Abstract
The major metabolite of the anticancer agent 5-fluorouracil (5-FU) is 5-fluorodeoxyuridine monophosphate (FdUMP), which is a potent inhibitor of thymidylate synthase (TS). Recently, we hypothesized that 5-FU-resistant colorectal cancer (CRC) cells have increased levels of TS protein relative to 5-FU-sensitive CRC cells and use a fraction of their TS to trap FdUMP, which results in resistance to 5-FU. In this study, we analyzed the difference between the regulation of the balance of the free, active form of TS and the inactive FdUMP-TS form in 5-FU-resistant HCT116 cells and parental HCT116 cells. Silencing of TYMS, the gene that encodes TS, resulted in greater enhancement of the anticancer effect of 5-FU in the 5-FU-resistant HCT116RF10 cells than in the parental HCT116 cells. In addition, the trapping of FdUMP by TS was more effective in the 5-FU-resistant HCT116RF10 cells than in the parental HCT116 cells. Our observations suggest that the regulation of the balance between the storage of the active TS form and the accumulation of FdUMP-TS is responsible for direct resistance to 5-FU. The findings provide a better understanding of 5-FU resistance mechanisms and may enable the development of anticancer strategies that reverse the sensitivity of 5-FU resistance in CRC cells.
Introduction
5-Fluorouracil (5-FU) is a key anticancer drug used for the chemotherapy of colorectal cancer (CRC).1,2 In the body, 5-FU is converted to 5-fluorodeoxyuridine monophosphate (FdUMP), which is a potent inhibitor of thymidylate synthase (TS).2−4 TS, encoded by the TYMS gene in humans, catalyzes the conversion of dUMP to dTMP using the co-substrate 5,10-methylenetetrahydrofolate (CH2-THF).5 FdUMP forms a covalent ternary complex with TS and CH2-THF.1,2,4,6−8 This covalent ternary complex inhibits TS, depletes the intracellular dTTP pool, and subsequently inhibits DNA synthesis.1−4 In addition, 5-FU can exert cytotoxic effects through its incorporation into DNA and RNA as fluorodeoxyuridine triphosphate (FdUTP) and fluorouridine triphosphate (FUTP), respectively.1−3
Cancer cells are known to acquire resistance to anticancer drugs through a variety of mechanisms. The common cancer resistance mechanisms include inactivation of drugs, enhancement of drug efflux, alteration of drug target molecules, utilization of bypass pathways, facilitation of DNA damage repair, and escaping cell death.1,2,9 Many studies have examined the mechanisms of resistance to 5-FU and its derivatives.1,2,9 The function and/or expression of TS and other enzymes related to the 5-FU anabolism or catabolism pathways are often altered, accelerating resistance to 5-FU.1,2,9−11 In addition, the known mechanisms of 5-FU resistance are perturbance of cell death and autophagy, altered epigenetic repression, and expression/functional changes in drug transporters and noncoding RNA (i.e., microRNA and long noncoding RNA).1,2,9 It is widely considered that TS is part of an important molecular mechanism that enhances 5-FU sensitivity and that targeting TS is an excellent strategy to reverse 5-FU resistance.1,2,12 Indeed, numerous studies have shown that the gene amplification of TYMS, leading to mRNA and protein overexpression is a major mechanism of resistance to 5-FU and its derivatives.12−15 In addition, we have shown that 5-FU-resistant CRC cells increase TYMS expression relative to 5-FU-sensitive CRC cells and use a fraction of TS to trap FdUMP, which results in resistance to 5-FU and its derivatives.16 We predict that the regulation of TS status, which refers to the balance between the active free-TS form and the inactive FdUMP-TS covalent complex, may confer 5-FU resistance.16
In this study, we investigated the anticancer sensitivity of the 5-FU-resistant HCT116 cells and the parental HCT116 cells to 5-FU after TYMS knockdown. In addition, we analyzed the difference in the regulation of the balance between the active free-TS form and the inactive FdUMP-TS form in 5-FU-resistant HCT116 cells and the parental HCT116 cells. We discussed the possibility of the FdUMP trapping by the TS protein as one of the mechanisms of 5-FU resistance.
