Abstract
Methicillin-resistant strains susceptible to gentamicin (Gms MRSA) have emerged since 1993 in several French hospitals. To study whether particular clones have spread in various French cities and whether some clones are related to gentamicin-resistant (Gmr) MRSA strains, various methods (antibiotyping, phage typing, determination of SmaI macrorestriction patterns before and after hybridization with IS256 transposase and aacA-aphD probes) were used to compare 62 Gms MRSA strains isolated from 1995 to 1997 in nine cities and 15 Gmr MRSA strains. Eighteen major SmaI genotypes were identified, of which 11 included only Gms MRSA strains and 5 included only Gmr MRSA strains. Each of the Gmr MRSA strains contained 6 to 13 SmaI fragments hybridizing with the insertion sequence IS256, of which a single band also hybridized with the aacA-aphD gene. No such hybridizing sequences were detected in 60 of the 62 Gms MRSA strains. Thus, the divergence between Gmr and Gms MRSA strains is revealed, not only by their distributions in distinct SmaI genotypes but also by the differences in hybridization patterns. Two of the 62 Gms MRSA strains had the uncommon feature of carrying several SmaI bands hybridizing with IS256, suggesting that they are possibly related to the Gmr MRSA strains grouped in the same SmaI genotype. Five of the 11 SmaI genotypes including only Gms MRSA strains contained strains from diverse cities, isolated during different years and with different antibiograms, suggesting that some clones have spread beyond their cities of origin and persisted.
Until 1992, at least 99% of the French methicillin-resistant Staphylococcus aureus (MRSA) strains sent to the Reference Center were resistant to gentamicin, kanamycin, and related aminoglycosides. All the strains tested carried at least one chromosomal copy of a Tn4001-like structure delimited by two inverted copies of the insertion sequence IS256 and including the aacA-aphD gene (10, 11, 17). Most of these strains also harbored multiple autonomous copies of IS256 on several SmaI fragments of the cellular DNA (9, 17, 18).
In 1993, gentamicin-susceptible (Gms) MRSA strains emerged in several French hospitals (1, 15, 16, 19). In these hospitals, their incidence has increased from 2 to 5 to 20 to 60%. In one of these hospitals (1), the 97 Gms MRSA strains isolated from 1992 to 1996 were distributed into three unrelated pulsotypes. In another hospital (15), 16 of the 26 Gms MRSA strains isolated from 1993 to 1996 clustered in the same pulsotype whereas the 10 other strains were distributed in different and distant pulsotypes. In each of these hospitals, the Gms strains were unrelated to the epidemic Gmr MRSA clones. We typed the Gms MRSA strains suspected to be responsible for very recent (1995) outbreaks in several other French hospitals (19); the spread of the same clone in each hospital was demonstrated.
In this study, we used several typing methods to analyze 62 Gms MRSA strains isolated from 1995 to 1997 in nine hospitals in nine French cities and 15 Gmr MRSA strains which were representative of the Gmr MRSA strains that spread in three of these hospitals in 1996 and in 1997. The aim of this study was to find out (i) whether some Gms clones are closely related to Gmr MRSA, (ii) whether particular Gms MRSA clones have spread and persisted in various French cities, as is found for Gmr MRSA strains (18), and (iii) if the strains grouped in the same pulsotype are easily traceable by antibiotyping and phage typing.
MATERIALS AND METHODS
Bacterial strains and plasmids.
A total of 77 MRSA strains isolated from 77 infected patients were studied (Table 1). They included 62 Gms strains isolated between 1995 and 1997 in nine hospitals in different French cities and 15 Gmr strains isolated in three of the hospitals in 1996 and in 1997.
TABLE 1.
