Abstract
A total of 422 Mycobacterium tuberculosis isolates from eight countries were subjected to IS6110 and IS1081 DNA fingerprinting by means of restriction fragment analysis to characterize M. tuberculosis strains from each country. Chinese, Mongolian, Hong Kong, Filipino, and Korean isolates had comparatively more copies of IS6110 (proportion with eight or more copies; 95% ± 5%), while Thai, Malaysian, and Vietnamese isolates had fewer copies (proportion with eight or more copies, 60% ± 4%). We found a number of novel IS1081 types in this study. One IS1081 type was present in 56% of Filipino isolates, had a specific 6.6-kb PvuII fragment in its IS6110 DNA fingerprint, and was termed the “Filipino family.” The IS1081 types of Thai isolates had interposing characteristics between the characteristics of northeastern Asian and southeastern Asian IS1081 types. A 1.3-kb single-copy IS6110 fragment was found only in Vietnamese M. tuberculosis isolates. Although M. tuberculosis isolates from each country had comparatively similar characteristics depending on the classification factor, each country's isolates showed characteristic DNA fingerprints and differed slightly from the isolates from the other countries in either the mode number of IS6110 copies or the distribution of IS1081 types.
Mycobacterium tuberculosis, the causative agent of human tuberculosis (TB), contains an insertion sequence in chromosome IS6110 which belongs to the IS3 family of enterobacterial insertion sequence (IS) elements (37). This insert is present in other species of the M. tuberculosis complex but not in the more distantly related mycobacteria (4). The presence of multiple copies of IS6110 within the chromosome of M. tuberculosis allows the easy detection of M. tuberculosis in tuberculous patients by PCR (12). Also, irregular insertion of IS6110 within the genome permits the discrimination of strains of M. tuberculosis species (4) by a DNA fingerprinting technique, restriction fragment length polymorphism (RFLP) analysis. This technique can be used to display variable polymorphisms after initial restriction enzyme digestion and probe hybridization.
DNA fingerprinting of M. tuberculosis provides a versatile tool for the identification of transmission (25, 28), the investigation of TB outbreaks (11, 17), the distinction between reactivation and reinfection (26), as well as proof of laboratory cross-contamination (1, 2, 6). IS6110 provides the most variable patterns yet discovered when it is used as a probe for RFLP analysis for the subdivision of M. tuberculosis strains (3, 5, 32). We used an IS6110 probe for the initial screening of M. tuberculosis isolates from China, Mongolia, Hong Kong, the Philippines, Korea, Thailand, Malaysia, and Vietnam. Another insertion sequence, IS1081, has five to seven repeats in all strains belonging to the M. tuberculosis complex (8). The putative transposase of IS1081 bears a high degree of resemblance to the products of the equivalent open reading frames of IS1245 and IS256 in Staphylococcus aureus (9, 27). Although IS1081 has recently been disregarded as a probe for DNA fingerprinting of M. tuberculosis because of its conserved copy number and low discriminatory power, it may be a useful subsidiary marker in epidemiological studies of TB (37) and has been used to differentiate wild-type M. bovis isolates (23, 34).
The polymorphic GC-rich repetitive sequence (PGRS), repeated multiple times in the genome of M. tuberculosis complex and in other mycobacteria such as M. kansasii, M. gastri, and M. szulgai, was cloned into plasmid pTBN12 (21). PGRS, which does not contain as many variations as IS6110, is useful as a secondary probe in subdividing clusters of M. tuberculosis isolates with fewer IS6110 copies (5) or in differentiating M. bovis isolates from animals (24). We also used PGRS as a subsidiary marker in subdividing IS6110 clusters in this study.
There have been some reports on the DNA fingerprinting analysis of M. tuberculosis strains isolated from the Western Pacific Region (WPR) (10, 13, 15, 18, 22, 30). The DNA fingerprinting information acquired in this study should allow a better understanding of the epidemiology of M. tuberculosis strains and aid in determining the transmission routes of M. tuberculosis in the WPR.
MATERIALS AND METHODS
Bacterial strains.
Sixty-four M. tuberculosis isolates from Henan Province in China, 25 isolates from Mongolia, 14 isolates from Hong Kong, 34 isolates from the Philippines, 49 isolates from Thailand, 50 isolates from Hanoi, Vietnam, 48 isolates from Malaysia, and 138 isolates from Korea were included in this study. Korean isolates consisted of 131 isolates cultured from the 7th Nationwide Tuberculosis Prevalence Survey in Korea in 1995 (14) and 7 isolates newly found in a follow-up survey. The subsequent survey was performed 18 months later for those patients who received a radiological diagnosis of suspected TB but whose TB was not proven bacteriologically in the 1995 survey. All isolates were supplied by the national tuberculosis reference laboratory of each country and were identified as M. tuberculosis by standard biochemical tests, including the niacin accumulation test, the nitrate reduction test, and the heat-labile catalase test (16).
Bacterial growth and chromosomal DNA isolation.
Isolates were grown in Löwenstein-Jensen medium for 4 weeks. Chromosomal DNA was isolated as described by van Soolingen et al. (33). In short, 1.5 ml of the concentrated culture was heated at 80°C for 30 min to kill the cells. After centrifugation, the cells were resuspended in 500 μl of TE buffer (0.01 M Tris-HCl, 0.001 M EDTA [pH 8.0]). Lysozyme was added to a final concentration of 1 mg/ml, and the tube was incubated for 1 h at 37°C. Seventy microliters of 10% sodium dodecyl sulfate and 6 μl of proteinase K (10 mg/ml) were added, and the mixture was incubated for 10 min at 65°C. An 80-μl volume of N-cetyl-N,N,N,-trimethyl ammonium bromide was added, and the mixture was vortexed briefly and then incubated for 10 min at 65°C. An equal volume of chloroform-isoamyl alcohol (24:1; vol/vol) was added, and the mixture was vortexed for 10 s. After centrifugation for 5 min, 0.6 volume of isopropanol was added to the supernatant to precipitate the DNA. After cooling for 30 min at −20°C and centrifugation for 15 min, the pellet was washed once with 70% ethanol. The air-dried pellet was redissolved in 20 μl of 0.1× TE buffer (0.001 M Tris-HCl, 0.0001 M EDTA [pH 8.0]).
