Abstract
The micromeres of the sea urchin embryo are distinct from other blastomeres. After they arise through an asymmetric cell division at the 8- to 16-cell stage, micromeres immediately function as organizers. They also commit themselves to specific cell fates such as larval skeletogenic cells and primordial germ cells, while other blastomeres remain plastic and uncommitted at the 16-cell stage. In the phylum Echinodermata, only the sea urchin (class Echinoidea) embryo forms micromeres that serve as apparent organizers during early embryogenesis. Therefore, it is considered that micromeres are the derived features and that modification(s) of the developmental system allowed evolutionary introduction of this unique cell lineage. In this chapter, we summarize the both historic and recent observations that demonstrate unique properties of micromeres and discuss how this lineage of micromeres may have arisen during echinoderm evolution.
1. Introduction
The sea urchin embryo has served as a model system in the field of embryology for over a century. In particular, the micromere, a cell type formed at the 16-cell stage in the embryo continues to draw the attention of many scientists because of its unique cell geometry, cell fate, function, and molecular properties. The micromere lineage arises through the first apparent asymmetric and unequal cell division at the 16-cell stage. This cell division is horizontal and produces unequal sized daughter cells, resulting in four macromeres and four micromeres in the vegetal hemisphere. Micromeres are positioned slightly tacked inward at the vegetal pole because of the oblique orientation of the spindle during asymmetric cell division, while animal blastomeres divide equally and vertically (Fig. 1A). Micromeres undergo another horizontal yet oblique unequal cell division at the 32-cell stage to form large and small micromeres that are also tacked toward the vegetal pole. These two lineages autonomously result in two specific cell fates: the large micromere develops to a singular fate of skeletogenic cells (Okazaki, 1975), and the small micromere gives rise to primordial germ cells (PGCs) (Yajima & Wessel, 2011a, 2012). The other embryonic blastomeres, on the other hand, do not become fully committed to specific cell fates during early embryogenesis.
Fig. 1.
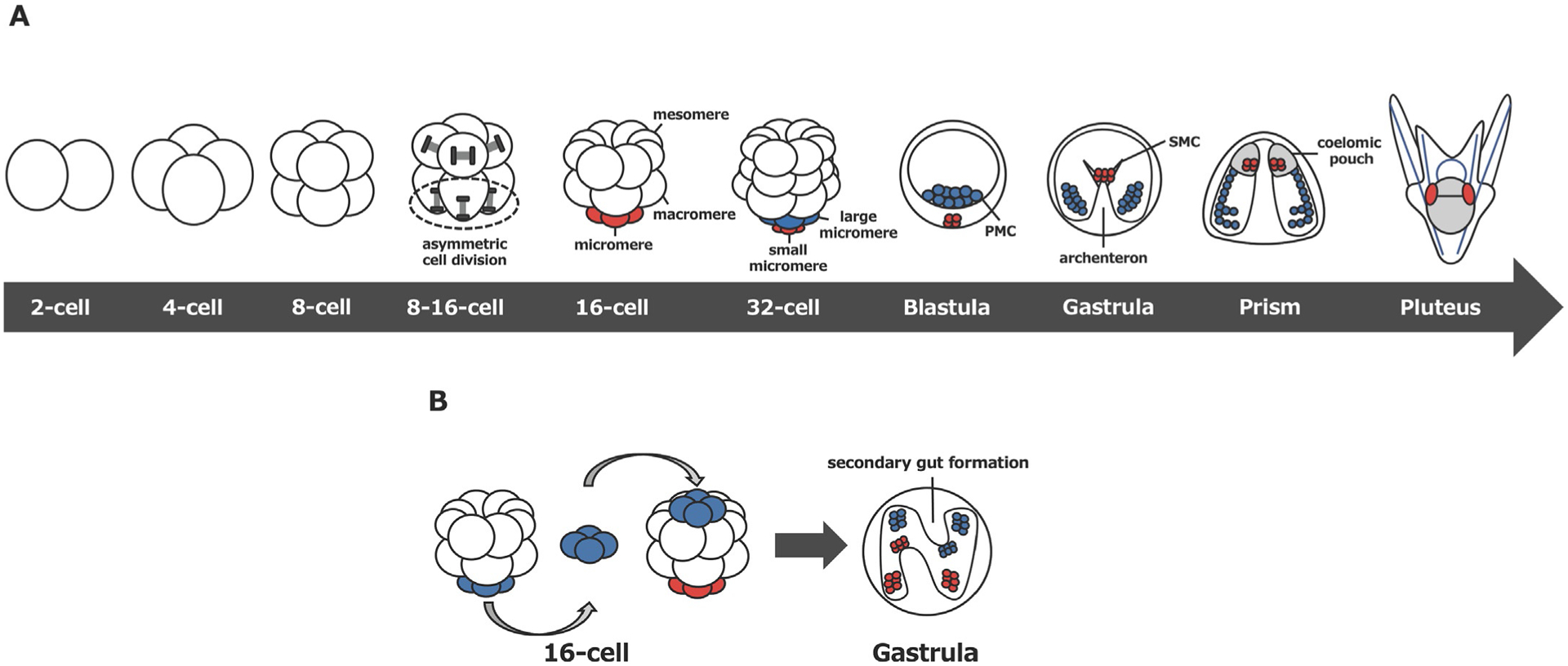
(A) Summary of early development of the sea urchin embryo. Small micromeres or large micromeres and their corresponding descendants are indicated in red or blue, respectively. PMC; primary mesenchyme cell, SMC; secondary mesenchyme cells. (B) Transplantation of extra micromeres to the animal cap at the 16-cell stage induces the secondary gut. Host micromeres or transplanted micromeres and their descendants are indicated in red or blue, respectively.
Micromeres are unique in their function. They are capable of signaling to adjacent, macromere-derived cells and induce the site of invagination. Therefore, micromeres are considered to be major organizers in the sea urchin embryo (Boveri, 1901; Hörstadius, 1928; Ransick & Davidson, 1993, 1995). This historical discovery was initially made around the time when Spemann’s organizer was discovered in the amphibian embryo (Spemann & Mangold, 1924). This calls extra attention to micromeres, which show similar inductive capability in the embryo of the sea urchin, an invertebrate animal. It is not yet entirely clear; however, if/how the mechanism of organizers’ functions is evolutionarily conserved or distinct between sea urchins and amphibians or among distantly related organisms.
Micromeres also show unique structural and chemical properties, such as denuded cortical membrane structure and an increased level of calcium (Dale, Yazaki and Tosti, 1997; Dan, Endo, & Uemura, 1983; Yazaki, 2001; Yazaki, Abe, Santella, & Koyama, 2004). However, the physiological significance of these specialized properties is yet to be determined. Lastly, micromeres are distinct at the molecular level. They selectively accumulate a variety of transcription-, translation- and signaling factors (e.g., delta, wnt8, pmar1, and vasa) by largely unknown mechanisms that promote development of each lineage (Angerer & Angerer, 2003; Cui, Siriwon, Li, Davidson, & Peter, 2014; Juliano et al., 2006; Materna & Davidson, 2012; Oliveri, Carrick, & Davidson, 2002; Swartz et al., 2014; Voronina et al., 2008; Wikramanayake et al., 2004; Yajima & Wessel, 2011b). For example, Vasa protein, a conserved germline marker, is present in all blastomeres until the 8-cell stage, and then becomes specifically enriched into micromeres at the onset of micromere formation and further into the small micromere lineage during the successive unequal cleavage (Voronina et al., 2008; Yajima & Wessel, 2011b). Importantly, when embryos are treated with SDS at the 8- to 16-cell stage, which randomizes axes of cell division and results in equal-sized daughter cells, the resultant embryos often fail in gastrulation (Tanaka, 1976) and Vasa is no longer asymmetric at the vegetal pole (Poon, Fries, Wessel, & Yajima, 2019). Therefore, unequal cell division appears to be important for micromere’s proper function as an organizer as well as for establishment of its molecular properties.
Collectively, micromere formation at the 16-cell stage appears to be an important event for embryogenesis of the sea urchin, yet the mechanism that makes micromeres so distinct from other cell types through a single asymmetric cell division is largely unknown. Further, in the phylum Echinodermata, micromere formation or early asymmetric cell division is seen only in sea urchins (class Echinoidea). The formation of micromeres is thus considered to be a derived feature among echinoderms, which appear to have also evolved organizer-dependent development specifically in the echinoid lineage. Asymmetric cell division during embryogenesis is critical for diversifying early cell fates in many organisms, and evolutionary changes in this process could impact the entire developmental program. Therefore, micromeres of the sea urchin have been an ideal model to study a fundamental biological question of how a new cell type emerges in the developmental program during evolution. Although many questions are yet to be answered, in this chapter, we summarize both historical and recent findings in relation to development of the micromere lineage in the sea urchin embryo.
2. Micromere formation in the sea urchin embryo
2.1. Early development of the sea urchin embryo
Upon fertilization, the sea urchin embryo divides symmetrically and yields both morphologically and molecularly equal blastomeres at the 4-cell stage through two vertical cell divisions (Fig. 1A). The embryo then undergoes the first asymmetric cell division through a horizontal cell division to produce an 8-cell stage embryo, which is morphologically symmetrical but has molecularly distinct vegetal and animal blastomeres. These first three cleavage patterns are highly similar among other echinoderms, yet the fourth cleavage is both morphologically and molecularly asymmetric and unique to the class Echinoidea (sea urchins) that includes euechinoid sea urchins (modern sea urchins), cidaroid sea urchins (pencil urchins) and sand dollars. In modern sea urchins and sand dollars, the four blastomeres in the animal hemisphere undergo symmetric and vertical cell divisions to generate eight intermediate size blastomeres (mesomeres), while the four vegetal blastomeres undergo asymmetric and horizontal cell division to yield four large and four small daughter cells (macromeres and micromeres, respectively). Micromeres undergo another asymmetric cell division at the fifth cleavage to form large and small micromeres, each of which gives rise exclusively to primary mesenchyme cells (PMCs)/skeletogenic cells and the germline, respectively.
