Abstract
IncI2 plasmids appear to have only recently become associated with resistance genes; however, their tendency to carry resistance to the antibiotics of last resort and their widespread distribution increase their relative importance. In this study, we describe lineages within this plasmid family that have an increased likelihood of acquisition of antimicrobial resistance genes. Globally distributed mcr-1-carrying IncI2 plasmids were found to cluster with other IncI2 plasmids carrying extended-spectrum beta-lactamase genes, and separately from the non-resistant IncI2 plasmids. In addition, insertion sequence (IS) elements with no direct association with the acquired resistance genes also clustered with the resistance plasmids in the phylogenetic tree. In recognition of the biased sequencing of resistant plasmids globally, the analysis was also performed on resistant and non-resistant IncI2 plasmids sequenced in the USA through government surveillance efforts that do not rely on antibiotic selection. This analysis confirmed a distinct clustering associated with both resistance and mobile elements and identified possible genomic changes in core genes that correlate with increased acquisition of foreign DNA. This work highlights a potential genetic mechanism for increased uptake of foreign DNA within this prevalent family of plasmids.
Keywords: mcr-1, IncI2, antibiotic resistance, insertion sequences, beta-lactamase, plasmids
1. Introduction
The family of IncI plasmids have been identified as the most frequent assigned incompatibility type among currently sequenced plasmids [1]. IncI2 plasmids in particular have recently garnered increased attention due to their association with extended-spectrum beta-lactamase (ESBL) genes [2,3] and the first mobile colistin resistance gene (mcr-1) [4]. The emergence of plasmid-mediated colistin resistance has highlighted the potentially devastating consequences of the ongoing dissemination of novel antibiotic resistance genes without detection [5,6]. Although colistin resistance had previously been documented in several pathogens (for a review, see [7]), the known mechanisms were chromosomally associated and therefore unlikely to become mobilized via plasmids among different bacterial lineages. However, the discovery by Liu et al. [4] of a novel resistance mechanism that was conferred by a single gene on a conjugative plasmid changed this paradigm and has led to an international effort to determine how widely this extra-chromosomal type of resistance has been circulating, and the factors that led to its development. Retrospective sampling indicates that mcr-1-containing plasmids have been present in food-producing animals for several decades in China, where it has been shown to be increasing in prevalence in recent years [8]. As has already been described by several researchers [9,10,11], ISApl1 mobilization of the mcr-1 cassette through the action of Tn6330 has facilitated its dissemination among different plasmid lineages, and stabilization has occurred through the loss of the ISApl1 in individual plasmids [12]. The mechanism by which the mcr-1 cassette has been acquired and stabilized may differ among the plasmid families, as demonstrated by patterns in the variability in the flanking sequences for specific plasmid families.
In the USA, detection of mcr-1-containing plasmids has been rare. Through the USDA Food Safety Inspection Service (FSIS) program, Enterobacteriaceae isolates obtained from turkey, swine, chicken and cattle samples acquired at slaughter were analyzed for the presence of the mcr-1 gene. From the screened samples, two isolates (both from swine) were found to be carrying colistin-resistant plasmids [13]. Both strains were fully sequenced [14,15] and mcr-1 genes were found to be carried on IncI2 plasmids with high similarity to the initial mcr-1 plasmid identified in China [4]. In a subsequent multi-locus sequence (MLST) study of a relatively small population of IncI2 plasmids (n = 81) [16] it was shown that multiple mechanisms were involved in remodeling this group of plasmids. These included transposons, homologous recombination, insertion/deletions and programmed re-assortment (specifically for pilV). In the current study herein, we extend the exploration to include single nucleotide polymorphism (SNP) analyses to see if this increases the resolution for characterization of these changes. In this paper, we evaluated the genetic diversity in globally distributed IncI2 plasmids to examine the emergence of IncI2 plasmids carrying antimicrobial resistance (AMR) genes. Recognizing the inherent bias in these sequenced isolates (due to increased global surveillance of AMR plasmids), we performed a separate analysis on the subset of IncI2 plasmids that have been sequenced in the USA to better understand the genetic changes that may have facilitated acquisition of antibiotic resistance genes within this plasmid family.