Results
Knockdown of TYMS Enhances the Anticancer Effect of 5-FU on 5-FU-Resistant HCT116RF10 Cells Compared with the Effect on Parental HCT116 Cells
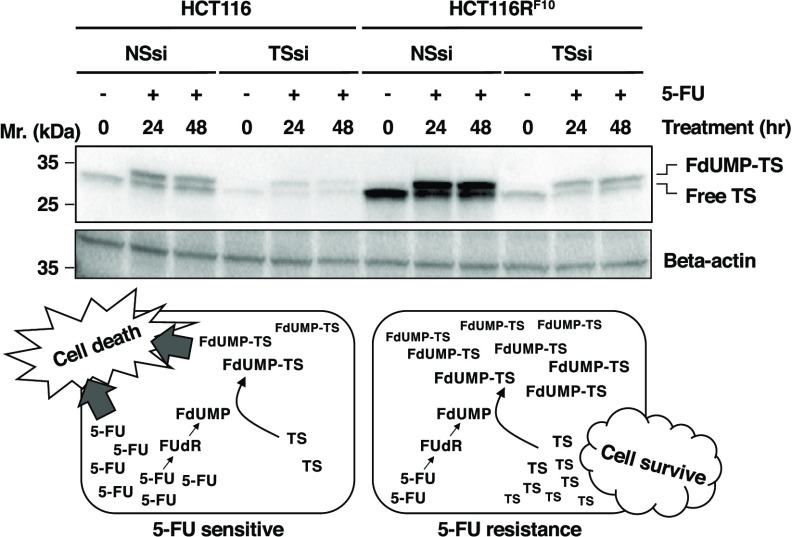
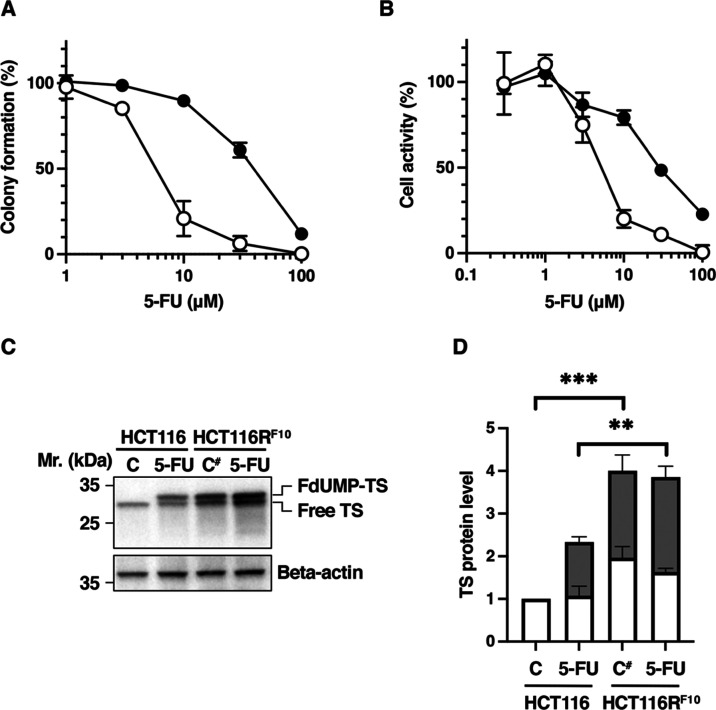
The main anticancer mechanism of 5-FU is inhibiting TS by FdUMP, an active metabolite of 5-FU.1,2,17 The fundamental mechanism for this activity, proposed by Santi in 1980,4 is that FdUMP forms a covalent ternary complex with TS and CH2-THF.4 We have investigated the mechanisms of resistance to 5-FU in human CRC cell models, 5-FU-resistant HCT116RF10 cells, and parental HCT116 cells, revealing their genetic background by exome analysis. The concentration that confers 50% efficacy (EC50) of 5-FU in the 5-FU-resistant HCT116RF10 and parental HCT116 cells in the colony formation and WST-8 assays is shown in Table 1 and Figures 1A,B. We recently hypothesized that 5-FU-resistant CRC cells have upregulated TYMS expression and use a fraction of their TS to trap FdUMP, resulting in 5-FU resistance.16 Indeed, the protein levels of free-TS, FdUMP-TS-CH2-THF covalent complex, and total TS were significantly higher in HCT116RF10 cells than in HCT116 cells under the passage culture conditions (Figures 1C,D). Additionally, the protein levels of free-TS (native enzyme), FdUMP-TS covalent complex (which we termed as FdUMP-TS), and total TS were individually about 1.6–1.8-fold higher in HCT116RF10 cells than in HCT116 cells after treatment with 100 μM 5-FU for 24 h. In these experiments, we tested 5-FU at a concentration of 100 μM, which has a sufficient anticancer effect in HCT116RF10 and HCT116 cells. Interestingly, the total TS and FdUMP-TS levels were upregulated about twofold in HCT116 cells but not in HCT116RF10 cells after treatment with 5-FU for 24 h compared with individual subculture conditions. These results indicate that the 5-FU-resistant HCT116RF10 cells may have a system that traps FdUMP with TS and removes FdUMP-TS as a resistance mechanism.
Table 1. Summary of 5-FU Sensitivities in the 5-FU-Resistant HCT116RF10 Cells and Parental HCT116 Cells.
| EC50 (μM) |
||
|---|---|---|
| cell line | colony formation | WST-8 |
| HCT116 | 5.5 | 5.1 |
| HCT116RF10 | 38.0 | 29.0 |
Figure 1.
Two TS protein forms, free-TS and FdUMP-TS, are higher in 5-FU-resistant HCT116RF10 cells than in 5-FU-sensitive parental HCT116 cells. (A) 5-FU sensitivity of HCT116 and HCT116RF10 cells using colony formation assay. The cells were treated with the indicated concentration of 5-FU and incubated for 10 d. Colony formation (%) represents the average of three independent experiments, with error bars showing the ±SE (standard error) of triplicates. Solid circle, HCT116RF10 cells; open circle, HCT116 cells. (B) Cells were tested for cell activity after 72 h of treatment with the indicated concentration of 5-FU. Results represent the averages of three independent experiments, with error bars showing the ±SE of triplicates. (C) Protein levels of TS and β-actin in HCT116RF10 and HCT116 cells. Whole-cell lysates were prepared from parental HCT116 and HCT116RF10 cells. The expression levels of β-actin were used as an internal control. The data represent at least three independent experiments. (D) Protein levels of two TS forms, free-TS and FdUMP-TS, in HCT116 and HCT116RF10 cells. TS protein levels in HCT116RF10 cells are shown by the ratio of TS density to β-actin density relative to the control value for HCT116 cells. Results represent the average of three independent experiments, with error bars showing the ±SE of triplicates. C, control; passage culture condition of parental HCT116 cells (no drug or solvent). C#, passage culture condition of 5-FU-resistant HCT116RF10 cells; the cells were continually treated with 10 μM 5-FU. 5-FU, the cells were treated with 100 μM 5-FU for 24 h. White bar, free-TS form; gray bar, FdUMP-TS form. Student’s t-test, **p < 0.01 and ***p < 0.001. One-way analysis of variance (ANOVA), p < 0.0001 (for each total TS and FdUMP-TS levels of all groups).