DNA probes.
Three different DNA probes were used in this study. The IS6110 probe was a 245-bp, PCR-amplified probe containing the sequence of the right arm of IS6110 from the M. tuberculosis H37Rv reference strain. Primers for the IS6110 probe were oligonucleotides INS-1 (5′-CGTGAGGGCATCGAGGTGGC-3′) and INS-2 (5′-GCGTAGGCGTCGGTGACAAA-3′) (29). The IS1081-specific DNA probe of 300 bp was amplified by PCR with oligonucleotides 1081-a (5′-TCGCGTGATCCTTCG-3′) and 1081-b (5′-CGCAGCTTGGGGATCGCGAC-3′) (34), which are based on the inverted repeat sequence from positions 333 to 347 and 613 to 632 of the IS1081 sequence, respectively (8). The PGRS probe was a 754-bp fragment amplified with oligonucleotide primers PGRS-F (5′-GAATCTCCGGCTGTGTCATT-3′) (GenBank accession no. MTV049, positions 33476 to 33495) and PGRS-R (5′-CAACTATTGGTTCGGCGATT-3′) (GenBank accession no. MTV049, positions 34210 to 34229), designed from one segment (GenBank accession no. MTV049) of the full genome library of M. tuberculosis H37Rv released through the Internet by the Sanger Center (7).
RFLP analysis.
DNA fingerprinting was performed by a standardized procedure as described by van Embden and colleagues (29, 31, 33), in which chromosomal DNA was restricted with PvuII for RFLP analysis with IS6110 as a probe (IS6110 RFLP analysis), and RFLP analysis with IS1081 as a probe (IS1081 RFLP analysis) and with AluI for RFLP analysis with PGRS as a probe (PGRS RFLP analysis) (32). The digested DNA was separated overnight by pulsed-field gel electrophoresis with a 0.8% agarose gel in a buffer containing 90 mM Tris base, 90 mM boric acid, and 2 mM EDTA (2.5 V/cm, 1.5 forward/backward). The separated DNA was transferred from the gel to a positively charged nylon membrane (Hybond N+; Amersham) by using a vacuum transfer device (Hybaid Corp). After hybridization for repetitive elements with labeled DNA probes, the bound probes were detected with an enhanced chemiluminescence direct nucleic acid detection system (Amersham) according to the manufacturer's recommendations.
Statistical analysis.
Data were analyzed with the software package SAS 6.12 (SAS Institute, Cary, N.C.). Categorical variables were compared by the χ2 test or Fisher's exact test.
RESULTS
IS6110-based DNA fingerprints.
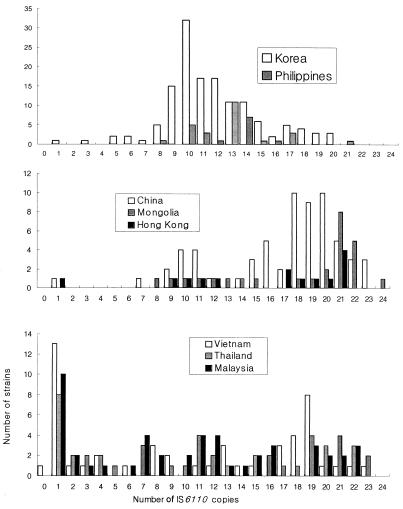
The results of IS6110 RFLP analysis of M. tuberculosis isolates from each country are summarized in Table 1. Chinese, Mongolian, Hong Kong, Filipino, and Korean isolates had comparatively more copies of IS6110 (proportion of isolates with eight or more copies, 95% ± 5%), while Thai, Malaysia, and Vietnamese isolates had fewer copies (proportion of isolates with eight or more copies, 60% ± 4%) (P < 0.005). On the basis of the distribution of the number of IS6110 copies of M. tuberculosis in the RFLP analysis, the eight regions could be categorized into three groups. The first group consisted of China, Mongolia, and Hong Kong, the second group consisted of Korea and the Philippines, and the third group consisted of Thailand, Vietnam, and Malaysia (Fig. 1). On the basis of RFLP analysis, most Chinese, Mongolian, and Hong Kong isolates were found to have about 20 copies of IS6110 (proportion with ≥17 copies, 68% ± 4%). The DNA fingerprints of Chinese isolates showed multiple peaks at 10, 16, and 20 copies of IS6110. Mongolian and Hong Kong isolates had similar distributions in terms of the number of IS6110 copies and the presence of 21 copies with single mode (Fig. 1). In many Chinese and Mongolian isolates, multiple fragments of less than 1.4 kb were found to be aggregated by IS6110 RFLP analysis.
TABLE 1.
Comparison of IS6110 RFLP analysis results for isolates from eight regions
| Country | No. of isolates | No. of clustersa | No. (%) of isolates with classified patternsb | No. (%) of isolates with more than eight copiesc | Mode no. of copiesd |
|---|---|---|---|---|---|
| China (Henan) | 64 | 4 (8) | 60 (93.8) | 62 (96.8) | 20 |
| Mongolia | 25 | 4 (9) | 20 (80.0) | 25 (100) | 21 |
| Hong Kong | 14 | 1 (2) | 13 (92.9) | 13 (92.8) | 21 |
| Korea | 138 | 6 (15) | 129 (93.5) | 131 (94.9) | 10 |
| Philippines | 34 | 1 (2) | 33 (97.1) | 34 (100) | 13 |
| Thailand | 49 | 2 (8) | 43 (87.8) | 31 (63.2) | 1 |
| Malaysia | 48 | 3 (12) | 39 (82.3) | 27 (56.2) | 1 |
| Vietnam (Hanoi) | 50 | 3 (14) | 39 (78.0) | 31 (62.0) | 1 |
Numbers in parentheses indicate the number of isolates within the clusters.