Specification of the micromere lineage occurs autonomously even in vitro (Okazaki, 1975; Yajima & Wessel, 2012), while other blastomeres (mesomeres and macromeres) remain unspecified and undergo conditional specification in response to inductive signals from micromeres. Under normal conditions, mesomeres and macromeres give rise to ectoderm and endomesoderm, respectively. However, when extra micromeres are transplanted to an ectopic position such as the animal pole, secondary gut formation occurs at that site (Fig. 1B) (Boveri, 1901; Hörstadius, 1928; Ransick & Davidson, 1993). This classical experiment demonstrates that micromeres have an inductive capability and are capable of changing the fate of adjacent cells to become endomesoderm. When micromeres are removed at the 16-cell stage, on the other hand, the resultant embryo often undergoes a severe developmental delay and gastrulation failure in S. purpuratus (Ransick & Davidson, 1995). Of note, in H. pulcherrimus, another sea urchin species, micromere-removal appears to cause only modest developmental delay, while randomizing left-right polarity in larvae (Kitazawa & Amemiya, 2007). Therefore, the level of micromere’s essentiality for gastrulation may vary for species and its signaling function appears to be dispensable through compensatory mechanisms that are not yet well identified (Voronina et al., 2008; Yajima & Wessel, 2015). Nonetheless, it is a consistent observation in multiple sea urchin species that micromeres provide an important role as organizers in normal development.
While signaling to adjacent blastomeres, micromeres commit themselves to specific fates at the 16-cell stage. Under normal development, one of the micromere descendants, the large micromere, gives rise to PMCs, which ingress into the blastocoel prior to gastrulation and form the skeleton of the larva. Another micromere descendant, the small micromere, invaginates together with secondary mesenchyme cells (SMCs) at the tip of archenteron during gastrulation and contributes to coelomic pouches that give rise to the adult rudiment. In addition to this unique location of small micromeres, based on the experimental outcomes of their molecular profiling and lineage tracing, small micromere descendants are proposed to contribute to the next generation as a germline (Yajima & Wessel, 2012).
2.2. Evolutionary introduction of micromeres during echinoid diversification
Echinoderms are deuterostomes and close relatives of chordates. Among echinoderms, only sea urchin embryos undergo asymmetric cell division at the 16-cell stage and form micromeres. Micromeres of the sea urchin are known to serve as an organizing center at the vegetal pole of the embryo, while other echinoderm embryos such as sea star embryos do not undergo asymmetric cell division or form an apparent signaling center during early embryogenesis. The sea star embryo lacks micromeres as well as PMCs and does not form a larval skeleton; therefore, it appears to completely lack the large micromere lineage. Furthermore, after fifth cleavage, small micromeres of the sea urchin start accumulating markers for primordial germ cells (PGCs) (e.g., vasa, nanos, piwi) (Juliano et al., 2006; Juliano, Yajima, & Wessel, 2010; Rodriguez et al., 2005; Voronina et al., 2008; Yajima & Wessel, 2011a, 2011b, 2012, 2015). In contrast, the sea star embryo enriches such germline markers in the tissue called posterior enterocoel (PE) only after the larval stage (Fresques, Zazueta-Novoa, Reich, & Wessel, 2013; Juliano & Wessel, 2009). These observations suggest that the evolutionary introduction of micromeres allowed early establishment of two unique cell lineages, the larval skeleton and the germline, in the sea urchin. Of note, the brittle star, another echinoderm, forms larval skeleton without forming micromeres. Transcriptomic analyses of the brittle star embryos, however, suggest that larval skeletogenesis likely evolved independently in two classes of echinoderms, sea urchins and brittle stars, potentially by co-opting each of the adult skeletogenic programs that are present in all echinoderms (Dylus, Czarkwiani, Blowes, Elphick, & Okiveri, 2018). Therefore, it is considered that brittle stars have acquired larval skeleton through a mechanism different from micromeres in the process of evolution, yet its exact mechanism remains unknown.
The pencil urchin, cidaroid sea urchin, is considered to be an ancestral type of the sea urchin and diverged around 252 million-years ago (Bottjer, Davidson, Peterson, & Cameron, 2006). Therefore, the pencil urchin is an evolutionary intermediate between the sea star and the sea urchin, and indeed demonstrates an intermediate type of development: the embryo forms 0–4 micromere-like cells of variable sizes. Importantly, a classic cell lineage experiment demonstrates that these micromeres of the pencil urchin contribute to the larval skeleton as do micromeres of the sea urchin (Wray & McClay, 1988). These observations support the contention that the micromere lineage is a derived trait of euechinoids. Further studies are awaited to determine if these micromere-like cells of the pencil urchin embryo have inductive capability; this would shed light on when the functional properties of micromeres were acquired during the evolutionary transition to modern sea urchins.
3. Unique properties of the micromere
3.1. Structural and chemical properties of the micromere
It has been reported that the micromere has a unique membrane composition. Transmission electron microcopy reveals that its plasma membrane is smooth and lacks cortical pigment granules. This is distinct from the other blastomeres, which have microvilli and pigment granules (Dale, Yazaki and Tosti, 1997; Dan, 1954). Furthermore, in the animal blastomere, a row of vesicles covers the cell surface, while the future micromere region of the vegetal blastomere lacks such vesicles (Dan et al., 1983). These observations suggest that micromeres may have different cellular signaling capability through its unique membrane, yet no direct functional study has been conducted to prove this point so far.
Several studies, however, support the view that micromeres have a unique intracellular signaling capability. For example, it has been reported that micromeres show a unique calcium ion elevation upon their formation. The calcium ion is an intracellular messenger in eukaryotic cells and plays critical roles in various cellular events including cell differentiation, proliferation and apoptosis across organisms (Berridge, Lipp, & Bootman, 2000). In the sea urchin embryo, it has been observed that intracellular calcium ion concentration elevates in the future micromere region during the fourth cell division (Yazaki, 2001; Yazaki et al., 2004). Subsequently, calcium oscillation occurs for about 10min, resulting in an increased concentration of the intracellular calcium ion in micromeres upon completion of cytokinesis (Yazaki et al., 2004). This calcium oscillation is proposed to be triggered by calcium influx caused by the activation of stretch-dependent calcium channels, followed by the release of calcium ions from the endoplasmic reticulum (Yazaki et al., 2004; Yazaki, Tosti, & Dale, 1995). These observations suggest a potential role of calcium oscillation in micromere formation and/or signaling.
Although the detailed functional significance of calcium elevation in micromeres has yet to be identified, in other organisms, it is known that phospholipase C (PLC) hydrolyses phosphatidylinositol 4,5-bisphosphate (PIP2) in the plasma membrane to produce 1,2-diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (IP3). IP3 subsequently induces the endoplasmic reticulum to release calcium ions. Calcium ions and DAG then activate conventional protein kinase C (cPKC), which mediates a wide range of biological functions such as cell shape regulation, proliferation and apoptosis. Therefore, in the sea urchin embryo, it has been proposed that a similar mechanism may be involved in calcium ion elevation in micromeres, and which may activate cPKC and controls a number of downstream events unique to micromeres. Supporting this idea, it has been reported that cPKC plays a critical role in embryonic axis formation of the sea urchin embryo (Yazaki et al., 2014). cPKC activator increases both nuclear and cytoplasmic localization of β-catenin in vegetal blastomeres, while depleting the nuclear β-catenin signal in animal blastomeres, enhancing the animal-vegetal (AV) gradient of β-catenin expression (Yazaki et al., 2014). A similar phenotype is caused by LiCl, a classically known vegetalizing agent (Livingston & Wilt, 1992). On the other hand, cPKC inhibitor induces nuclear localization of β-catenin in all blastomeres, diminishing the AV gradient in the embryo (Yazaki et al., 2014).
It is not yet identified how cPKC functions in the AV axis formation, but its inhibitor appears to alter a distribution of actin filaments by shifting their location from the plasma membrane to the cytoplasm. This suggests cPKC’s potential role in cytoskeletal control. Similar to cPKC inhibitor, GdCl3, an inhibitor for stretch-activated ion channels, also suppresses calcium ion influx in micromeres and delays gastrulation (Yazaki et al., 2004, 2014). Taken together, Yazaki et al. hypothesizes that calcium ion influx may be mediated by membrane-stretch activity of cPKC in micromeres and that cPKC may be not only a downstream but also an upstream factor of calcium ion dynamics, contributing to calcium homeostasis in the cell. It is, however, not yet directly tested if calcium ion influx actually activates cPKC in micromeres nor if cPKC indeed stretches the membrane and controls calcium ion dynamics in the cell. Additionally, it will be important to identify in the future if/how the activated cPKC signaling leads to what downstream pathways to control micromere signaling and AV axis formation. These remaining questions may be addressed by visualizing and manipulating dynamics of cPKC signaling, calcium ion influx, and β-catenin nuclear localization specifically in micromeres in the future.
3.2. Specification of the micromere lineage
Micromeres are committed to two specific cell fates at the 16-cell stage. Through another asymmetric cell division at the 16–32-cell stage, micromeres autonomously accumulate and segregate cell fate determinants important for skeletogenic and germline specification into large and small micromeres, respectively. In 1975, Okazaki reported this unique feature of micromeres at the first time by observing isolated micromeres in a culture dish. She observed that even under in vitro culture conditions, micromeres give rise to PMCs, and these PMCs migrate and form a larval skeleton through much the same process as endogenous PMCs in the embryo. In this isolated culture condition, micromeres still undergo asymmetric cell division on time to form large and small micromeres. Small micromeres then asymmetrically enrich Vasa, a germline marker, during this asymmetric cell division in an autonomous manner (Yajima & Wessel, 2012). In addition, these isolated small micromeres in a dish activate another germline marker, nanos, and rarely undergo cell division and remain quiescent, all of which are typical traits of PGCs. On the contrary, large micromeres undergo up to five cell divisions from day 3 to 5 after fertilization, which is also a typical trait of PMCs (Yajima & Wessel, 2012). These observations further suggest that the micromere lineage is autonomously specified by the 16-cell stage, yet the detailed mechanism that drives this autonomous process still remains largely unknown.