2. Results
2.1. Global IncI2 Diversity
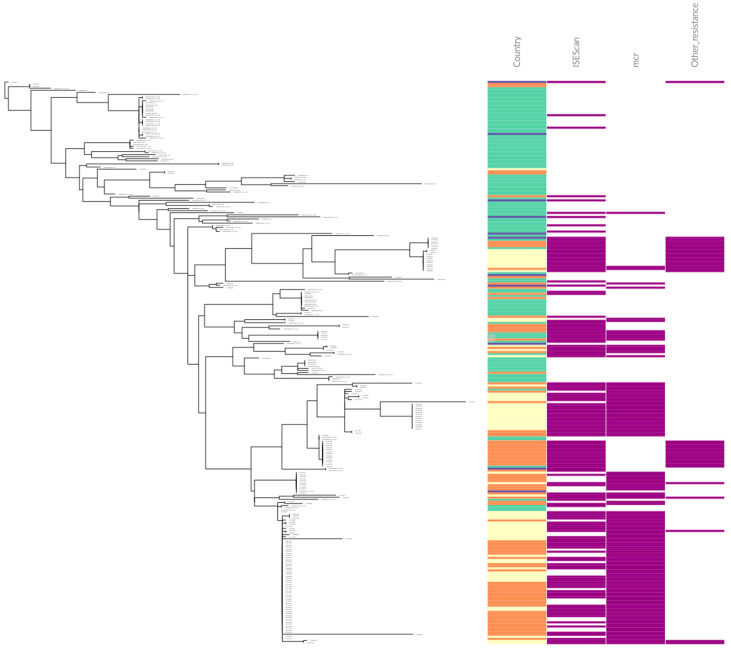
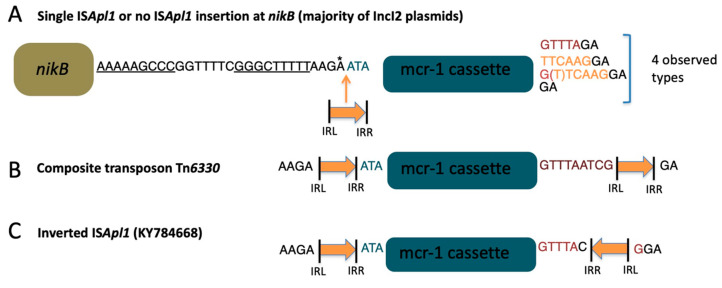
To investigate the relationship between globally distributed IncI2 plasmids, a total of 261 plasmids were extracted from public databases including NCBI’s GenBank database and USA government surveillance efforts (see methods and Supplemental Table S1). Within the IncI2 plasmids carrying an identified mcr-1 gene (n = 121), the vast majority were isolated from E. coli (72%) or Shigella (18%) followed by Salmonella (7%) with the remaining 3% spread between diverse strains (Supplemental Table S1). The core genome phylogenetic tree (see methods) was rooted by the original IncI2 plasmid described (AP002527) and indicated that the majority of mcr-1-containing plasmids formed a highly related clade (Figure 1), as previously described [16]. The remainder of the mcr-1-containing plasmids, and IncI2 plasmids carrying other resistance genes, also occur in the portions of the tree most distant from the chosen root; however, whether the clustering of these plasmids was artificially influenced by biased sequencing of resistant plasmids could not be determined. For this reason, we selected a subset of IncI2 plasmids for further analysis (see next section). Consistent with other reports, the mcr-1 insertion has occurred almost exclusively in a single location (downstream of the nikB gene) suggesting either that these plasmids have descended from a common ancestor or that there is some form of targeted insertion for this location. Examination of the adjacent sequence revealed a conserved palindrome (Figure 2A) immediately downstream of nikB that could play a role in a targeted insertion in this location [16]. The majority of IncI2 plasmids carrying mcr-1 have either a single copy of ISApl1, or no copy, and variability in the flanking sequences illustrates multiple losses or rearrangements of the original composite transposon (Figure 2B,C). Consistent with the interpretation of a targeted insertion, analysis of an IncI2 plasmid carrying mcr-7 (MG267386) revealed that this resistance gene is also inserted directly adjacent to the palindrome downstream of the nikB gene. Although there is limited information available on the mechanisms for this mobilization [17], the lack of sequence similarity between mcr-1 and mcr-7 could indicate independent acquisition events in this location.
Figure 1.
Phylogenetic tree of global IncI2 plasmids based on alignment of core genes (see methods). Country designations are USA (teal), China (beige), UK (blue) and other (orange). The remaining columns are presence or absence of IS elements, mcr genes, or other resistance genes, respectively. Supplemental Table S1 contains all associated metadata in the order presented in this figure.
Figure 2.
