Abstract
Mitochondrial Ca2+-independent phospholipase A2γ (iPLA2γ/PNPLA8) was previously shown to be directly activated by H2O2 and release free fatty acids (FAs) for FA-dependent H+ transport mediated by the adenine nucleotide translocase (ANT) or uncoupling protein 2 (UCP2). The resulting mild mitochondrial uncoupling and consequent partial attenuation of mitochondrial superoxide production lead to an antioxidant effect. However, the antioxidant role of iPLA2γ in the brain is not completely understood. Here, using wild-type and iPLA2γ-KO mice, we demonstrate the ability of tert-butylhydroperoxide (TBHP) to activate iPLA2γ in isolated brain mitochondria, with consequent liberation of FAs and lysophospholipids. The liberated FA caused an increase in respiratory rate, which was fully inhibited by carboxyatractyloside (CATR), a specific inhibitor of ANT. Employing detailed lipidomic analysis, we also demonstrate a typical cleavage pattern for TBHP-activated iPLA2γ, reflecting cleavage of glycerophospholipids from both sn-1 and sn-2 positions releasing saturated FAs, monoenoic FAs, and predominant polyunsaturated FAs. The acute antioxidant role of iPLA2γ-released FAs is supported by monitoring both intramitochondrial superoxide and extramitochondrial H2O2 release. We also show that iPLA2γ-KO mice were more sensitive to stimulation by pro-inflammatory lipopolysaccharide, as reflected by the concomitant increase in protein carbonyls in the brain and pro-inflammatory IL-6 release in the serum. These data support the antioxidant and anti-inflammatory role of iPLA2γ in vivo. Our data also reveal a substantial decrease of several high molecular weight cardiolipin (CL) species and accumulation of low molecular weight CL species in brain mitochondria of iPLA2γ-KO mice. Collectively, our results support a key role of iPLA2γ in the remodeling of lower molecular weight immature cardiolipins with predominantly saturated acyl chains to high molecular weight mature cardiolipins with highly unsaturated PUFA acyl chains, typical for the brain.
Keywords: mitochondria, phospholipase iPLA2γ/PNPLA8, adenine nucleotide translocase, redox homeostasis, cardiolipin remodeling
1. Introduction
Cellular redox homeostasis can be defined as a dynamic steady state maintained by metabolic fluxes and redox feedback, in which oxidants (electrophiles) produced by aerobic life are reduced by antioxidant mechanisms, which reestablish nucleophilic tone [1,2]. While a spatiotemporal transient increase in oxidant production provides a means to essential regulatory redox signaling, a sustained deviation from the steady-state set point refers to physiological oxidative stress [3].
The brain is one of the main organs in the body with the highest metabolic demand and requires tight regulation of the surrounding environment [4,5]. The brain produces various reactive oxygen species (ROS) in enzymatic and non-enzymatic reactions as a byproduct of metabolism [6,7,8]. Both redox signaling and oxidative stress are recognized to be involved in all aspects of the central nervous system development, function, aging, and disease [9]. The brain deliberately produces electrophilic species to transmit redox signals in order to regulate critical functions of all its major cell types, namely glial cells (oligodendrocytes, astrocytes, microglia) and neurons [10]. Neurons are especially susceptible to harmful activities of oxidants due to their poor antioxidative equipment, and many of the reasons why the brain is vulnerable to oxidative stress remain obscure [11].
Similar to other tissues, mitochondria belong to the most important source of ROS in the brain [10,11]. There are at least eleven distinct mitochondrial sites that are known to leak electrons to oxygen to produce superoxide (O2●−) and downstream oxidants under different bioenergetic conditions [12]. Within the mitochondria, O2●− is dismutated to H2O2 by mitochondrial matrix manganese superoxide dismutase (MnSOD) [13]. In the brain, the MnSOD activity is essential as MnSOD deficiency causes increased susceptibility to oxidative mitochondrial injury in central nervous system neurons, including severe neurodegeneration and mitochondrial oxidative damage [14,15,16]. Mitochondria also possess a multilevel network of both enzymatic and non-enzymatic antioxidant systems for the detoxification of H2O2 [17,18,19,20]. The expression and contributions of enzymatic systems for H2O2 detoxification vary widely between tissues and include catalase, glutathione/glutathione peroxidase (GSH/GPx), and the thioredoxin/peroxiredoxin systems [21]. While the expression of catalase in brain mitochondria is negligible, and the GSH/GPx shows minimal contribution [20], brain mitochondria were shown to remove H2O2 mainly in a unique respiration-dependent manner primarily via the thioredoxin 2/peroxiredoxin 3 and 5 system [20].
The role of fatty acids (FAs), notably polyunsaturated FAs (PUFAs), in the maintenance of redox homeostasis in the brain is still unclear. Similarly, the metabolism of fatty acids in the brain is still not completely understood, especially in relation to complex brain morphology [10,22,23] and the accumulation of FAs in brain tissue is associated with some inherited neurological disorders [10,11]. In addition, high unsaturated lipid content defines a cause of oxidative stress because of fatty acid-associated deleterious effects (the so-called lipotoxicity) and also the susceptibility of PUFAs to lipid peroxidation [11,24].
There are several physiological and pathological sources of elevated FA in brain tissue, including the uptake of exogenous FAs from the blood and the liberation of endogenous FAs by intracellular phospholipase A2 [10,25,26]. The phospholipase A2 (PLA2) superfamily contains more than 50 enzymes in mammals that are subdivided into several distinct families on a structural and biochemical basis, including group VI, also termed patatin-like phospholipase domain-containing lipases (PNPLAs) [26]. The phospholipases iPLA2β (group VIA PLA2, PNPLA9) and iPLA2γ (group VIB, PNPLA8) were both shown to be located to mitochondria; however, only iPLA2γ is known to contain the N-terminal mitochondrial localization sequence [26,27]. Like all PNPLAs, the mitochondrial iPLA2γ catalyzes the cleavage of acyl groups from glycerophospholipids from both sn-1 and sn-2 positions and is predominantly an sn-1 lipase for phospholipids containing a PUFA chain at the sn-2 position [28]. iPLA2γ is also a major mediator releasing oxidized aliphatic chains from cardiolipin, and iPLA2γ together with cardiolipin play integrated roles in lipid second-messenger production and mitochondrial bioenergetics during oxidative stress [29]. In addition, studies utilizing iPLA2γ-knockout (KO) mice identified the obligatory role of iPLA2γ in neuronal mitochondrial structure and function [30]. iPLA2γ-KO mice are characterized by significant changes in hippocampal lipids, including cardiolipin content and molecular species composition, and the presence of increased levels of oxidized lipid molecular species [30]. In addition, decreasing iPLA2γ activity increases lipid oxidative stress accompanied by mitochondrial disorders in the brain [31]. Thus, neurodegeneration is strongly related to iPLA2γ activity [32], and upregulating iPLA2γ activity can restore mitochondrial membrane and function [31,33].
Our previous results revealed a non-canonical role for iPLA2γ in participating in cellular antioxidant protection. We showed that the iPLA2γ is directly stimulated by hydroperoxides and hence can be activated physiologically by redox signaling or oxidative stress [34,35,36]. The tert-butylhydroperoxide (TBHP) or H2O2-activated iPLA2γ liberates FAs, which interact with mitochondrial uncoupling proteins (UCPs) and adenine nucleotide translocases (ANTs). This induces FA-mediated H+ transport, leading to a mild dissipation of the mitochondrial protonmotive force (also termed mild mitochondrial uncoupling) [37,38,39,40,41,42,43,44]. This leads to the attenuation of mitochondrial superoxide formation ([34,35,36], reviewed in [45,46]). Therefore, iPLA2γ is a crucial component of a feedback regulation, where partial ROS increase is subsequently eliminated by mitochondrial uncoupling due to activation of the iPLA2γ-UCP(ANT) antioxidant synergy [45,46].
Here, we tested the hypothesis that mitochondrial iPLA2γ participates in the maintenance of redox homeostasis in the brain due to the interaction of iPLA2γ-released FAs with brain UCPs or ANT. Using wild-type (WT) and iPLA2γ-KO mice, we demonstrate the ability of TBHP to activate iPLA2γ in isolated brain mitochondria, with consequent liberation of FAs and lysophospholipids. The liberated FA caused an increase in respiratory rate, which was fully inhibited by carboxyatractyloside (CATR), a specific inhibitor of ANT. Employing detailed lipidomic analysis, we also demonstrate a typical cleavage pattern for TBHP-activated iPLA2γ, reflecting cleavage of glycerophospholipids from both sn-1 and sn-2 positions releasing free saturated FAs, monoenoic FAs, and predominant PUFAs. The acute antioxidant role of iPLA2γ is supported by monitoring both intramitochondrial superoxide and extramitochondrial H2O2 release and is further supported by the measurement of protein carbonyl content in brain tissue and IL-6 concentration in the serum. Moreover, iPLA2γ participation in remodeling of cardiolipins was suggested by the predominance of lower-molecular-weight cardiolipins containing relatively saturated acyl chains in mitochondria of iPLA2γ-KO mice, reflecting stalled deacylation of immature cardiolipins and their subsequent reacylation/transacylation into mature cardiolipins containing long PUFA acyl chains.
2. Materials and Methods
2.1. Chemicals and Reagents
r-Bromenol lactone (r-BEL) and carboxyatractyloside (CATR) were obtained from Cayman Chemical (Ann Arbor, MI, USA). Amplex Red and MitoSOX RedTM were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Other reagents were obtained from Sigma-Aldrich (Darmstadt, Germany).
2.2. Creation of iPLA2γ/PNPLA8 Knockout Mice
The iPLA2γ/PNPLA8 knockout mice were generated using transcription activator-like effector nucleases (TALENs) as described previously [47], starting with C57Bl6/N mice. TALENs were designed to target the Pnpla8 exon-3 to eliminate the XbaI restriction site (Supplemental Figure S1). DNA from tails of young mice was analyzed by the PCR restriction-fragment-length polymorphism. Purified PCR products were digested with XbaI (ThermoFisher, Waltham, MA, USA). To further select appropriate mice for breeding, the PCR products were subcloned into the pGEM®-T Easy Vector System (Promega, Madison, WI, USA) and sequenced using M13 reverse and forward sequencing primers. For further breeding, mice bearing a 13 base-pairs long deletion in exon-3, which resulted in the formation of a premature stop codon in the fourth exon, were selected. The wild-type (WT) mice used were those backcrossed >10 generations into the iPLA2γ/PNPLA8-knockout mice background. Animals were housed in open cages and kept at standard housing conditions with free access to water and a normal chow diet (NCD; Altromin 1314 Forti, Postfach, Germany). All animal studies were ethically reviewed and performed in accordance with European Directive 2010/63/EU, complying with the NIH Publication No.85–23 (revised 1996) and the ARRIVE guidelines; and were approved by the Czech Central Commission for Animal Welfare.
2.3. Isolation of Mitochondria
Typically, 2–4 mice 20–24 weeks old were sacrificed by cervical dislocation. The brains were removed and washed in ice-cold isolation medium (225 mM of mannitol, 75 mM of sucrose, 5 mM of HEPES, 1 mM of EGTA, 0.1 mM of EDTA, pH 7.4 by TRIS) supplemented with 0.5% bovine serum albumin (BSA) and with 5 mM of N-acetylcysteine (NAC), where indicated. Brain mitochondria were isolated following the procedure of Stepanova et al. [48], combining differential centrifugation and digitonin treatment. Briefly, after homogenization with a Dounce homogenizer, homogenates were centrifuged at 2000× g for 10 min at 4 °C. Pellets were discarded, and 0.02% digitonin was added to supernatants for synaptosome lysing. Supernatants were centrifuged at 10,000× g for 10 min at 4 °C, pellets were resuspended in the isolation medium lacking BSA and NAC, and were centrifuged as above. The final pellet was resuspended to an approximate concentration of 20 mg protein/mL in the isolation medium lacking BSA. The exact protein concentration was determined by the BCA method (Sigma).
2.4. High-Resolution Respirometry
Mitochondrial oxygen consumption was monitored using an Oxygraph 2k high-resolution respirometer (Oroboros, Innsbruck, Austria) after air calibration and background correction. The respiratory medium included KCl (125 mM), HEPES (20 mM), EGTA (0.2 mM), KH2PO4 (2 mM), MgCl2 (2 mM), pH 7.4 at 30 °C. The respiratory medium routinely contained both Complex I- and Complex II-linked substrates, glutamate (5 mM), malate (1 mM), and succinate (5 mM), referred to as GMS, and ATP synthase inhibitor oligomycin (1 μg/mL), where applicable. A low non-saturating concentration of BSA (typically titrated to 0.5 μM) was also used during the experiments to minimize the effect of residual contaminating FAs.