Several studies, on the other hand, have characterized the molecular properties of micromeres after their formation at 16-cell stage. A set of gene products accumulates in this lineage after the 16-cell stage, further facilitating a lineage segregation of micromeres. For example, β-catenin, a protein critical for micromere specification, is uniformly present in the cytoplasm until the 8-cell stage (Miller & McClay, 1997; Yazaki et al., 2014). Yazaki et al. (2014) reported that β-catenin initially translocates into the nucleus of macromeres at the 16-cell stage and then into that of micromeres after the 32-cell stage. Another study, however, did not observe this initial nuclear translocation of β-catenin in macromeres at 16-cell stage (Logan, Miller, Ferkowicz, & McClay, 1999). Although the reason for this slight inconsistency is unclear, both studies report that nuclear translocation of β-catenin takes place in large and small micromeres at the fifth cleavage division (Logan et al., 1999; Yazaki et al., 2014). Nuclear β-catenin is known to play critical roles in the Wnt signaling in a variety of cells and organisms. In the sea urchin embryo, nuclear β-catenin facilitates zygotic wnt8 expression in micromeres, which contributes to gastrulation and endomesoderm specification (Cui et al., 2014; Wikramanayake et al., 2004). Nuclear β-catenin is more stable in micromeres than in animal blastomeres due to the presence of Disheveled (Dsh). Dsh is a component of the Wnt signaling pathway and is maternally localized to the vegetal cortex of the fertilized egg (Leonard & Ettensohn, 2007; Weitzel et al., 2004). Overexpression of the DIX domain of Dsh, which acts as a dominant negative, prevents β-catenin nuclear accumulation in the vegetal blastomeres, indicating that Dsh contributes to the stability of β-catenin in the micromere lineage (Weitzel et al., 2004). In animal blastomeres, on the other hand, β-catenin is phosphorylated by Glycogen Synthase Kinase-3-β (GSKβ) in the absence of Dsh, resulting in β-catenin degradation through the ubiquitin-proteasome pathway (Emily-Fenouil, Ghiglione, Lhomond, Lepage, & Gache, 1998; Weitzel et al., 2004). Nuclear β-catenin in the micromere lineage acts as a transcription coactivator with TCF to facilitate downstream gene expression (Huang et al., 2000; Vonica, Weng, Gumbiner, & Venuti, 2000). Therefore, the β-catenin/TCF complex plays a central role in regulating the molecular and functional properties of micromere descendants after the 16-cell stage.
Small micromeres arise at the fifth cleavage division. Based on their unique cellular behavior and molecular properties, it is considered that the small micromeres constitute the definitive PGC lineage. For example, small micromeres are slow in cell cycling due to having a longer S phase, which is a conserved feature of PGCs. Indeed, they divide only twice during early embryogenesis up to the late gastrula stage (Tanaka & Dan, 1990). The molecular properties of small micromeres are also similar to those of PGCs in other organisms. For example, germline markers such as Vasa, Nanos and Piwi that are conserved across animals, are expressed in the small micromere lineage (Juliano et al., 2006, 2010; Rodriguez et al., 2005; Voronina et al., 2008; Yajima & Wessel, 2012, 2015). Expression of these molecules appears to occur autonomously in this lineage by yet unknown mechanisms. Furthermore, adult sea urchins derived from small micromere-depleted embryos have smaller gonads that lack gametes (Yajima & Wessel, 2011a). Based on these studies, small micromeres are considered to contribute to the germline in the sea urchin, yet further lineage tracing studies with genetically marked small micromeres will be useful to understand if small micromeres contribute solely to the germline or both to the germline and somatic structures of the adult sea urchin.
3.3. The gene regulatory network of micromeres
Gene regulatory networks (GRNs) are especially well characterized in sea urchins (Davidson et al., 2002). Network models are constructed through gene knockdowns using a morpholino antisense oligonucleotide for each gene of interest followed by quantitative gene expression profiling to identify changes in the expression levels of downstream genes. In the large micromere GRN, the β-catenin/TCF pathway is a central element that regulates Notch signaling, and which is critical for the inductive function of micromeres (Fig. 2). Maternally deposited β-catenin/TCF activates pmar1 repressor, which blocks transcription of a target gene hesC (Oliveri et al., 2002; Revilla-i-Domingo, Oliveri, & Davidson, 2007; Vonica et al., 2000). Since HesC is a transcriptional repressor, it suppresses expression of downstream factors. Therefore, this mechanism is called a double-negative gate and important for micromere specification (Fig. 2B). The double-negative gate promotes expression of not only skeletogenic regulators (e.g., alx1, tbr, ets1) important for PMC specification, but also expression of delta, a ligand of the Notch signaling needed for SMC specification (Fig. 2B) (Oliveri et al., 2002; Revilla-i-Domingo et al., 2007). At the blastula stage, the closest endomesoderm cells to the large micromeres express a Notch receptor and receive the Delta signal and differentiate into nonskeletogenic mesoderm, including pigment cells, blastocoelar cells and muscle cells (Materna & Davidson, 2012; Sherwood & McClay, 1997; Sherwood & McClay, 1999; Sweet, Hodor, & Ettensohn, 1999). Moreover, Delta from the large micromeres activates small micromere specific foxY, which is considered to be critical for multipotency of coelomic pouch cells, suggesting a possible role of the Notch signaling for small micromere specification in addition to mesoderm specification (Materna & Davidson, 2012; Materna, Swartz, & Smith, 2013; Song & Wessel, 2012).
Fig. 2.
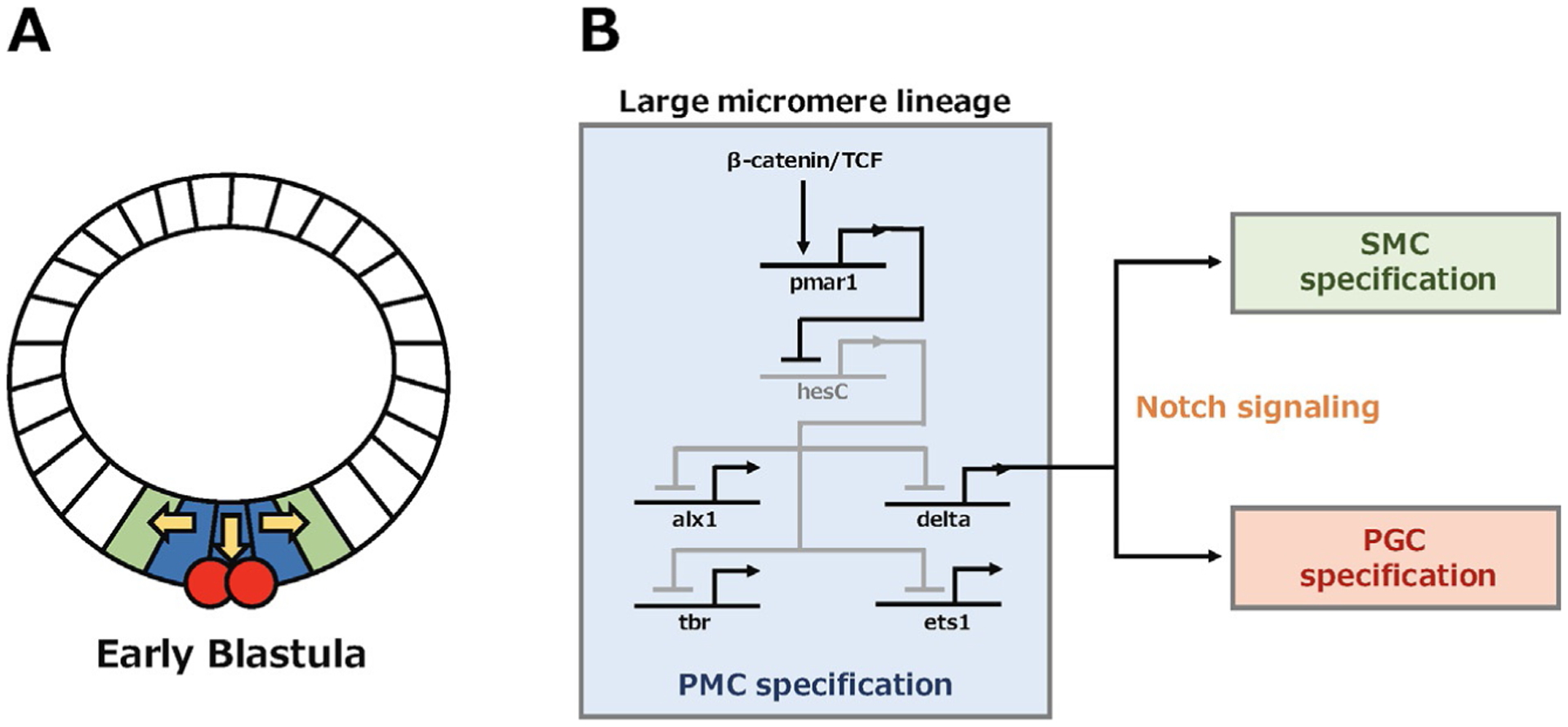
(A) Sea urchin embryo drawing at the early blastula stage. The large micromere lineage (blue) signals to adjacent endomesoderm (green) and small micromere (red) to specify SMC and PGC, respectively. (B) The GRNs model in the large micromere to regulate PMC specification and organize fates of neighbor cells through the Notch signaling.
A recent new technology, single cell RNA-seq, further expands our knowledge of sea urchin GRNs (Foster, Oulhen, & Wessel, 2021; Massri et al., 2021; Perillo et al., 2020). With this technology, Massri et al. were able to computationally track the progression of cell lineages during embryogenesis and found that micromere descendants, i.e., the PMCs and germline, molecularly diverge from the rest of cells in the embryo as early at the 32-cell stage, while non-micromere cells remain molecularly in an intermediate state until the gastrula stage. This finding further supports the contention that micromeres independently undergo early cell differentiation.
3.4. Modification of the micromere GRNs during evolution
Using the sea urchin GRNs as a foundation, an effort has been recently made to elucidate the GRNs of other echinoderms. This comparative approach allows researchers to understand how GRNs have been modified or pre-served during species diversification. In the pencil urchin, skeletogenic genes such as alx1, tbr and ets1 are expressed at different locations and/or at different times than in sea urchins (Erkenbrack & Davidson, 2015). Although alx1 is expressed in the micromere lineage of the pencil urchin, the timing of its expression is shifted to a later stage compared to that of the sea urchin. In contrast, tbr and ets1 expressions are expanded to non-skeletogenic mesoderm and not restricted in the micromere lineage. Further, hesC, which is repressed in the micromeres of the euechinoid sea urchins, is expressed in that those of the pencil urchin, which suggests the pencil urchin may not have the double-negative gate controlled by pmar1 gene. In fact, pmar1 gene was first reported to be absent in both genome and transcriptome databases of the pencil urchin, E. tribuloides (Erkenbrack & Davidson, 2015). Recently, in another pencil urchin P. baculosa, pmar1-like gene was identified to be present and lowly expressed in the vegetal region during early embryogenesis, yet promoting endomesodermal specification in a hesC-independent manner (Yamazaki et al., 2020). Based on these two independent studies, it appears that pmar1 gene may be present but the double-negative gate is absent in the pencil urchins. This raises a possibility that micromeres of the pencil urchin embryos may not have inductive capabilities as organizers, yet which still needs to be experimentally tested in near future.
