System Collaborating Laboratories. Detection and characterization of novel rotavirus strains in the United States. J Clin Microbiol. 1998;36:3223–3229. doi: 10.1128/jcm.36.11.3223-3229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saitou N, Nei M. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 35.Santos N, Gouvea V, Timenetsky M C, Clark H F, Riepenhoff-Talty M, Garbarg-Chenon A. Comparative analysis of VP8* sequences from rotaviruses possessing M37-like VP4 recovered from children with and without diarrhoea. J Gen Virol. 1994;75:1775–1780. doi: 10.1099/0022-1317-75-7-1775. [DOI] [PubMed] [Google Scholar]
- 36.Santos N, Riepenhoff-Talty M, Clark H F, Offit P, Gouvea V. VP4 genotyping of human rotavirus in the United States. J Clin Microbiol. 1994;32:205–208. doi: 10.1128/jcm.32.1.205-208.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steele A D, Garcia D, Sears J, Gerna G, Nakagomi O, Flores J. Distribution of VP4 gene alleles in human rotaviruses by using probes to the hyperdivergent region of the VP4 gene. J Clin Microbiol. 1993;31:1735–1740. doi: 10.1128/jcm.31.7.1735-1740.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steele A D, van Niekerk M C, Mphahlele M J. Geographic distribution of human rotavirus VP4 genotypes and VP7 serotypes in five South African regions. J Clin Microbiol. 1995;33:1516–1519. doi: 10.1128/jcm.33.6.1516-1519.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Timenetsky M C S T, Santos N, Gouvea V. Survey of rotavirus G and P types associated with human gastroenteritis in São Paulo, Brazil, from 1986 to 1992. J Clin Microbiol. 1994;32:2622–2624. doi: 10.1128/jcm.32.10.2622-2624.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Urasawa S, Hasegawa A, Urasawa T, Taniguchi K, Wakasugi F, Suzuki H, Inouye S, Pongprot B, Supawadee J, Suprasert S, Rangsiyanond P, Tonusin S, Yamazi Y. Antigenic and genetic analyses of human rotaviruses in Chian Mai, Thailand: evidence for a close relationship between human and animal rotaviruses. J Infect Dis. 1992;166:227–234. doi: 10.1093/infdis/166.2.227. [DOI] [PubMed] [Google Scholar]
- 42.Wyatt R, James W, Bohl E, Theil K, Saif L, Kalica A, Greenberg H, Kapikian A, Chanock R. Human rotavirus type 2: cultivation in vitro. Science. 1980;207:189–191. doi: 10.1126/science.6243190. [DOI] [PubMed] [Google Scholar]