Abstract
Accumulating evidence suggests that intestinal bacteria play an important role in the pathogenesis of colorectal cancer (CRC). Due to the complexity of the intestinal microbiome, identification of the specific causative microbial agents in CRC remains challenging, and the search for the causative microbial agents is intense. However, whether bacteria or their products can induce inflammation that results in tumorigenesis or directly causes CRC in humans is still not clear. This review will mainly focus on the progress of bacterial infection and CRC, and introduce the microbial contribution to the hallmarks of cancer. This article uses Salmonella and its chronic infection as an example to investigate a single pathogen and its role in the development of CRC, based on laboratory and epidemiological evidence. The bacterial infection leads to an altered intestinal microbiome. The review also discusses the dysfunction of the microbiome and the mechanism of host–microbial interactions, for example, bacterial virulence factors, key signaling pathways in the host, and microbial post-translational modifications in the tumorigenesis. Colonic carcinogenesis involves a progressive accumulation of mutations in a genetically susceptible host leading to cellular autonomy. Moving forward, more human data are needed to confirm the direct roles of bacterial infection in CRC development. Insights into the inhibiting infection will help to prevent cancer and develop strategies to restore the balance between host and microorganisms.
Keywords: Adenomatous polyposis coil, Beta-catenin, Colorectal cancer, Microbiome, Post-translational modifications, Salmonella
Introduction
Colorectal cancer (CRC) is one of the cancers that have a major impact on the lives of the world population. It accounts for >0.57 million deaths worldwide.[1] It is the third most common cancer in the world and the second most common cancer in China. CRC progresses in the stepwise process which begins when healthy tissue develops into the precancerous polyps or adenoma in the colon.[2] The colon is the natural habitat for a dynamic and highly competitive bacterial community and contains up to 100 trillion organisms. The intestinal microbiome is the entire habitat in the intestine, including the microorganisms (bacteria, viruses, archaea, and fungi), their genomes (ie, genes), and the surrounding environmental conditions. Dysfunction of the microbiome in the intestinal microbiome could foster chronic inflammation and the production of carcinogenic metabolites, leading to neoplasia.[3,4] However, the links between microbial colonization and CRC risk are starting to be unraveled. Evidence to support a direct link of intestinal bacteria and its virulence factors to human sporadic CRC is still limited.
About 20% of human cancers are linked to infection by virus, bacterial, or parasites.[5] However, the majority of the research papers and books focused on viral infection and cancer; limited topics of bacterial infection in cancer are mainly Helicobacter pylori and gastric cancer. Gram-negative Salmonella enterica is an intracellular pathogen to both humans and animals, posing a major public health concern worldwide. It is reported that antibody levels against Salmonella flagellin were higher in CRC and pre-cancer cases than controls in two distinct populations in the US and the Netherlands.[6] This study has shown that smoking and dietary intake (ie, iron) is one of the mediating factors, suggesting a possible link of Salmonella to CRC. We have discovered that Salmonella with bacterial AvrA-expression increased the incidence of colorectal tumors in a mouse CRC model.[7,8] Furthermore, our data showed that bacterial AvrA is more often found in human tumor-adjacent mucosa than in colorectal mucosa of non-cancer patients.[7,8]
The current review will highlight the progress of infection and the emerging roles of the microbiome in the pathophysiology of CRC. This review uses Salmonella infection as a sample to discuss the microbial contribution to the development of CRC and the mechanism of host–microbial interactions in the tumorigenesis. In cancer treatment, inhibiting infection, inhibiting chronic inflammation, and reversing malnutrition are reasonable strategies to restore the balance between host and microorganisms. A better understanding of the role of bacteria in inflammation and cancer will lead to more targeted strategies in which specific pathways of mammalian cells are selectively affected.
Microbial Contribution to the Hallmarks of Cancer
The hallmarks constitute an organizing principle for rationalizing the complexities of cancer. Hanahan and Weinberg[9] reviewed the hallmarks of cancer in 2000. These comprise six biological capabilities acquired during the multistep development of tumors. They include sustaining proliferative signaling, evading growth suppressors, resisting cell death, enabling replicative immortality, inducing angiogenesis, and activating invasion and metastasis. In 2011, Hanahan and Weinberg[10] further summarized the emerging hallmarks of cancer and added two emerging hallmarks of potential generality to this list-reprogramming of energy metabolism and evading immune destruction. Underlying these hallmarks is genome instability, which generates the genetic diversity that expedites their acquisition and inflammation, which fosters multiple hallmark functions. Tumors contain a repertoire of recruited, ostensibly normal cells that contribute to the acquisition of hallmark traits by creating the “tumor microenvironment.”[10] Interestingly, microbes contribute to the multistep development of tumors and can make the microenvironment in favor of tumor growth. We can detect enhanced bacteria (eg, Bacteroides fragilis) in the tumors of colon cancer patients and in the experimental animal model.[11]
Colonic carcinogenesis involves a progressive accumulation of mutations in a genetically susceptible host leading to eventual cellular autonomy.[12] Many common cancers develop as a consequence of chronic inflammation and/or infectious. Patients with inflammatory bowel disease (IBD) are at increased risk of developing CRC. The presence of bacteria in the gut appears to be of crucial importance in the pathogenesis of CRC. However, whether bacteria can induce inflammation that results in tumorigenesis or directly causes CRC in humans is not known. The nature and regulation of host–bacterial interactions in the gut are areas of intense scientific and clinical interest. The pathogens have used bacterial effectors and/or microbial metabolites to manipulate the signaling pathways in the host for their benefit. The pathways involved include the p53, adenomatous polyposis coil (APC)/β-catenin, and nuclear factor kappa-B (NF-κB).