Similar to the pencil urchin, the sea star has a pmar1-related gene, phb, and lacks the double-negative gate (Yamazaki et al., 2020). In the sea star embryo, hesC and delta are co-expressed at the vegetal pole until the mid-gastrula stage (Cary et al., 2020; Dylus et al., 2016). Inhibition of Notch signaling in the sea star embryo appears to decrease hesC expression and increase delta expression in the vegetal pole, suggesting a reciprocal relationship between Notch signaling and hesC expression (Cary et al., 2020). Notch signaling is known to mediate a patterning process called lateral inhibition, whereby signaling cells instruct adjacent cells to adopt a different fate than the instructing cell. Therefore, Cary et al. have proposed that this reciprocal regulation of Notch and HesC expression inhibits mesoderm cell differentiation, resulting in a gradual partitioning of endomesoderm through lateral inhibition during gastrulation (Cary et al., 2020). In contrast, at the blastula stage of the sea urchin embryo, suppression of hesC by pmar1 unilaterally activates the downstream Notch signaling in large micromere descendants, which induces adjacent cells to differentiate into mesodermal SMCs in the sea urchin. Therefore, although Notch signaling is conserved among echinoderms, the location and the timing of its function appears to be modified during evolution. Based on these observations, it has been proposed that acquisition of pmar1 function in hesC suppression facilitated formation of the β-catenin/TCF-mediated double-negative gate, modifying the entire micromere GRNs to be unique to the sea urchin. Although what facilitated a change of pmar1 function in the sea urchin has yet to be determined, a small modification of the GRNs may be sufficient to create a new cell type such as micromeres. Further comparative GRNs study in various echinoderms and other taxa will be important to identify key biological events and mechanisms that ultimately contributes to speciation of animals.
4. Mechanism of micromere formation through asymmetric cell division
4.1. Conserved molecular mechanism of asymmetric cell division
Asymmetric cell division produces two daughter cells with different fates and is a primary mechanism to generate cell diversity during development in a variety of organisms. The fundamental steps and basic molecular mechanisms of asymmetric cell division appear to be broadly conserved across organisms. Typically, the cell first sets up a polarity, enriches fate determinants toward a specific subcellular region and orients the mitotic spindle along the cell polarity axis (Betschinger & Knoblich, 2004). Of note, not all asymmetric cell division creates daughter cells of different sizes but micromeres are formed through unequal cell division. In the case of unequal cell division, polarity factors in the cell pull the spindle toward one side, resulting in asymmetric positioning of the spindle and unequal cleavage. In the sea urchin embryo, micromeres are formed through the following steps, (1) localization of polarity factors at the vegetal cortex by the 8-cell stage, prior to micromere formation; (2) asymmetrical positioning of the nucleus and spindle at the 8- to 16-cell stage, during micromere formation; (3) enrichment of fate determinants, during micromere formation; and (4) completion of unequal cell division followed by further expressions of micromere-specific molecules after micromere formation.
At the molecular level, in various organisms, a set of conserved polarity factors such as Partitioning-defective protein (Par) complex (Par3/Par6/aPKC), Activator of G-protein (AGS), Gα and Nuclear Mitotic Apparatus (NuMA in mammal, Mud in Drosophila) control cell polarity and spindle orientation and/or positioning (Fig. 3) (Ajduk & Zernicka-Goetz, 2016; Wavreil & Yajima, 2020). In the sea urchin, several G protein subunits and AGS are expressed throughout embryonic development (Voronina & Wessel, 2004, 2006). A yeast two-hybrid screen demonstrates that among the G protein subunits, only Gαi partners with AGS (Voronina & Wessel, 2006). AGS/Gαi localizes at the vegetal cortex from 4- to 8-cell stage well before micromere formation in the embryo (Poon et al., 2019). AGS knockdown appears to disturb nuclear translocation to the vegetal cortex and alter the spindle orientation in vegetal blastomeres in the 8- to 16-cell embryo. This causes a failure of unequal cleavage and of enrichment of cell fate determinants such as Vasa in resultant vegetal cells (Poon et al., 2019). Indeed, AGS-knockdown causes gastrulation failure, suggesting a loss of the micromere signaling in these knockdown embryos (Poon et al., 2019). Gαi-knockdown, on the other hand, causes effects similar to, but milder than AGS-knockdown, suggesting AGS may be the major factor that controls micromere formation. Collectively, AGS appears to be crucial not only for facilitating an unequal cell division but also for micromere function as an organizer in the sea urchin embryo.
Fig. 3.
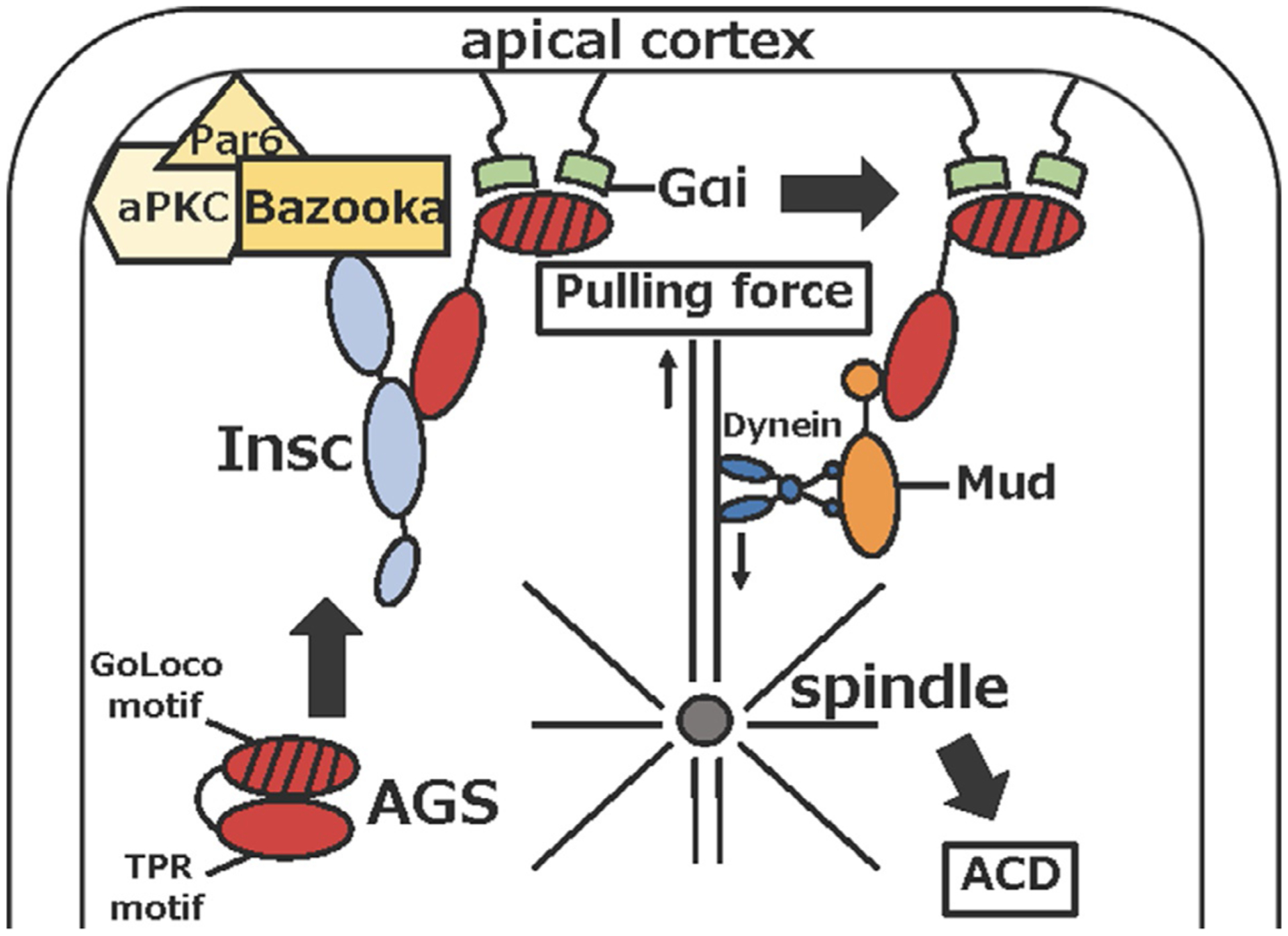
Molecular mechanism of asymmetric cell division in Drosophila neuroblast.
What recruits AGS/Gαi to the vegetal cortex prior to micromere formation has yet to be determined, but asymmetric cell division often starts from cell polarization mediated by the Inscuteable (Insc) and Par complex in embryogenesis of other organisms (Ajduk & Zernicka-Goetz, 2016; Henrique & Schweisguth, 2003). For example, in Drosophila embryonic neuroblast, Insc recruits AGS-family protein Pins to the apical pole to bind Gαi, and further connects Pins/Gαi to the Bazooka (Par3 homolog) through Insc (Schaefer, Shevchenko, Shevchenko, & Knoblich, 2000; Schober, Schaefer, & Knoblich, 1999; Wodarz, Ramrath, Kuchinke, & Knust, 1999; Yu, Morin, Cai, Yang, & Chia, 2000). In mammalian stem cells, LGN (mammalian AGS ortholog) is reported to interact with Par3 via Insc to control asymmetric cell division and cell fate in the similar manner to Drosophila neuroblast cells (Culurgioni et al., 2018; Izaki, Kamakura, Kohjima, & Sumimoto, 2006; Williams, Ratliff, Postiglione, Knoblich, & Fuchs, 2014; Zhu et al., 2011). Based on these observations, the Par complex appears to play a role in enriching AGS/Gαi localization at the subcortical region through Insc, and which contributes to cell fate regulation of the daughter cells.