2.5. Intramitochondrial Detection of Superoxide Formation
Surplus superoxide released into the mitochondrial matrix was monitored by increasing fluorescence of MitoSOX RedTM (Thermo Fisher), which accumulates in the mitochondrial matrix [35,36,49]. To support a uniform distribution of MitoSOX Red within mitochondria, the isolated brain mitochondria at a concentration of 0.2 mg/mL were preincubated with 1 μM of MitoSOX and 5 mM of NAC in respiration buffer containing respiratory substrates (only 5 mM of glutamate, 1 mM of malate) for 45 min. After this preincubation, 0.5% BSA was added for another 5 min to remove any residual released FAs. Mitochondria were centrifuged at 10,000× g for 10 min at 4 °C, and pellets were resuspended in the isolation medium. Fluorescence was monitored with an RF 5301 PC spectrofluorometer (Shimadzu, Duisburg, Germany) at excitation of 510 nm and emission of 590 nm at 30 °C.
2.6. Extramitochondrial Detection of H2O2 Release
The H2O2 released out of mitochondria was detected with AmplexTM UltraRed Reagent (ThermoFisher), which allows for a highly sensitive and selective quantitative enzymatic detection of H2O2 [50]. Mitochondria were used at a concentration of 0.2 mg/mL protein in a standard respiration medium at 30 °C, and the monitoring of H2O2 release was initiated by adding the Amplex UltraRed probe (10 μM) in the presence of horseradish peroxidase (5 U/mL). The fluorescence signal was calibrated for each experimental condition by sequential additions of H2O2 aliquots. Fluorescence intensity was detected using excitation and emission wavelengths of 572 nm and 580 nm on an RF 5301 PC spectrofluorometer (Shimadzu) at 30 °C.
2.7. LC–MS-Based Lipidomic Profiling
Extraction was carried out using a biphasic solvent system of cold methanol, methyl tert-butyl ether (MTBE), and water. In more detail, 0.4 mg of the mitochondrial pellet was homogenized (1.5 min) with 275 µL of methanol and 275 µL of 10% methanol using a grinder. Then, 1 mL of MTBE was added, and the tubes were shaken (1 min) and centrifuged (16,000 rpm, 5 min, 4 °C). For lipidomic profiling, 500 µL of upper organic phase was collected, evaporated, and resuspended using methanol containing 12-[[(cyclohexylamino) carbonyl]amino]-dodecanoic acid (CUDA) internal standard, shaken (30 s), centrifuged (16,000 rpm, 5 min, 4 °C), and used for LC-MS analysis in negative electrospray ion mode.
The LC-MS analysis systems consisted of a Vanquish UHPLC System (Thermo Fisher) coupled to a QExactive Plus mass spectrometer (Thermo Fisher). Experimental conditions are described in detail in [51]. A sample volume of 5 μL was used for injection. LC-MS instrumental files from lipidomic profiling were processed through MS-DIAL 4.70 software [52], including lipid annotations. Exported raw data were filtered using blank samples, serial dilution samples, and quality control (QC) pool samples with relative standard deviation (RSD) < 30%, normalized using the LOESS approach by means of QC pool samples, regularly injected between 10 actual samples. Samples were randomized across the platform run. Only lipid species with a signal intensity 10-fold higher than in corresponding blank samples were considered for further data analysis.
2.8. Agent/Drug Application to Mice
Where indicated, mice were injected with lipopolysaccharides (LPS) (6 mg/kg body weight) or with iPLA2γ inhibitor r-BEL (1 mg/kg body weight) and LPS in parallel. LPS was administered one time intraperitoneally (i.p.) 6 h before sacrifice, while r-BEL was subcutaneously applied to mice in 24-h periods, three times during 72 h [33]. Animals treated with vehicle (phosphate-buffered saline and/or DMSO) under the same conditions were used as the control group. Mice were sacrificed, selected tissues were collected, and aliquots of each tissue were immediately frozen in liquid nitrogen and stored at −80 °C until the analysis was performed. Interleukin-6 (IL-6) concentrations were determined in mouse sera. The protein carbonyl content was established from tissue homogenates. Typically, one group contained 3–4 mice, gender-mixed.
2.9. Tissue Homogenates
The selected tissues were minced into small pieces in the mitochondrial isolation medium and homogenized using a Potter-Elvegen tissue grinder. The homogenates were centrifuged at 10,000× g for 10 min at 4 °C, and the supernatants were stored in aliquots at −80 °C. For protein carbonylation, the supernatant absorbance at 280 and 260 nm was checked to determine the possible presence of nucleic acids in the sample. As the ratio of 280/260 nm was less than 1, samples were incubated with streptomycin sulfate at a final concentration of 1% in the sample [53]. After 15 min of incubation at room temperature, the samples were centrifuged at 6000× g for 10 min at 4 °C, and the supernatants were used for determining protein carbonyl content.
2.10. Quantification of Interleukin-6 Levels
IL-6 levels were determined from collected sera using a commercially available kit (IL-6 Mouse ProQuantum Immunoassay Kit, Invitrogen, Waltham, MA, USA). The high-sensitivity immunoassay utilizes proximity ligation assay technology to combine antibody-antigen binding specificity and amplification capabilities of real-time PCR. Briefly, mouse sera were incubated with antibody conjugated to a DNA oligonucleotide for 1 h at room temperature according to manufacturer instructions. After incubation, ligase was added to the samples for the creation of a template strand for amplification. A qPCR reaction was performed on the Bio-Rad CFX96 instrument (Bio-Rad, Hercules, CA, USA).
2.11. Quantification of Protein Carbonyls
The samples were prepared using the method developed by Levine et al. [53] with modifications. Briefly, samples with protein concentrations in the range of 1–10 mg/mL of tissue homogenates were treated with 0.8 mL of 20 mM 2,4-dinitrophenylhydrazine (DNPH) in 2.5 M of HCl for 60 min at room temperature in the dark. The reaction was stopped by adding 1 mL of 20% trichloroacetic acid (TCA), and the TCA-treated samples were incubated on ice for 5 min and centrifuged at 10,000× g for 10 min at 4 °C. The pellets were resuspended in 1 mL of 10% TCA, incubated on ice for 5 min, and then centrifuged as above. The precipitates were washed three times in 1 mL of 1:1 ethanol: ethyl acetate solution and centrifuged as above. The final pellet was dissolved in 0.5 mL of 6 M guanidine hydrochloride. The total protein carbonyl content was measured at 25 °C following the absorbance at 370 nm and determined using the molar absorption coefficient of 22,000 M−1 cm−1.
2.12. Statistical Analysis
The lipidomic data were normalized to total-ion current (TIC) before subsequent analysis. The multivariate analyses were performed using MetaboAnalyst 5.0 [54]. Hierarchical clustering analysis (Euclidean distance) was performed using ComplexHeatmap (Bioconductor) package by RStudio version 1.4. [55] with log10-transformed data scaled with Pareto scaling [56]. The remaining statistical analysis was performed using the GraphPad Prism software version 5.01. The difference between two groups was analyzed using the unpaired Student’s T-test, while two-way analysis of variance (ANOVA) followed by Bonferroni’s post-test was used for comparison of multiple groups. Differences with p < 0.05 were considered statistically significant.
3. Results
3.1. Fatty Acid-Induced Increase in Respiration following Redox Activation of iPLA2γ
3.1.1. Uncovering the Redox Activation of iPLA2γ in Isolated Brain Mitochondria
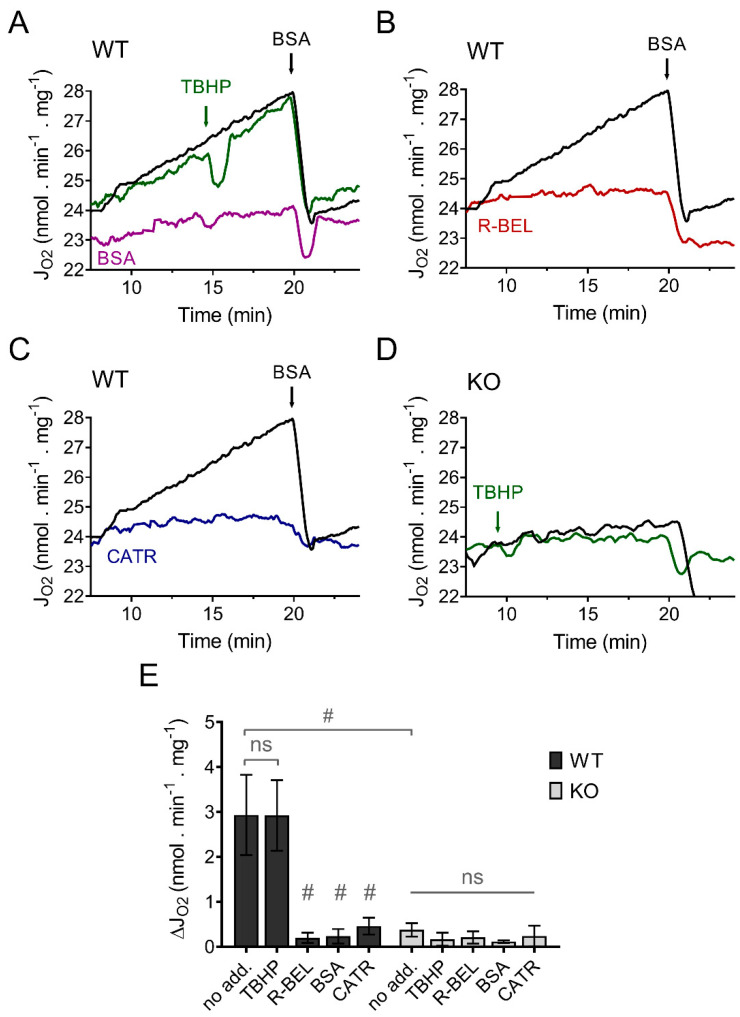
In contrast to our previous results using mitochondria isolated from the heart, lung, and spleen, which revealed a TBHP-dependent increase in respiration due to activation of iPLA2γ [34,36], isolated murine brain mitochondria displayed a steady increase in the rate of basal respiration without the obligatory requirement for TBHP (Figure 1A). This increase in respiratory rate was entirely prevented by an excess of bovine serum albumin (Figure 1A), by a selective iPLA2γ inhibitor r-BEL (Figure 1B), and also by CATR (Figure 1C). On the contrary, no such increase in respiration was observed in brain mitochondria isolated from iPLA2γ-KO mice (Figure 1D). These results indicate that the observed increase in the respiratory rate can be attributed to FAs being gradually liberated by iPLA2γ during the experiment and that iPLA2γ in isolated brain mitochondria is intrinsically active. The CATR inhibition also shows that the liberated FAs interact with ANT to increase the H+ conductance and, consequently, the respiratory rate.
Figure 1.
iPLA2γ is intrinsically active in brain mitochondria isolated from wild-type mice. Representative traces are shown for state-4 respiration rates (JO2) evolving in time in the presence of 5 mM of glutamate, 1 mM of malate, and 5 mM of succinate (GMS). (A) Spontaneous increase in the rate of respiration (black trace) was not sensitive to TBHP (green trace) but was prevented by BSA (purple trace). (B) The effect of r-BEL (red trace). (C) Effect of CATR (blue trace). (D) The basal state-4 respiration rate (black trace) and the effect of TBHP (green trace) in mitochondria isolated from iPLA2γ-KO mice. (E) The effects of various reagents on the respiration rates of brain mitochondria isolated from WT (black bars) or iPLA2γ-KO mice (grey bars) monitored by changes (Δ) in O2 flux (JO2). Reagent concentrations: TBHP, 50 µM; BSA, 5 µM; r-BEL, 2 µM; CATR, 2 µM. All inhibitors were added before the addition of TBHP. Values are plotted as means ± standard deviations. The number of replicates and isolations was at least 4. # p < 0.001 compared to no addition (no add). ns, No significant differences were found between the rates in mitochondria isolated from iPLA2γ-KO mice.