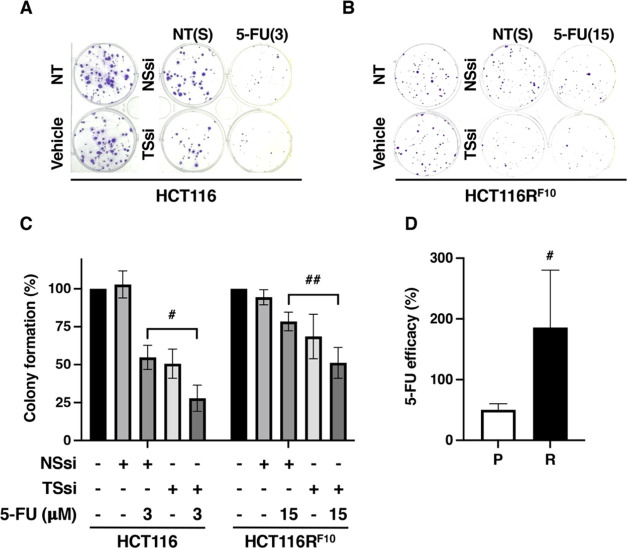
First, to elucidate the relationship between 5-FU resistance and TYMS expression, we analyzed the anticancer activity of 5-FU in the 5-FU-resistant HCT116RF10 cells and parental HCT116 cells transfected with TYMS-targeted siRNA. HCT116 and HCT116RF10 cells were treated with the indicated concentration of 5-FU (EC20 values: 3 μM for HCT116 cells; 15 μM for HCT116RF10 cells), respectively. Additionally, the knockdown of TYMS enhanced the anticancer activity of 5-FU in both types of CRC cells (Figure 2A–C). In the parental HCT116 cells, the percentage of colony formation following 5-FU treatment was lower when the cells were transfected with TYMS-targeted siRNA (28%) than with nonsilencing siRNA (55%) (Figures 2A,C). Similarly, in 5-FU-resistant HCT116RF10 cells (Figures 2B,C), the percentage of colony formation after 5-FU treatment was lower after transfection with TYMS-targeted siRNA (51%) than with nonsilencing siRNA (79%). The enhancement of the anticancer effect of 5-FU cytotoxicity by TYMS knockdown was stronger in HCT116RF10 cells (186%) than in parental HCT116 cells (50%) (Figure 2D). There are numerous reports that the phenotype of 5-FU sensitivity and resistance is influenced by the levels of TS protein and enzymatic activity in cancer cells.13−15,18 These observations suggest that the TS protein’s intracellular abundance, status, and function are important for the phenotypic characteristics of sensitivity and resistance to 5-FU in cancer cells.
Figure 2.
TYMS knockdown results in stronger enhancement of the anticancer activity of 5-FU in HCT116RF10 cells than in HCT116 cells. (A) Image of colony formation in HCT116 cells. (B) Image of colony formation in HCT116RF10 cells. Anticancer activity of 5-FU in HCT116RF10 cells and HCT116 cells, measured using the colony formation assay. HCT116RF10 cells and HCT116 cells were transfected with TYMS-targeted siRNA or nonsilencing siRNA. Then, both types of cells were treated with the indicated concentration of 5-FU and incubated for 9 days. NT, nontreatment; vehicle, lipofectamine RNAiMax alone; NT(S), solvent (dimethyl sulfoxide (DMSO)); NSsi, nonsilencing siRNA; TSsi, TYMS-targeted siRNA; 5-FU(3), 5-FU 3 μM; 5-FU(15), 5-FU 15 μM. (C) Colony formation (%) represents the average of three independent experiments, each performed in duplicate, with error bars showing the SE of triplicate experiments. Student’s t-test, # p = 0.0840, ## p = 0.0828, and one-way ANOVA, p < 0.0001 (for all groups). (D) 5-FU efficacy (%) indicates the enhancement of 5-FU efficacy in HCT116RF10 cells and HCT116 cells, respectively. 5-FU efficacy was calculated using the values for colony formation: 5-FU efficacy (%) = (NSsi + 5-FU – TSsi + 5-FU)/(NSsi alone – TSsi alone) × 100. White bar, P: parental HCT116 cells; black bar, R: 5-FU-resistant HCT116RF10 cells. Student’s t-test, # p = 0.2273 (vs P) and F-test p = 0.0219 (vs P).