One cluster containing several isolates was regarded as one pattern.
Proportion of isolates from China, Mongolia, Hong Kong, Korea, and the Philippines with eight or more copies, 95% ± 5%; proportion of isolates from Thailand, Malaysia, and Vietnam with eight or more copies, 60% ± 4%.
The eight countries could be divided into three groups by mode number of copies.
FIG. 1.
Comparison of IS6110-based DNA fingerprints of M. tuberculosis strains isolated from the WPR.
The number of IS6110 copies in the 138 Korean M. tuberculosis isolates ranged from 1 to 20, with the majority (74.6%) having from 9 to 14 copies (Table 1). Plotting of the results displayed an almost normal distribution curve, with a mode of 10 copies (23.2%). Although the Filipino IS6110 copy number histogram overlapped the Korean histogram more than it overlapped the others, the mode copy number for Filipino isolates was slightly shifted to higher copy numbers and was bimodal, with the minor mode being 10 copies (14.7%) of IS6110 and the major mode being 13 copies (32.4%) of IS6110. Filipino isolates had peculiar IS6110 RFLP patterns, with many common PvuII DNA fragments, especially those of 4.4, 2.8, 2.3, and 2.0 kb (Fig. 2). Despite their similarity, Filipino isolates could easily be distinguished from one another, with most isolates (97.1%) showing distinct IS6110 RFLP types (Table 1).
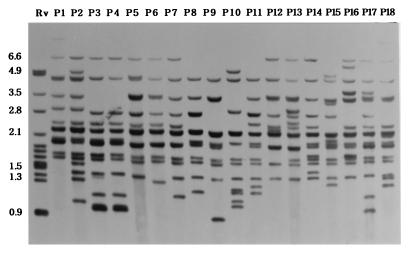
FIG. 2.
IS6110 DNA fingerprints of M. tuberculosis strains isolated from the Philippines. Most strains showed similar patterns because of the common 4.4-, 2.8-, 2.3-, and 2.0-kb fragments. Strains containing a 6.6-kb fragment by IS6110 fingerprinting were classified as type J. Lane 1, M. tuberculosis H37Rv. Numbers on the left are in kilobases.
Comparatively lower proportions (60% ± 4%) of Thai, Vietnamese, and Malaysian M. tuberculosis isolates had greater than eight IS6110 copies compared with the proportions for isolates from northeastern Asian countries, but higher proportions of isolates had a single copy of IS6110 (21% ± 5%) (P < 0.005). Even though isolates from these three countries all had high proportions of single copies of IS6110, their distributions differed: among Vietnamese isolates, 10 had 1.3-kb (20.0%), 2 had 1.5-kb (4.0%), and 1 (2.0%) had 4.5-kb fragments; among Malaysian isolates, 7 had 1.5-kb (14.6%) and 3 had 4.5-kb (6.3%) fragments; and among Thai isolates 6 had 1.5-kb (12.2%) and 2 had 4.5-kb (4.1%) fragments. The 1.3-kb single-copy IS6110 fragment was found only in Vietnamese isolates (Fig. 3). The northeastern Asian (China, Korea, Hong Kong) strains did not have the 1.3- or 1.5-kb IS6110 fragments but, instead, contained a 4.5-kb single-copy fragment. The 1.3- and 4.5-kb single-copy fragments identified by IS6110 RFLP analysis were commonly type A by IS1081 RFLP analysis, while the 1.5-kb single-copy fragment were of variable types.
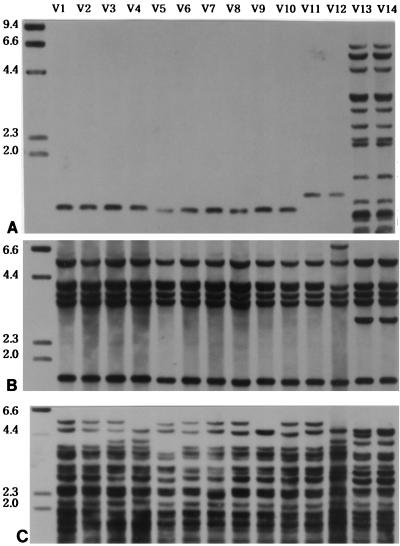
FIG. 3.
Clusters of Vietnamese M. tuberculosis strains (strains V1 to V14) hybridized with IS6110 (A), IS1081 (B), and PGRS (C). Clusters of isolates with the same IS6110 fingerprints were typed as VA (V1 to V10), VB (V11 and V12), or VC (V13 and V14). Four strains of type VA with a 1.3-kb PvuII fragment, two strains of type VB with a 1.5-kb fragment, and two strains of type VC were differentiated with the PGRS probe. Numbers on the left are in kilobases.
Analysis of IS1081 DNA fingerprints.
Six novel IS1081 types were identified: J and K types in Filipino isolates, M and N types in Malaysian isolates, and O and P types in Vietnamese isolates (Fig. 4; Table 2). Filipino isolates had very peculiar IS1081 patterns, with the novel J type, designated the “Filipino family,” as the mode (55.9%). Strains with IS6110 fingerprints of the J type had 6.6-kb PvuII DNA fragments in common (Fig. 2). Filipino strains had a higher proportion (17.6%) of type I fingerprints than strains from other countries but a lower proportion of type C fingerprints (17.6%; Table 2). The two Filipino strains with type K fingerprints had 10 and 13 copies of IS6110, respectively. The distribution pattern of IS1081 types among Mongolian strains was interposed between those among Chinese and Korean isolates. It was similar to that for Korean isolates with respect to the proportions of C and A types but was also similar to that for Chinese isolates with respect to the proportions of H and I types (Table 2). An especially high proportion (21.4%) of Hong Kong isolates had type B IS1081 fingerprints.