Sequences surrounding the mcr-1 cassette in the IncI2 plasmids characterized in this study. The mcr-1 cassette includes the mcr-1 gene and the PAP-2 hypothetical protein, as well as flanking sequences that are conserved among all the plasmids in this study. IRL and IRR refer to the left and right inverted repeats respectively for ISApl1 (orange arrow). (A) Palindromic sequence (7 bp central core flanked by 9 bp inverted repeats underlined in figure) found downstream of the nikB gene and adjacent to most mcr-1 insertions in IncI2 plasmids. The final ‘A’ in this sequence (denoted with a star) is present in cases with ISApl1, and absent when ISApl1 is absent. The sequences in blue appear to have inserted with the mcr-1 cassette. The upstream sequence in black is present in the IncI2 plasmids without mcr-1. Downstream of the mcr-1 cassette shows greater variability. The sequences in red correspond with the original sequence from the composite transposon (shown in B), sequences in orange are consistent with the end of the IRR in the composite transposon and may represent a remnant after loss of the downstream ISApl1. (B) The composite transposon Tn6330 consists of the mcr-1 cassette flanked by two copies of ISApl1. The sequences highlighted in red occur between the end of the mcr-1 cassette and the inverted repeat of the ISApl1 element located downstream of the cassette (relative to mcr-1 transcription). (C) Only one composite transposon has an inverted ISApl1 at the downstream end, and this inversion appears to have centered around the ‘GTTTA’ sequence between the mcr-1 cassette and the inverted repeat for ISApl1.
Within the phylogenetic tree, the IncI2 plasmids did not segregate according to collection date, country of origin, or isolation source of the strain. However, some trends emerged, such as the observation that the majority of IncI2 plasmids have been isolated from either China or the USA (Figure 1) and that almost half of the IncI2 plasmids containing the mcr-1 gene were isolated in China (54/121, 45%). Notably, all of the isolates sequenced from China (with one exception—CP010220 isolated from mouse feces) were carrying some form of antibiotic resistance gene, presumably due to sample selection bias through extensive research into antibiotic resistance prevalence there (for a review, see [18]). Conversely, most strains containing IncI2 plasmids lacking the mcr-1 gene (100/140, 71%) were isolated in the USA through government surveillance programs. These programs perform whole-genome sequencing projects of potential pathogenic importance, and those containing IncI2 plasmids were therefore not specifically chosen based on phenotypic resistance.
In examining the collection of IncI2 plasmids that were not carrying resistance genes, we were intrigued by the finding that a substantial proportion (99/121, 82%) also did not carry any IS elements as determined by our analysis (Table 1). Although this is an expected finding for the IS elements associated with the resistance genes observed (for example IS30, IS1182 and IS1380), there were additional IS elements (e.g., IS200/605, IS1) that were over-represented in IncI2 plasmids carrying resistance genes.
Table 1.
Distribution of IS elements in global collection of IncI2 plasmids correlated with carriage of resistance genes.
| ISEScan | No AMR | mcr-1 | mcr-7.1_1 | aadA, dfrA | bla CMY-2_1 | bla CTX-M-132_1 | bla CTX-M-15_1 | bla CTX-M-199_1 | bla CTX-M-55_1 | bla CTX-M-64_1 | blaKPC-3, aac(6’)-Ib, aadA, blaOXA-9, blaTEM-1 | blaKPC-3, blaTEM-1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No IS found | 99 | 32 | - | - | - | - | - | - | - | - | - | - |
| IS1 | - | 2 | - | - | - | - | - | - | - | - | - | - |
| IS1, IS200/IS605 | - | 13 | - | - | - | - | - | - | - | - | - | - |
| IS1, IS30, IS91 | - | 2 | - | - | - | - | - | - | - | - | - | - |
| IS1, IS5 | - | 1 | - | - | - | - | - | - | - | - | - | - |
| IS110, IS3, IS30 | - | 1 | - | - | - | - | - | - | - | - | - | - |
| IS1182, IS21 | - | - | - | - | - | - | - | - | - | - | 3 | 2 |
| IS1182, IS21, IS91 | - | - | - | - | - | - | - | - | - | - | 1 | - |
| IS1380 | - | 3 | 1 | - | 13 | 2 | - | 1 | 6 | 3 | - | - |
| IS1380, IS3, IS30 | - | 1 | - | - | - | - | - | - | 1 | - | - | - |
| IS1380, IS3, IS91 | - | - | - | - | - | - | 1 | - | - | - | - | - |
| IS1380, IS30 | - | 2 | - | - | - | - | - | - | 2 | - | - | - |
| IS200/IS605 | 15 | 11 | - | - | - | - | - | - | - | - | - | - |
| IS200/IS605, IS3 | - | 1 | - | - | - | - | - | - | - | - | - | - |