Trapping of FdUMP by the TS Protein is More Effective in 5-FU-Resistant HCT116RF10 Cells than in Parental HCT116 Cells
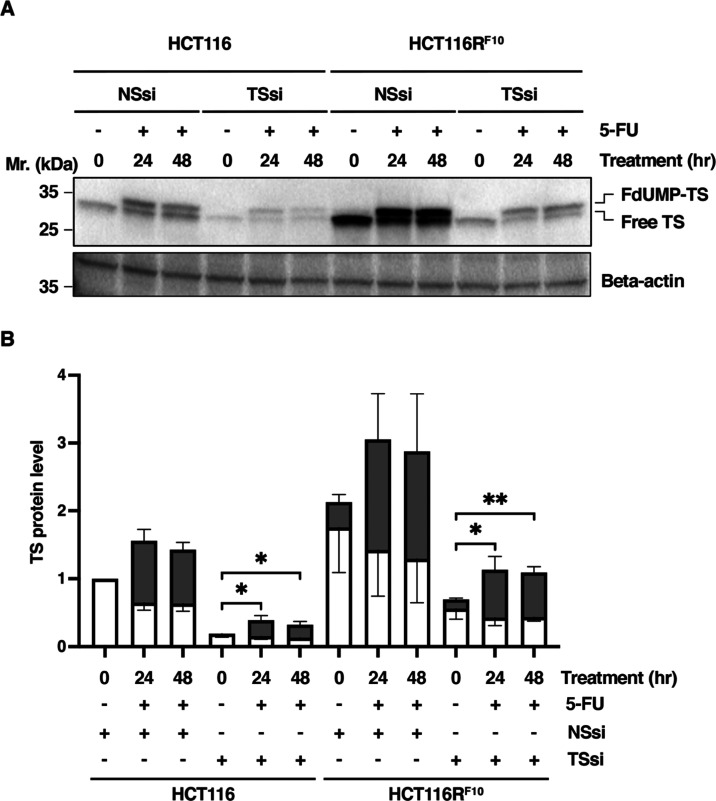
We tested the hypothesis that the TS protein is utilized to trap FdUMP, which results in resistance to 5-FU. As shown in Figure 3A,B, the expression of TYMS in untreated and 5-FU-treated parental HCT116 cells and 5-FU-resistant HCT116RF10 cells was suppressed by transfection of TYMS-targeted siRNA. In the untreated stage, the knockdown efficacies of the TS protein were 86% in HCT116 cells and 63% in HCT116RF10 cells transfected with TYMS-targeted siRNA compared to that in both cells transfected with nonsilencing siRNA, respectively. The other control experiment, in which nonsilencing siRNA was transfected, showed no effect on the expression of TS and β-actin in either cell type. Similarly, the transfection of TYMS-targeted siRNA in both types of cells showed no impact on the expression of β-actin. These control experiments showed similar protein levels of TS and β-actin in HCT116 cells and HCT116RF10 cells under both the passage culture condition and 5-FU-treated condition (Figure 1D). In both types of nonsilencing siRNA-transfected TS, i.e., total TS, appears to be overproduced in HCT116RF10 cells compared with the parental HCT116 cells with and without 5-FU treatment. The same results were observed when both cell types were transfected with TYMS-targeted siRNA. The induction of TS after treatment with 5-FU for 24 h was higher in parental HCT116 cells (1.7-fold increase in NSsi-transfected cells and 2.1-fold increase in TSsi-transfected cells) than in the 5-FU-resistant HCT116RF10 cells (1.4-fold increase in NSsi-transfected cells and 1.5-fold increase in TSsi-transfected cells). Furthermore, the accumulation of the FdUMP-TS protein after 5-FU for 24 h was dramatically increased in HCT116RF10 cells (1.8–3.0-fold higher) compared with HCT116 cells transfected with nonsilencing siRNA or TYMS-targeted siRNA. It is known that the FdUMP-TS protein band, indicating the FdUMP-covalent form, represents TS in ternary complexes and is correlated with the intracellular concentration of FdUMP.19−22 Similarly, the storage of active free-TS protein after 5-FU for 24 h was significantly increased in HCT116RF10 cells (2.5–2.9-fold higher) compared with HCT116 cells after transfection of nonsilencing siRNA or TYMS-targeted siRNA. Notably, the expression of free-TS protein in 5-FU-resistant HCT11RF10 cells was decreased (19% at 24 h and 26% at 48 h in NSsi-transfected cells; 23% at 24 h and 23% at 48 h in TSsi-transfected cells) by 5-FU treatment compared with no treatment after transfection of TYMS-targeted siRNA or nonsilencing siRNA, respectively. Similarly, the expression of free-TS protein in parental HCT116 cells was decreased (36% at 24 h and 37% at 48 h in NSsi-transfected cells; 21% at 24 h and 32% at 48 h in TSsi-transfected cells) by 5-FU treatment compared with the untreated control after transfection TYMS-targeted siRNA or nonsilencing siRNA. These observations indicate that the regulation of the balance between the storage of active free-TS and the accumulation of FdUMP-TS is a leading cause of direct resistance to 5-FU.
Figure 3.
Trapping efficiency of FdUMP by TS is higher in HCT116RF10 cells than in parental HCT116 cells. (A) Dynamics of the TS protein in TYMS-silenced HCT116RF10 cells and HCT116 cells after treatment with 5-FU. At 48 h after transfection with TYMS-targeted siRNA or nonsilencing siRNA, the cells were treated with 5-FU 100 μM for the indicated treatment time, and whole-cell lysates were prepared. The protein expression of TS and β-actin was measured by Western blotting analysis. The data are representative of at least three independent experiments. NSsi, nonsilencing siRNA; TSsi, TYMS-targeted siRNA; (B) TS protein level in HCT116RF10 cells and HCT116 cells. The levels of total TS, i.e., the active free-TS form and the inactive FdUMP-TS form, are indicated by the ratio of TS density to β-actin density for each treatment relative to the value for the NSsi-transfected parental HCT116 cells without 5-FU. The results represent the average of three independent experiments and the error bars show the ±SE of triplicate experiments. White bar, free-TS form; gray bar, FdUMP-TS form. Student’s t-test, * p < 0.05 and ** p < 0.01, one-way ANOVA, p < 0.001 (for total TS levels of all groups), and p < 0.05 (for FdUMP-TS levels of all groups).
Discussion
TS, which is encoded by the TYMS gene in humans, catalyzes the conversion of dUMP to dTMP using the co-substrate CH2-THF as a methyl donor.5 The TS enzyme is believed to exist in two forms, a monomer and a dimer, which are in monomer–homodimer equilibrium.5 The TS dimer is essential for its catalytic activity. It is known that binding of TS, in its dimeric form, to its own mRNA leads to the formation of an autoregulatory feedback loop that represses the translation of TYMS mRNA.19,23−26 Many mechanisms have been proposed to explain 5-FU resistance in cancer cells. One important mechanism is the disruption of the autoregulatory feedback loop for the repression of translation. TS ligands, such as 5-FU, disrupt the binding of the TS enzyme with TYMS mRNA, leading to translational derepression and overproduction of the TS enzyme.19,25,26 In addition to translational derepression, enzyme stabilization has been suggested as the primary mechanism of TS induction by fluoropyrimidines in human CRC and ovarian cancer cell lines.27−29 Furthermore, it is proposed that fluoropyrimidine-mediated increases in TS levels are induced by its effect on TS enzyme stability, with no effect on TYMS mRNA.28,30,31 The amplification of TYMS, leading to the overproduction of TYMS mRNA and TS protein, is another mechanism of resistance to fluoropyrimidines like 5-FU and its derivatives.12 These observations indicated that an understanding of translational derepression, enzyme stabilization, and gene amplification as the process of TS induction can help to elucidate the mechanism of the acquisition of 5-FU resistance. These findings clearly suggest that the mechanisms of 5-FU resistance are a complex and serious problem.