An additional detailed analysis of the coupling states and respiration rates of the mitochondria isolated from WT and iPLA2γ-KO mice, performed in the presence of BSA in excess (0.1%), revealed that both behave identically under the given experimental conditions (Supplemental Figure S2). Specifically, both mitochondrial isolations have nearly identical rates of basal respiration following the addition of substrates, have identical state-3 respiration determined by the addition of ADP (OXPHOS capacity), and have similar maximal respiratory rates obtained by titration of the uncoupler FCCP (maximal respiratory chain capacity) (Supplemental Figure S2). Moreover, the addition of 0.5 μM of linoleic acid to the respiring isolated brain mitochondria also led to an identical increase in respiration in both preparations from WT and iPLA2γ-KO mice, and this increase was abolished by BSA and CATR (Supplemental Figure S3). Altogether, these results show that brain mitochondria isolated from WT and iPLA2γ-KO mice are similar in respect to their respiratory properties and the ability of FAs to interact with ANT and increase H+ conductance.
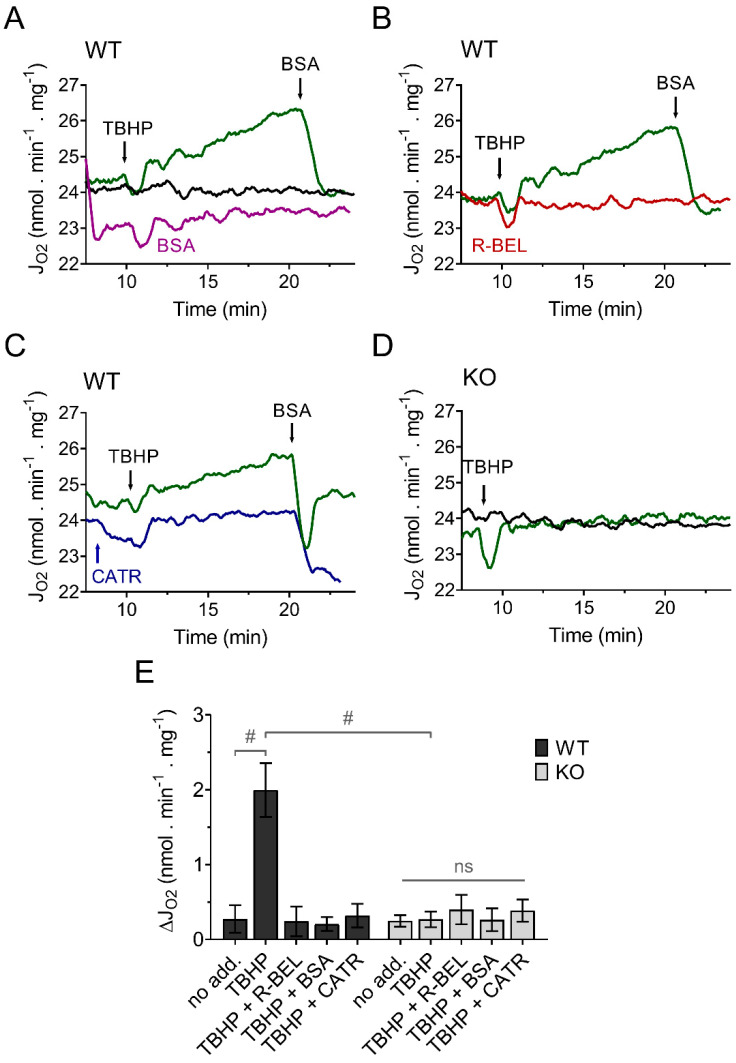
Because we previously demonstrated that the recombinant reconstituted iPLA2γ is sensitive to thiol-reducing compounds [35], we expected that the presence of a membrane-permeable thiol reducing compound, such as N-acetyl cysteine (NAC), during mitochondrial isolation, could reduce the intrinsically active brain mitochondrial iPLA2γ and thus restore the ability of iPLA2γ to respond to and be activated by external or internal oxidants. The results shown in Figure 2. support our prediction. Brain mitochondria isolated from WT mice in the presence of NAC show a negligible increase in the basal respiratory rate, but the addition of TBHP led to a steady increase in the rate of basal respiration (Figure 2A), comparable to the increase seen in the absence of NAC. Analogously, this increase in respiratory rate was completely prevented by a BSA excess (Figure 2A), by a selective iPLA2γ inhibitor r-BEL (Figure 2B), and also by CATR (Figure 2C). Again, no such increase in respiration was observed in brain mitochondria isolated from iPLA2γ-KO mice (Figure 2D). These results are consistent with the ability of iPLA2γ to be reversibly inhibited by thiol reducing compounds, either directly or indirectly. They support the hypothesis of redox activation of iPLA2γ by oxidative posttranslational modification of accessible protein thiols [46].
Figure 2.
TBHP activates iPLA2γ in brain mitochondria isolated in the presence of N-acetyl cysteine. Representative traces are shown for state-4 respiration rates (JO2) evolving in time in the presence of 5 mM glutamate, 1 mM malate, and 5 mM succinate. (A) The basal respiration rate (black trace) was increased by TBHP (green trace), and the TBHP-induced increase in the respiration rate was prevented by BSA (purple trace). (B) The effect of r-BEL (red trace). (C) Effect of CATR (blue trace). (D) The basal state-4 respiration rate (black trace) and the effect of TBHP (green trace) in mitochondria isolated from iPLA2γ-KO mice. (E) The effects of various reagents on the respiration rate of brain mitochondria isolated from WT (black bars) or iPLA2γ-KO mice (grey bars) monitored by changes (Δ) in O2 flux (JO2). Reagent concentrations were identical to those described in Figure 1. The number of replicates and isolations was at least 4 for both WT and iPLA2γ-KO. # p < 0.001 compared to no addition (no add). ns, No significant differences between the rates were found in mitochondria isolated from iPLA2γ-KO mice.
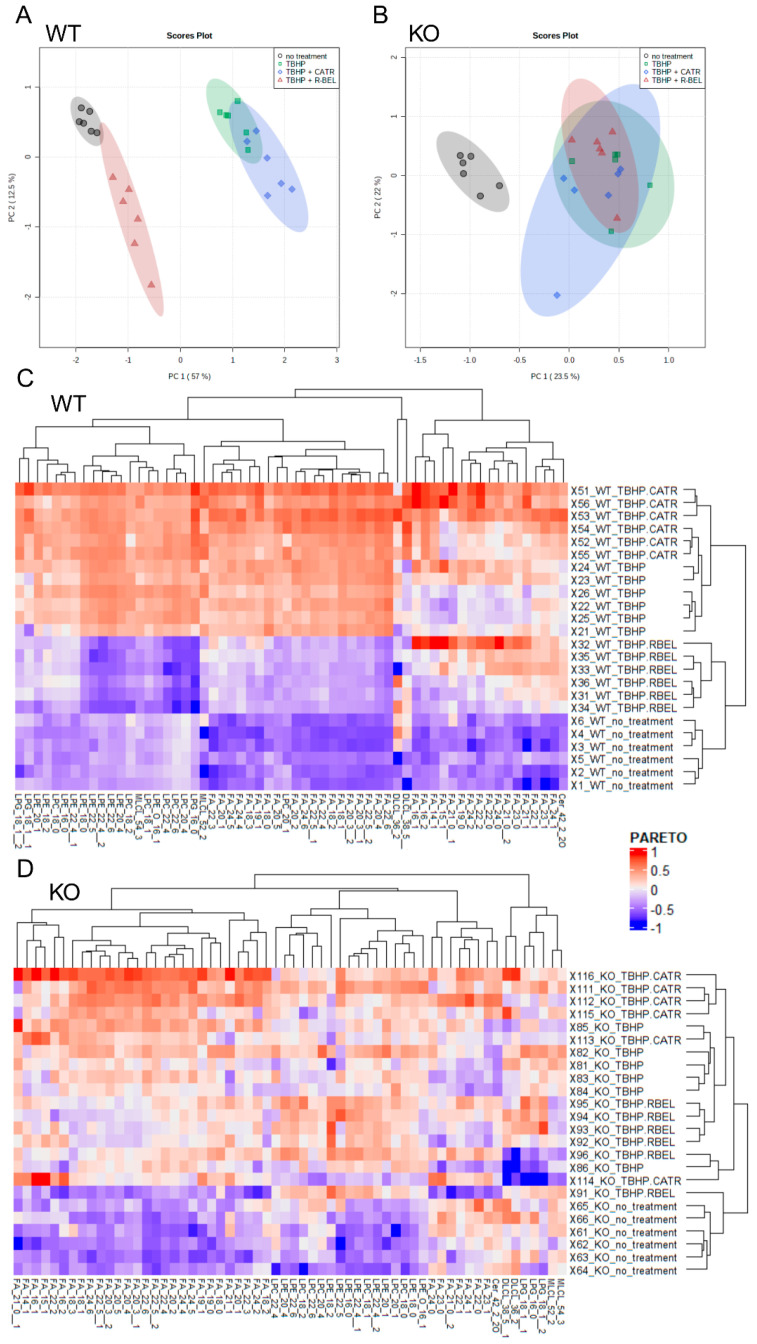
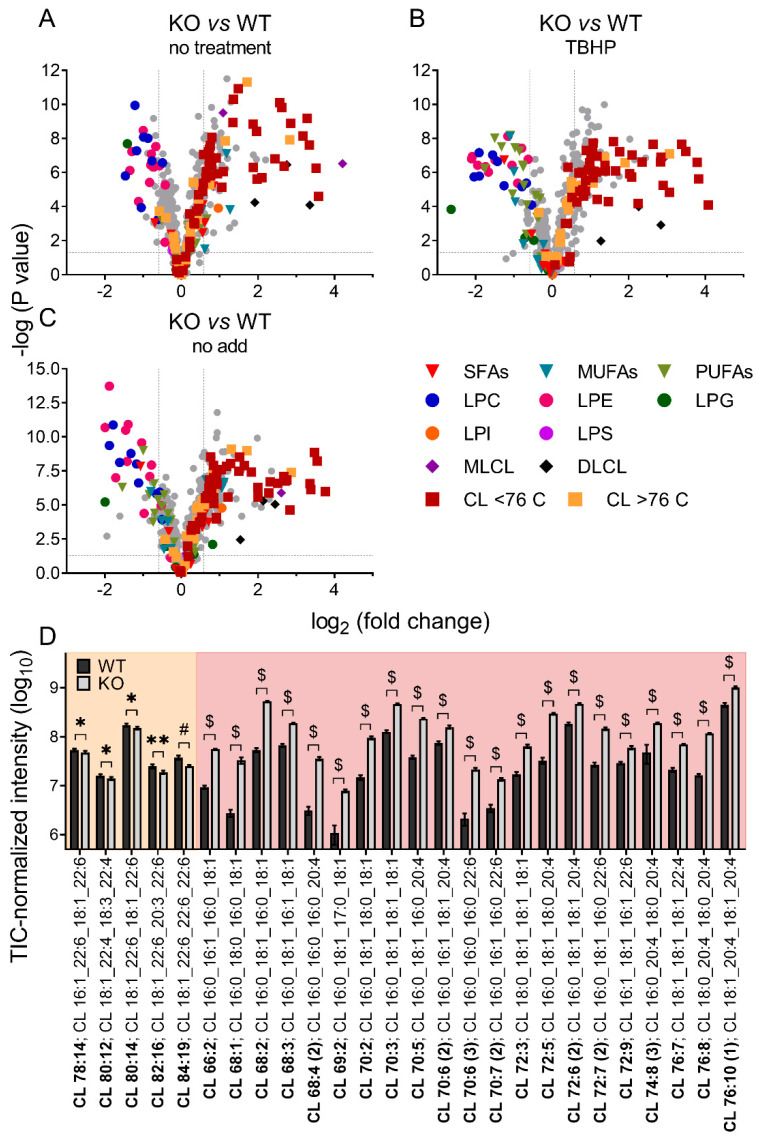
Next, to obtain a complete reaction pattern of TBHP-stimulated catalytic activities of iPLA2γ, mitochondria isolated in the presence of NAC were allowed to respire for 1 h under the conditions described in Figure 2, and the subsequently isolated mitochondrial pellets were subjected to unbiased LC-MS lipidomic profiling. First, to reveal patterns in the data, we performed a multivariate principal component analysis (PCA) based on amounts of lipid species normalized to total ion current (TIC) for all experimental conditions (Figure 3A,B). The score plots show the similarity of phenotypically similar groups and the variability of remote groups, and in this case, active vs. inactive iPLA2γ. In WT brain mitochondria, the result of this analysis formed two related clusters. As shown in Figure 3A,C, the group of “TBHP + r-BEL” clustered together with the “no treatment” group (mitochondria frozen immediately following the isolation and not subjected to the TBHP treatment), which indicates that both groups demonstrate a common metabolic phenotype, where iPLA2γ is not activated. On the contrary, the groups of “TBHP” and “TBHP + CATR” formed another cluster, which indicates that these two groups represent metabolic phenotype following the iPLA2γ activation. Simply, they represent cleaved products of iPLA2γ, liberated FAs, and lysophospholipids. Furthermore, no such clustering was found in iPLA2γ-KO brain mitochondria (Figure 3B,D). The PCA analysis also revealed a separation of WT and iPLA2γ-KO samples in PC1 (Supplemental Figure S4), which indicates that there exists a long-term effect of the iPLA2γ ablation on the overall lipid profile besides the changes initiated by the respective acute treatments (see Section 3.2 below).