| IS200/IS605, IS30 | 1 | 9 | - | - | - | - | - | - | - | - | - | - |
| IS200/IS605, IS91 | 1 | 1 | - | - | - | - | - | - | - | - | - | - |
| IS3 | 3 | 3 | - | 1 | - | - | - | - | - | - | - | - |
| IS3, IS30 | - | 1 | - | - | - | - | - | - | - | - | - | - |
| IS3, IS91 | - | 1 | - | - | - | - | - | - | - | - | - | - |
| IS30 | 1 | 20 | - | - | - | - | - | - | - | - | - | - |
| IS30, IS5 | - | 1 | - | - | - | - | - | - | - | - | - | - |
| IS30, IS630 | - | 1 | - | - | - | - | - | - | - | - | - | - |
| IS30, IS66 | - | 2 | - | - | - | - | - | - | - | - | - | - |
| IS30, IS91 | - | 1 | - | - | - | - | - | - | - | - | - | - |
| IS4 | - | 4 | - | - | - | - | - | - | - | - | - | - |
| IS66 | 1 | 1 | - | - | - | - | - | - | - | - | - | - |
| IS91 | - | 6 | - | - | - | - | - | - | - | - | - | - |
In addition to our phylogenetic analysis based on core genes, MOB-suite was used to perform whole-sequence-based assignment to MOB clustering groups. This revealed 6 sub-clusters (secondary nodes AI345-AI350) within the IncI2 collection and confirmed that the mcr-1 plasmids were a closely related cluster as they were primarily associated with the sub-node designated AI350 (92% of mcr plasmids were within this node). IS element distribution was also found to differ according to the 6 designated nodes (Supplemental Table S2). AI345 and AI346 had less than 10% of their plasmids associated with IS elements (0/16 and 3/38, respectively) and 34% (17/49) of plasmids in node AI347 were associated with IS elements (primarily IS200/605 alone or in combination with IS30-mcr-1). AI348 had a higher percentage of plasmids carrying IS elements (65%, 17/26) but notably all the plasmids carrying IS elements in this node were associated with beta-lactamase genes. Likewise, although 75% of the representatives for AI349 were carrying IS elements, this is not considered representative as there were only 8 plasmids assigned to this group. Finally, 70% of the plasmids associated with node AI350 (86/123) were found to carry IS elements, although the high abundance of mcr-1 carrying plasmids in this group biases this result. Notably, of the plasmids in node AI350 carrying mcr-1, 27 of these did not carry any IS elements consistent with previous reports of the ISApl1 element being lost subsequent to mcr-1 insertion.
2.2. Comparison of IncI2 Plasmids from USA Surveillance
The plasmids sequenced through USA surveillance efforts represented a greater diversity of plasmids lacking resistance genes (97/106) and therefore provided an opportunity to test the hypothesis that acquisition of mobile elements (including but not limited to those carrying antibiotic resistance genes) was related to genetic differences in the core genes of the IncI2 plasmids. This collection was also balanced in terms of bacterial hosts, with 56% of the plasmids isolated from Salmonella (59/106) and 42% from E. coli (44/106). There were also two plasmids isolated from Klebsiella and a single plasmid from Shigella in this collection.
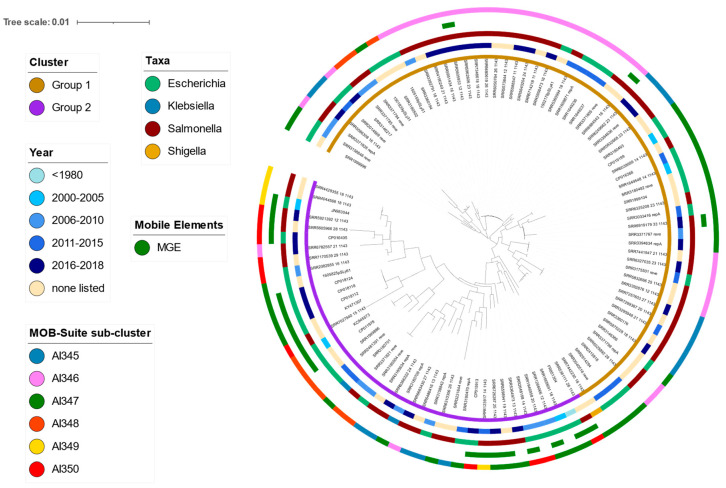
The pangenome analysis of USA IncI2 plasmids illustrated that they had similar overall diversity to the full IncI2 collection analyzed previously. There were 29 core genes identified in the USA dataset (Supplemental Table S3), and a total of 260 unique genes identified (compared to 27 core and 264 total genes for the full IncI2 collection). Most of the plasmids in the USA collection did not contain any IS elements (Table 2) and all except three plasmids carrying IS elements clustered together based on SNP phylogeny of core genes and will be referred to as Group 2 (Figure 3). Linkage analysis confirmed that the clade of plasmids in Group 2 shows greater linkages among alleles, particularly in relation to genes involved in conjugation and pilus formation (Supplemental Table S4). Although many of the plasmid submissions did not include isolation dates, it should be noted that the oldest plasmid in the collection (FR851304, isolation date 1979) is found within Group 2, suggesting that this branch represents a separate trajectory of plasmids as opposed to a recent divergence. Among the accessory genes strongly correlated with this clade were the molecular chaperone dnaJ as well as a primosomal protein dnaT. Of note, the addiction gene relE was also found solely within this clade of plasmids (with one exception—SRR3158846), which may be important for understanding the host range and stability of this subset of plasmids.