Relevant characteristics of the 77 MRSA strains typed by several methods
| Genotypesa
|
Characteristics of MRSA strains
|
|||||
|---|---|---|---|---|---|---|
| SmaI | SmaI/IS256 | Source | Yr of isolation | No. of strains | Phages used at RTD and giving at least 5 PFU | Drug resistance markersb |
| Ia | A | Clichy | 1995 | 3 | 616 | Pc Ox Nm Sp MLSc Pf (Sm Rf Fm) |
| Ib | B | Paris | 1997 | 1 | NTc | Gm Pc Ox Nm Sp MLSc Pf Fm |
| II | C | Blois | 1996 | 1 | 53, 77, 85 | Pc Ox Nm Pf Fm |
| Montreuil | 1995 | 1 | 84, 616 | Pc Ox Nm Sm Sp MLSi Pf | ||
| Toulouse | 1997 | 1 | 53, 83A, 620, 622, 629, 630 | Pc Ox Sm Sp Pf | ||
| III* | D# | Toulouse | 1997 | 1 | 84, 616 | Pc Ox Nm Sp MLSc Pf Fm |
| IV* | E# | Blois | 1996 | 1 | 616 | Pc Ox Nm Sm Sp MLSc Pf Fm Rf |
| Toulouse | 1996 | 1 | 84, 616 | Pc Ox Nm Sm Sp MLSc Pf Fm | ||
| V | F | Paris | 1997 | 1 | NT | Gm Pc Ox Nm Sp MLSc Pf Fm |
| VIa† | G1 | Paris | 1997 | 1 | 85 | Pc Ox Nm Sm Pf |
| VIb† | G2 | Evry | 1995 | 2 | 84, 616, 622 | Pc Ox Nm Sm Sp Pf |
| Colombes | 1995 | 7 | 77, 84, 85, 616, 622 | Pc Ox Nm Sm Sp Pf | ||
| Clichy | 1995 | 1 | NT | Pc Ox Nm Sm Sp MLSc Pf Rf | ||
| Toulouse | 1997 | 4 | NT | Pc Ox Nm Pf (Sm) | ||
| Toulouse | 1997 | 1 | 77 | Pc Ox Nm Sm Pf Rf Fa | ||
| SaintGermain | 1995 | 1 | NT | Pc Ox Nm Sm Sp Lc SgA Pf | ||
| VIc† | G3 | Toulouse | 1997 | 1 | 84, 85, 616, 622 | Pc Ox Nm Sm Sp Pf |
| VId† | G4 | Blois | 1996 | 1 | 77, 616, 622 | Pc Ox Nm Sp MLSc Pf Fm |
| Toulouse | 1997 | 1 | 77, 84, 85, 616, 622, 625 | Pc Ox Nm Spe MLSc Pf | ||
| VIIa† | H | Paris | 1997 | 1 | 616 | Pc Ox Nm Sp MLSc SgA Pf Fm Tp |
| VIIa† | I | Paris | 1997 | 1 | 84, 616 | Pc Ox Nm Sp MLSc Pf Fm |
| VIIb† | J | Paris | 1977 | 1 | 84, 616 | Gm Pc Ox Nm Sp MLSc Pf Fm |
| VIIc† | K1 | Toulouse | 1997 | 1 | 84, 616 | Pc Ox Nm Sm Sp MLSc Pf Fm |
| VIId† | K2 | Nîmes | 1995 | 9 | 616 | Pc Ox Nm Sm Sp MLSc Pf Fm |
| 1 | 84, 616 | Pc Ox Nm Sm Sp MLSc Pf Fm | ||||
| 3 | NT | Pc Ox Nm Sm Sp MLSc Pf Fm | ||||
| Toulouse | 1997 | 4 | 84, 616 | Pc Ox Nm Sm Sp MLSc (Pf SgA) | ||
| 1 | 84, 616, 622 | Pc Ox Nm Sm Sp MLSc Pf | ||||
| 1 | 616 | Pc Ox Nm Sm Sp MLSc Pf | ||||
| 1 | 616, 625 | Pc Ox Nm Sm Sp MLSc Pf | ||||
| Clichy | 1995 | 1 | 84, 616 | Pc Ox Nm Sm Sp MLSc Pf | ||
| Blois | 1996 | 1 | 616 | Pc Ox Nm Sp MLSc Pf Fm | ||
| VIIe† | L‡ | Toulouse | 1997 | 1 | 54, 630 | Gm Pc Ox Nm Sm Sp MLSc Pf Rf Tc Mn |
| VIIf† | M‡ | Paris | 1997 | 1 | 77 | Gm Pc Ox Nm Sm Sp MLSc Pf Rf |
| VIII | N | Blois | 1996 | 2 | 47, 54, 75, 77, 83A, 85, 616, 618, 620, 622, 623, 629 | Gm Pc Ox Nm Sm Sp MLSc Pf Rf |
| Blois | 1996 | 1 | NT | Gm Pc Ox Nm Sm Sp MLSc Pf Rf | ||
| IX§ | O∥ | Paris | 1997 | 1 | 84 | Pc Ox Nm MLSi Pf |
| X§ | P∥ | Paris | 1997 | 1 | NT | Pc Ox Nm MLSi Pf |
| XI | Q | Toulouse | 1997 | 1 | 84, 616, 622, 630 | Pc Ox Nm Sm Sp MLSc Pf |
| XII | R | Blois | 1996 | 1 | 3A, 3C, 6, 42E, 47, 54, 75, 77, 84, 94, 96, 81, 616, 618, 620, 622, 623, 629 | Pc Ox Lc SgA Pf |
| 1 | 29, 6, 42E, 47, 53, 54, 75, 77, 83A, 84, 85, 81, 616, 620, 622, 623, 625, 626, 629, 630 | Pc Ox Lc SgA | ||||