Figure 3.
Lipidomic analysis of isolated WT and iPLA2γ-KO mouse mitochondria. Principal component analysis scores plots (PCA) of TIC-normalized log10-transformed intensities for WT (A), iPLA2γ-KO (B). (C,D) Heatmap based on hierarchical clustering presented in dendrograms according to the Euclidean distance. Data are log10-transformed and scaled by Pareto scaling. Lipids with loadings values in PC1 > 0.05 in WT (C) and iPLA2γ-KO (D) mouse brain mitochondria. Isomers are marked with the number after double underscores. Lipidomic data were collected from n = 6 replicates.
3.1.2. Detailed Lipidomic Analyses Reveal the Main Products of the TBHP-Activated iPLA2γ in Brain Mitochondria
Subsequently, we analyzed metabolites, which were responsible for sample clustering within PC1 (Supplemental Figure S5). Hierarchical clustering analysis of metabolites with loading values in PC1 > 0.05 is presented in Figure 3C,D. Importantly, upregulated metabolites in the treatment groups in WT samples distinguished clusters of free FAs and several groups of lysophospholipids, which were absent following the r-BEL treatment (Figure 3C). This type of clustering was completely missing in iPLA2γ-KO samples (Figure 3D). Therefore, the WT pattern “TBHP” and “TBHP + CATR” should indicate the iPLA2γ-specific products.
Moreover, several saturated FAs were liberated after the TBHP treatment, such as 18:0, 19:0, 23:0, 24:0, 22:0, and 21:0 (ordered by descending PC1 loadings values) (Supplemental Figure S4). These data independently support the conclusion that the phospholipase cleaving these FAs by its PLA1 activity is a member of the PNPLA family of group VI because this group possesses the ability to cleave sn-1 positions of the glycerol backbone occupied typically with saturated FAs. We conclude that the identified species of lysophosphatidylcholines (LPCs), lysophosphatidylethanolamines (LPEs), lysophosphatidylglycerols (LPGs), monolysocardiolipins (MLCLs), and dilysocardiolipins (DLCLs) are explicitly formed by the TBHP-induced activity of iPLA2γ in brain mitochondria.
3.1.3. Univariate Analyses of Lipidomic Data Demonstrate a More Detailed Pattern of iPLA2γ Reaction Products
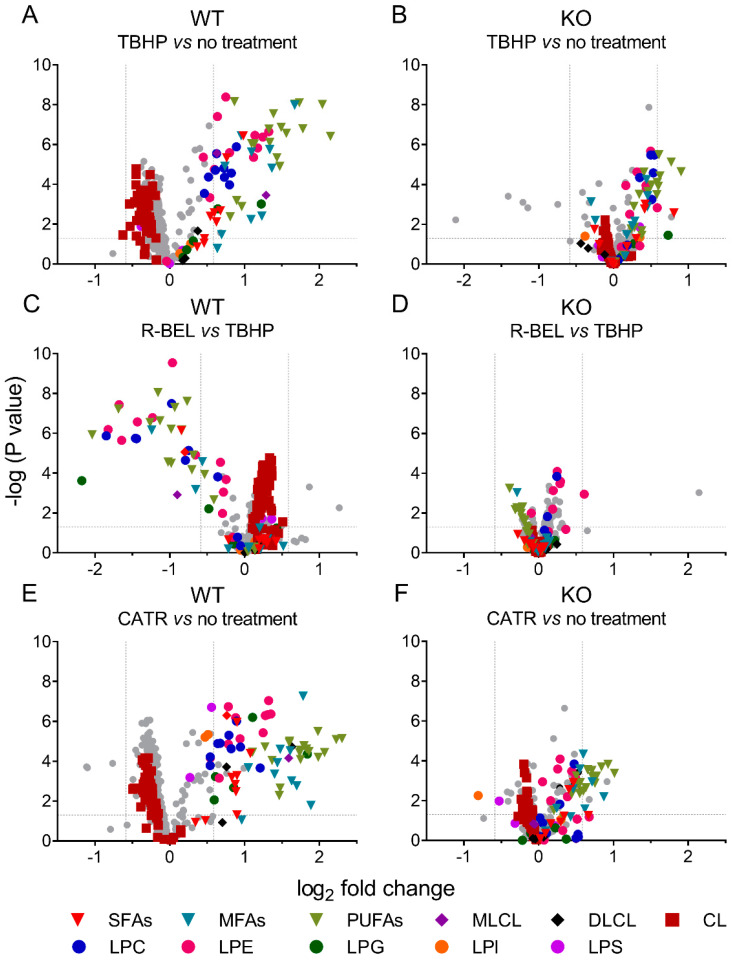
Either considering multivariate analyses with PC1 loadings > 0.05 (Supplemental Figure S4) or comparing two selected groups using univariate analyses, we further examined the differences between mitochondria isolated from WT and iPLA2γ-KO mice in more detail (Figure 4). In agreement with the multivariate analyses, the addition of TBHP significantly increased the relative concentration of free saturated, monoenoic, and polyunsaturated FAs, MLCL, and lysophospholipids (LPC, LPG, LPE), while it decreased the relative levels of cardiolipins (CL) (Figure 4A). The addition of r-BEL inhibited the TBHP-dependent iPLA2γ activation in WT mitochondria (Figure 4C), resulting in a similar pattern to that obtained with iPLA2γ-KO mice (Figure 4B,D). Surprisingly, more products, including DLCL and lysophospholipids LPS and LPI, were detected when CATR was used together with TBHP in WT brain mitochondria (Figure 4E). This pattern ceased for CATR plus TBHP treatment of iPLA2γ-KO samples (Figure 4F).
Figure 4.
Volcano plot analysis of TBHP-induced, iPLA2γ-dependent cleavage of phospholipids. Brain mitochondria isolated from WT mice and respiring in the presence of TBHP are related to mitochondria with no treatment frozen immediately after isolation (A) and to mitochondria treated with TBHP + r-BEL (C). Analogously, brain mitochondria isolated from iPLA2γ-KO mice and respiring in the presence of TBHP are related to mitochondria with no treatment frozen immediately after isolation (B) and to mitochondria treated with TBHP + r-BEL (D). TBHP + CATR treatment is related to mitochondria with no treatment frozen immediately after isolation in WT (E) and iPLA2γ-KO (F) mice. Abbreviations: SFAs, saturated FAs; MFAs, monoenoic FAs; PUFA, polyunsaturated FAs; MLCL, monolysocardiolipin; DLCL, dilysocardiolipin; CL, cardiolipin; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; LPG, lysophosphatidylglycerol; LPI, lysophosphatidylinositol; LPS, lysophosphatidylserine. Lipidomics data were collected from n = 6 replicates.
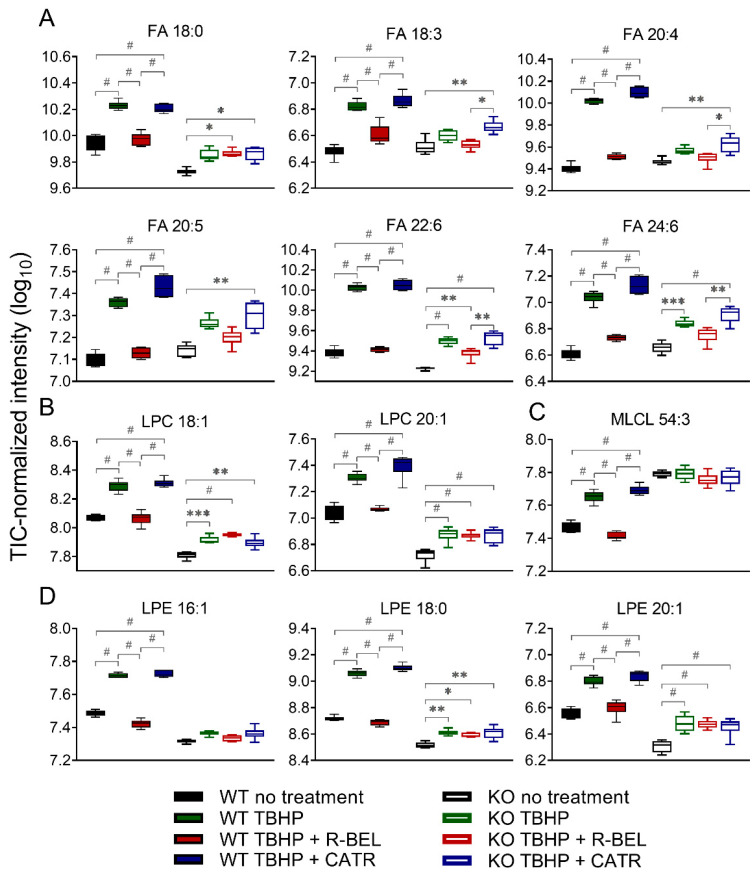
Next, we focused on those lipid species in the WT group, which were consistently downregulated in the presence of r-BEL and were not affected by r-BEL treatment in the iPLA2γ-KO group. In Figure 5, we present 12 of the most prominent lipid metabolites that fulfilled such requirements and were positively correlated with the TBHP-induced iPLA2γ activity in brain mitochondria. These include stearic acid (C18:0), α-linolenic acid (18:3), arachidonic acid (C20:4), eicosapentaenoic acid (C20:5), docosahexaenoic acid (C22:6), and tetracosahexaenoic acid (C24:6) (Figure 5A). Specifically, the addition of TBHP resulted in an approximate 4.8-fold increase in C22:6 (p < 0.0001, n = 6), a 4.5-fold increase in C20:4, while C24:6, C22:4, C20:5, and C18:0 were increased more than 3-fold. Again, the presence of cleaved stearic acid, an unsaturated FA, is consistent with the unique ability of PNPLAs, i.e., group VI phospholipases, to cleave the sn-1 side chain of phospholipids. The overall pattern of FAs released from both sn-1 (predominantly SFAs) plus sn-2 cleaved positions (predominantly PUFAs) is consistent with the lipid signatures typical to brain mitochondria [57].
Figure 5.
The changes in selected fatty acids and lysophospholipids of isolated WT and iPLA2γ-KO mouse mitochondria. (A) Fatty acids, (B) lysophosphatidylcholines, (C) monolysocardiolipin, (D) lysophosphatidylethanolamines. * p < 0.05; ** p < 0.01; *** p < 0.005; # p < 0.001. Lipidomic data are collected from n = 6.
The corresponding parallel increase in certain lysophospholipids species was also found, namely lysophosphatidylcholines (LPC 18:1, LPC 20:1, Figure 5B), monolysocardiolipin (MLCL 54:3, Figure 5C), and lysophosphatidylethanolamines (LPE 16:1, LPE 18:0, and LPE 20:1, Figure 5D), which, again, reflect the pattern of iPLA2γ preferential activity. In brain mitochondria isolated from iPLA2γ-KO mice, the changes in the selected FAs and lysophospholipids were much less prominent, or they were absent altogether (Figure 5 A–D). These residual TBHP-induced changes observed in iPLA2γ-KO may reflect activities of other mitochondria-located phospholipases, such as iPLA2β, or may also possibly reflect the presence of contaminant cytosolic phospholipases.