Table 2.
Distribution of IS elements within USA IncI2 plasmids analyzed. Please note that IS30, IS1182 and IS1380 are the only IS elements directly associated with antibiotic resistance genes in these plasmids.
| ISEScan Result | No AMR | mcr-1.1 | bla CMY-2 | blaKPC-3, aac(6′)-Ib, aadA, blaOXA-9, blaTEM-1 |
|---|---|---|---|---|
| No IS found | 84 | - | - | - |
| IS1182, IS21 | - | - | - | 2 |
| IS1380 | - | - | 1 | - |
| IS200/IS605 | 8 | 2 | - | - |
| IS200/IS605, IS30 | 1 | 3 | - | - |
| IS200/IS605, IS91 | - | 1 | - | - |
| IS3 | 2 | - | - | - |
| IS30 | 1 | - | - | - |
| IS66 | 1 | - | - | - |
Figure 3.
Phylogenetic analysis of the core gene SNP analysis for the USA subset of IncI2 plasmids. Rings (starting from inner to outer) correspond with: (1) designation of Group 1 or 2 based on core gene phylogeny, (2) year of isolation, (3) taxa, (4) presence or absence of mobile genetic elements and, (5) sub-cluster assigned using MOB-suite. All colors are indicated in the legends. Figure created using iTOL [19]. Analysis using MOB-suite confirmed that representatives from all 6 sub-nodes identified in the full dataset were also present in the smaller USA dataset. As the mcr-1 plasmids were primarily associated with AI350, it was not surprising that the USA plasmids were less prevalently found in the AI350 node (n = 10) in this dataset and were instead more commonly assigned to the AI345-347 nodes (n = 15, 33 and 35 respectively). Notably, IS elements continued to be absent from plasmids assigned to node AI345 and AI346, consistent with the results of the larger dataset. The plasmids carrying mcr-1 were divided between AI350 and AI347, suggesting separate acquisition/introduction events for mcr-1 plasmids in the USA. An examination of the association between the core SNP phylogeny and the assigned MOB nodes indicated that changes within the core genes could be occurring broadly within IncI2 plasmids, as opposed to being isolated to a closely related node. Similar to the pattern observed for IS elements, nodes AI348-350 were primarily associated with Group 2 (67%, 100% and 100%, respectively). Of the plasmids assigned to AI347, 46% were associated with Group 2, which is slightly higher than the observed association of IS elements with this node in the full dataset (35%). Despite a large number of representatives, only 6% of plasmids in node AI346 (2/33) were associated with Group 2. Interestingly, 27% of AI345 plasmids (4/15) were associated with Group 2, although there were no plasmids from this node found to carry any IS elements in either dataset.
Analysis using MOB-suite confirmed that representatives from all 6 sub-nodes identified in the full dataset were also present in the smaller USA dataset. As the mcr-1 plasmids were primarily associated with AI350, it was not surprising that the USA plasmids were less prevalently found in the AI350 node (n = 10) in this dataset and were instead more commonly assigned to the AI345-347 nodes (n = 15, 33 and 35 respectively). Notably, IS elements continued to be absent from plasmids assigned to node AI345 and AI346, consistent with the results of the larger dataset. The plasmids carrying mcr-1 were divided between AI350 and AI347, suggesting separate acquisition/introduction events for mcr-1 plasmids in the USA. An examination of the association between the core SNP phylogeny and the assigned MOB nodes indicated that changes within the core genes could be occurring broadly within IncI2 plasmids, as opposed to being isolated to a closely related node. Similar to the pattern observed for IS elements, nodes AI348-350 were primarily associated with Group 2 (67%, 100% and 100%, respectively). Of the plasmids assigned to AI347, 46% were associated with Group 2, which is slightly higher than the observed association of IS elements with this node in the full dataset (35%). Despite a large number of representatives, only 6% of plasmids in node AI346 (2/33) were associated with Group 2. Interestingly, 27% of AI345 plasmids (4/15) were associated with Group 2, although there were no plasmids from this node found to carry any IS elements in either dataset.