| XIII | S | Toulouse | 1997 | 1 | 77, 620, 630 | Gm Pc Ox Nm Sm Sp MLSc Pf Rf Tc Mn Fm |
| XIV | T | Blois | 1996 | 1 | 54, 77, 85, 630, 56B | Gm Pc Ox Nm Sm Sp MLSc Pf Rf Tc Mn Fm |
| XV | U | Blois | 1996 | 1 | NT | Pc Ox Sm Sp Pf |
| XVI | V | Toulouse | 1997 | 4 | 29, 47, 54, 75, 77, 83A, 84, 85, 616, 620, 622, 623, 626, 629, 630 | Gm Pc Ox Nm Sm Sp MLSi Pf Rf Tc Mn Fm Fa |
| 1 | 54 | Gm Pc Ox Nm Sm Sp MLSi Pf Rf Tc Mn Fm Fa | ||||
| XVII | W | Paris | 1997 | 1 | 616 | Pc Ox Nm Sp MLSc Pf Fm |
| XVIII | X | Paris | 1997 | 1 | NT | Pc Ox Nm MLSc Tc Mn |
Genotypes which include strains which are possibly related (less than seven band differences) are marked with same symbols (*, †, ‡, §, ∥, and #).
All the strains resistant to neomycin or to gentamicin were also resistant to tobramycin and kanamycin. Antibiotic resistance markers in boldface are common to all the strains; those in parentheses vary between strains. Abbreviations: Fa, fusidic acid; Fm, fosfomycin; Gm, gentamicin; Km, kanamycin; Lc, lincosamides; MLSc, constitutive resistance to macrolides-lincosamides-streptogramin B; MLSi, inducible resistance to macrolides-lincosamides-streptogramin B; Mn, minocycline; Nm, neomycin; Ox, oxacillin; Pc, penicillinase production; Pf, pefloxacin; Rf, rifampin; SgA, streptogramin A; Sm, streptomycin; Sp, spectinomycin; Tc, tetracycline; Tp, trimethoprim.
NT, not typeable.
pSF815A (12) and pIP1551 (9) were used as probes. pSF815A is pUC8 plus a 1.5-kb AluI insert carrying aacA-aphD, which encodes the bifunctional enzyme, AAC6′-APH2", that inactivates gentamicin, kanamycin, and related antibiotics. pIP1551 consists of pUC18 carrying a 468-bp insert from the transposase gene of insertion sequence IS256 (4).
Susceptibilities to antibiotics.
Susceptibilities to antibiotics were determined by a disk diffusion assay (5) with commercially available antibiotic disks (Diagnostics Pasteur). Additional disks prepared in our laboratory contained pristinamycin IIA (20 μg) or pristinamycin IB (40 μg).
Phage typing.
The strains were phage typed by the standard method of Blair and Williams (3) with the international set of 23 phages and an additional set of 9 phages: 616, 618, 620, 622, 623, 625, 626, 629, 630 (21). These phages were used at routine test dilution (RTD).
Pulsed-field gel electrophoresis of macrorestriction fragments and comparative analysis of banding patterns.
The protocol used for the determination of SmaI restriction patterns was described previously (7). Concatemeric bacteriophage lambda DNA molecules (48.5 kb; Bio-Rad) and SmaI fragments of the cellular DNA from S. aureus NCTC8325 were used as size standards. Macrorestriction fingerprints were compared visually and were scanned with GelCompar software (Applied Maths, Kortrijk, Belgium). A similarity matrix was created by using the band-based Dice similarity coefficient (8). The unweighted pair group method with average linkages was used to cluster the strains on the basis of the SmaI patterns.