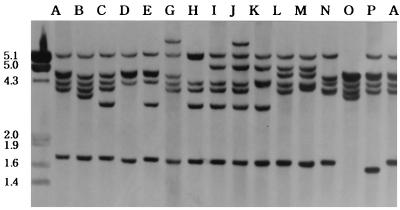
FIG. 4.
Various types of IS1081 DNA fingerprints of M. tuberculosis isolated from the WPR. The type F IS1081 fingerprint was not detected in this study, but novel IS1081 types were found: J and K types in the Philippines, M and N types in Malaysia, and O and P types in Vietnam. Numbers on the left are in kilobases.
TABLE 2.
Distribution of IS1081 types of M. tuberculosis isolates in each regiona
| Country | No. (%) of isolates of the following type:
|
Total | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | ||
| China | 5 (7.8) | 50 (78.1) | 1 (1.6) | 6 (9.4) | 2 (3.2) | 64 (100.0) | |||||||||||
| Mongolia | 6 (24.0) | 13 (52.0) | 1 (4.0) | 4 (16.0) | 1 (4.0) | 25 (100.0) | |||||||||||
| Hong Kong | 1 (7.1) | 3 (21.4) | 9 (64.3) | 1 (7.1) | 14 (100.0) | ||||||||||||
| Korea | 37 (26.8) | 2 (1.4) | 96 (69.6) | 1 (0.7) | 2 (1.4) | 138 (100.0) | |||||||||||
| Philippines | 6 (17.6) | 1 (2.9) | 6 (17.6) | 19 (55.9) | 2 (5.9) | 34 (100.0) | |||||||||||
| Thailand | 16 (32.7) | 1 (2.0) | 24 (49.0) | 1 (2.0) | 5 (10.2) | 2 (4.1) | 49 (100.0) | ||||||||||
| Malaysia | 25 (52.1) | 2 (4.2) | 16 (33.3) | 1 (2.1) | 1 (2.1) | 1 (2.1) | 1 (2.1) | 1 (2.1) | 48 (100.0) | ||||||||
| Vietnam | 24 (48.0) | 1 (2.0) | 19 (38.0) | 2 (4.0) | 1 (2.0) | 1 (2.0) | 1 (2.0) | 1 (2.0) | 50 (100.0) | ||||||||
Type C was found to be dominant in China, Mongolia, Hong Kong, and Korea, while the novel type J was dominant in the Philippines. Thai strains showed characteristics of both the C and the A types. In Malaysia and Vietnam, type A was dominant.
The distribution of IS1081 types among Korean isolates was different from that among the Chinese isolates with respect to the distribution of A and H types (Table 2). The distribution pattern of IS1081 types among Thai isolates revealed traits of the patterns for both Chinese and Vietnamese isolates because it had a relatively high proportion of isolates with type H fingerprints, similar to the proportion among Chinese isolates, but the proportions of isolates with type A and C fingerprints were also similar to those among Vietnamese isolates. Malaysian and Vietnamese isolates had similar IS1081 types, with higher proportions of type A fingerprints than type C fingerprints (Table 2).
Analysis of IS6110 clusters.
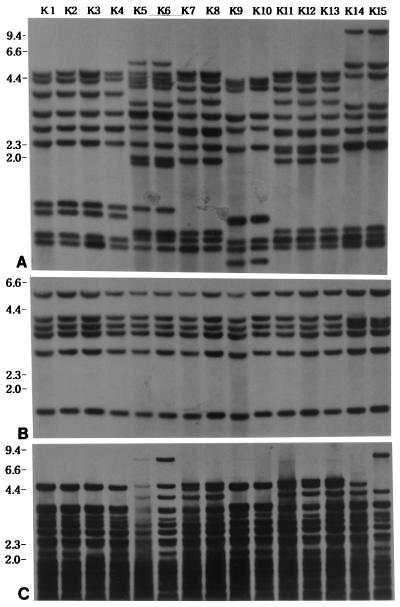
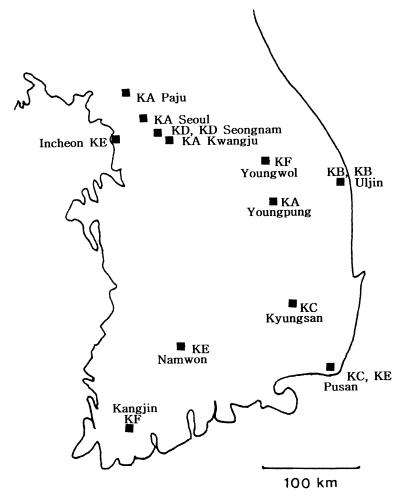
The 138 Korean isolates had 129 distinct IS6110 patterns, including six clusters that consisted of two clusters with 10 copies, two clusters with 12 copies, one cluster with 14 copies, and one cluster with 16 copies. One IS6110 cluster with 12 copies was differentiable with the PGRS probe (Fig. 5). Epidemiologic connections were ascertained by having patients fill out a questionnaire concerning residence history, frequency of hospital visits, and personal acquaintances. We were able to elucidate the epidemiologic connections of two clusters of four Korean patients with pulmonary TB; one consisted of patients infected through household contact (cluster type KD, patients K9 and K10) and the other consisted of patients infected through neighborhood contact (cluster type KB, patients K5 and K6) (Table 3; Fig. 6). The infections in patients K5 and K6 in cluster type KB and patient K9 in cluster type KD were left undiagnosed until the 1995 nationwide survey. In view of their extent of infection, as shown by chest radiography (patient K5, who had a moderately advanced infection; patient K6, who had minimal infection) and sputum examinations (Table 3), it was evident that patient K5 in cluster type KB had infected patient K6. In the case of cluster type KD, patient K9 was the son of patient K10, who had not been a patient in 1995. In the 1996 follow-up survey, however, patient K10 was found to have developed TB. The remaining patients within clusters were found to have no personal contacts with each other or to be neighbors with one another. The two strains from patients in cluster type KC had different drug susceptibilities (Table 3).