In the sea urchin embryo, Par6 and its partner Cdc42, a member of the Rho family of small GTPases, localize to the cortex to outline the entire embryo as early as the 2-cell stage (Alford, Ng, & Burgess, 2009; Moorhouse, Gudejko, McDougall, & Burgess, 2015; Prulière, Cosson, Chevalier, Sardet, & Chenevert, 2011; Shiomi & Yamaguchi, 2008). Embryos that lack Cdc42 activity by knockdown or inhibitor treatment fail in PMC migration and alignment, and overall skeletogenesis (Sepúlveda-Ramírez, Toledo-Jacobo, Henson, & Shuster, 2018). Similarly, down-regulation of Par6 causes defects in skeletogenesis in resultant embryos (Shiomi, Yamazaki, Kagawa, Kiyomoto, & Yamaguchi, 2012). These observations suggest that Cdc42 and Par6 are important for skeletogenesis that is controlled by PMCs, the large micromere progeny after blastula stage. However, it is yet to be tested if Cdc42 and Par6 are also involved in micromere formation at the 8- to 16-cell stage. Furthermore, aPKC, another conserved polarity factor that is known to work with Cdc42 and Par6, is also expressed during embryogenesis of the sea urchin. Two reports differ, however, in their conclusions regarding the expression pattern of aPKC. One report suggests that aPKC outlines the entire cortex but is excluded from the vegetal cortex at the 16- to 64-cell stage (Prulière et al., 2011). Another report suggests that aPKC outlines the entire cortex including the vegetal cortex and that an inhibitor of aPKC has no impact on micromere formation (Moorhouse et al., 2015). Overall, these observations are all still too preliminary to allow solid conclusions. Direct functional studies are needed to determine if/how these conserved polarity factors such as Par6, Cdc42 and aPKC are involved in cell polarity establishment during micromere formation.
Once cell polarity is established, the spindle becomes positioned and oriented along this polarity axis in the cell, which generates spindle pulling force and leads to cytokinesis. When this cell polarity is asymmetric, that causes asymmetric spindle position and/or orientation, resulting in asymmetric cell division. In Drosophila and mammals, it is known that microtubule motor proteins such as dynein and kinesin control spindle orientation and microtubule-pulling force via interaction with AGS. The linker region of AGS is phosphorylated during early M-phase, and which facilitates binding of the membrane-associated Discs large (Dlg). This further recruits kinesin to attach astral microtubule to the cortex (Asaba, Hanada, Takeuchi, & Chishti, 2003; Bellaıche et al., 2001; Hanada, Lin, Tibaldi, Reinherz, & Chishti, 2000; Johnston, Hirono, Prehoda, & Doe, 2009; Mauser & Prehoda, 2012; Siegrist & Doe, 2005; Yamada, Hanada, & Chishti, 2007). Subsequently, during late mitosis, Mud (in Drosophila) or NuMA (in mammal) forms a complex with AGS and pulls the spindle through dynein toward the cortex (Culurgioni, Alfieri, Pendolino, Laddomada, & Mapelli, 2011; Izumi, Ohta, Hisata, Raabe, & Matsuzaki, 2006; Johnston et al., 2009; Mauser & Prehoda, 2012; Siller, Cabernard, & Doe, 2006; Takayanagi et al., 2019; Zhu et al., 2011). In the sea urchin embryo, immunoprecipitation experiments show that AGS/Gαi interacts with NuMA and dynein as well as other fate determinants such as Vasa (Poon et al., 2019), suggesting that a similar mechanism of AGS-NuMA-Dynein-dependent spindle orientation and microtubule pulling may be employed in micromere formation. Furthermore, the phosphorylation site of the AGS linker region appears to be also conserved in sea urchin AGS, but its functional involvement in kinesin dynamics is yet to be determined. Further studies are required to reveal how similar the mechanism of asymmetric cell division used in micromere formation is to the other model systems that have been used to study asymmetric cell division.
4.2. Evolutionary introduction of the micromere through modifications of the AGS protein
AGS appears to be a key regulator of micromere formation, which is unique to the sea urchin embryo. Importantly, introduction of sea urchin AGS into the sea star embryo leads to formation of micromere-like cells, while non-functional version of sea urchin AGS does not appear to induce such cells (Poon et al., 2019). This suggests that sea urchin AGS but not sea star AGS may have an ability to induce micromere-like cells in the embryo. Based on these observations, Poon et al. have proposed that sea urchin AGS protein has been modified during the process of evolution and has obtained a distinct function necessary for formation of a new cell type, the micromere. In this initial study, however, overexpression of sea star AGS was not performed, which is critical to directly compare functional differences of sea urchin and sea star AGS in micromere formation.
AGS protein is consisted of two major functional domains: the N-terminal tetratricopeptide repeats (TPR) motifs and the C-terminal G-protein regulatory (GoLoco) motifs. AGS in general stays in a closed form via intermolecular binding between the TPR and GoLoco motifs. AGS switches to the open state when Gα binds to GoLoco motifs and disrupts the TPR-GoLoco interaction, thereby allowing the TPR motifs to associate with other proteins necessary for asymmetric cell division (Du & Macara, 2004; Johnston et al., 2009; Nipper, Siller, Smith, Doe, & Prehoda, 2007; Pan et al., 2013). The number of TPR and GoLoco motifs appears to vary across taxa, even within the same phylum (Waldron & Yajima, 2020). In echinoderms, the N-terminal TPR motifs are rather similar in actual sequence among various echinoderm AGS proteins, and their evolutionary modifications and/or roles in micromere formation are yet to be determined. In contrast, the C-terminal AGS sequence are highly varied among echinoderms. For example, echinoids, including sand dollars and sea urchins, possess three to four GoLoco motifs, while the sea star possesses only two GoLoco motifs (Fig. 4). Importantly, sea urchin AGS that lacks the first GoLoco motif fails to induce formation of micromere-like cells in the sea star embryo (Poon et al., 2019), suggesting the first GoLoco motif is required for micromere formation. Based on these observations, it is proposed that addition of an extra GoLoco motif in the sea urchin AGS protein has facilitated an asymmetric cell division at the 8- to 16-cell stage, which ultimately contributed to formation of micromeres in the modern sea urchin. Futher experiments such as introducing chimeric sea star AGS that is inserted with the first GoLoco motif of the sea urchin into the sea star embryo may be helpful to test this hypothesis in the future.
Fig. 4.
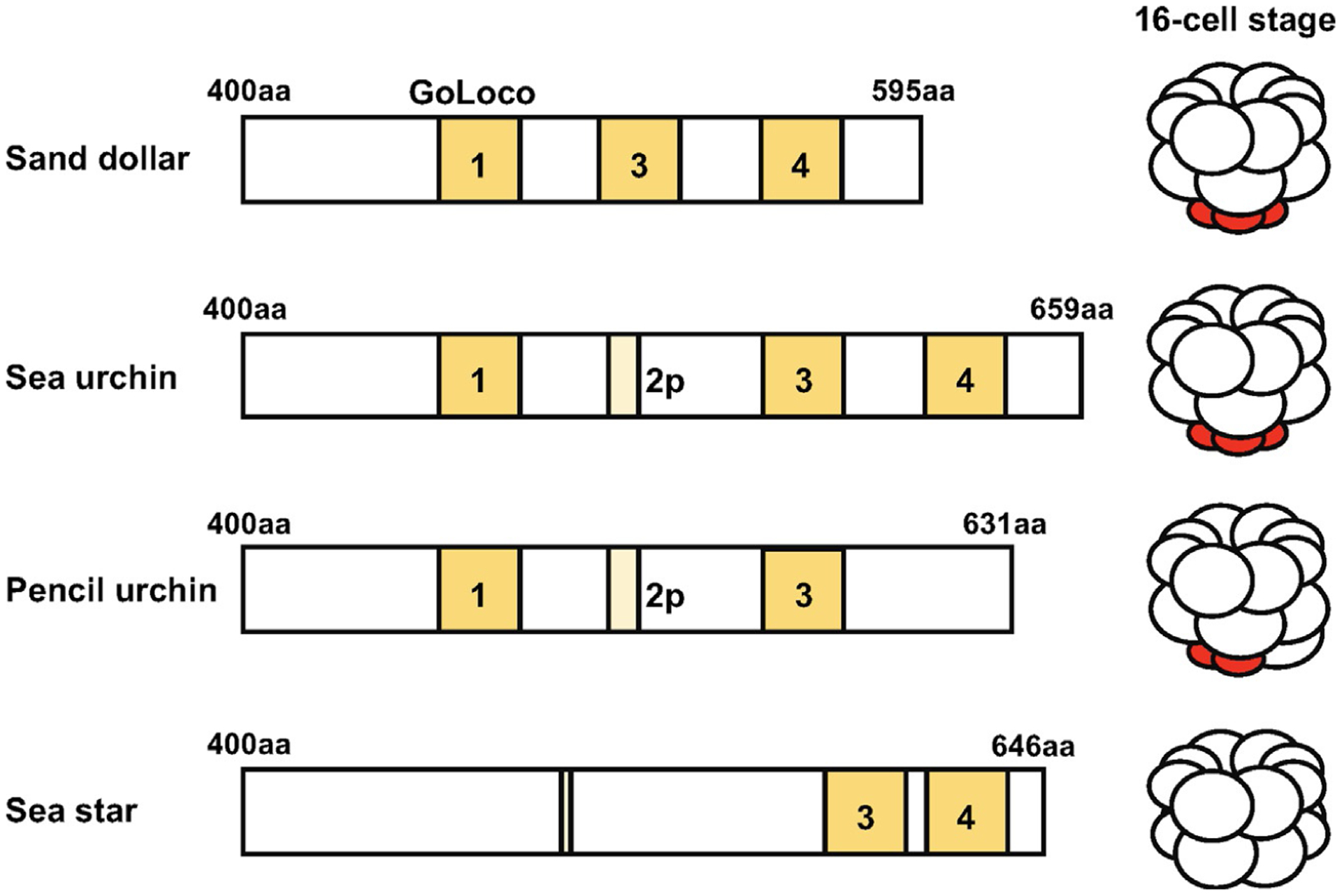
Predicted C-terminal GoLoco motifs of each AGS protein among echinoderms based on NCBI blast search results (Poon et al., 2019). Conserved GoLoco motifs are highlighted in yellow. Less conserved or partial motifs are colored in light yellow. Micromeres are indicated in red.