If the dendrograms revealed clusters that included strains with percentages of similarity of at least 80, the patterns of the strains in the same cluster were compared visually on the same gel. The strains were clustered according to the following criteria proposed by Tenover et al. (24): (i) strains were grouped in the same major genotype if their patterns differed by no more than three bands (such strains were considered to be closely related and monoclonal); (ii) if the number of band differences between patterns was between four and six, the strains were scored as possibly related but were, nevertheless, classified into distinct genotypes to discriminate them clearly from the unambiguously closely related strains; and (iii) differences between patterns involving at least seven bands indicated different or unrelated strains. Major genotypes are designated by roman numerals. Strains with indistinguishable patterns were classified within the same subtype. Subtypes are designated by roman numerals with letter suffixes.
Within each major SmaI genotype, the SmaI patterns were also compared by analysis of the differences revealed by hybridization with an IS256 probe (18). SmaI bands of the same apparent mobilities but differing by the presence or absence of hybridizing nucleotide sequences were considered to show an additional difference. The criteria used to cluster the strains into major genotypes and subtypes were the same as those used after analysis of SmaI patterns. The strains having the same SmaI pattern after hybridization with the IS256 probe belong to the same subtype, and the patterns of the strains clustered in the same major genotype differ by three or fewer bands (bands of different sizes or distinguishable after hybridization with IS256). The genotypes characterized by IS256 probing are designated by capital letters, and the subtypes are designated by capital letters with arabic numbers.
Blotting and hybridization.
DNA was transferred from agarose gels to Hybond N+ membranes (Amersham) with a Trans-Vac TE80 vacuum blotter (Hoefer Scientific Instruments, San Francisco, Calif.). Prehybridization and hybridization were performed under high-stringency conditions, as previously described (6). The plasmid used as a probe was labeled with [α-32P]dCTP (110 TBq · mmol−1) by random priming with the Megaprime DNA-labeling system (Amersham). The blots were exposed to Hyperfilm (Amersham) at −80°C.
Preparation of oligonucleotides and PCR.
The following oligonucleotides were prepared by the phosphoramidite method with an Applied Biosystems model 380 B DNA synthesizer: oligonucleotide A, 5′-GGGATCATAGCGTCATTATTC-3′; oligonucleotide B, 5′-AACGATTGTGACACGATAGCC-3′. Oligonucleotides A and B were used as primers in PCR to amplify a 529-bp sequence (20) from within the mecA gene (23). The forward primer (A) corresponds to nucleotide positions 516 to 536, and the reverse primer (B) corresponds to the complement of positions 1024 to 1044. The PCR was performed with a precycle of 5 min at 94°C and 5 min at 60°C; this was followed by 30 cycles of 1 min at 72°C, 1 min at 94°C, and 1 min at 60°C.
RESULTS
Analysis of antibiotypes.
The 62 Gms MRSA strains studied were distributed into 19 antibiotypes (Table 1). Five antibiotypes were distinguished among the 15 Gmr MRSA strains. The Gms strains exhibited heterogeneous and low-level resistance to oxacillin. This resistance was more easily detectable at 30°C than at 37°C and after a prolongation of the incubation time from 24 to 48 h. Fourteen of the 15 Gmr strains exhibited high-level resistance to oxacillin, i.e., they grew at the contact of the disks containing 5 μg of oxacillin after 24 h of incubation at 37°C. All the strains exhibiting a low-level resistance to oxacillin were screened for the presence of the mecA gene by PCR, and an amplicon of the expected size (529 bp) was detected in all cases (results not shown).
There were several examples of strains from hospitals in different cities having the same antibiotype (Table 1).
Analysis of phage types.
Sixty-two of the 77 MRSA strains could be typed with the phages used at RTD (Table 1). Despite the use of 9 phages in addition to the international collection of 23 phages routinely used to type S. aureus strains, 42 of the 62 typeable strains were susceptible to no more than three phages (RTD). Strains isolated in a single hospital in 1995 were all grouped in the same phage type. In 1996 and 1997, strains of diverse phage types were isolated in single hospitals. Note that some single phage types contained strains isolated in different cities between 1995 and 1997 and having different antibiograms.
Analysis of the SmaI patterns.