FIG. 5.
Clusters of Korean M. tuberculosis strains hybridized with IS6110 (A), IS1081 (B), and PGRS (C). Clusters of patients infected with isolates with the same IS6110 fingerprints were typed as KA (patients K1 to K4), KB (patients K5 and K6), KC (patients K7 and K8), KD (patients K9 and K10), KE (patients K11 to K13), and KF (patients K14 to K15). Two strains from patients in cluster type KF were discriminated with the PGRS probe. Numbers on the left are in kilobases.
TABLE 3.
Comparison of Korean patients whose isolates were clustered by IS6110 fingerprinting
| Patients | Sexa/age (yr) | No. of IS6110 copies | Cluster type | Date of collectionb | Direct smear resultc | Culture resultd | Drug susceptibilitye | Personal acquaintancef |
|---|---|---|---|---|---|---|---|---|
| K1 | M/61 | 10 | KA | 5/15/1995 | − | + | All − | No |
| K2 | F/21 | 10 | KA | 6/22/1995 | + | ++ | All − | No |
| K3 | M/75 | 10 | KA | 9/27/1995 | + | ++ | All − | No |
| K4 | M/27 | 10 | KA | 7/18/1995 | − | + | All − | No |
| K5 | M/56 | 16 | KB | 5/25/1995 | +++ | ++++ | All − | Neighbor |
| K6 | F/84 | 16 | KB | 6/9/1995 | − | + | All − | Neighbor |
| K7 | M/37 | 12 | KC | 6/7/1995 | +++ | +++ | I/P/E/R/T | No |
| K8 | M/59 | 12 | KC | 7/7/1995 | − | + | All − | No |
| K9 | M/25 | 10 | KD | 6/15/1995 | +++ | +++ | All − | Family |
| K10 | M/61 | 10 | KD | 12/11/1996 | − | + | All − | Family |
| K11 | M/71 | 14 | KE | 6/23/1995 | − | +2 | All − | No |
| K12 | M/39 | 14 | KE | 7/13/1995 | − | +1 | All − | No |
| K13 | F/20 | 14 | KE | 10/4/1996 | − | + | All − | No |
| K14 | F/62 | 12 | KF | 5/5/1995 | + | + | All − | No |
| K15 | F/49 | 12 | KF | 6/26/1995 | + | + | All − | No |
M, male; F, female.
Month/day/year.
−, 0 in 300 fields; +, 1 to 10 in 100 fields; ++, 1 to 10 in 10 fields; +++, 1 to 10 in 1 field.
−, no colonies; +1, one colony; +2, two colonies; +, 50 to 100 colonies; ++, 100 to 200 colonies; +++, 200 to 500 colonies; ++++, more than 500 colonies.
All −, the strain was susceptible to all drugs tested. I/P/E/R/T, the strain was resistant to isoniazid, p-aminosalicylic acid, ethambutol, rifampin, and prothionamide.
With the exception of two clusters, patients contained within the same clusters had no personal acquaintance with one another.
FIG. 6.
Geographical distribution of patients in six Korean IS6110 clusters (clusters KA and KF). The patients in cluster type KD contracted TB by household contact while those in KB contracted TB by neighborhood contact.
Clusters of Thai, Vietnamese, and Malaysian isolates with single copies of IS6110 appeared. Most Vietnamese isolates in clusters had a 1.3-kb PvuII DNA fragment, while the Thai and Malaysian isolates in clusters had a 1.5-kb fragment (Table 4; Fig. 3). The majority of isolates in clusters with a single copy of IS6110 could be subdivided by RFLP analysis with PGRS. In particular, all Thai isolates in clusters with a single copy of IS6110 could be subdivided with the PGRS probe (Table 4). Among the 70 isolates in clusters containing a single copy of IS6110, the IS1081 probe could subdivide only seven isolates (10%), while the PGRS probe was able to subdivide 28 isolates (40%). Distinction of clusters of isolates with the same numbers of copies of IS6110 with the PGRS probe was possible for isolates with multiple copies of IS6110 (15%) as well as isolates with single copies of IS6110 (77.3%). We did not search for epidemiologic connections of clusters of isolates with the same number of copies of IS6110 found in countries other than Korea.
TABLE 4.
Analysis of IS6110 clusters
| Type of IS6110 clusters in each countrya | No. of IS6110 copiesb | No. of isolatesc | No. of isolates subclassified with:
|
|
|---|---|---|---|---|
| IS1081 | PGRS | |||
| CA of China | 11 | 2 | None | None |
| CB of China | 11 | 2 | None | None |
| CC of China | 10 | 2 | None | None |
| CD of China | 20 | 2 | None | 2 |
| MGA of Mongolia | 22 | 3 | None | None |
| MGB of Mongolia | 21 | 2 | None | None |
| MGC of Mongolia | 21 | 2 | None | None |
| MGD of Mongolia | 21 | 2 | None | None |
| HA of Hong Kong | 21 | 2 | None | None |
| KA of Korea | 10 | 4 | None | None |
| KB of Korea | 16 | 2 | None | None |
| KC of Korea | 12 | 2 | None | None |
| KD of Korea | 10 | 2 | None | None |
| KE of Korea | 14 | 3 | None | None |
| KF of Korea | 12 | 2 | None | 2 |
| PA of Philippines | 13 | 2 | None | None |
| TA of Thailand | 1 (1.5 kb) | 6 | 2 | 6 |
| TB of Thailand | 1 (4.5 kb) | 2 | None | 2 |
| MA of Malaysia | 1 (1.5 kb) | 7 | 3 | 7 |
| MB of Malaysia | 1 (4.5 kb) | 3 | None | None |
| MC of Malaysia | 11 | 2 | None | None |
| VA of Vietnam | 1 (1.3 kb) | 10 | None | 5 |
| VB of Vietnam | 1 (1.5 kb) | 2 | 2 | 2 |
| VC of Vietnam | 18 | 2 | None | 2 |
The first letter is the initial of the country; the second one is a serial indication of clusters.