Recently, we established a 5-FU-resistant cell line, HCT116RF10 cells, from parental human CRC HCT116 cells and analyzed the resistance mechanisms of 5-FU.16 In previous findings, HCT116RF10 cells were weakly sensitive to SN-38, the active metabolite of irinotecan, and cisplatin compared with the parental HCT116 cells.16 The sensitivity of SN-38 and cisplatin was 1.4-fold (EC50 = 3 nM in HCT116RF10 cells; 4.2 nM in HCT116 cells) and 1.2-fold (EC50 = 4.5 μM in HCT116RF10 cells; 5.2 μM in HCT116 cells) higher in HCT116RF10 cells than in parental HCT116 cells, respectively.16 Additionally, the parental HCT116 cells grow with a doubling time of approximately 18 h. In contrast, 5-FU-resistant HCT116RF10 cells grow with a doubling time of approximately 27 h in both passage culture conditions with and without 10 μM 5-FU. Interestingly, the 5-FU-resistant HCT116RF10 cells exhibited a lower ability to form colonies and tumor spheres compared with parental HCT116 cells in colony formation and three-dimensional cell culture experiments.16 We consider that the difference of proliferation capacity and clonogenicity may be less relevant to anticancer drug sensitivity in HCT116RF10 cells and HCT116 cells. Further, we previously reported that 5-FU-resistant HCT116RF10 cells have increased TYMS expression relative to 5-FU-sensitive parental HCT116 cells and they use a fraction of TS to trap FdUMP, thereby resulting in resistance to 5-FU and its derivative fluorodeoxyuridine.16
In this study, we demonstrated that the regulation of the balance between the storage of active free-TS and the accumulation of inactive FdUMP-TS is responsible for the resistance to 5-FU. Our findings suggest that the TS enzyme in 5-FU-resistant HCT116RF10 cells can actively and efficiently trap FdUMP. Notably, several studies have shown that 5-FU treatment enhances TS enzyme induction, mainly the ternary complex among TS, FdUMP, and CH2-THF in various human CRC cells and tissues.1,2,12,32−34 Indeed, the expression levels of TYMS mRNA and TS protein are molecular biomarkers predicting tumor sensitivity to 5-FU.1,2 Additionally, 5-FU resistance is associated with the level of TS protein and enzymatic activity in several human CRC cells and tumors.1,2,12,32 The numerous findings may support the hypothesis that the trapping of FdUMP by TS enzyme confers resistance to 5-FU and its derivatives, in that several CRC cells and patients with high TS levels are less sensitive to 5-FU. However, it is critical that many studies to date have not discussed the relationship between the FdUMP trapping capacity by TS enzyme, i.e., FdUMP-TS level at total TS level, and the anticancer sensitivity to 5-FU in human CRC cells. Previously, many researchers understood that 5-FU exerts its anticancer effects through inhibition of TS by its active metabolite FdUMP and incorporation of 5-FU’s metabolites, i.e., FUMP and FdUMP, into RNA and DNA, respectively. In particular, we realize that the main anticancer mechanism of 5-FU is inhibiting TS by its active metabolite, FdUMP. In this study, particularly, our findings suggest that the TS enzyme, which is the target of FdUMP, acts as a resistance factor that traps FdUMP in 5-FU-resistant HCT116RF10 cells. We think that additional studies in several 5-FU-resistant human CRC cells are needed to understand the mechanisms of 5-FU resistance utilizing the trap of FdUMP by the TS enzyme. We also consider that the regulatory mechanisms of monomeric and dimeric TS protein form differ between 5-FU-resistant HCT116RF10 cells and the 5-FU-sensitive parental HCT116 cells. We further investigated the relationship between the regulation of TS protein status, i.e., the balance between active form of free-TS and the inactive TS form (FdUMP-TS–CH2-THF), and that the potential regulators of 5-FU resistance include TS-interacting proteins, mRNAs, and noncoding RNAs.
Conclusions
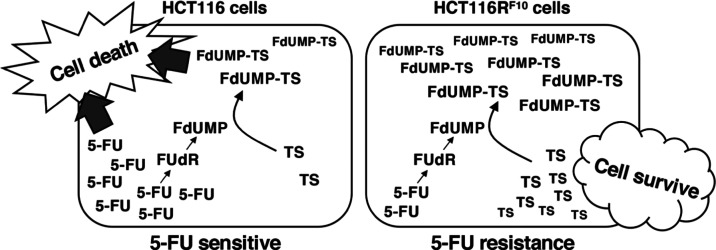
Collectively, we demonstrated that the trapping of FdUMP by its target enzyme TS confers resistance to 5-FU. In addition, we showed that 5-FU-resistant HCT116RF10 cells became resistant to 5-FU by regulating the balance between the storage of the active TS protein and the accumulation of FdUMP-TS protein. In contrast, parental HCT116 cells are sensitized to 5-FU by the depletion of TS, which is due to the formation of the FdUMP-TS complex (Figure 4). Our findings provide a better understanding of the mechanisms of 5-FU resistance and may lead to the development of anticancer strategies to reverse sensitivity to 5-FU and its derivatives.