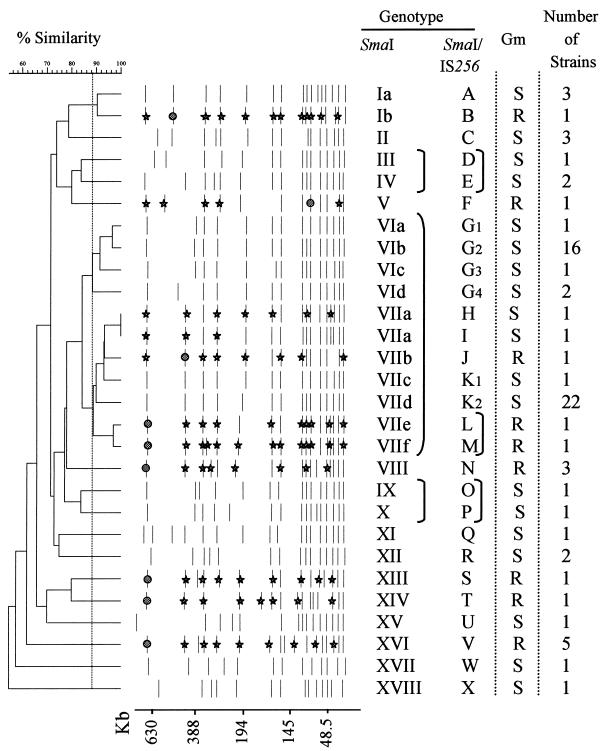
There were 19 different SmaI patterns among the 62 Gms strains and 9 patterns among the 15 Gmr MRSA strains. The patterns of eight of these strains are shown in Fig. 1, and the schematic representation of the 28 different patterns is reported in Fig. 2. A dendrogram of all SmaI patterns was constructed on the basis of the levels of similarity (Fig. 2). For patterns with no more than three fragment differences, the percentages of similarity were 88 to 100. An 88% similarity cutoff value gave 18 major SmaI genotypes of which 2 (genotypes I and VII [Fig. 2]) included both Gms and Gmr MRSA strains. An 82% similarity cutoff value enabled the delineation of 15 SmaI genotypes, each including putatively related strains whose SmaI patterns differed by six or fewer bands. The 82% cutoff value was used to group the strains belonging to the following SmaI genotypes: III and IV, VI and VII, and IX and X (Fig. 2). Forty-five of the 62 Gms strains, including those of diverse origins and antibiograms, were grouped in SmaI genotypes VI and VII.
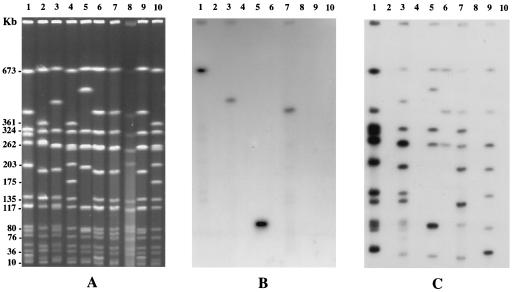
FIG. 1.
Pulsed-field gel electrophoresis of SmaI-digested total DNA from eight MRSA strains. (A) SmaI macrorestriction patterns; (B) hybridization patterns with the aacA-aphD-probe; (C) hybridization patterns with the IS256 probe. Lanes 1 to 3 and 5 to 9, SmaI-digested DNA from MRSA strains belonging to SmaI genotypes (Table 1) VIIf, VIa, Ib, V, VIIa, VIIb, XVII, and VIIg, respectively; lanes 4 and 10, SmaI-digested cellular DNA from strain NCTC8325 used as a standard.
FIG. 2.
Classification and schematic representation of the SmaI patterns of the 77 MRSA strains studied. The SmaI and SmaI/IS256 genotypes assigned to the strains are reported, and the brackets linking some of them indicate that the strains, although in different genotypes, are possibly related (less than seven band differences between their patterns). The bands marked with a circle are those hybridizing with aacA-aphD and IS256 probes, and the bands marked with a star are those hybridizing with the IS256 probe only.
Analysis of SmaI patterns after hybridization with aacA-aphD and IS256 probes.
All the Gmr MRSA strains hybridized with the aacA-aphD probe, whereas none of the Gms MRSA strains gave any detectable signal (Fig. 2). In the 15 Gmr strains tested, the hybridizing nucleotide sequences were on a single SmaI fragment of ≈100 kb (1 strain) or >400 kb (14 strains).
Six to 13 SmaI bands hybridizing with IS256 were present for each of the 15 Gmr strains; these bands included the single band hybridizing with aacA-aphD (Fig. 2). Only 2 of the 62 Gms MRSA strains hybridized with IS256, and the hybridizing sequences were on three or seven SmaI bands (Fig. 2).