Most clusters in Thailand, Malaysia, and Vietnam had a single copy of IS6110. Numbers in parentheses indicate the fragment size in the clusters.
Number of isolates belonging to each cluster.
DISCUSSION
TB is a major health problem in developing countries. The 181 countries reporting to the World Health Organization provided notification of a total of 3,368,879 cases of TB in 1997, with 834,583 cases (25%) in the WPR (36). The global proportion of cases in the WPR has gradually been increasing: 20% in 1985, 22% in 1990, 24% in 1995, and 25% in 1997 (20, 35, 36). The seven countries included in this study (excluding Thailand, which belongs to the Southeast Asia Region) covered 91% of the cases of TB in the WPR in 1997 (36), making TB a major concern in this region.
Korea has performed nationwide TB prevalence surveys at 5-year intervals since 1965, with the most recent one conducted in 1995. The prevalence of pulmonary TB per 100,000 population in Korea has decreased in the last 30 years: the prevalence of direct smear-positive TB has gone from 686 per 100,000 in 1965 to 93 per 100,000 in 1995, and the prevalence of smear- and/or culture-positive TB has gone from 940 per 100,000 in 1965 to 219 per 100,000 in 1995 (14). Despite this successful reduction, the prevalence of TB in Korea still remains high. By better characterizing M. tuberculosis strains from developing countries (including Korea), we hope to enlarge our understanding of the dynamics of the epidemiology of TB and develop improved strategies to reduce the prevalence of TB in this region.
van Soolingen et al. (30) reported that strains of the Beijing family were found to dominate in China and surrounding countries, including Korea, Mongolia, and Thailand, whereas Beijing strains were rarer in other countries in Middle East Asia, Africa, South America, and Europe. van Soolingen et al. (30) defined Beijing family strains as containing only 9 of the 43 spacer sequences by spoligotyping and a 3.6-kb PvuII fragment by IS1081 fingerprinting. We did not study the number of spacer sequences in this study, but a 3.6-kb PvuII fragment was present in strains of the C, E, H, I, J, and K types by IS1081 fingerprinting (Fig. 4). The proportions of Beijing family strains with IS1081 types containing only a 3.6-kb PvuII fragment in each country were 100% in the Philippines, 92% in China, 76% in Mongolia, 72% in Korea, 71% in Hong Kong, 63% in Thailand, 40% in Vietnam, and 35% in Malaysia. Although in our study the proportion of Beijing family strains was higher than that in the report of van Soolingen et al. (30) (86% in China, 50% in Mongolia, 43% in Korea, 37% in Thailand), our results confirmed that China and surrounding countries had higher proportions than countries such as Malaysia or Vietnam. We also found that China and surrounding countries showed slight distinctions in the proportion of Beijing family strains, IS1081 types, and mode number of IS6110 copies. With that in mind, we tried in this study to further subdivide these countries according to a number of factors.
For the Korean isolates, we obtained a more normalized distribution curve for the number of IS6110 copies than that obtained in a previous study (15). The range of IS6110 copies was wider than the 1 to 13 copies reported previously, but the mode was identical at 10 copies (15). The IS6110 RFLP pattern of Korean isolates greatly resembled that of Japanese isolates, ranging from 1 to 19 copies, with the majority having 8 to 15 copies (22), quite unlike those of China or Mongolia, with modes of about 20 copies.
Despite the small number, the IS6110 RFLP histogram for the 14 Hong Kong strains was similar to that of Das et al. (10). In this study the proportion of 1.3-kb fragments (76.9%) among Vietnamese strains with a single copy of IS6110 was also similar to the proportion (80.0%) of Lilly et al. (18). The high proportion of 1.3-kb fragments in the IS6110 fingerprints of the Vietnamese isolates differed from the proportion of such fragments among Malaysian and Thai isolates in this study. The IS6110 RFLP pattern of Thai strains, 16.3% of which contained a single copy of IS6110, was similar to that (20.3%) of Palittapongarnpim et al. (19), and the Thai strains could preferentially be subgrouped with Malaysian or Vietnamese strains rather than Chinese, Mongolian, or Korean strains. Even though M. tuberculosis isolates from the eight regions could be subgrouped according to each factor, we found that isolates from each country had their own peculiar DNA fingerprinting traits.
With respect to IS1081 fingerprinting, G, H, I, and L types were previously reported by other investigators, with the H type found for Mongolian and Korean isolates (15, 30), the G type found for Mongolian isolates (30), the I type found for Korean isolates (15), and the L type found for Indian and New Zealand isolates (9, 32). The F type, which had previously been found for M. bovis isolates (34), was not found in this study. We found some novel IS1081 types in this study, namely, Filipino J and K types, Malaysian M and N types, and Vietnamese O and P types. In the case of the highly prevalent Filipino J type (55.9%), its existence was assumed to be caused by evolutionary isolation because the Philippines is relatively isolated geographically. More in-depth analysis of M. tuberculosis isolates from various parts of the whole country are needed to properly characterize the Filipino strains. Isolates with these novel IS1081 types were found only in countries surrounding the South China Sea. It is remarkable that so many isolates with novel IS1081 types could be found in this region, considering the relatively high degree of stability of IS1081, which lacks the variable patterns of IS6110 (32, 34, 37). Unfortunately, this particular study was too limited to explain their presence, whether they were merely chromosomal mutations in the pool of M. tuberculosis strains or whether there was a specific geographical or racial cause for adaptation of M. tuberculosis. The identification of so many novel types of M. tuberculosis strains in this study is a reflection of the prior lack of IS1081 fingerprinting because of its perceived low discriminative power. We suggest that further DNA fingerprinting studies involving IS1081 may yield better characterization and evolutionary understanding of M. tuberculosis isolates, especially in developing countries with a high prevalence of TB.