In addition to the TPR and GoLoco motifs, studies in other organisms suggest the functional importance of a linker region that connects the TPR and GoLoco motifs of AGS. In Drosophila S2 cells, the Pins linker domain is phosphorylated by Aurora kinase A, which causes Dlg and kinesin to associate with the microtubule, controlling the spindle orientation (Johnston et al., 2009). In MDCK cells, the same linker domain is phosphorylated by aPKC, which removes the sub-cellular localization of AGS from the cortex, thus controlling spindle orientation (Hao et al., 2010). However, a possible functional contribution of the AGS linker domain is yet to be tested in echinoderms. The molecular dissection of sea urchin AGS will be useful to fully understand the mechanism of AGS-mediated micromere formation in the sea urchin embryo.
5. Unique transcriptional and translational activity of the micromere and its descendants
5.1. Unique activity of the small micromere
Transcriptional and translational activities become broadly suppressed in small micromeres after their formation at fifth cleavage. It appears that only maternally deposited factors function to maintain the pluripotency nature of PGCs in this lineage (Oulhen, Swartz, Laird, Mascaro, & Wessel, 2017; Swartz et al., 2014). Maternal transcripts are ubiquitously expressed in eggs and early embryos of the sea urchin, yet their major turn-over occurs during the blastula-to-gastrula stage in all cells except the small micromere lineage (Swartz et al., 2014). At this maternal to zygotic transition, CNOT, a CCR4-related deadenylase, is responsible for degrading maternal mRNAs in the cells. In the small micromere lineage, however, RNA-binding protein Nanos is present and degrades cnot6 mRNA, resulting in the protection of maternal transcripts in this lineage. Indeed, mis-localization of CNOT6 in the small micromeres compromises the stable expression of germline factors such as Vasa (Swartz et al., 2014). These findings suggest that specification of the small micromere lineage may be largely dependent on the maternal factors. This further supports the contention that micromere descendants undergo autonomous specification in culture (Yajima & Wessel, 2012).
Although it is not yet well understood in the sea urchin, Nanos may also contribute to the unusually long cell cycle of small micromeres (Tanaka & Dan, 1990). Nanos is known to target cyclinB and suppresses its expression in the germline of several organisms such as Drosophila and Xenopus (Kadyrova, Habara, Lee, & Wharton, 2007; Lai, Zhou, Luo, Fox, & King, 2011). Therefore, Nanos may also contribute to suppression of cell cycle regulators in small micromeres of the sea urchin, although this needs to be experimentally tested in the future. Overall, these observations illustrate that unique mechanisms of transcriptional and translational regulation operate in the small micromere lineage. Further studies that identify the targets and the mechanism of Nanos and other germline factors will be helpful to understand the developmental process of this unique lineage.
5.2. Hypothesis: Asymmetric segregation of a translational regulator Vasa and its possible involvement in micromere specification
In the sea urchin embryo, protein synthesis occurs throughout the cell cycle including M-phase (Gross & Fry, 1966). Furthermore, a subset of mRNAs, polyribosomes and polyA-binding proteins is enriched on the mitotic apparatus (Hamill, Davis, Drawbridge, & Suprenant, 1994; Suprenant, 1993). Based on these observations, it has been hypothesized that localized translation on the mitotic apparatus may occur and contribute to mitotic regulation during early embryogenesis of several organisms (Waldron & Yajima, 2020). Indeed, more recent studies in the Xenopus embryo or lysates prepared from the embryo suggest that translation of cyclinB on the mitotic apparatus is required for proper cell cycle progression (Groisman et al., 2000). Although it is yet to be proven if such a process operates during sea urchin embryogenesis, this mechanism could explain the rapid lineage segregation of the micromere lineage through an asymmetric cell division.
A DEAD-box RNA helicase, Vasa, is one potential candidate that may mediate a process of localized translation on the mitotic apparatus. Vasa is known to be associated with the spindle during M-phase and becomes enriched into micromeres through an asymmetric cell division at the 8- to 16-cell stage (Yajima & Wessel, 2011b). Vasa is also required for the translation of cell cycle regulators such as cyclinB and retinoblastoma1 during sea urchin development (Fernandez-Nicolas, Xu, & Yajima, 2019; Yajima & Wessel, 2015), pointing to its function as a translational regulator. Although the detailed mechanism of Vasa function in sea urchin embryogenesis is still largely unknown, its localization mechanism to the micromere has been partly revealed. Gustavus (Gus), an ECS-type E3 ubiquitin ligase, is proposed to have dual functions and control Vasa enrichment in micromeres: Gus degrades Vasa in nonmicromere cells yet protects it in micromeres, resulting in increased Vasa enrichment in the micromere lineage (Gustafson, Yajima, Juliano, & Wessel, 2011). The functional significance of this Vasa enrichment in the micromere is not yet clear. Vasa may be passively segregated into the PGC precursor cells through asymmetric cell divisions. Or, one may also speculate that Vasa is needed for its immediate function on the mitotic apparatus during micromere formation. For example, Vasa may directly target mRNAs responsible for asymmetric cell division. In Drosophila neuroblasts, Abstrakt, another DEAD-box helicase, is known to directly bind and translate insc mRNA that is apically localized in the cell (Irion et al., 2004; Knirr, Breuer, Paululat, & Renkawitz-Pohl, 1997). During Drosophila oogenesis, Vasa is required for proper localization of bcb, osk and grk mRNAs that are important for establishing embryonic polarity and for piRNA pathway regulation (Tomancak, Guichet, Zavorszky, & Ephrussi, 1998) in germ cells (Xiol et al., 2014). These observations suggest a potential role of Vasa in controlling localization, translation and/or degradation of mRNAs important for asymmetric cell division or cell fate specification. Future studies that identify Vasa’s target mRNAs and its function on the mitotic apparatus during asymmetric cell division will be essential to determine its possible role in micromere formation.
6. Conclusions and perspectives
Micromeres display distinctive molecular features compared to the rest of the blastomeres in the sea urchin embryo. These features include an elevation of calcium levels, the presence of a unique GRNs circuit, and distinct spatial and temporal regulation of cell fate determinants. Besides the topics mentioned above, other elements may further make this lineage unique. For example, mitochondrial activity and pH are reported to be reduced in small micromere descendants (Oulhen et al., 2017). Reduction of mitochondria activity could change the metabolism from oxidative phosphorylation to glycolytic regulation, which may induce acidification of the cytoplasm. Although a number of unique properties of the micromere and its descendants has been reported, if/how each of these elements cross-talks to form this unique lineage remains largely unknown. Therefore, future studies could include investigating the functional interactions between these unique properties of the micromere, which will provide a comprehensive view of how this unique lineage is regulated.
The evolutionary introduction of asymmetric cell division into the early developmental program of the sea urchin have allowed formation of a new cell type, the micromere. It appears that modifications of AGS at least in part contributed to introduction of the micromere. However, what modifications of AGS triggered formation of the micromere is yet to be known. Further, it has not been tested if other polarity factors associated with AGS have also undergone evolutionary modifications to contribute to formation of the micromere. Further molecular dissection of AGS and its partner proteins both in the sea urchin embryo and in other echinoderm embryos will be critical to understand the mechanism and the evolutionary process of micromere formation in the sea urchin.
Acknowledgment
Our research on this topic was funded by the National Institute of General Medical Sciences (1R01GM126043-01) and the National Science Foundation (IOS-1940975) to M.Y.
References
- Ajduk A, & Zernicka-Goetz M (2016). Polarity and cell division orientation in the cleavage embryo: From worm to human. Molecular Human Reproduction, 22, 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alford LM, Ng MM, & Burgess DR (2009). Cell polarity emerges at first cleavage in sea urchin embryos. Developmental Biology, 330, 12–20. [DOI] [PubMed] [Google Scholar]
- Angerer LM, & Angerer RC (2003). Patterning the sea urchin embryo: Gene regulatory networks, signaling pathways, and cellular interactions. Current Topics in Developmental Biology, 53, 159–198. [DOI] [PubMed] [Google Scholar]
- Asaba N, Hanada T, Takeuchi A, & Chishti AH (2003). Direct interaction with a kinesin-related motor mediates transport of mammalian discs large tumor suppressor homologue in epithelial cells. The Journal of Biological Chemistry, 278, 8395–8400. [DOI] [PubMed] [Google Scholar]
- Bellaıche Y, Radovic A, Woods DF, Hough CD, Parmentier ML, O’Kane CJ, et al. (2001). The partner of Inscuteable/discs-large complex is required to establish planar polarity during asymmetric cell division in drosophila. Cell, 106, 355–366. [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Lipp P, & Bootman MD (2000). The versatility and universality of calcium signalling. Nature Reviews Molecular Cell Biology, 1, 11–21. [DOI] [PubMed] [Google Scholar]
- Betschinger J, & Knoblich JA (2004). Dare to be different: Asymmetric cell division in Drosophila C. elegans and vertebrates. Current Biology, 14, R674–R685. [DOI] [PubMed] [Google Scholar]
- Bottjer DJ, Davidson EH, Peterson KJ, & Cameron RA (2006). Paleogenomics of echinoderms. Science, 314, 956–960. [DOI] [PubMed] [Google Scholar]
- Boveri T (1901). Über die Polarität des Seeigeleies. Verhandlungen der Physikalisch-Medizinischen Gesellschaft zu Würzburg, 34, 145–176. [Google Scholar]