3. Discussion
The first discovery of mcr-1 [4] set off an intensive search for the gene in many parts of the world. It was found fairly frequently in Southeast Asia and less often in the rest of the world but widely scattered in all the continents save Antartica [20]. Most of the isolations came from farm animals, notably pigs. At the time of the first isolation the use of colistin was not regulated in much of Southeast Asia and was commonly used for promoting growth [20] and for prophylaxis for gastrointestinal disease during weaning [21]. It is apparent that the use of colistin created selective pressure favoring bacteria carrying mcr-1. The gene was most commonly found on IncI2 plasmids [22].
IncI2 plasmids appear to have only recently become associated with resistance genes; however, their tendency to carry resistance to the antibiotics of last resort and their widespread distribution increase their relative importance. Both the β-lactamase- and mcr-1-containing plasmids clustered together in the IncI2 phylogenetic analysis, and this cluster of plasmids was also more likely to be carrying mobile genetic elements (MGEs), including MGEs that were not directly related to acquisition of the resistance genes. Although the clustering may have resulted from strong bias in the representative isolates that have been sequenced, the clustering of plasmids containing increased accessory genes was also evident when examining the collection of IncI2 plasmids sequenced through multiple surveillance networks within the USA. Our results illustrate that the IncI2 plasmids that were part of the resistance cluster had a greater diversity of accessory genes and shared several mobile elements that were not specifically associated with the resistance genes that they carried, and that were missing from the other clade of IncI2 plasmids. Identifying the source of these shared mobile elements (other co-resident plasmids vs. direct acquisition through DNA uptake) could aid in identifying potential routes of resistance gene acquisition in this family of plasmids.
In conclusion, we have demonstrated increased prevalence of IS elements and AMR genes within a subset of IncI2 plasmids and that mcr gene acquisition may be associated with a short palindrome sequence directly downstream of the nikB gene. In addition, our core gene SNP analysis identified strong linkages of allelic variants that could indicate evolution towards increased acquisition of MGEs within the IncI2 family of plasmids. Future work aimed at determining whether the acquisition of certain genes or specific mutations within core genes has facilitated increased gene flexibility within this subset of plasmids, would be pertinent. Our work suggests that mutational changes in conjugation-related genes, or specific gene acquisitions such as relE, could influence host range or plasmid stability and therefore impact the evolutionary path of this family of plasmids. As IncI2 plasmids are among the most prevalent mcr-1-carrying plasmids globally and are also found in the greatest diversity of bacterial hosts and geographic locations [23], understanding the selective pressures and evolution of this subset of IncI2 plasmids could potentially direct future research and identify key bacterial hosts, isolation sources, or co-occurring plasmids that have influenced the acquisition of mobile elements in these highly disseminated plasmids.
4. Materials and Methods
4.1. Phylogenetic Reconstruction of IncI2 Plasmids
A total of 261 plasmids (listed in Table S1) were collected from public databases and government surveillance programs in the USA. Genes were annotated using Prokka v1.14.6 [24] and a general time reversible phylogenetic tree was made in FastTree v2.1.11 [25] using the core genome alignment produced by Roary v3.13.0 [26] with the original described IncI2 plasmid (AP002527) designated as the root. Visualizations of constructed trees and pangenome content were created using Phandango [27] and iTOL [19]. MOB-suite v3.0.3 was used to reconstruct plasmid content from each assembly. MOB-recon was used to analyze plasmid sequences, which includes MOB_typer to perform relaxase and replicon typing of plasmids, as well as generate MOB-cluster codes and host range information.
4.2. Analysis of USA Plasmids
We undertook a separate phylogenetic analysis of USA plasmids due to the extent of isolates available through government surveillance programs that did not specifically rely on antibiotic selection for isolate acquisition. A total of 106 plasmids with the source designations of “US”, “USA”, “Pulsenet”, “GenomeTrakr”, “FSIS” and “NARST” were analyzed separately as the USA cohort. Alignment of the USA cohort was performed using the MAFFT v7.308 [28] in Geneious, and the same phylogenetic analysis as described above was performed using the Compute Canada Graham cluster. To determine linkage between loci among the plasmids, the linkage allele table described by Meinersmann [16] was extended to include the new plasmids of interest. The table was imported into Arlequin v3.5.2.2 [29]. The exact test of linkage disequilibrium [30] and a locus-by-locus AMOVA with population specific Fst’s [31] were calculated.