ACKNOWLEDGMENTS
We deeply appreciate Dick van Soolingen's help in setting up the RFLP techniques. We also thank Yeun Kim for technical assistance.
REFERENCES
- 1.Bauer J, Thomsen V, Poulsen S, Andersen Å B. False-positive results from cultures of Mycobacterium tuberculosis due to laboratory cross-contamination confirmed by restriction fragment length polymorphism. J Clin Microbiol. 1997;35:988–991. doi: 10.1128/jcm.35.4.988-991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharya M, Dietrich S, Mosher L, Siddiqui F, Reisberg B E, Paul W S, Warren J R. Cross-contamination of specimens with Mycobacterium tuberculosis, clinical significance, causes, and prevention. Microbiol Infect Dis. 1997;109:324–330. doi: 10.1093/ajcp/109.3.324. [DOI] [PubMed] [Google Scholar]
- 3.Braden C R, Temleton G L, Cave M D, Valway S, Onorato I M, Castro K G, Moers D, Yang Z, Stead W W, Bates J H. Interpretation of restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolates from a state with a large rural population. J Infect Dis. 1997;175:1446–1452. doi: 10.1086/516478. [DOI] [PubMed] [Google Scholar]
- 4.Cave M D, Eisenach K D, McDermott P F, Bates J H, Crawford J T. IS6110: conservation of sequence in the Mycobacterium tuberculosis complex and its utilization in DNA fingerprinting. Mol Cell Probes. 1991;5:73–80. doi: 10.1016/0890-8508(91)90040-q. [DOI] [PubMed] [Google Scholar]
- 5.Chaves F, Yang Z, Hajj H E, Alonso M, Burman W J, Eisenach K D, Dronda F, Bates J H, Cave M D. Usefulness of the secondary probe pTBN12 in DNA fingerprinting of Mycobacterium tuberculosis. J Clin Microbiol. 1996;34:1118–1123. doi: 10.1128/jcm.34.5.1118-1123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohn D L, Brien R J. The use of restriction fragment length polymorphism (RFLP) analysis for epidemiological studies of tuberculosis in developing countries. Int J Tuberc Lung Dis. 1998;2:16–26. [PubMed] [Google Scholar]
- 7.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 8.Collins D M, Stephens D M. Identification of an insertion sequence, IS1081, in Mycobacterium bovis. FEMS Microbiol Lett. 1991;83:11–16. doi: 10.1016/0378-1097(91)90435-d. [DOI] [PubMed] [Google Scholar]
- 9.Collins D M, Erasmuson S K, Stephens D M, Yates G F, de Lisle G W. DNA fingerprinting of Mycobacterium bovis strains by restriction fragment analysis and hybridization with insertion elements IS1081 and IS6110. J Clin Microbiol. 1993;31:1143–1147. doi: 10.1128/jcm.31.5.1143-1147.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das S, Chan S L, Allen B W, Mitchison D A, Lowrie D B. Application of DNA fingerprinting with IS986 to sequential mycobacterial isolates obtained from pulmonary tuberculosis patients in Hong Kong before, during and after short-course chemotherapy. Tuberc Lung Dis. 1993;74:47–51. doi: 10.1016/0962-8479(93)90068-9. [DOI] [PubMed] [Google Scholar]
- 11.Daley C L, Small P M, Schecter G F, Schoolnik G K, McAdam R A, Jacobs W R, Jr, Hopewell P C. An outbreak of tuberculosis with accelerated progression among persons infected with the human immunodeficiency virus. An analysis using restriction-fragment-length polymorphisms. N Engl J Med. 1992;326:231–235. doi: 10.1056/NEJM199201233260404. [DOI] [PubMed] [Google Scholar]
- 12.Eisenach K D, Cave M D, Bates J H, Crawford J T. Polymerase chain reaction amplification of a repetitive DNA sequence specific for Mycobacterium tuberculosis. J Infect Dis. 1990;161:977–981. doi: 10.1093/infdis/161.5.977. [DOI] [PubMed] [Google Scholar]
- 13.Fomukong N G, Tang T H, Al-Maamary S, Ibrahim W A, Ramayah S, Yates M, Zainuddin Z F. Insertion sequence typing of Mycobacterium tuberculosis: characterization of a widespread subtype with a single copy of IS6110. Tuberc Lung Dis. 1994;75:435–440. doi: 10.1016/0962-8479(94)90117-1. [DOI] [PubMed] [Google Scholar]
- 14.Hong Y P, Kim S J, Lew W J, Lee E K, Han Y C. The seventh nationwide tuberculosis prevalence survey in Korea, 1995. Int J Tuberc Lung Dis. 1998;2:27–36. [PubMed] [Google Scholar]
- 15.Huh Y J, Ahn D I, Kim S J. Limited variation of DNA fingerprints (IS6110 and IS1081) in Korean strains of Mycobacterium tuberculosis. Tuberc Lung Dis. 1995;76:324–329. doi: 10.1016/s0962-8479(05)80031-0. [DOI] [PubMed] [Google Scholar]
- 16.Kent P T, Kubica G P. Public health mycobacteriology. A guide for the level III laboratory. Atlanta, Ga: Centers for Disease Control; 1985. pp. 80–96. [Google Scholar]
- 17.Lemaitre N, Sougakoff W, Truffot-Pernot C, Cambau E, Derenne J P, Bricaire F. Use of DNA fingerprinting for primary surveillance of nosocomial tuberculosis in a large urban hospital: detection of outbreaks in homeless people and migrant workers. Int J Tuberc Lung Dis. 1998;2:390–396. [PubMed] [Google Scholar]