3.2. Differences in Brain Mitochondrial Lipid Composition of iPLA2γ- KO Mice
Mitochondrial iPLA2γ Participates in Cardiolipin Remodeling
Unbiased lipidomics also revealed profound differences in the cardiolipin species profile for mitochondria of WT and iPLA2γ-KO mice. Figure 6 shows a detailed analysis of CL species where the iPLA2γ-KO mouse brain mitochondria were related to WT under several experimental conditions. First, we analyzed the CL species profile of isolated brain mitochondria that were not subjected to any further treatment and were frozen immediately after the isolation (Figure 6A). We further analyzed the CL species profile following the respiration of NAC-treated mitochondria and inducing the iPLA2γ activity by TBHP (Figure 6B), and following the respiration of mitochondria without the NAC treatment, containing the intrinsically active iPLA2γ (Figure 6C). In all tested conditions, numerous specific species of C66 to C76 cardiolipins were elevated in mitochondria isolated from iPLA2γ-KO mice. Detailed compositions of CL acyl chains of isolated iPLA2γ-KO brain mitochondria that were not subjected to any further treatment show a substantial decrease of five high molecular weight CL species (CL78:14, CL80:12, CL80:14, CL82:16, and CL84:19) and, in contrast, several-fold accumulation of many species of lower molecular weight CLs (Figure 6D). The existence of these elevated cardiolipin species in iPLA2γ-KO relative to WT mouse brain mitochondria could be explained by a stalled specific deacylation of CL acyl chains due to the iPLA2γ ablation. Since CL68:1 is likely to be synthesized from phosphatidylglycerol PG {18:0,18:1} and 1,2-dihexadecanoyl-sn-glycero-3-cytidine-5′-diphosphate (CDP-DG {16:0,16:0}), or from PG {16:0,18:1} and CDP-DG {18:0,16:0}, without activity of iPLA2γ, cardiolipin is no longer deacylated in its C18:0 and C16:0 acyl chains and remains unmodified in the brain tissues of iPLA2γ-KO mice.
Figure 6.
Differences in cardiolipin composition for brain mitochondria from iPLA2γ-KO mice vs. WT. (A) Isolated mitochondria without further treatment, frozen instantly after isolation, (B) mitochondria isolated in the presence of NAC and respiring for 1 h in the presence of TBHP, and (C) mitochondria isolated in the absence of NAC and respiring for 1 h in the absence of TBHP. (D) The changes in cardiolipin composition of individual cardiolipin species of isolated WT and iPLA2γ-KO mitochondria, without further treatment, frozen instantly after isolation (D). Isomers are marked with the number in parentheses. * p < 0.05; ** p < 0.01; # p < 0.001; $ p < 0.000001. Lipidomic data are collected from n = 6 replicates.
These data suggest that phospholipase iPLA2γ participates in the deacylation of lower molecular weight immature cardiolipins with predominantly saturated acyl chains to high molecular weight mature cardiolipins with highly unsaturated PUFA acyl chains, typical for the brain.
3.3. Antioxidant Role of iPLA2γ-Released Fatty Acids in Isolated Brain Mitochondria
3.3.1. Mitochondrial Superoxide Release into the Matrix Decreases following Redox Activation of iPLA2γ
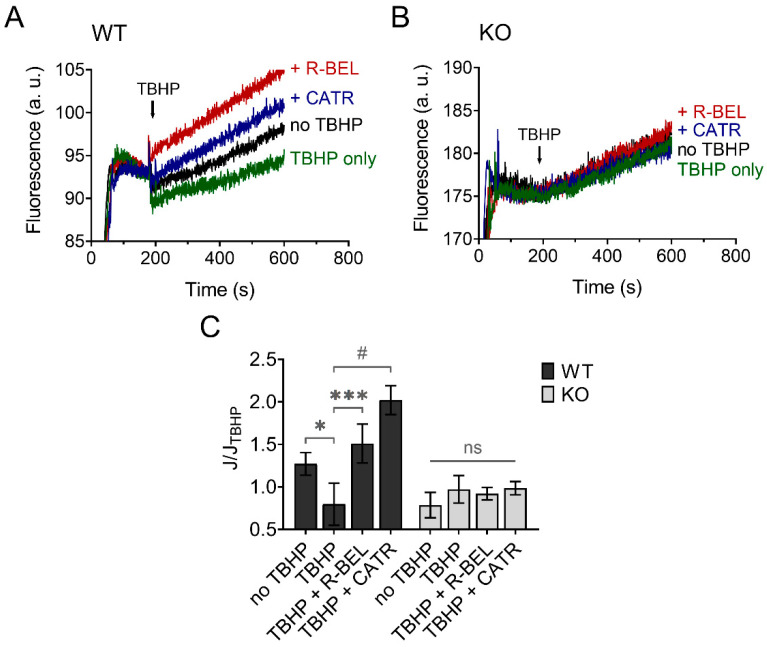
The results presented so far are consistent with hydroperoxide-dependent activation of iPLA2γ, leading to the release of free FAs, which then interact with ANT. This causes an increase in the proton conductance of the inner mitochondrial membrane [40], monitored as an increase in the rate of respiration (Figure 2) [34,36]. Next, we tested whether this mechanism mediates an antioxidant function of iPLA2γ in the brain. Figure 7 shows traces of fluorescence of mitochondria-targeted, superoxide-selective probe MitoSOX following the respiration of isolated brain mitochondria. The addition of TBHP led to a decrease in the rate of MitoSOX fluorescence compared to the baseline (no TBHP), and this effect of TBHP was completely reversed in the presence of a selective iPLA2γ inhibitor r-BEL or by ANT inhibitor CATR (Figure 7A). No such qualitative changes were observed in brain mitochondria isolated from iPLA2γ-KO mice (Figure 7B).
Figure 7.
TBHP-dependent activation of iPLA2γ leads to decreased superoxide formation. Representative traces of MitoSOX fluorescence in isolated brain mitochondria respiring in the presence of 5 mM of glutamate, 1 mM of malate, and 5 mM of succinate. (A) The basal rate of superoxide release (black trace) was decreased by TBHP (green trace), and the TBHP-induced effect was reversed by r-BEL (red trace) and CATR (blue trace). (B) The basal rate of superoxide release (black trace), the effect of TBHP (green trace), r-BEL (red trace), and CATR (blue trace) in mitochondria isolated from iPLA2γ-KO mice. (C) The effects of tested compounds on the rates of superoxide release (J) relative to the rate observed in the presence of TBHP (JTBHP) of brain mitochondria isolated from WT (black bars) or iPLA2γ-KO mice (grey bars). Reagent concentrations were identical to those described in Figure 1. The number of replicates was at least 6 for both WT and iPLA2γ-KO mice. * p < 0.05; *** p < 0.005; # p < 0.001 compared to JTBHP. ns, No significant differences between the rates were found in mitochondria isolated from iPLA2γ-KO mice.
3.3.2. Extramitochondrial H2O2 Release Decreases following Redox Activation of iPLA2γ
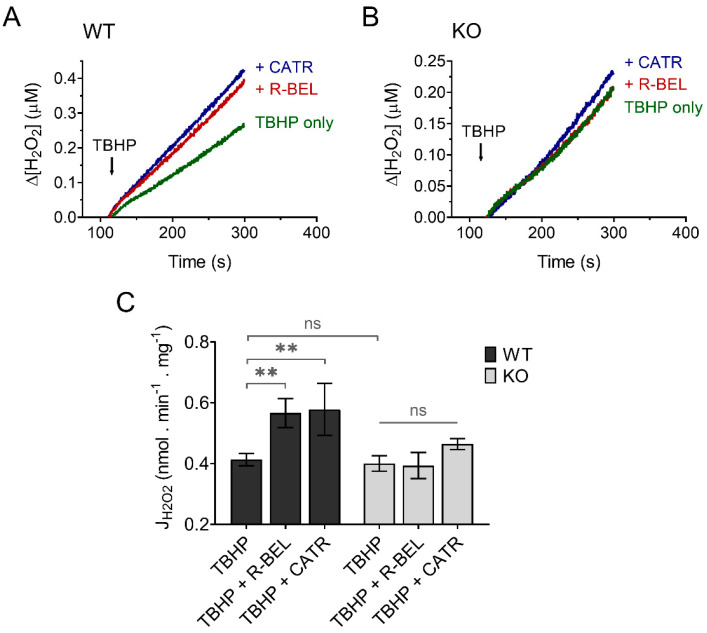
Since H2O2 readily penetrates mitochondrial membranes, it is routinely assayed quantitatively utilizing the extramitochondrial florescent probe Amplex Ultra Red in the presence of horseradish peroxidase [50]. To minimize the effect of matrix consumers of H2O2 and thus maximize the amount of H2O2 released out of mitochondria, we performed our experiments in the presence of the thioredoxin-reductase inhibitor auranofin, which was shown previously to increase the apparent H2O2 efflux rates [58]. Indeed, under these experimental conditions utilizing both Complex I- and Complex II-linked substrate GMS, the H2O2 release from mitochondria was relatively fast (Figure 8). The addition of TBHP to brain mitochondria isolated from WT mice resulted in a steady increase in H2O2 release, which was accelerated in the presence of r-BEL, a selective iPLA2γ inhibitor (Figure 8A), which is consistent with the redox-dependent activation of iPLA2γ and its simultaneous antioxidant activity. The presence of CATR caused an acceleration of H2O2 release comparable to that seen in the presence of r-BEL (Figure 8A,C). Again, no such qualitative differences were observed in brain mitochondria isolated from iPLA2γ-KO mice (Figure 8B,C).
Figure 8.
TBHP-dependent activation of iPLA2γ leads to decreased H2O2 formation. Representative traces of extramitochondrial H2O2 release determined from calibrated resorufin fluorescence in isolated brain mitochondria respiring in the presence of 5 mM of glutamate, 1 mM of malate, 5 mM of succinate (GMS), and 2 μM of auranofin. (A) The rate of H2O2 release in the presence of TBHP (green trace) was inhibited by r-BEL (red trace) and CATR (blue trace). (B) The rate of H2O2 release in the presence of TBHP (green trace), r-BEL (red trace), and CATR (blue trace) in mitochondria isolated from iPLA2γ-KO mice. (C) The effects of tested compounds on the rates of H2O2 release of brain mitochondria isolated from WT (black bars) or iPLA2γ-KO mice (grey bars). The reagent concentrations were identical to those described in Figure 1. The jump in fluorescence due to the interaction of TBHP with AmplexRed was omitted for clarity. The number of replicates was at least 4 for both WT and iPLA2γ-KO. ** p < 0.01. ns, No significant differences were found between the rates in mitochondria isolated from iPLA2γ-KO mice.
3.4. Antioxidant Role of iPLA2γ In Vivo
3.4.1. iPLA2γ—Dependent Antioxidant Function Leads to Decreased Protein Carbonyl Content in the Brain
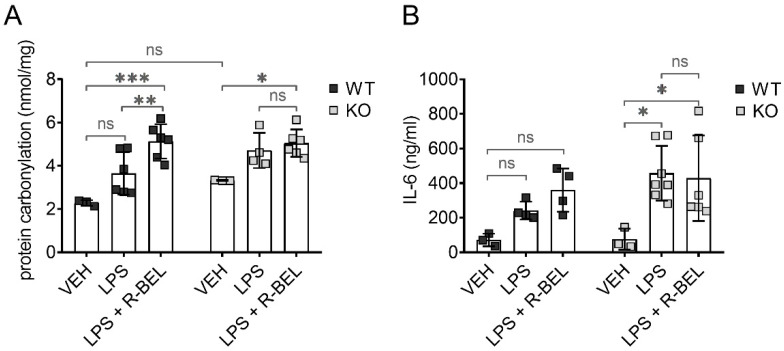
To test whether iPLA2γ may also play an antioxidant role in vivo, we utilized our previously developed protocols [59] and administered lipopolysaccharides (LPS) to WT and iPLA2γ-KO mice together with iPLA2γ inhibitor r-BEL. LPS is widely used to study the acute inflammatory response together with acute oxidative stress and causes a dose-dependent increase in protein carbonyl content, which is the most widely used marker of oxidative modification of proteins and a footprint of oxidative stress [60]. Figure 9A shows the levels of protein carbonyls in brain homogenates from WT and iPLA2γ-KO mice. The addition of LPS did not lead to statistically significant changes in the protein carbonyl levels in brain homogenates from WT mice, indicating a protective role of iPLA2γ. In contrast, LPS treatment caused about a 1.5-fold increase in protein carbonyl levels in iPLA2γ-KO mice. The simultaneous administration of LPS and the selective iPLA2γ inhibitor r-BEL increased the protein carbonyl content of WT mice to a level similar to KO mice, whereas no further effect of r-BEL administration was observed in homogenates of KO mice. These results support the hypothesis that iPLA2γ plays a critical role in the regulation of cellular redox homeostasis and oxidative stress in brain tissues.