A comparative analysis of the 77 MRSA strains tested enabled the detection of 24 SmaI/IS256 genotypes (Fig. 2). None of these genotypes included both Gmr and Gms MRSA strains. The strains belonging to genotypes D and E, L and M, or O and P were possibly related because their patterns differed by six or fewer bands.
Clustering of Gms MRSA strains on the basis of various typing methods.
The seven strains isolated in 1995 in the hospital of Colombes were grouped in the same type, whatever the typing method used (phage typing, antibiotyping, comparison of SmaI patterns) (Table 1). The same was also observed for the 2 strains isolated in Evry in 1995 and for 10 of the 13 strains isolated in 1995 in Nîmes hospital. The three other strains isolated in Nîmes hospital had the same antibiotype and the same SmaI genotype, but they were not phage typeable. In the three hospitals, the Gms strains analyzed were those collected during the 6 months following their first emergence.
In contrast, for the strains isolated in the Toulouse (19 strains in 1997), Blois (7 strains in 1996), Paris (7 strains in 1997), and Clichy (4 strains in 1995) hospitals, there was no correlation between the clusterings obtained by using the different typing methods. In each of these four hospitals, in which the first Gms strains were isolated in 1993 to 1994, the strains were distributed in two to seven unrelated SmaI genotypes (seven or more band differences between the SmaI patterns).
DISCUSSION
None of the Gms strains tested contained nucleotide sequences hybridizing with the aacA-aphD gene. Thus, it is unlikely that these strains result from a modification in this gene abolishing its expression. Consequently, gentamicin and netilmicin or any related aminoglycosides may be used for therapy. The divergence between Gms and Gmr strains was revealed not only by their distribution into distant SmaI genotypes, but also by their differences in hybridization with the IS256 probe. As shown in this study and other papers (9, 17, 26), the presence of multiple autonomous copies of IS256 is common in Gmr staphylococci but is rare in Gms staphylococci. The detection, in 2 of the 62 Gms MRSA strains studied, of three and seven SmaI bands hybridizing with the IS256 probe suggested that these strains are possibly related to the Gmr MRSA strains grouped in the same SmaI genotype (VII).
Gms MRSA strains considered closely or possibly related on the basis of the comparison of SmaI patterns were isolated in different French cities and at time intervals of up to 2 years (1995 to 1997). This suggests a capacity to disseminate beyond a single hospital or city and to persist. This phenomenon was most clearly observed among the strains in the SmaI genotypes VI and VII, which include possibly related strains (less than seven band differences). It would be worth looking for strains belonging to these two genotypes in other European countries to assess whether Gms MRSA strains have disseminated similarly to Gmr MRSA strains (18). Unfortunately, phage typing and antibiotyping are not appropriate to trace such strains, not only because of the diversity observed among strains in the same genotype but also because some strains having the same phage type and/or antibiotype belong to distant genotypes. However, at the beginning of an outbreak in a hospital, these two phenotypic markers may be useful to screen closely related strains. Indeed, for such strains, our study revealed a good correlation between the results obtained by different typing methods.
This study and those previously published (1, 15) show that a multiplicity of unrelated Gms strains are isolated in hospitals. Diversity within a single hospital is mostly observed when the strains are collected several years after the emergence of the first Gms strains. In these cases, the diversity may be a result of rearrangements in persistent clones, which become endemic. However, we also detected unrelated clones among strains which have recently emerged in different hospitals or in the same hospital. Therefore, the increased incidence of Gms MRSA strains in several hospitals is not only attributable to the spread of derivatives of the same clone but also to the emergence of unrelated clones. The emergence of such clones in France coincided, in several hospitals, with a change in antibiotic prescribing patterns, including a decrease in gentamicin consumption, and with infection control programs (13, 15). The result has been a decrease in the proportion of Gmr MRSA strains (15) and in particular of the endemic phage type 77 Gmr MRSA strains (18).