Figure 4.
Predictive model of the regulation of TS status by balancing the accumulation of the inactive FdUMP-TS form and the storage of the active free-TS form in the 5-FU-resistant HCT116RF10 cells and parental HCT116 cells. We show that the trapping of FdUMP by TS enzyme is more effective in 5-FU-resistant HCT116RF10 cells than in parental HCT116 cells. In addition, we predict that the regulation of the balance between the storage of the active TS form and the accumulation of FdUMP-TS is responsible for direct resistance to 5-FU. 5-FU, 5-fluorouracil; FUdR, 5-fluorodeoxyuridine; FdUMP, 5-fluorodeoxyuridine monophosphate; TS, thymidylate synthase.
Materials and Methods
Reagents
5-FU was purchased from FUJIFILM Wako Pure Chemical (Osaka, Japan) and stored as a 100 mM stock in dimethyl sulfoxide (DMSO; Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) at −25 °C. The TYMS-targeted siRNA (Hs_TYMS_3 FlexiTube siRNA, catalog number: SI00021616, sequence: unpublished) and nonsilencing siRNA (AllStars negative control siRNA, catalog number: 1027280, sequence: unpublished) were obtained from QIAGEN (Dusseldorf, Germany) and stored as a 20 μM stock solution in RNase-free water at −25 °C. Invitrogen Lipofectamine RNAiMax reagent was purchased from Thermo Fisher Scientific (Waltham, MA).
Cell Lines and Cell Culture
The human CRC cell line HCT116 was obtained from the American Type Culture Collection (Manassas, VA). 5-FU-resistant HCT116 (HCT116RF10) cells were produced in accordance with a previously described method.16 The parental HCT116 and 5-FU-resistant HCT116RF10 cell lines were then cultured as previously described.16 Both the parental HCT116 cells and the 5-FU-resistant HCT116RF10 cells were grown in Dulbecco’s modified Eagle’s medium (D-MEM, Cat#:043-30085, FUJIFILM Wako Pure Chemical). The culture medium contained 10% heat-inactivated fetal bovine serum, 100 units/mL penicillin, and 100 μg/mL streptomycin.
Transfection
The transfection of TYMS-targeted siRNA (TSsi) or nonsilencing siRNA (NSsi) was performed using the Lipofectamine RNAiMax reagent (Thermo Fisher Scientific) in accordance with the manufacturer’s protocol. Briefly, cells were seeded into six-well plates (5 × 104 cells/well) and then incubated overnight. Prior to transfection, the culture medium was exchanged for 1 mL/well Opti-MEM (Thermo Fisher Scientific). The cells were transfected with TSsi or NSsi (each at 10 nM final concentration). At 4–6 h after transfection, the medium was removed and replaced with an antibiotic-free culture medium.
Colony Formation Assay
The colony formation assay was performed in accordance with a previously described method.16,35,36 The cells were detached using Accutase, suspended in medium, inoculated into six-well plates (200 cells/well), and incubated overnight. Experiments were performed in triplicate. The cells were treated with various concentrations of 5-FU or with solvent (i.e., DMSO) as the negative control. After incubation for 10 days, the cells were fixed with 4% formaldehyde solution, stained with 0.1% (w/v) crystal violet, and the number of colonies in each well was counted. In the transfection experiments, the cells were transfected with TSsi or NSsi (10 nM, as above). After incubation for 24 h, the cells were treated with various concentrations of 5-FU or with DMSO. After incubation for 9 days, the colonies were fixed, stained, and counted.
Cell Activity by WST-8 Assay
Cell activity assays were performed as previously described.16 Cell activity was determined using the Cell Counting Kit-8 (WST-8) cell proliferation assay (Dojindo, Tokyo, Japan).
Western Blotting
Western blotting analysis was performed as previously described.16,35 The antibodies used were rabbit anti-thymidylate synthase (D5B3) monoclonal antibody (9045S, 1:1000, Cell Signaling Technologies, Massachusetts), mouse anti-DPYD (A-5) monoclonal antibody (sc-376712, 1:1000, Santa Cruz Biotechnology, Texas), mouse anti-β-actin monoclonal antibody (A19178-200UL, 1:20 000, Sigma-Aldrich), horseradish peroxidase-linked antirabbit IgG (1:20 000, GE Healthcare, Connecticut), and horseradish peroxidase-linked whole-antibody antimouse IgG (1:20 000, GE Healthcare).
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 9 software. The data are presented as the mean ± standard error. Significant differences among groups were evaluated using Student’s t-test, F-test, and one-way analysis of variance (ANOVA). A p value of <0.05 was considered to indicate statistical significance.
Acknowledgments
We thank Dr. Hikoya Hayatsu (Okayama University) and Dr. Yusuke Wataya (Okayama University) for their helpful discussions.
Author Contributions
A.S. conceived and designed the project. C.K., N.N., Y.O., and A.S. acquired the data. C.K., Y.O., and A.S. analyzed and interpreted the data. A.S. wrote the paper.
The authors declare no competing financial interest.