- Cary GA, McCauley BS, Zueva O, Pattinato J, Longabaugh W, & Hinman VF (2020). Systematic comparison of sea urchin and sea star developmental gene regulatory networks explains how novelty is incorporated in early development. Nature Communications, 11, 6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M, Siriwon N, Li E, Davidson EH, & Peter IS (2014). Specific functions of the Wnt signaling system in gene regulatory networks throughout the early sea urchin embryo. Proceedings of the National Academy of Sciences of the United States of America, 111, E5029–E5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culurgioni S, Alfieri A, Pendolino V, Laddomada F, & Mapelli M (2011). Inscuteable and NuMA proteins bind competitively to Leu-Gly-Asn repeat-enriched protein (LGN) during asymmetric cell divisions. Proceedings of the National Academy of Sciences of the United States of America, 108, 20998–21003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culurgioni S, Mari S, Bonetti P, Gallini S, Bonetto G, Brennich M, et al. (2018). Insc:LGN tetramers promote asymmetric divisions of mammary stem cells. Nature Communications, 9, 1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale B, Yazaki I, & Tosti E (1997). Polarized distribution of L-type calcium channels in early sea urchin embryos. American Journal of Physiology, 273, 822–825. [DOI] [PubMed] [Google Scholar]
- Dan K (1954). The cortical movement in Arbacia punctulata eggs through cleavage cycles. Embrologia, 2, 115–122. [Google Scholar]
- Dan K, Endo S, & Uemura I (1983). Studies on unequal cleavage in sea urchins II. Surface differentiation and the direction of nuclear migration. Development, Growth and Differentiation, 25, 227–237. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, et al. (2002). A genomic regulatory network for development. Science, 295, 1669–1678. [DOI] [PubMed] [Google Scholar]
- Du Q, & Macara IG (2004). Mammalian pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell, 119, 503–516. [DOI] [PubMed] [Google Scholar]
- Dylus DV, Czarkwiani A, Blowes LM, Elphick MR, & Okiveri P (2018). Developmental transcriptomics of the brittle star Amphiura filiformis reveals gene regulatory network rewiring in echinoderm larval skeleton evolution. Genome Biology, 19, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dylus DV, Czarkwiani A, Stångberg J, Ortega-Martinez O, Dupont S, & Oliveri P (2016). Large-scale gene expression study in the ophiuroid Amphiura filiformis provides insights into evolution of gene regulatory networks. EvoDevo, 7, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emily-Fenouil F, Ghiglione C, Lhomond G, Lepage T, & Gache C (1998). GSK3β/shaggy mediates patterning along the animal-vegetal axis of the sea urchin embryo. Development, 125, 2489–2498. [DOI] [PubMed] [Google Scholar]
- Erkenbrack EM, & Davidson EH (2015). Evolutionary rewiring of gene regulatory network linkages at divergence of the echinoid subclasses. Proceedings of the National Academy of Sciences of the United States of America, 112, E4075–E4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Nicolas A, Xu D, & Yajima M (2019). A tumor suppressor Retinoblastoma1 is essential for embryonic development in the sea urchin. Developmental Dynamics, 248, 1273–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster S, Oulhen N, & Wessel G (2021). Correction: A single cell RNA sequencing resource for early sea urchin development. Development, 148, dev199412. [DOI] [PubMed] [Google Scholar]
- Fresques T, Zazueta-Novoa V, Reich A, & Wessel GM (2013). Selective accumulation of germ-line associated gene products in early development of the sea star and distinct differences from germ-line development in the sea urchin. Developmental Dynamics, 243, 568–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman I, Huang Y-S, Mendez R, Cao Q, Theurkauf W, & Richter JD (2000). CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: Implications for local translational control of cell division. Cell, 103, 435–447. [DOI] [PubMed] [Google Scholar]
- Gross PR, & Fry BJ (1966). Continuity of protein synthesis through cleavage metaphase. Science, 153, 749–751. [DOI] [PubMed] [Google Scholar]
- Gustafson EA, Yajima M, Juliano CE, & Wessel GM (2011). Post-translational regulation by Gustavus contributes to selective vasa protein accumulation in multipotent cells during embryogenesis. Developmental Biology, 349, 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill D, Davis J, Drawbridge J, & Suprenant KA (1994). Polyribosome targeting to microtubules: Enrichment of specific mRNAs in a reconstituted microtubule preparation from sea urchin embryos. The Journal of Cell Biology, 127, 973–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T, Lin L, Tibaldi EV, Reinherz EL, & Chishti AH (2000). GAKIN, a novel kinesin-like protein associates with the human homologue of the drosophila discs large tumor suppressor in T lymphocytes. The Journal of Biological Chemistry, 275, 28774–28784. [DOI] [PubMed] [Google Scholar]
- Hao Y, Du Q, Chen X, Zheng Z, Balsbaugh JL, Maitra S, et al. (2010). Par3 controls epithelial spindle orientation by aPKC-mediated phosphorylation of apical pins. Current Biology, 20, 1809–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrique D, & Schweisguth F (2003). Cell polarity: The ups and downs of the Par6/aPKC complex. Current Opinion in Genetics & Development, 13, 341–350. [DOI] [PubMed] [Google Scholar]
- Hörstadius S (1928). Über die determination des keimes bei echinodermen. Acta Zoologica, 9, 1–191. [Google Scholar]
- Huang L, Li X, El-Hodiri HM, Dayal S, Wikramanayake AH, & Klein WH (2000). Involvement of Tcf/Lef in establishing cell types along the animal-vegetal axis of sea urchins. Development Genes & Evolution, 210, 73–81. [DOI] [PubMed] [Google Scholar]
- Irion U, Leptin M, Siller K, Fuerstenberg S, Cai Y, Doe CQ, et al. (2004). Abstrakt, a DEAD box protein, regulates Insc levels and asymmetric division of neural and mesodermal progenitors. Current Biology, 14, 138–144. [DOI] [PubMed] [Google Scholar]
- Izaki T, Kamakura S, Kohjima M, & Sumimoto H (2006). Two forms of human Inscuteable-related protein that links Par3 to the pins homologues LGN and AGS3. Biochemical and Biophysical Research Communications, 341, 1001–1006. [DOI] [PubMed] [Google Scholar]
- Izumi Y, Ohta N, Hisata K, Raabe T, & Matsuzaki F (2006). Drosophila pins-binding protein mud regulates spindle-polarity coupling and centrosome organization. Nature Cell Biology, 8, 586–593. [DOI] [PubMed] [Google Scholar]
- Johnston CA, Hirono K, Prehoda KE, & Doe CQ (2009). Identification of an Aurora-a/PinsLINKER/Dlg spindle orientation pathway using induced cell polarity in S2 cells. Cell, 138, 1150–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano CE, Voronina E, Stack C, Aldrich M, Cameron AR, & Wessel GM (2006). Germ line determinants are not localized early in sea urchin development, but do accumulate in the small micromere lineage. Developmental Biology, 300, 406–415. [DOI] [PubMed] [Google Scholar]
- Juliano CE, & Wessel GM (2009). An evolutionary transition of vasa regulation in echinoderms. Evolution & Development, 11, 560–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano CE, Yajima M, & Wessel GM (2010). Nanos functions to maintain the fate of the small micromere lineage in the sea urchin embryo. Developmental Biology, 337, 220–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrova LY, Habara Y, Lee TH, & Wharton RP (2007). Translational control of maternal cyclin B mRNA by Nanos in the drosophila germline. Development, 134, 1519–1527. [DOI] [PubMed] [Google Scholar]
- Kitazawa C, & Amemiya S (2007). Micromere-derived signal regulates larval left-right polarity during sea urchin development. Journal of Experimental Zoology, 307A, 249–262. [DOI] [PubMed] [Google Scholar]
- Knirr S, Breuer S, Paululat A, & Renkawitz-Pohl R (1997). Somatic mesoderm differentiation and the development of a subset of pericardial cells depend on the not enough muscles (nem) locus, which contains the inscuteable gene and the intron located gene, skittles. Mechanism of Development, 67, 69–81. [DOI] [PubMed] [Google Scholar]
- Lai F, Zhou Y, Luo X, Fox J, & King ML (2011). Nanos1 functions as a translational repressor in the Xenopus germline. Mechanisms of Development, 128, 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard JD, & Ettensohn CA (2007). Analysis of dishevelled localization and function in the early sea urchin embryo. Developmental Biology, 306, 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston BT, & Wilt FH (1992). Phorbol esters alter cell fate during development of sea urchin embryos. The Journal of Cell Biology, 119, 1641–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CY, Miller JR, Ferkowicz MJ, & McClay DR (1999). Nuclear β-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development, 126, 345–357. [DOI] [PubMed] [Google Scholar]
- Massri AJ, Greenstreet L, Afanassiev A, Berrio A, Wray GA, Schiebinger G, et al. (2021). Developmental single-cell transcriptomics in the Lytechinus variegatus sea urchin embryo. Development, 148(19). 10.1242/dev.198614, dev198614. https://pubmed.ncbi.nlm.nih.gov/34463740/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materna SC, & Davidson EH (2012). A comprehensive analysis of Delta signaling in pre-gastrular sea urchin embryos. Developmental Biology, 364, 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materna SC, Swartz SZ, & Smith J (2013). Notch and nodal control forkhead factor expression in the specification of multipotent progenitors in sea urchin. Development, 140, 1796–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauser JF, & Prehoda KE (2012). Inscuteable regulates the pins-mud spindle orientation pathway. PLoS One, 7, e29611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JR, & McClay DR (1997). Changes in the pattern of adherens junction-associated β-catenin accompany morphogenesis in the sea urchin embryo. Developmental Biology, 192, 310–322. [DOI] [PubMed] [Google Scholar]
- Moorhouse KS, Gudejko HFM, McDougall A, & Burgess DR (2015). Influence of cell on early development of the sea urchin embryo. Developmental Dynamics, 244, 1469–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nipper RW, Siller KH, Smith NR, Doe CQ, & Prehoda KE (2007). Gαi generates multiple pins activation states to link cortical polarity and spindle orientation in drosophila neuroblasts. Proceedings of the National Academy of Sciences of the United States of America, 104, 14306–14311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki K (1975). Spicule formation by isolated micromeres of the sea urchin embryo. American Zoologist, 15, 567–581. [Google Scholar]
- Oliveri P, Carrick DM, & Davidson EH (2002). A regulatory gene network that directs micromere specification in the sea urchin embryo. Developmental Biology, 246, 209–228. [DOI] [PubMed] [Google Scholar]
- Oulhen N, Swartz SZ, Laird J, Mascaro A, & Wessel GM (2017). Transient translational quiescence in primordial germ cells. Development, 144, 1201–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z, Zhu J, Shang Y, Wei Z, Jia M, Xia C, et al. (2013). An autoinhibited conformation of LGN reveals a distinct interaction mode between GoLoco motifs and TPR motifs. Structure, 21, 1007–1017. [DOI] [PubMed] [Google Scholar]