Acknowledgments
We are grateful to John Nash, James Robertson and Justin Schonfeld for supplying SRA sequence assemblies and for helpful discussions throughout the project. NR would also like to thank Adina Howe and the members of GERMSlab, as well as Julian Trachsel and members of the USDA Bioinformatics club for helpful scripts and computational assistance.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics11020181/s1, Table S1: Metadata for global collection of IncI2 plasmids; Table S2: Mob-suite distribution for IncI2 plasmids in this study; Table S3: Core genes identified in the USA plasmids; Table S4: Linkage analysis.
Author Contributions
Conceptualization, N.R. and R.J.M.; Data curation, N.R. and E.W.; Formal analysis, N.R., G.C. and R.J.M.; Methodology, N.R. and R.J.M.; Supervision, H.K.A.; Validation, R.J.M.; Visualization, N.R. and G.C.; Writing—original draft, N.R.; Writing—review and editing, G.C., H.K.A. and R.J.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was undertaken thanks in part to funding from the Canada First Research Excellence Fund and was enabled in part by support by Compute Ontario (www.computeontario.ca) and Compute Canada (www.computecanada.ca). This research was supported in part by an appointment to the Agricultural Research Service (ARS) Research Participation Program administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and the U.S. Department of Agriculture (USDA). ORISE is managed by ORAU under DOE contract number DE-AC05-06OR23100. This research used resources provided by the SCINet project and the AI Center of Excellence of the USDA Agricultural Research Service, ARS project number 0500-00093-001-00-D and funds from ARS project number 6040-32000-009-00-D. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S Department of Agriculture. USDA is an equal opportunity provider and employer.
Data Availability Statement
All sequences used in this study were acquired through publicly available databases.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Douarre P.E., Mallet L., Radomski N., Felten A., Mistou M.Y. Analysis of COMPASS, a New Comprehensive Plasmid Database Revealed Prevalence of Multireplicon and Extensive Diversity of IncF Plasmids. Front. Microbiol. 2020;11:483. doi: 10.3389/fmicb.2020.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong M.H., Liu L., Yan M., Chan E.W., Chen S. Dissemination of IncI2 Plasmids That Harbor the blaCTX-M Element among Clinical Salmonella Isolates. Antimicrob. Agents Chemother. 2015;59:5026–5028. doi: 10.1128/AAC.00775-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bortolaia V., Hansen K.H., Nielsen C.A., Fritsche T.R., Guardabassi L. High diversity of plasmids harbouring bla(CMY-2) among clinical Escherichia coli isolates from humans and companion animals in the upper Midwestern USA. J. Antimicrob. Chemother. 2014;69:1492–1496. doi: 10.1093/jac/dku011. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y.Y., Wang Y., Walsh T.R., Yi L.X., Zhang R., Spencer J., Doi Y., Tian G., Dong B., Huang X., et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 5.Rhouma M., Letellier A. Extended-spectrum β-lactamases, carbapenemases and the mcr-1 gene: Is there a historical link? Int. J. Antimicrob. Agents. 2017;49:269–271. doi: 10.1016/j.ijantimicag.2016.11.026. [DOI] [PubMed] [Google Scholar]
- 6.Tartor Y.H., Abd El-Aziz N.K., Gharieb R.M.A., El Damaty H.M., Enany S., Soliman E.A., Abdellatif S.S., Attia A.S.A., Bahnass M.M., El-Shazly Y.A., et al. Whole-Genome Sequencing of Gram-Negative Bacteria Isolated From Bovine Mastitis and Raw Milk: The First Emergence of Colistin mcr-10 and Fosfomycin fosA5 Resistance Genes in Klebsiella pneumoniae in Middle East. Front. Microbiol. 2021;12:770813. doi: 10.3389/fmicb.2021.770813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baron S., Hadjadj L., Rolain J.M., Olaitan A.O. Molecular mechanisms of polymyxin resistance: Knowns and unknowns. Int. J. Antimicrob. Agents. 2016;48:583–591. doi: 10.1016/j.ijantimicag.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 8.Shen Z., Wang Y., Shen Y., Shen J., Wu C. Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect. Dis. 2016;16:293. doi: 10.1016/S1473-3099(16)00061-X. [DOI] [PubMed] [Google Scholar]
- 9.Li R., Xie M., Zhang J., Yang Z., Liu L., Liu X., Zheng Z., Chan E.W., Chen S. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J. Antimicrob. Chemother. 2017;72:393–401. doi: 10.1093/jac/dkw411. [DOI] [PubMed] [Google Scholar]