- 18.Lilly K W Y, Ross B C, Jackson K M, Dwyer B. Characterization of Mycobacterium tuberculosis strains from Vietnamese patients by Southern blot hybridization. J Clin Microbiol. 1993;31:1615–1618. doi: 10.1128/jcm.31.6.1615-1618.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palittapongarnpim P, Luangsook P, Tansuphaswadikul S, Chuchottaworn C, Prachaktam R, Sathapatayavongs B. Restriction fragment length polymorphism study of Mycobacterium tuberculosis in Thailand using IS6110 as probe. Int J Tuberc Lung Dis. 1997;1:370–376. [PubMed] [Google Scholar]
- 20.Raviglione M C, Snider D E, Jr, Kochi A. Global epidemiology of tuberculosis. JAMA. 1995;273:220–226. [PubMed] [Google Scholar]
- 21.Ross B C, Raios K, Jackson K, Dwyer B. Molecular cloning of a highly repeated DNA element from Mycobacterium tuberculosis and its use as an epidemiological tool. J Clin Microbiol. 1992;30:942–946. doi: 10.1128/jcm.30.4.942-946.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi M, Kazumi Y, Fukasawa Y, Hirano K, Mori T, Dale J W, Abe C. Restriction fragment length polymorphism analysis of epidemiologically related Mycobacterium tuberculosis isolates. Microbiol Immunol. 1993;37:289–294. doi: 10.1111/j.1348-0421.1993.tb03212.x. [DOI] [PubMed] [Google Scholar]
- 23.Skuce R A, Brittain D, Hughes M S, Beck L A, Neill S D. Genomic fingerprinting of Mycobacterium bovis from cattle by restriction fragment length polymorphism analysis. J Clin Microbiol. 1994;32:2387–2392. doi: 10.1128/jcm.32.10.2387-2392.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skuce R A, Brittain D, Hughes M S, Beck L A, Neill S D. Differentiation of Mycobacterium bovis isolates from animals by DNA typing. J Clin Microbiol. 1996;34:2469–2474. doi: 10.1128/jcm.34.10.2469-2474.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Small P M, Hopewell P C, Singh S P, Paz A, Parsonnet J, Ruston D C, Schecter G F, Daley C L, Schoolnik G K. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N Engl J Med. 1994;330:1703–1709. doi: 10.1056/NEJM199406163302402. [DOI] [PubMed] [Google Scholar]
- 26.Small P M, Shafer R W, Hopewell P C, Singh S P, Murphy M J, Desmond E, Sierra M F, Schoolnik G K. Exogenous reinfection with multidrug-resistant Mycobacterium tuberculosis in patients with advanced HIV infection. N Engl J Med. 1993;328:1137–1144. doi: 10.1056/NEJM199304223281601. [DOI] [PubMed] [Google Scholar]
- 27.Stanley J, Saunders N. DNA insertion sequences and the molecular epidemiology of Salmonella and Mycobacterium. J Med Microbiol. 1996;45:236–251. doi: 10.1099/00222615-45-4-236. [DOI] [PubMed] [Google Scholar]
- 28.van Deutekom H, Gerritsen J J J, van Soolingen D, van Amijden E J C, van Embden J D A. A molecular epidemiological approach to studying the transmission of tuberculosis in Amsterdam. Clin Infect Dis. 1997;25:1071–1077. doi: 10.1086/516072. [DOI] [PubMed] [Google Scholar]
- 29.van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, Mcadam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Soolingen D, Qian L, de Haas P E W, Douglas J T, Traore H, Portaels F, Huang Z Q, Enkhsaikan D, Nymadawa P, van Embden J D A. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J Clin Microbiol. 1995;33:3234–3238. doi: 10.1128/jcm.33.12.3234-3238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Soolingen D, de Haas P E W, Hermans P W M, van Embden J D A. DNA fingerprinting of Mycobacterium tuberculosis. Methods Enzymol. 1993;235:196–205. doi: 10.1016/0076-6879(94)35141-4. [DOI] [PubMed] [Google Scholar]
- 32.van Soolingen D, de Haas P E W, Hermans P W M, Groenen P M A, van Embden J D A. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J Clin Microbiol. 1993;31:1987–1995. doi: 10.1128/jcm.31.8.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Soolingen D, Hermans P W M, de Haas P E W, Soll D R, van Embden J D A. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains; evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Soolingen D, Hermans P W M, de Haas P E W, van Embden J D A. Insertion element IS1081-associated restriction fragment length polymorphisms in Mycobacterium tuberculosis complex species: a reliable tool for recognizing Mycobacterium bovis BCG. J Clin Microbiol. 1992;30:1772–1777. doi: 10.1128/jcm.30.7.1772-1777.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.World Health Organization. Global tuberculosis control. 1997. pp. 3–16. . Publication no. WHO/TB/97.225. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 36.World Health Organization. Global tuberculosis control. 1999. pp. 2–34. . Publication no. WHO/CDC/TB/99.259. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 37.Yang Z H, de Haas P E W, van Soolingen D, van Embden J D A, Andersen A B. Restriction fragment length polymorphism of Mycobacterium tuberculosis strains isolated from Greenland during 1992: evidence of tuberculosis transmission between Greenland and Denmark. J Clin Microbiol. 1994;32:3018–3025. doi: 10.1128/jcm.32.12.3018-3025.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]