Figure 9.
Antioxidant role of iPLA2γ in the cytosol and in vivo. Differences in protein carbonyl levels in brain homogenates (A) and interleukin-6 levels in serum (B) from WT mice and iPLA2γ-KO mice. Where indicated, mice were injected with LPS (5 mg/kg body weight) or iPLA2γ inhibitor r-BEL (1 mg/kg body weight) and LPS in parallel. * p < 0.05; ** p < 0.01; *** p < 0.005. ns, No significant differences were found between the rates in mitochondria isolated from iPLA2γ-KO mice.
3.4.2. iPLA2γ–ANT Antioxidant Synergy Decreases the Levels of Inflammatory Marker IL-6
Following LPS injection, the concentrations of IL-6 increased significantly and were higher in iPLA2γ-KO mice compared to age-matched WT controls. Moreover, r-BEL, the selective inhibitor of iPLA2γ, further elevated IL-6 concentrations in WT mice to the IL-6 levels comparable to those obtained with iPLA2γ-KO mice, while the inhibitor had no further effect in iPLA2γ-KO mice (Figure 9B). These data demonstrate that the inhibition or ablation of iPLA2γ leads to an increase in IL-6 serum levels and are consistent with a proposed antioxidant role of iPLA2γ in vivo.
4. Discussion
In this work, we demonstrate several consequences of catalytic activities of redox-activated mitochondrial phospholipase iPLA2γ in the brain. We show that (1) FAs liberated by mitochondrial phospholipase iPLA2γ interact with the ANT and play an antioxidant role, (2) iPLA2γ participates in cardiolipin post-biosynthetic remodeling (3) iPLA2γ releases a range of very long-chain PUFAs which may serve as precursors of unique mitochondria-derived lipid mediators of cellular redox signaling.
The molecular mechanism explaining the antioxidant role of iPLA2γ is supported by the ability of ANT to mediate FA-dependent H+ transport across the inner mitochondrial membrane, which leads to a mild increase in respiration, corresponding dissipation of the mitochondrial protonmotive force, and consequent attenuation of mitochondrial superoxide formation by the mitochondrial electron transfer system. The decrease in mitochondrial superoxide formation is followed by a decrease in the formation of H2O2 and consequent oxidants. The antioxidant mechanism based on the iPLA2γ-ANT synergy may have consequences extending beyond the respective mitochondrial compartments. This is because mitochondrially-produced oxidants, namely H2O2 and membrane-permeable electrophilic lipids, can diffuse to cytosolic subcellular compartments and increase the electrophilic tone in the cytosol. This increase has to be compensated by the cytosolic antioxidant mechanisms, reestablishing the redox steady-state. Altogether, the ability of mitochondria to regulate the production of superoxide by the dissipation of the protonmotive force substantiates an antioxidant effect of mitochondria within the intracellular environment [17,45,46].
The conclusions of this study are in contrast to previous data on the putative roles of Ca2+-independent phospholipases A2 on mitochondrial ROS production in rat brain mitochondria. It has been concluded that iPLA2 does not have a role in the modulation of ROS production in the brain [61]. However, that study probed the effect of externally added DHA (C24:6 PUFA), as the presumed primary product of iPLA2 activity, in isolated respiring mitochondria with the attempt to mimic the role of iPLA2-liberated fatty acids on the mitochondrial ROS generation. The addition of DHA induced a CATR-sensitive increase in oxygen consumption, which is consistent with ANT-mediated DHA-dependent uncoupling and compatible with our data. However, DHA diminished mitochondrial H2O2 release only in the presence of succinate, a complex II-linked substrate, but stimulated ROS production in the presence of complex I-linked substrates, which verifies the ability of externally supplied DHA to interact with complex I of the mitochondrial electron transfer system directly, leading to effects associated with lipotoxicity [10]. The results of our study are based on acute activation of iPLA2γ, which leads to a local release of fatty acids and thus may avoid the lipotoxicity associated with externally added DHA. In addition, our data show that the profile of iPLA2γ-liberated fatty acids is broad and is not limited to DHA. Our conclusions are further supported by the lack of the iPLA2γ-dependent antioxidant mechanism in parallel control experiments using iPLA2γ-KO mice.
Consistent with previous studies [61], our data show that iPLA2γ is intrinsically active in routinely isolated brain mitochondria without the need for added oxidants. We also demonstrate that the presence of NAC during mitochondrial isolation restores the hydroperoxide-induced activation of brain iPLA2γ, which may reflect a plausible in vivo situation when the thiol-dependent redox status of the respective mitochondrial compartments controls the activity of iPLA2γ. This idea is supported by our previous finding showing that thiol-reducing agents prevent TBHP-induced activation of recombinant reconstituted iPLA2γ [35] and further supported by the data obtained in the Oximouse database (https://oximouse.hms.harvard.edu/, accessed on 20 December 2021). The comprehensive and quantitative mapping of the mouse cysteine redox proteome in vivo provided by the database reveals that Cys553 and Cys708 of iPLA2γ are oxidized in several tissues, including the brain, suggesting a reversible modification of the respective cysteine residues by hydroperoxides or other electrophiles compatible with oxidizing accessible thiol groups. Coincidentally, a recent study showed that incubation of isolated brain mitochondria with NAC restores the redox state of the glutathione system and increases the levels of S-glutathionylated proteins [62]. Altogether, the results support our hypothesis that iPLA2γ contains cysteine residues accessible to reversible oxidative posttranslational modifications, and oxidation of these residues leads to reversible activation of iPLA2γ.
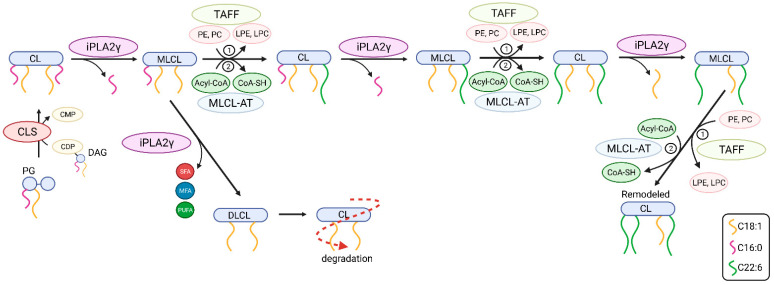
Moreover, our data are consistent with the key role of iPLA2γ in brain mitochondrial lipid biogenesis. Specifically, the obtained lipidomic data suggest the participation of iPLA2γ in cardiolipin post-biosynthetic remodeling (Figure 10). The CL acyl chain composition is tissue-specific, and unlike other tissues, the CL profile in the mammalian brain is temporally dependent, highly diversified, and contains a complex repertoire of CL molecular species [63,64]. iPLA2γ is considered, together with iPLA2β, a major mitochondrial enzyme involved in cardiolipin remodeling [63]. In addition, iPLA2γ has been shown to play a significant role in the generation of MLCL by hydrolysis of short-chain, relatively saturated FAs in the Tafazzin-deficient brain and is involved in the hydrolysis of oxidized CL following traumatic brain injury [33]. Our data show a substantial decrease of five high molecular weight CL species (CL78:14, CL80:12, CL80:14, CL82:16, and CL84:19), and, in contrast, up to a 10-fold accumulation of low molecular weight cardiolipins, namely CL68:1, CL68:2, CL68:4, CL70:6, and CL72:5. These data indicate that iPLA2γ aids deacylation of lower molecular weight cardiolipins with predominantly saturated side chains. Our data support an indispensable role of iPLA2γ in the remodeling of immature CL to mature species containing high molecular weight and highly unsaturated acyl chains, typical for the brain (Figure 10).
Figure 10.
The putative roles of iPLA2γ in post-biosynthetic cardiolipin remodeling and degradation. Shown are steps from newly synthesized nascent cardiolipin (CL) to mature CL. The iPLA2γ-catalyzed deacylation of CL yields fatty acid (depicted as squiggle) and monolysoCL (MLCL) and is followed by reacylation catalyzed by either (1) Tafazzin transacylase (TAFF) or by (2) MLCL acyltransferase (MLCL-AT). Abbreviations: PG, phosphatidylglycerol; CLS, CL synthase; DAG, diacylglycerol; CDP, cytidine diphosphate; CMP, cytidine monophosphate; SFA, saturated fatty acid; MFA, monoenoic fatty acid; PUFA, polyunsaturated fatty acid; PE, phosphatidylethanolamine; PC, phosphatidylcholine; DLCL, dilysoCL.
In addition, our detailed lipidomic analyses accurately illustrate the recognized substrate specificity and product pattern of iPLA2γ reaction complying with the documented specificity for the PNPLA type of phospholipases [28,65]. At first, we established that in isolated brain mitochondria, iPLA2γ liberates both saturated and (poly)unsaturated fatty acids from glycerophospholipids, which is consistent with iPLA2γ hydrolyzing both the sn-1 and sn-2 positions of the glycerol backbone. Second, in the particular case of cardiolipin cleavage, the activation of iPLA2γ led primarily to the accumulation of MLCL species, which is consistent with the role of iPLA2γ in cardiolipin remodeling. However, we also detected a moderate increase in DLCL, which indicates that iPLA2γ can hydrolyze up to two aliphatic chains from cardiolipins. This result is consistent with the finding of Kiebish et al. [66], who detected DLCL in the hearts of mice that were created by crossing strain null for iPLA2γ with inducible Taffazin knockdown mice. Those results indicate that iPLA2γ plays a critical role in the initial and rapid production of DLCL for cardiolipin remodeling in an acyl-chain-specific manner [66]. From the available lipid and CDP-DG sources within the brain, we conclude that iPLA2γ is essential in the synthesis of the high-molecular-weight cardiolipins containing PUFAs in all four acyl chains, which is further documented by our lipidomic analysis of iPLA2γ-KO mice.
Our data also demonstrate the ability of iPLA2γ to release a range of very long-chain PUFAs, including C24:6, C22:6, C20:5, and C20:4. These PUFAs are metabolically active, participate in the regulation of peripheral immune function, and have been shown to regulate microglia activation in the brain [67]. Specifically, arachidonic acid (C20:4, AA), the main n-6 PUFA, and docosahexaenoic acid, the main n-3 PUFA, are suggested to regulate the molecular signaling of microglia, especially in the context of neuroinflammation and behavior [67]. In addition, both AA and DHA are substrates of cyclooxygenases and lipoxygenases in the brain, resulting in an array of lipid signaling mediators, including fatty acid-derived electrophilic species. These oxidizing compounds are generally considered for their pro-inflammatory properties, but they also have an impact on numerous redox-dependent cell signaling pathways [68]. Previous studies suggested a role for iPLA2β in the release of DHA in the brain [69,70,71]. However, the ability of iPLA2γ to mediate the release of AA and DHA in the brain has not been fully explored, and suggests an intriguing role for iPLA2γ in mitochondria-produced lipid-dependent intracellular signaling in the brain.
Finally, we show that mice with ablated iPLA2γ were more sensitive to stimulation by LPS, as reflected by the concomitant increase in protein carbonyls in brain tissue homogenates and pro-inflammatory IL-6 release detected in the serum. These data support the link between the prevention of pro-oxidative and pro-inflammatory consequences of these treatments and the antioxidant and anti-inflammatory role of iPLA2γ in vivo. Then, the proposed iPLA2γ-dependent antioxidant mechanism supported by our study may be part of a broad spectrum of physiological functions which iPLA2γ plays in the brain, including a causative effect of decreased/compromised activity of iPLA2γ leading to neurodegenerative disorders.
5. Conclusions
Our data support an antioxidant mechanism in the brain utilizing the release of fatty acids catalyzed by the redox-activated mitochondrial phospholipase iPLA2γ. The interaction of iPLA2γ-released fatty acids with adenine nucleotide translocase increases proton conductance of the inner mitochondrial membrane and consequently decreases mitochondrial protonmotive force and mitochondrial ROS formation. Our data further indicate a feedback loop in which pro-oxidant milieu or increased oxidative stress activates mitochondrial iPLA2γ and initiates iPLA2γ-dependent, ANT-mediated antioxidant protection. Moreover, we found an indispensable role of iPLA2γ in the remodeling of lower molecular weight immature cardiolipins with predominantly saturated acyl chains to high molecular weight mature cardiolipins with highly unsaturated PUFA acyl chains, typical for the brain.