- 10.Poirel L., Kieffer N., Nordmann P. In Vitro Study of ISApl1-Mediated Mobilization of the Colistin Resistance Gene mcr-1. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.00127-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snesrud E., He S., Chandler M., Dekker J.P., Hickman A.B., McGann P., Dyda F. A Model for Transposition of the Colistin Resistance Gene mcr-1 by ISApl1. Antimicrob. Agents Chemother. 2016;60:6973–6976. doi: 10.1128/AAC.01457-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snesrud E., McGann P., Chandler M. The Birth and Demise of the ISApl1-mcr-1-ISApl1 Composite Transposon: The Vehicle for Transferable Colistin Resistance. MBio. 2018;9 doi: 10.1128/mBio.02381-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meinersmann R.J., Ladely S.R., Plumblee J.R., Cook K.L., Thacker E. Prevalence of mcr-1 in the Cecal Contents of Food Animals in the United States. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.02244-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meinersmann R.J., Ladely S.R., Bono J.L., Plumblee J.R., Hall M.C., Genzlinger L.L., Cook K.L. Complete Genome Sequence of a Colistin Resistance Gene (mcr-1)-Bearing Isolate of Escherichia coli from the United States. Genome Announc. 2016;4:e01283-16. doi: 10.1128/genomeA.01283-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meinersmann R.J., Ladely S.R., Plumblee J.R., Hall M.C., Simpson S.A., Ballard L.L., Scheffler B.E., Genzlinger L.L., Cook K.L. Colistin Resistance mcr-1-Gene-Bearing Escherichia coli Strain from the United States. Genome Announc. 2016;4:e00898-16. doi: 10.1128/genomeA.00898-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meinersmann R.J. The biology of IncI2 plasmids shown by whole-plasmid multi-locus sequence typing. Plasmid. 2019;106:102444. doi: 10.1016/j.plasmid.2019.102444. [DOI] [PubMed] [Google Scholar]
- 17.Yang Y.Q., Li Y.X., Lei C.W., Zhang A.Y., Wang H.N. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2018;73:1791–1795. doi: 10.1093/jac/dky111. [DOI] [PubMed] [Google Scholar]
- 18.Qiao M., Ying G.G., Singer A.C., Zhu Y.G. Review of antibiotic resistance in China and its environment. Environ. Int. 2018;110:160–172. doi: 10.1016/j.envint.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 19.Letunic I., Bork P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2007;23:127–128. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valiakos G., Kapna I. Colistin Resistant mcr Genes Prevalence in Livestock Animals (Swine, Bovine, Poultry) from a Multinational Perspective. A Systematic Review. Vet. Sci. 2021;8:265. doi: 10.3390/vetsci8110265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhouma M., Fairbrother J.M., Beaudry F., Letellier A. Post weaning diarrhea in pigs: Risk factors and non-colistin-based control strategies. Acta Vet. Scand. 2017;59:31. doi: 10.1186/s13028-017-0299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S., Abbas M., Rehman M.U., Wang M., Jia R., Chen S., Liu M., Zhu D., Zhao X., Gao Q., et al. Updates on the global dissemination of colistin-resistant Escherichia coli: An emerging threat to public health. Sci. Total Environ. 2021;799:149280. doi: 10.1016/j.scitotenv.2021.149280. [DOI] [PubMed] [Google Scholar]
- 23.Li X.P., Sun R.Y., Song J.Q., Fang L.X., Zhang R.M., Lian X.L., Liao X.P., Liu Y.H., Lin J., Sun J. Within-host heterogeneity and flexibility of mcr-1 transmission in chicken gut. Int. J. Antimicrob. Agents. 2020;55:105806. doi: 10.1016/j.ijantimicag.2019.09.010. [DOI] [PubMed] [Google Scholar]
- 24.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 25.Price M.N., Dehal P.S., Arkin A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Page A.J., Cummins C.A., Hunt M., Wong V.K., Reuter S., Holden M.T., Fookes M., Falush D., Keane J.A., Parkhill J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hadfield J., Croucher N.J., Goater R.J., Abudahab K., Aanensen D.M., Harris S.R. Phandango: An interactive viewer for bacterial population genomics. Bioinformatics. 2018;34:292–293. doi: 10.1093/bioinformatics/btx610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Excoffier L., Lischer H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 30.Slatkin M. Linkage disequilibrium in growing and stable populations. Genetics. 1994;137:331–336. doi: 10.1093/genetics/137.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Excoffier L., Smouse P.E., Quattro J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences used in this study were acquired through publicly available databases.