The levels of resistance to β-lactams due to expression of the mecA gene carried by all the French Gms MRSA strains isolated since 1992 (this study) and before the emergence of the first Gmr MRSA strain in 1975 (14) are very low and are expressed heterogeneously. Several genes other than mecA, including those of the mecA regulatory region (mecI, mecR-I) and promoter, are involved in the expression of mecA (2, 27). It would be valuable to compare the SmaI restriction patterns and the regulatory regions of mecA of the early and recent French Gms MRSA strains to determine whether they are related. If they are, it would mean that some of the old Gms MRSA clones have persisted with a low prevalence in the hospitals and have emerged with an increased frequency as the selective pressure exerted by the use of gentamicin has decreased. Data from each hospital about the consumption of antibiotics and the incidence of each antibiotic resistance marker among staphylococci would help in the evaluation of the contribution of the selective pressure exerted by antibiotics on the spread of clones. Note that other factors such as those involved in virulence and colonization (13, 22, 25) and/or resistance to environmental stress may also contribute to the spread and the persistence of some clones.
ACKNOWLEDGMENTS
We thank the microbiologists who sent us the strains and C. Tran for secretarial assistance.
REFERENCES
- 1.Aubry-Damon H, Legrand P, Brun-Buisson C, Astier A, Soussy C-J, Leclercq R. Reemergence of gentamicin-susceptible strains of methicillin-resistant Staphylococcus aureus: roles of an infection control program and changes in aminoglycoside use. Clin Infect Dis. 1997;25:647–653. doi: 10.1086/513749. [DOI] [PubMed] [Google Scholar]
- 2.Berger-Bächi B. Resistance not mediated by β-lactamase (methicillin-resistance) In: Crossley K B, Archer G L, editors. The Staphylococci in human disease. New York, N.Y: Churchill Livingstone; 1997. pp. 158–174. [Google Scholar]
- 3.Blair J E, Williams R E O. Phage typing of staphylococci. Bull W H O. 1961;24:771–784. [PMC free article] [PubMed] [Google Scholar]
- 4.Byrne M E, Rouch D A, Skurray R A. Nucleotide sequence analysis of IS256 from the Staphylococcus aureus gentamicin-tobramycin-kanamycin-resistance transposon Tn4001. Gene. 1989;81:361–367. doi: 10.1016/0378-1119(89)90197-2. [DOI] [PubMed] [Google Scholar]
- 5.Chabbert Y A. Sensibilité bactérienne aux antibiotiques. In: Le Minor L, Veron M, editors. Bactériologie Médicale. Paris, France: Flammarion, Médecine Science; 1982. pp. 204–212. [Google Scholar]
- 6.Chesneau O, Allignet J, El Solh N. Thermonuclease gene as a target nucleotide sequence for specific recognition of Staphylococcus aureus. Mol Cell Probes. 1993;7:301–310. doi: 10.1006/mcpr.1993.1044. [DOI] [PubMed] [Google Scholar]
- 7.Derbise A, Dyke K G H, El Solh N. Characterization of a Staphylococcus aureus transposon Tn5405, located within Tn5404 and carrying the aminoglycoside resistance genes, aphA-3 and aadE. Plasmid. 1996;35:174–188. doi: 10.1006/plas.1996.0020. [DOI] [PubMed] [Google Scholar]
- 8.Dice L R. Measures of the amount of ecological association between species. Ecology. 1945;26:297–302. [Google Scholar]
- 9.Dyke K G H, Aubert S, El Solh N. Multiple copies of IS256 in staphylococci. Plasmid. 1992;28:235–246. doi: 10.1016/0147-619x(92)90055-f. [DOI] [PubMed] [Google Scholar]
- 10.El Solh N, Fouace J M, Pillet J, Chabbert Y A. Plasmid DNA content of multiresistant Staphylococcus aureus strains. Ann Inst Pasteur Microbiol. 1981;132B:131–156. [PubMed] [Google Scholar]
- 11.El Solh N, Moreau N, Ehrlich S D. Molecular cloning and analysis of Staphylococcus aureus chromosomal aminoglycoside resistance genes. Plasmid. 1986;15:104–118. doi: 10.1016/0147-619x(86)90047-8. [DOI] [PubMed] [Google Scholar]