Acknowledgments
The authors would like to acknowledge the Metabolomics Core Facility at the Institute of Physiology of the Czech Academy of Sciences for conducting LC–MS-based lipidomic profiling, and the Czech Centre of Phenogenomics, Institute of Molecular Genetics of the Czech Academy of Sciences for creating the iPLA2γ ablated mice. We are grateful to Fariha Ansari for help in the preparation of this manuscript.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antiox11020198/s1, Figure S1: Creation of iPLA2γ/PNPLA8 knockout mice and the knockout verification. Figure S2: Respiratory control analysis of brain mitochondria isolated from WT and iPLA2γ-KO mice. Figure S3: Linoleic acid-dependent increase in respiration in brain mitochondria isolated from WT and iPLA2γ-KO mice. Figure S4: Lipidomic analysis of isolated WT and iPLA2γ-KO mouse brain mitochondria. Figure S5: Metabolites responsible for sample clustering.
Author Contributions
Conceptualization, P.P., P.J., A.G. and M.J.; methodology, P.P., K.G., K.S., L.A., B.H., J.T., P.J., A.G. and M.J.; formal analysis, P.P., K.G., K.S., P.J. and M.J.; writing—original draft preparation, P.P., P.J. and M.J.; writing—review and editing, P.P., K.G., K.S., A.G., P.J. and M.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Czech Ministry of Education Inter-Excellence program grant LTA USA 17174 and The Czech Science Foundation GA22-17173S (to M.J.) and by NIH RO1NS112381 and R21NS125466 (to A.G.).
Institutional Review Board Statement
The animal study protocol was approved by the Institutional Review Board and Ethics Committee of the Czech Academy of Sciences (protocol code 11388/2019-MZE-17214, date of approval 25 May 2019).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data will be available from the corresponding author upon a reasonable request. The data are not publicly available due to there are still numerous raw data that were collected during the experiments and are not included in this manuscript, because they were considered superfluous. These could be available upon request.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ursini F., Maiorino M., Forman H.J. Redox homeostasis: The Golden Mean of healthy living. Redox Biol. 2016;8:205–215. doi: 10.1016/j.redox.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sies H., Jones D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020;21:363–383. doi: 10.1038/s41580-020-0230-3. [DOI] [PubMed] [Google Scholar]
- 3.Sies H. Oxidative eustress: On constant alert for redox homeostasis. Redox Biol. 2021;41:101867. doi: 10.1016/j.redox.2021.101867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rinaldi C., Donato L., Alibrandi S., Scimone C., D’Angelo R., Sidoti A. Oxidative Stress and the Neurovascular Unit. Life. 2021;11:767. doi: 10.3390/life11080767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee K.H., Cha M., Lee B.H. Crosstalk between Neuron and Glial Cells in Oxidative Injury and Neuroprotection. Int. J. Mol. Sci. 2021;22:13315. doi: 10.3390/ijms222413315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vinokurov A.Y., Stelmashuk O.A., Ukolova P.A., Zherebtsov E.A., Abramov A.Y. Brain region specificity in reactive oxygen species production and maintenance of redox balance. Free Radic. Biol. Med. 2021;174:195–201. doi: 10.1016/j.freeradbiomed.2021.08.014. [DOI] [PubMed] [Google Scholar]
- 7.Woo J., Cho H., Seol Y., Kim S.H., Park C.J., Yousefian-Jazi A., Hyeon S., Lee J., Ryu H. Power Failure of Mitochondria and Oxidative Stress in Neurodegeneration and Its Computational Models. Antioxidants. 2021;10:229. doi: 10.3390/antiox10020229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantó-Santos J., Grau-Junyent J.M., Garrabou G. The Impact of Mitochondrial Deficiencies in Neuromuscular Diseases. Antioxidants. 2020;9:964. doi: 10.3390/antiox9100964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franco R., Vargas M.R. Redox Biology in Neurological Function, Dysfunction, and Aging. Antioxid. Redox Signal. 2018;28:1583–1586. doi: 10.1089/ars.2018.7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schönfeld P., Reiser G. How the brain fights fatty acids’ toxicity. Neurochem. Int. 2021;148:105050. doi: 10.1016/j.neuint.2021.105050. [DOI] [PubMed] [Google Scholar]
- 11.Cobley J.N., Fiorello M.L., Bailey D.M. 13 reasons why the brain is susceptible to oxidative stress. Redox Biol. 2018;15:490–503. doi: 10.1016/j.redox.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong H.-S., Dighe P.A., Mezera V., Monternier P.-A., Brand M.D. Production of superoxide and hydrogen peroxide from specific mitochondrial sites under different bioenergetic conditions. J. Biol. Chem. 2017;292:16804–16809. doi: 10.1074/jbc.R117.789271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Case A.J., Tian J., Zimmerman M.C. Increased mitochondrial superoxide in the brain, but not periphery, sensitizes mice to angiotensin II-mediated hypertension. Redox Biol. 2017;11:82–90. doi: 10.1016/j.redox.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lebovitz R.M., Zhang H., Vogel H., Cartwright J., Jr., Dionne L., Lu N., Huang S., Matzuk M.M. Neurodegeneration, myocardial injury, and perinatal death in mitochondrial superoxide dismutase-deficient mice. Proc. Natl. Acad. Sci. USA. 1996;93:9782–9787. doi: 10.1073/pnas.93.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh S.S., Sullivan K.A., Wilkinson J.E., Backus C., Hayes J.M., Sakowski S.A., Feldman E.L. Neurodegeneration and early lethality in superoxide dismutase 2-deficient mice: A comprehensive analysis of the central and peripheral nervous systems. Neuroscience. 2012;212:201–213. doi: 10.1016/j.neuroscience.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinerfeld D., Traini M.D., Weinberger R.P., Cochran B., Doctrow S.R., Harry J., Melov S. Endogenous mitochondrial oxidative stress: Neurodegeneration, proteomic analysis, specific respiratory chain defects, and efficacious antioxidant therapy in superoxide dismutase 2 null mice. J. Neurochem. 2004;88:657–667. doi: 10.1046/j.1471-4159.2003.02195.x. [DOI] [PubMed] [Google Scholar]
- 17.Ježek P., Plecitá-Hlavatá L. Mitochondria in homeostasis of reactive oxygen species in cell, tissues, and organism. Int. J. Biochem. Cell Biol. 2005;37:2478–2503. doi: 10.1016/j.biocel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 18.Ježek P., Plecitá-Hlavatá L. Mitochondrial reticulum network dynamics in relation to oxidative stress, redox regulation, and hypoxia. Int. J. Biochem. Cell Biol. 2009;41:1790–1804. doi: 10.1016/j.biocel.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 19.Ježek P., Holendová B., Plecitá-Hlavatá L. Redox Signaling from Mitochondria: Signal Propagation and Its Targets. Biomolecules. 2020;10:93. doi: 10.3390/biom10010093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drechsel D.A., Patel M. Respiration-dependent H2O2 Removal in Brain Mitochondria via the Thioredoxin/Peroxiredoxin System. J. Biol. Chem. 2010;285:27850–27858. doi: 10.1074/jbc.M110.101196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren X., Zou L., Zhang X., Branco V., Wang J., Carvalho C., Holmgren A., Lu J. Redox Signaling Mediated by Thioredoxin and Glutathione Systems in the Central Nervous System. Antioxid. Redox Signal. 2017;27:989–1010. doi: 10.1089/ars.2016.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romano A., Koczwara J.B., Gallelli C.A., Vergara D., Micioni Di Bonaventura M.V., Gaetani S., Giudetti A.M. Fats for thoughts: An update on brain fatty acid metabolism. Int. J. Biochem. Cell Biol. 2017;84:40–45. doi: 10.1016/j.biocel.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 23.Schönfeld P., Reiser G. Brain energy metabolism spurns fatty acids as fuel due to their inherent mitotoxicity and potential capacity to unleash neurodegeneration. Neurochem. Int. 2017;109:68–77. doi: 10.1016/j.neuint.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 24.Niki E. Lipid peroxidation: Physiological levels and dual biological effects. Free Radic. Biol. Med. 2009;47:469–484. doi: 10.1016/j.freeradbiomed.2009.05.032. [DOI] [PubMed] [Google Scholar]
- 25.Wolf M.J., Izumi Y., Zorumski C.F., Gross R.W. Long-term potentiation requires activation of calcium-independent phospholipase A2. FEBS Lett. 1995;377:358–362. doi: 10.1016/0014-5793(95)01371-7. [DOI] [PubMed] [Google Scholar]
- 26.Murakami M., Sato H., Taketomi Y. Updating phospholipase A2 biology. Biomolecules. 2020;10:1457. doi: 10.3390/biom10101457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mancuso D.J., Jenkins C.M., Gross R.W. The Genomic Organization, Complete mRNA Sequence, Cloning, and Expression of a Novel Human Intracellular Membrane-associated Calcium-independent Phospholipase A2. J. Biol. Chem. 2000;275:9937–9945. doi: 10.1074/jbc.275.14.9937. [DOI] [PubMed] [Google Scholar]
- 28.Yan W., Jenkins C.M., Han X., Mancuso D.J., Sims H.F., Yang K., Gross R.W. The Highly Selective Production of 2-Arachidonoyl Lysophosphatidylcholine Catalyzed by Purified Calcium-independent Phospholipase A2γ: Identification of a novel enzymatic mediator for the generation of a key branch point intermediate in eicosanoid signali. J. Biol. Chem. 2005;280:26669–26679. doi: 10.1074/jbc.M502358200. [DOI] [PubMed] [Google Scholar]
- 29.Liu G.-Y., Moon S.H., Jenkins C.M., Li M., Sims H.F., Guan S., Gross R.W. The phospholipase iPLA2γ is a major mediator releasing oxidized aliphatic chains from cardiolipin, integrating mitochondrial bioenergetics and signaling. J. Biol. Chem. 2017;292:10672–10684. doi: 10.1074/jbc.M117.783068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mancuso D.J., Kotzbauer P., Wozniak D.F., Sims H.F., Jenkins C.M., Guan S., Han X., Yang K., Sun G., Malik I., et al. Genetic Ablation of Calcium-independent Phospholipase A2γ Leads to Alterations in Hippocampal Cardiolipin Content and Molecular Species Distribution, Mitochondrial Degeneration, Autophagy, and Cognitive Dysfunction. J. Biol. Chem. 2009;284:35632–35644. doi: 10.1074/jbc.M109.055194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chao H., Liu Y., Fu X., Xu X., Bao Z., Lin C., Li Z., Liu Y., Wang X., You Y., et al. Lowered iPLA2γ activity causes increased mitochondrial lipid peroxidation and mitochondrial dysfunction in a rotenone-induced model of Parkinson’s disease. Exp. Neurol. 2018;300:74–86. doi: 10.1016/j.expneurol.2017.10.031. [DOI] [PubMed] [Google Scholar]
- 32.Shukla A., Saneto R.P., Hebbar M., Mirzaa G., Girisha K.M. A neurodegenerative mitochondrial disease phenotype due to biallelic loss-of-function variants in PNPLA8 encoding calcium-independent phospholipase A2γ. Am. J. Med. Genet. Part A. 2018;176:1232–1237. doi: 10.1002/ajmg.a.38687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chao H., Anthonymuthu T.S., Kenny E.M., Amoscato A.A., Cole L.K., Hatch G.M., Ji J., Kagan V.E., Bayır H. Disentangling oxidation/hydrolysis reactions of brain mitochondrial cardiolipins in pathogenesis of traumatic injury. JCI Insight. 2018;3:e97677. doi: 10.1172/jci.insight.97677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ježek J., Jabůrek M., Zelenka J., Ježek P. Mitochondrial phospholipase A2 activated by reactive oxygen species in heart mitochondria induces mild uncoupling. Physiol. Res. 2010;59:737–747. doi: 10.33549/physiolres.931905. [DOI] [PubMed] [Google Scholar]