References
- Longley D. B.; Harkin D. P.; Johnston P. G. 5-Fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- Blondy S.; David V.; Verdier M.; Mathonnet M.; Perraud A.; Christou N. 5-Fluorouracil resistance mechanisms in colorectal cancer: from classical pathways to promising processes. Cancer Sci. 2020, 111, 3142–3154. 10.1111/cas.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberger C. Fluorinated pyrimidines. Prog. Nucleic Acid Res. Mol. Biol. 1965, 4, 1–50. [DOI] [PubMed] [Google Scholar]
- Santi D. V. Perspective on the design and biochemical pharmacology of inhibitors of thymidylate synthetase. J. Med. Chem. 1980, 23, 103–111. 10.1021/jm00176a001. [DOI] [PubMed] [Google Scholar]
- Garg D.; Henrich S.; Salo-Ahen O. M.; Myllykallio H.; Costi M. P.; Wade R. C. Novel approaches for targeting thymidylate synthase to overcome the resistance and toxicity of anticancer drugs. J. Med. Chem. 2010, 53, 6539–6549. 10.1021/jm901869w. [DOI] [PubMed] [Google Scholar]
- Santi D. V.; McHenry C. S. 5-Fluoro-2′-deoxyuridylate: covalent complex with thymidylate synthetase. Proc. Natl. Acad. Sci. U.S.A. 1972, 69, 1855–1857. 10.1073/pnas.69.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi D. V.; McHenry C. S.; Sommer H. Mechanism of interaction of thymidylate synthetase with 5-fluorodeoxyuridylate. Biochemistry 1974, 13, 471–481. 10.1021/bi00700a012. [DOI] [PubMed] [Google Scholar]
- Sommer H.; Santi D. V. Purification and amino acid analysis of an active site peptide from thymidylate synthetase containing covalently bound 5-fluoro-2′-deoxyuridylate and methylenetetrahydrofolate. Biochem. Biophys. Res. Commun. 1974, 57, 689–695. 10.1016/0006-291X(74)90601-9. [DOI] [PubMed] [Google Scholar]
- Sethy C.; Kundu C. N. 5-Fluorouracil (5-FU) resistance and the new strategy to enhance the sensitivity against cancer: implication of DNA repair inhibition. Biomed. Pharmacother. 2021, 137, 111285 10.1016/j.biopha.2021.111285. [DOI] [PubMed] [Google Scholar]
- Ishibiki Y.; Kitajima M.; Sakamoto K.; Tomiki Y.; Sakamoto S.; Kamano T. Intratumoural thymidylate synthase and dihydropyrimidine dehydrogenase activities are good predictors of 5-fluorouracil sensitivity in colorectal cancer. J. Int. Med. Res. 2003, 31, 181–187. 10.1177/147323000303100303. [DOI] [PubMed] [Google Scholar]
- Popat S.; Matakidou A.; Houlston R. S. Thymidylate synthase expression and prognosis in colorectal cancer: a systematic review and meta-analysis. J. Clin. Oncol. 2004, 22, 529–536. 10.1200/JCO.2004.05.064. [DOI] [PubMed] [Google Scholar]
- Peters G. J.; Backus H. H.; Freemantle S.; van Triest B.; Codacci-Pisanelli G.; van der Wilt C. L.; Smid K.; Lunec J.; Calvert A. H.; Marsh S.; McLeod H. L.; Bloemena E.; Meijer S.; Jansen G.; van Groeningen C. J.; Pinedo H. M. Induction of thymidylate synthase as a 5-fluorouracil resistance mechanism. Biochim. Biophys. Acta, Mol. Basis Dis. 2002, 1587, 194–205. 10.1016/S0925-4439(02)00082-0. [DOI] [PubMed] [Google Scholar]
- Copur S.; Aiba K.; Drake J. C.; Allegra C. J.; Chu E. Thymidylate synthase gene amplification in human colon cancer cell lines resistant to 5-fluorouracil. Biochem Pharmacol 1995, 49, 1419–1426. 10.1016/0006-2952(95)00067-A. [DOI] [PubMed] [Google Scholar]
- Johnston P. G.; Lenz H. J.; Leichman C. G.; Danenberg K. D.; Allegra C. J.; Danenberg P. V.; Leichman L. Thymidylate synthase gene and protein expression correlate and are associated with response to 5-fluorouracil in human colorectal and gastric tumors. Cancer Res. 1995, 55, 1407–1412. [PubMed] [Google Scholar]
- Wang W.; Marsh S.; Cassidy J.; McLeod H. L. Pharmacogenomic dissection of resistance to thymidylate synthase inhibitors. Cancer Res. 2001, 61, 5505–5510. [PubMed] [Google Scholar]
- Kurasaka C.; Ogino Y.; Sato A. Molecular mechanisms and tumor biological aspects of 5-fluorouracil resistance in HCT116 human colorectal cancer cells. Int. J. Mol. Sci. 2021, 22, 2916. 10.3390/ijms22062916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N.; Yin Y.; Xu S. J.; Chen W. S. 5-Fluorouracil: mechanisms of resistance and reversal strategies. Molecules 2008, 13, 1551–1569. 10.3390/molecules13081551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.; McLeod H. L.; Cassidy J.; Collie-Duguid E. S. Mechanisms of acquired chemoresistance to 5-fluorouracil and tomudex: thymidylate synthase dependent and independent networks. Cancer Chemother. Pharmacol. 2007, 59, 839–845. 10.1007/s00280-006-0384-5. [DOI] [PubMed] [Google Scholar]
- Chu E.; Koeller D. M.; Johnston P. G.; Zinn S.; Allegra C. J. Regulation of thymidylate synthase in human colon cancer cells treated with 5-fluorouracil and interferon-gamma. Mol. Pharmacol. 1993, 43, 527–533. [PubMed] [Google Scholar]
- Drake J. C.; Allegra C. J.; Johnston P. G. Immunological quantitation of thymidylate synthase-FdUMP-5,10-methylenetetrahydrofolate ternary complex with the monoclonal antibody TS 106. Anticancer Drugs 1993, 4, 431–435. 10.1097/00001813-199308000-00002. [DOI] [PubMed] [Google Scholar]