- 12.Ferretti J J, Gilmore K S, Courvalin P. Nucleotide sequence analysis of the gene specifying the bifunctional 6′-aminoglycoside-acetyltransferase 2"-aminoglycoside-phosphotransferase enzyme in Streptococcus faecalis and identification and cloning of gene regions specifying the two activities. J Bacteriol. 1986;167:631–638. doi: 10.1128/jb.167.2.631-638.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frénay H M E, Theelen J P G, Schouls L M, Vandenbrouke-Grauls C M J E, Verhoef J, van Leeuwen W J, Mooi F R. Discrimination of epidemic and nonepidemic methicillin-resistant Staphylococcus aureus strains on the basis of protein A gene polymorphism. J Clin Microbiol. 1994;32:846–847. doi: 10.1128/jcm.32.3.846-847.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leclercq R. Methicillin-resistant Staphylococcus aureus in France: evolution and present status. In: Brun-Buisson C, El Solh N, Casewell M W, Régnier B, editors. Methicillin resistant staphylococci. Paris, France: Médecine-Sciences, Flammarion; 1995. pp. 92–99. [Google Scholar]
- 15.Le Maitre N, Sougakoff W, Masmoudi A, Fievet M H, Bismuth R, Jarlier V. Characterization of gentamicin-susceptible strains of methicillin-resistant Staphylococcus aureus involved in nosocomial spread. J Clin Microbiol. 1998;36:81–85. doi: 10.1128/jcm.36.1.81-85.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller G H, Sabatelli F J, Naples L, Hare S R, Shaw K J, Group A R S. The most frequently occurring aminoglycoside resistance mechanisms—combined results of surveys in eight regions of the world. J Chemother. 1995;7:17–30. [PubMed] [Google Scholar]
- 17.Monzon-Moreno C, Aubert S, Morvan A, El Solh N. Usefulness of three probes in typing isolates of methicillin-resistant Staphylococcus aureus (MRSA) J Med Microbiol. 1991;35:80–88. doi: 10.1099/00222615-35-2-80. [DOI] [PubMed] [Google Scholar]
- 18.Morvan A, Aubert S, Godard C, El Solh N. Contribution of a typing method based on IS256 probing of SmaI-digested cellular DNA to discrimination of European phage type 77 methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol. 1997;35:1415–1423. doi: 10.1128/jcm.35.6.1415-1423.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morvan A, El Solh N. Société Française de Microbiologie (ed.), Proceedings of the 8th International Symposium on Staphylococci and Staphylococcal Infections. Aix-les-Bains, France: Société Française de Microbiologie; 1996. Analysis of the gentamicin-susceptible methicillin-resistant Staphylococcus aureus strains which have recently emerged with increased incidence in several French hospitals; p. 278. [Google Scholar]
- 20.Prédari S C, Ligozzi M, Fontana R. Genotypic identification of methicillin-resistant coagulase-negative staphylococci by polymerase chain reaction. Antimicrob Agents Chemother. 1991;35:2568–2573. doi: 10.1128/aac.35.12.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richardson J F, Chittasobhon N, Marples R R. Supplementary phages for the investigation of strains of methicillin-resistant Staphylococcus aureus. J Med Microbiol. 1988;25:67–74. doi: 10.1099/00222615-25-1-67. [DOI] [PubMed] [Google Scholar]
- 22.Roberts J I S, Gaston M A. Protein A and coagulase expression in epidemic and nonepidemic Staphylococcus aureus. J Clin Pathol. 1987;40:837–840. doi: 10.1136/jcp.40.8.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song M D, Wachi M, Doi M, Ishino F, Matsuhashi M. Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett. 1987;221:167–171. doi: 10.1016/0014-5793(87)80373-3. [DOI] [PubMed] [Google Scholar]
- 24.Tenover F C, Arbeit R, Archer G, Biddle J, Byrne S, Goering R, Hancock G, Hébert G A, Hill B, Hollis R, Jarvis W R, Kreiswirth B, Eisner W, Maslow J, McDougal L K, Miller J M, Mulligan M, Pfaller M A. Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J Clin Microbiol. 1994;32:407–415. doi: 10.1128/jcm.32.2.407-415.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Wamel W J B, Fluit A C, Wadström T, van Dijk H, Verhoef J, Vandenbroucke-Grauls C M J E. Phenotypic characterization of epidemic versus sporadic strains of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 1995;33:1769–1774. doi: 10.1128/jcm.33.7.1769-1774.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walcher-Salesse S, Monzon-Moreno C, Aubert S, El Solh N. An epidemiological assessment of coagulase-negative staphylococci from an intensive care unit. J Med Microbiol. 1992;36:321–331. doi: 10.1099/00222615-36-5-321. [DOI] [PubMed] [Google Scholar]
- 27.Weller T M A. The distribution of mecA, mecR1 and mecI and sequence analysis of mecI and the mec promoter region in staphylococci expressing resistance to methicillin. J Antimicrob Chemother. 1999;43:15–22. doi: 10.1093/jac/43.1.15. [DOI] [PubMed] [Google Scholar]