- 35.Ježek J., Dlasková A., Zelenka J., Jabůrek M., Ježek P. H2O2-Activated Mitochondrial Phospholipase iPLA2γ Prevents Lipotoxic Oxidative Stress in Synergy with UCP2, Amplifies Signaling via G-Protein–Coupled Receptor GPR40, and Regulates Insulin Secretion in Pancreatic β-Cells. Antioxid. Redox Signal. 2015;23:958–972. doi: 10.1089/ars.2014.6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaburek M., Jezek J., Zelenka J., Ježek P. Antioxidant activity by a synergy of redox-sensitive mitochondrial phospholipase A2 and uncoupling protein-2 in lung and spleen. Int. J. Biochem. Cell Biol. 2013;45:816–825. doi: 10.1016/j.biocel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 37.Jaburek M., Garlid K.D. Reconstitution of Recombinant Uncoupling Proteins. UCP1, -2, and -3 have similar affinities for ATP and are unaffected by coenzyme Q10. J. Biol. Chem. 2003;278:25825–25831. doi: 10.1074/jbc.m302126200. [DOI] [PubMed] [Google Scholar]
- 38.Jabůrek M., Varecha M., Gimeno R.E., Dembski M., Jezek P., Zhang M., Burn P., Tartaglia L.A., Garlid K.D. Transport Function and Regulation of Mitochondrial Uncoupling Proteins 2 and 3. J. Biol. Chem. 1999;274:26003–26007. doi: 10.1074/jbc.274.37.26003. [DOI] [PubMed] [Google Scholar]
- 39.Jaburek M., Miyamoto S., Di Mascio P., Garlid K., Ježek P. Hydroperoxy Fatty Acid Cycling Mediated by Mitochondrial Uncoupling Protein UCP2. J. Biol. Chem. 2004;279:53097–53102. doi: 10.1074/jbc.M405339200. [DOI] [PubMed] [Google Scholar]
- 40.Bertholet A.M., Chouchani E.T., Kazak L., Angelin A., Fedorenko A., Long J.Z., Vidoni S., Garrity R., Cho J., Terada N., et al. H+ transport is an integral function of the mitochondrial ADP/ATP carrier. Nature. 2019;571:515–520. doi: 10.1038/s41586-019-1400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bertholet A.M., Kazak L., Chouchani E.T., Bogaczyńska M.G., Paranjpe I., Wainwright G.L., Bétourné A., Kajimura S., Spiegelman B.M., Kirichok Y. Mitochondrial Patch Clamp of Beige Adipocytes Reveals UCP1-Positive and UCP1-Negative Cells Both Exhibiting Futile Creatine Cycling. Cell Metab. 2017;25:811–822.e4. doi: 10.1016/j.cmet.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wojtczak L., Wiȩckowski M.R. The mechanisms of fatty acid-induced proton permeability of the inner mitochondrial membrane. J. Bioenerg. Biomembr. 1999;31:447–455. doi: 10.1023/A:1005444322823. [DOI] [PubMed] [Google Scholar]
- 43.Wojtczak L., Wiȩckowski M.R., Schönfeld P. Protonophoric Activity of Fatty Acid Analogs and Derivatives in the Inner Mitochondrial Membrane: A Further Argument for the Fatty Acid Cycling Model. Arch. Biochem. Biophys. 1998;357:76–84. doi: 10.1006/abbi.1998.0777. [DOI] [PubMed] [Google Scholar]
- 44.Brustovetsky N., Klingenberg M. The reconstituted ADP/ATP carrier can mediate H+ transport by free fatty acids, which is further stimulated by mersalyl. J. Biol. Chem. 1994;269:27329–27336. doi: 10.1016/S0021-9258(18)46989-X. [DOI] [PubMed] [Google Scholar]
- 45.Ježek P., Holendova B., Garlid K.D., Jaburek M. Mitochondrial Uncoupling Proteins: Subtle Regulators of Cellular Redox Signaling. Antioxid. Redox Signal. 2018;29:667–714. doi: 10.1089/ars.2017.7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jabůrek M., Průchová P., Holendová B., Galkin A., Ježek P. Antioxidant Synergy of Mitochondrial Phospholipase PNPLA8/iPLA2γ with Fatty Acid–Conducting SLC25 Gene Family Transporters. Antioxidants. 2021;10:678. doi: 10.3390/antiox10050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kašpárek P., Krausova M., Haneckova R., Kriz V., Zbodakova O., Korinek V., Sedlacek R. Efficient gene targeting of theRosa26locus in mouse zygotes using TALE nucleases. FEBS Lett. 2014;588:3982–3988. doi: 10.1016/j.febslet.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 48.Stepanova A., Kahl A., Konrad C., Ten V., Starkov A., Galkin A. Reverse electron transfer results in a loss of flavin from mitochondrial complex I: Potential mechanism for brain ischemia reperfusion injury. J. Cereb. Blood Flow Metab. 2017;37:3649–3658. doi: 10.1177/0271678X17730242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Plecitá-Hlavatá L., Engstová H., Holendová B., Tauber J., Špaček T., Petrásková L., Křen V., Špačková J., Gotvaldová K., Ježek J., et al. Mitochondrial Superoxide Production Decreases on Glucose-Stimulated Insulin Secretion in Pancreatic β Cells Due to Decreasing Mitochondrial Matrix NADH/NAD+Ratio. Antioxid. Redox Signal. 2020;33:789–815. doi: 10.1089/ars.2019.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou M., Diwu Z., Panchuk-Voloshina N., Haugland R.P. A Stable Nonfluorescent Derivative of Resorufin for the Fluorometric Determination of Trace Hydrogen Peroxide: Applications in Detecting the Activity of Phagocyte NADPH Oxidase and Other Oxidases. Anal. Biochem. 1997;253:162–168. doi: 10.1006/abio.1997.2391. [DOI] [PubMed] [Google Scholar]
- 51.Janovska P., Melenovsky V., Svobodova M., Havlenova T., Kratochvilova H., Haluzik M., Hoskova E., Pelikanova T., Kautzner J., Monzo L., et al. Dysregulation of epicardial adipose tissue in cachexia due to heart failure: The role of natriuretic peptides and cardiolipin. J. Cachexia Sarcopenia Muscle. 2020;11:1614–1627. doi: 10.1002/jcsm.12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsugawa H., Ikeda K., Takahashi M., Satoh A., Mori Y., Uchino H., Okahashi N., Yamada Y., Tada I., Bonini P., et al. A lipidome atlas in MS-DIAL 4. Nat. Biotechnol. 2020;38:1159–1163. doi: 10.1038/s41587-020-0531-2. [DOI] [PubMed] [Google Scholar]
- 53.Levine R.L., Garland D., Oliver C.N., Amici A., Climent I., Lenz A.G., Ahn B.W., Shaltiel S., Stadtman E.R. Determination of carbonyl content in oxidatively modified proteins. Methods Enzymol. 1990;186:464–478. doi: 10.1016/0076-6879(90)86141-h. [DOI] [PubMed] [Google Scholar]
- 54.Xia J., Psychogios N., Young N., Wishart D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. Spec. Publ. 2009;37:W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 56.Grace S.C., Hudson D.A. Metabolomics—Fundamentals and Applications. IntechOpen; 2016. [(accessed on 27 December 2021)]. Processing and Visualization of Metabolomics Data Using R. Available online: https://www.intechopen.com/chapters/52527. [DOI] [Google Scholar]
- 57.Pollard A.K., Ortori C.A., Stöger R., Barrett D., Chakrabarti L. Mouse mitochondrial lipid composition is defined by age in brain and muscle. Aging. 2017;9:986–998. doi: 10.18632/aging.101204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Munro D., Banh S., Sotiri E., Tamanna N., Treberg J.R. The thioredoxin and glutathione-dependent H2O2 consumption pathways in muscle mitochondria: Involvement in H2O2 metabolism and consequence to H2O2 efflux assays. Free Radic. Biol. Med. 2016;96:334–346. doi: 10.1016/j.freeradbiomed.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 59.Jaburek M., Jezek J., Ježek P. Cytoprotective activity of mitochondrial uncoupling protein-2 in lung and spleen. FEBS Open Bio. 2018;8:692–701. doi: 10.1002/2211-5463.12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stadtman E.R., Levine R.L. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids. 2003;25:207–218. doi: 10.1007/s00726-003-0011-2. [DOI] [PubMed] [Google Scholar]
- 61.Nordmann C., Strokin M., Schönfeld P., Reiser G. Putative roles of Ca2+ -independent phospholipase A2 in respiratory chain-associated ROS production in brain mitochondria: Influence of docosahexaenoic acid and bromoenol lactone. J. Neurochem. 2014;131:163–176. doi: 10.1111/jnc.12789. [DOI] [PubMed] [Google Scholar]
- 62.Semenovich D.S., Plotnikov E.Y., Titko O.V., Lukiyenko E.P., Kanunnikova N.P. Effects of Panthenol and N-Acetylcysteine on Changes in the Redox State of Brain Mitochondria under Oxidative Stress In Vitro. Antioxidants. 2021;10:1699. doi: 10.3390/antiox10111699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheng H., Mancuso D.J., Jiang X., Guan S., Yang J., Yang K., Sun G., Gross R.W., Han X. Shotgun Lipidomics Reveals the Temporally Dependent, Highly Diversified Cardiolipin Profile in the Mammalian Brain: Temporally Coordinated Postnatal Diversification of Cardiolipin Molecular Species with Neuronal Remodeling. Biochemistry. 2008;47:5869–5880. doi: 10.1021/bi7023282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oemer G., Koch J., Wohlfarter Y., Alam M.T., Lackner K., Sailer S., Neumann L., Lindner H.H., Watschinger K., Haltmeier M., et al. Phospholipid Acyl Chain Diversity Controls the Tissue-Specific Assembly of Mitochondrial Cardiolipins. Cell Rep. 2020;30:4281–4291.e4. doi: 10.1016/j.celrep.2020.02.115. [DOI] [PubMed] [Google Scholar]
- 65.Hayashi D., Mouchlis V.D., Dennis E.A. Each phospholipase A2 type exhibits distinct selectivity toward sn-1 ester, alkyl ether, and vinyl ether phospholipids. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids. 2022;1867:159067. doi: 10.1016/j.bbalip.2021.159067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kiebish M.A., Yang K., Liu X., Mancuso D.J., Guan S., Zhao Z., Sims H.F., Cerqua R., Cade W.T., Han X., et al. Dysfunctional cardiac mitochondrial bioenergetic, lipidomic, and signaling in a murine model of Barth syndrome. J. Lipid Res. 2013;54:1312–1325. doi: 10.1194/jlr.M034728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Layé S., Nadjar A., Joffre C., Bazinet R.P. Anti-Inflammatory Effects of Omega-3 Fatty Acids in the Brain: Physiological Mechanisms and Relevance to Pharmacology. Pharmacol. Rev. 2018;70:12–38. doi: 10.1124/pr.117.014092. [DOI] [PubMed] [Google Scholar]
- 68.Yang B., Fritsche K.L., Beversdorf D.Q., Gu Z., Lee J.C., Folk W.R., Greenlief C.M., Sun G.Y. Yin-Yang Mechanisms Regulating Lipid Peroxidation of Docosahexaenoic Acid and Arachidonic Acid in the Central Nervous System. Front. Neurol. 2019;10:642. doi: 10.3389/fneur.2019.00642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Green J.T., Orr S.K., Bazinet R.P. The emerging role of group VI calcium-independent phospholipase A2 in releasing docosahexaenoic acid from brain phospholipids. J. Lipid Res. 2008;49:939–944. doi: 10.1194/jlr.R700017-JLR200. [DOI] [PubMed] [Google Scholar]
- 70.Cheon Y., Kim H.-W., Igarashi M., Modi H.R., Chang L., Ma K., Greenstein D., Wohltmann M., Turk J., Rapoport S.I., et al. Disturbed brain phospholipid and docosahexaenoic acid metabolism in calcium-independent phospholipase A2-VIA (iPLA2β)-knockout mice. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids. 2012;1821:1278–1286. doi: 10.1016/j.bbalip.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Blanchard H., Taha A.Y., Cheon Y., Kim H.-W., Turk J., Rapoport S.I. iPLA2β Knockout Mouse, a Genetic Model for Progressive Human Motor Disorders, Develops Age-Related Neuropathology. Neurochem. Res. 2014;39:1522–1532. doi: 10.1007/s11064-014-1342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be available from the corresponding author upon a reasonable request. The data are not publicly available due to there are still numerous raw data that were collected during the experiments and are not included in this manuscript, because they were considered superfluous. These could be available upon request.