- Mori R.; Futamura M.; Tanahashi T.; Tanaka Y.; Matsuhashi N.; Yamaguchi K.; Yoshida K. 5FU resistance caused by reduced fluoro-deoxyuridine monophosphate and its reversal using deoxyuridine. Oncol. Lett. 2017, 14, 3162–3168. 10.3892/ol.2017.6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu T.; Mori R.; Futamura M.; Fukada M.; Tanaka H.; Yasufuku I.; Sato Y.; Iwata Y.; Imai T.; Imai H.; Tanaka Y.; Okumura N.; Matsuhashi N.; Takahashi T.; Yoshida K. Mechanism of acquired 5FU resistance and strategy for overcoming 5FU resistance focusing on 5FU metabolism in colon cancer cell lines. Oncol. Rep. 2021, 45, 27 10.3892/or.2021.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu E.; Koeller D. M.; Casey J. L.; Drake J. C.; Chabner B. A.; Elwood P. C.; Zinn S.; Allegra C. J. Autoregulation of human thymidylate synthase messenger RNA translation by thymidylate synthase. Proc. Natl. Acad. Sci. U.S.A. 1991, 88, 8977–8981. 10.1073/pnas.88.20.8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu E.; Voeller D.; Koeller D. M.; Drake J. C.; Takimoto C. H.; Maley G. F.; Maley F.; Allegra C. J. Identification of an RNA binding site for human thymidylate synthase. Proc. Natl. Acad. Sci. U.S.A. 1993, 90, 517–521. 10.1073/pnas.90.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyomarsi K.; Samet J.; Molnar G.; Pardee A. B. The thymidylate synthase inhibitor, ICI D1694, overcomes translational detainment of the enzyme. J. Biol. Chem. 1993, 268, 15142–15149. 10.1016/S0021-9258(18)82448-6. [DOI] [PubMed] [Google Scholar]
- Chu E.; Allegra C. J. The role of thymidylate synthase as an RNA binding protein. Bioessays 1996, 18, 191–198. 10.1002/bies.950180306. [DOI] [PubMed] [Google Scholar]
- Kitchens M. E.; Forsthoefel A. M.; Barbour K. W.; Spencer H. T.; Berger F. G. Mechanisms of acquired resistance to thymidylate synthase inhibitors: the role of enzyme stability. Mol. Pharmacol. 1999, 56, 1063–1070. 10.1124/mol.56.5.1063. [DOI] [PubMed] [Google Scholar]
- Kitchens M. E.; Forsthoefel A. M.; Rafique Z.; Spencer H. T.; Berger F. G. Ligand-mediated induction of thymidylate synthase occurs by enzyme stabilization. Implications for autoregulation of translation. J. Biol. Chem. 1999, 274, 12544–12547. 10.1074/jbc.274.18.12544. [DOI] [PubMed] [Google Scholar]
- Marverti G.; Ligabue A.; Paglietti G.; Corona P.; Piras S.; Vitale G.; Guerrieri D.; Luciani R.; Costi M. P.; Frassineti C.; Moruzzi M. S. Collateral sensitivity to novel thymidylate synthase inhibitors correlates with folate cycle enzymes impairment in cisplatin-resistant human ovarian cancer cells. Eur. J. Pharmacol. 2009, 615, 17–26. 10.1016/j.ejphar.2009.04.062. [DOI] [PubMed] [Google Scholar]
- Washtien W. L. Increased levels of thymidylate synthetase in cells exposed to 5-fluorouracil. Mol. Pharmacol. 1984, 25, 171–177. [PubMed] [Google Scholar]
- Abdel Mohsen A.-W.; Aull J. L.; Payne D. M.; Daron H. H. Ligand-induced conformational changes of thymidylate synthase detected by limited proteolysis. Biochemistry 1995, 34, 1669–1677. 10.1021/bi00005a023. [DOI] [PubMed] [Google Scholar]
- Peters G. J.; van der Wilt C. L.; van Triest B.; Codacci-Pisanelli G.; Johnston P. G.; van Groeningen C. J.; Pinedo H. M. Thymidylate synthase and drug resistance. Eur. J. Cancer 1995, 31, 1299–1305. 10.1016/0959-8049(95)00172-F. [DOI] [PubMed] [Google Scholar]
- van Triest B.; Pinedo H. M.; van Hensbergen Y.; Smid K.; Telleman F.; Schoenmakers P. S.; van der Wilt C. L.; van Laar J. A.; Noordhuis P.; Jansen G.; Peters G. J. Thymidylate synthase level as the main predictive parameter for sensitivity to 5-fluorouracil, but not for folate-based thymidylate synthase inhibitors, in 13 nonselected colon cancer cell lines. Clin. Cancer Res. 1999, 5, 643–654. [PubMed] [Google Scholar]
- Peters G. J.; van Triest B.; Backus H. H.; Kuiper C. M.; van der Wilt C. L.; Pinedo H. M. Molecular downstream events and induction of thymidylate synthase in mutant and wild-type p53 colon cancer cell lines after treatment with 5-fluorouracil and the thymidylate synthase inhibitor raltitrexed. Eur. J. Cancer 2000, 36, 916–924. 10.1016/S0959-8049(00)00026-5. [DOI] [PubMed] [Google Scholar]
- Ogino Y.; Sato A.; Uchiumi F.; Tanuma S. I. Cross resistance to diverse anticancer nicotinamide phosphoribosyltransferase inhibitors induced by FK866 treatment. Oncotarget 2018, 9, 16451–16461. 10.18632/oncotarget.24731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino Y.; Sato A.; Uchiumi F.; Tanuma S. I. Genomic and tumor biological aspects of the anticancer nicotinamide phosphoribosyltransferase inhibitor FK866 in resistant human colorectal cancer cells. Genomics 2019, 111, 1889–1895. 10.1016/j.ygeno.2018.12.012. [DOI] [PubMed] [Google Scholar]