- Perillo M, Oulhen N, Foster S, Spurrell M, Calestani C, & Wessel G (2020). Regulation of dynamic pigment cell states at single-cell resolution. eLife, 9, e60388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon J, Fries A, Wessel GM, & Yajima M (2019). Evolutionary modification of AGS protein contributes to formation of micromeres in sea urchins. Nature Communications, 10, 3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prulière G, Cosson J, Chevalier S, Sardet C, & Chenevert J (2011). Atypical protein kinase C controls sea urchin ciliogenesis. Molecular Biology of the Cell, 22, 2042–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransick A, & Davidson EH (1993). A complete second gut induced by transplanted micromeres in the sea urchin embryo. Science, 259, 1134–1138. [DOI] [PubMed] [Google Scholar]
- Ransick A, & Davidson EH (1995). Micromeres are required for normal vegetal plate specification in sea urchin embryos. Development, 121, 3215–3222. [DOI] [PubMed] [Google Scholar]
- Revilla-i-Domingo R, Oliveri P, & Davidson EH (2007). A missing link in the sea urchin embryo gene regulatory network: hesC and the double-negative specification of micromeres. Proceedings of the National Academy of Sciences of the United States of America, 104, 12383–12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AJ, Seipel SA, Hamill DR, Romancino DP, Carlo MD, Suprenant KA, et al. (2005). Seawi—A Sea urchin piwi/argonaute family member is a component of MT-RNP complexes. RNA, 11, 646–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Shevchenko A, Shevchenko A, & Knoblich JA (2000). A protein complex containing Inscuteable and the Gα-binding protein pins orients asymmetric cell divisions in drosophila. Current Biology, 10, 353–362. [DOI] [PubMed] [Google Scholar]
- Schober M, Schaefer M, & Knoblich JA (1999). Bazooka recruits Inscuteable to orient asymmetric cell divisions in drosophila neuroblasts. Nature, 402, 548–551. [DOI] [PubMed] [Google Scholar]
- Sepúlveda-Ramírez SP, Toledo-Jacobo L, Henson JH, & Shuster CB (2018). Cdc42 controls primary mesenchyme cell morphogenesis in the sea urchin embryo. Developmental Biology, 437, 140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood DR, & McClay DR (1997). Identification and localization of a sea urchin notch homologue: Insights into vegetal plate regionalization and notch receptor regulation. Development, 124, 3363–3374. [DOI] [PubMed] [Google Scholar]
- Sherwood DR, & McClay DR (1999). LvNotch signaling mediates secondary mesenchyme specification in the sea urchin embryo. Development, 126, 1703–1713. [DOI] [PubMed] [Google Scholar]
- Shiomi K, & Yamaguchi M (2008). Expression patterns of three par-related genes in sea urchin embryos. Gene Expression Patterns, 8, 323–330. [DOI] [PubMed] [Google Scholar]
- Shiomi K, Yamazaki A, Kagawa M, Kiyomoto M, & Yamaguchi M (2012). Par6 regulates skeletogenesis and gut differentiation in sea urchin larvae. Development Genes and Evolution, 222, 269–278. [DOI] [PubMed] [Google Scholar]
- Siegrist SE, & Doe CQ (2005). Microtubule-induced pins/Gai cortical polarity in drosophila neuroblasts. Cell, 123, 1323–1335. [DOI] [PubMed] [Google Scholar]
- Siller KH, Cabernard C, & Doe CQ (2006). The NuMA-related mud protein binds pins and regulates spindle orientation in drosophila neuroblasts. Nature Cell Biology, 8, 594–600. [DOI] [PubMed] [Google Scholar]
- Song JL, & Wessel GM (2012). The forkhead transcription factor FoxY regulates Nanos. Molecular Reproduction & Development, 79, 680–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spemann H, & Mangold H (1924). Über induktion von embryonalanlagen durch implantation artfremder organisatoren. Archiv für mikroskopische Anatomie und Entwicklungsmechanik, 100, 599–638. [Google Scholar]
- Suprenant KA (1993). Microtubules, ribosomes, and RNA: Evidence for cytoplasmic localization and translational regulation. Cell Motility and the Cytoskeleton, 25, 1–9. [DOI] [PubMed] [Google Scholar]
- Swartz SZ, Reich AM, Oulhen N, Raz T, Milos PM, Campanale JP, et al. (2014). Deadenylase depletion protects inherited mRNAs in primordial germ cells. Development, 141, 3134–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet HC, Hodor PG, & Ettensohn CA (1999). The role of micromere signaling in notch activation and mesoderm specification during sea urchin embryogenesis. Development, 126, 5255–5265. [DOI] [PubMed] [Google Scholar]
- Takayanagi H, Hayase J, Kamakura S, Miyano K, Chishiki K, Yuzawa S, et al. (2019). Intramolecular interaction in LGN, an adaptor protein that regulates mitotic spindle orientation. The Journal of Biological Chemistry, 294, 19655–19666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y (1976). Effects of the surfactants on the cleavage and further development of the sea urchin embryos. 1. The inhibition of micromere formation at the fourth cleavage. Development Growth & Differentiation, 18, 113–122. [DOI] [PubMed] [Google Scholar]
- Tanaka S, & Dan K (1990). Study of the lineage and cell cycle of small micromeres in embryos of the sea urchin, Hemicentrotus pulcherrimus. Development Growth & Differentiation, 32, 145–156. [DOI] [PubMed] [Google Scholar]
- Tomancak P, Guichet A, Zavorszky P, & Ephrussi A (1998). Oocyte polarity depends on regulation of gurken by vasa. Development, 125, 1723–1732. [DOI] [PubMed] [Google Scholar]
- Vonica A, Weng W, Gumbiner BM, & Venuti JM (2000). TCF is the nuclear effector of the β-catenin signal that patterns the sea urchin animal–vegetal axis. Developmental Biology, 217, 230–243. [DOI] [PubMed] [Google Scholar]
- Voronina E, Lopez M, Juliano CE, Gustafson E, Song JL, Extavour C, et al. (2008). Vasa protein expression is restricted to the small micromeres of the sea urchin, but is inducible in other lineages early in development. Developmental Biology, 314, 276–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina E, & Wessel GM (2004). Regulatory contribution of heterotrimeric G-proteins to oocyte maturation in the sea urchin. Mechanisms of Development, 121, 247–259. [DOI] [PubMed] [Google Scholar]
- Voronina E, & Wessel GM (2006). Activator of G-protein signaling in asymmetric cell divisions of the sea urchin embryo. Development Growth & Differentiation, 48, 549–557. [DOI] [PubMed] [Google Scholar]
- Waldron A, & Yajima M (2020). Localized translation on the mitotic apparatus: A history and perspective. Developmental Biology, 468, 55–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wavreil FDM, & Yajima M (2020). Diversity of activator of G-protein signaling (AGS)-family proteins and their impact on asymmetric cell division across taxa. Developmental Biology, 465, 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzel HE, Illies MR, Byrum CA, Xu R, Wikramanayake AH, & Ettensohn CA (2004). Differential stability of β-catenin along the animal-vegetal axis of the sea urchin embryo mediated by dishevelled. Development, 131, 2947–2956. [DOI] [PubMed] [Google Scholar]
- Wikramanayake AH, Peterson R, Chen J, Huang L, Bince JM, McClay DR, et al. (2004). Nuclear β-catenin-dependent wnt8 signaling in vegetal cells of the early sea urchin embryo regulates gastrulation and differentiation of endoderm and mesodermal cell lineages. Genesis, 39, 194–205. [DOI] [PubMed] [Google Scholar]
- Williams SE, Ratliff LA, Postiglione MP, Knoblich JA, & Fuchs E (2014). Par3–mInsc and Gαi3 cooperate to promote oriented epidermal cell divisions through LGN. Nature Cell Biology, 16, 758–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A, Ramrath A, Kuchinke U, & Knust E (1999). Bazooka provides an apical cue for Inscuteable localization in drosophila neuroblasts. Nature, 402, 544–547. [DOI] [PubMed] [Google Scholar]
- Wray GA, & McClay DR (1988). The origin of spicule-forming cells in a ‘primitive’ sea urchin (Eucidaris tribuloides) which appears to lack primary mesenchyme cells. Development, 103, 305–315. [DOI] [PubMed] [Google Scholar]
- Xiol J, Spinelli P, Laussmann MA, Homolka D, Yang Z, Cora E, et al. (2014). RNA clamping by vasa assembles a piRNA amplifier complex on transposon transcripts. Cell, 157, 1698–1711. [DOI] [PubMed] [Google Scholar]
- Yajima M, & Wessel GM (2011a). Small micromeres contribute to the germline in the sea urchin. Development, 138, 237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M, & Wessel GM (2011b). The DEAD-box RNA helicase vasa functions in embryonic mitotic progression in the sea urchin. Development, 138, 2217–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M, & Wessel GM (2012). Autonomy in specification of primordial germ cells and their passive translocation in the sea urchin. Development, 139, 3786–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M, & Wessel GM (2015). Essential elements for translation: The germline factor vasa functions broadly in somatic cells. Development, 142, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada KH, Hanada T, & Chishti AH (2007). The effector domain of human Dlg tumor suppressor acts as a switch that relieves autoinhibition of kinesin-3 motor GAKIN/KIF13B. Biochemistry, 46, 10039–10045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki A, Morino Y, Urata M, Yamaguchi M, Minokawa T, Furukawa R, et al. (2020). pmar1/phb homeobox genes and the evolution of the double-negative gate for endomesoderm specification in echinoderms. Development, 147, dev182139. [DOI] [PubMed] [Google Scholar]
- Yazaki I (2001). Ca2+ in specification of vegetal cell fate in early sea urchin embryos. The Journal of Experimental Biology, 204, 823–834. [DOI] [PubMed] [Google Scholar]
- Yazaki I, Abe M, Santella L, & Koyama Y (2004). Mechanisms of calcium elevation in the micromeres of sea urchin embryos. Biology of the Cell, 96, 153–167. [DOI] [PubMed] [Google Scholar]
- Yazaki I, Tosti E, & Dale B (1995). Cytoskeletal elements link calcium channel activity and the cell cycle in early sea urchin embryos. Development, 121, 1827–1831. [Google Scholar]
- Yazaki I, Tsurugaya T, Santella L, Chun JT, Amore G, Kusunoki S, et al. (2014). Ca2+ influx-linked protein kinase C activity regulates the β-catenin localization, micromere induction signalling and the oral–aboral axis formation in early sea urchin embryos. Zygote, 23, 426–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Morin X, Cai Y, Yang X, & Chia W (2000). Analysis of partner of inscuteable, a novel player of drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell, 100, 399–409. [DOI] [PubMed] [Google Scholar]
- Zhu J, Wen W, Zheng Z, Shang Y, Wei Z, Xiao Z, et al. (2011). LGN/mInsc and LGN/NuMA complex structures suggest distinct functions in asymmetric cell division for the Par3/mInsc/LGN and Gai/LGN/NuMA pathways. Molecular Cell, 43, 418–431. [DOI] [PMC free article] [PubMed] [Google Scholar]