Our recent book entitled “Inflammation, infection, and microbiome in cancers: evidence, mechanisms, and implications” has discussed the progress in bacterial infection and microbiome in various cancers.[13] In this book, Bleich and Arthur[14] have a chapter on the microbiome and hallmarks of cancer, which discusses mechanisms for how microbiota promote tumorigenesis in the intestine through crosstalk with the host, interactions between other microbes, and the role of microbial localization in relation to carcinogenesis, using the hallmarks of cancer as a framework.[14] Briefly, host–microbe interactions and the microbe–microbe interactions influence host physiology and microbial community dynamics. Microbes act as a community and influence one another both through physical (proximity) and chemical (metabolite) interactions and signaling. There is also constant communication and feedback between members of this community and the host.[14] In the following sections, we will mainly discuss the roles and mechanisms of bacteria, such as Salmonella, B. fragilis, and C. rodentium, in the development of CRC.
Salmonella infection and gastrointestinal cancer
More than 1 million people in the US acquire Salmonella infection annually as a foodborne illness mainly from eggs, meats, dairy, and other contaminated non-animal foods.[15] Widespread use of antibiotics in recent years[16] is likely to contribute to this high infection rate.[15]Salmonella belongs to the class of Gammaproteobacteria that includes the Enterobacteriaceae family, similar to Escherichia coli. It is an intracellular pathogen that infects both humans and a wide range of animals. Outcomes of Salmonella infection vary widely, ranging from mild self-limiting gastroenteritis to a severe systemic infection that can be fatal. Some of these acute infections result in a chronic carrier state with excretion of bacteria in stool and urine without symptoms, which represents another transmission mechanism to other humans.[15] In an animal model, recurrent infection with Salmonella (S.) enterica results in a progressive decline in protection against intestinal inflammation through the accelerated molecular aging of a protective host enzyme.[17]
Seroepidemiological studies based on in-house immunoassays have revealed that the incidence of non-typhoid Salmonella infection is much higher (∼600 times) than actually reported,[18] ranging from 56 per 1000 person-years in Finland to 547 in Poland during the years 2003–2008[19] and rising from 13 per 1000 person-years to 217 in Denmark from 1983 to 1999.[20] Furthermore, salmonellosis, primarily caused by its major serotypes, Salmonella serotype (S. ser.) Typhimurium and S. ser. Enteritidis, has also been implicated in the development of various chronic sequelae, including irritable bowel syndrome[21] and IBD.[22]
Two studies from Scandinavian countries have found that the probability of new IBD diagnosis following an episode of non-typhoid Salmonella infection (all subspecies combined) particularly within the first 10 years, significantly increases (2–3 folds) compared with the general population.[23,24]S. ser. Typhi carrier status is well recognized to increase the risk of gallbladder cancer. A meta-analysis of 22 studies by Koshiol et al[25] demonstrated the summary odds ratios of 4.6 and 5 for gallbladder cancer according to serology and culture, respectively. Moreover, antibody titers to Salmonella ser. Typhimurium were higher in colorectal tumor cases than controls in the US and the Netherlands and that smoking and dietary iron is one of the mediating factors, suggesting a possible link of non-typhoid Salmonella infection to intestinal tumorigenesis.[6] Impotently, this observation was recently confirmed by an independent population-based linkage study in the Netherlands. The investigators analyzed cancer registry and public health surveillance data and found an increased incidence of CRC, compared with the general population, among residents who were reported for enteric (not systemic) salmonellosis caused by Salmonella Enteritidis.[26] In this nationwide registry study, age difference and location of tumors are also reported. The increased risk of CRC was observed among patients who had reported (severe) Salmonella infection between 20 years and 60 years of age when compared with the baseline CRC risk in the Dutch population.[26] This increased risk was significant following infection with S. Enteritidis and for the proximal part of the colon. Moreover, it was shown that among CRC patients, the risk of having had a previously notified Salmonella infection was higher for individuals with pre-infectious IBD.[26] A recent study conducted a population-based cohort study using four health registries in Denmark.[27] Person-level demographic data of all residents were linked to laboratory-confirmed non-typhoidal salmonellosis and to CRC diagnoses in 1994 to 2016. Hazard ratios for cancer (overall/proximal/distal) associated with salmonellosis were estimated. However, it found no increased risk of CRC >/=1-year post-infection. Although stratifying by serovar, there was a significantly increased risk of proximal CRC >/=1-year post-infection with serovars other than Enteritidis and Typhimurium. CRC risk was significantly increased in the first-year post-infection, which could be explained by increased stool testing around the time of cancer diagnosis. The association between proximal CRC and non-Enteritidis/non-Typhimurium serovars is unclear. Thus, epidemiological evidence for Salmonella infections warrants further investigation, maybe depending on the studied populations.
In the animal models, we have established an S. enterica serotype Typhimurium (S. Typhimurium) chronic infection model by only gavaging mice once with S. Typhimurium and observing the chronic role of bacterial infection in the host.[28] This model allows us to examine the correlation between chronic infection and cancer development. Our data have shown that infection in the wild-type mice rising in a specific pathogen-free facility will not develop CRC post-chronic Salmonella infection.[28] However, using an azoxymethane (AOM)/dextran sodium sulfate (DSS) mouse model of inflammatory CRC model[7] or an APC-deficient model (unpublished data), we have revealed that colorectal tumor incidence indeed significantly increased in the Salmonella-infected mice, compared with mice without bacterial infection.[7] In support of this observation, we have demonstrated that Salmonella protein AvrA was immunohistochemically detectable in human colorectal mucosa from CRC patients more often than that from patients with no colorectal pathologies.[29] A higher risk of colon carcinoma formation after infection with wild-type Salmonella strains in APC+/− mice genetically predisposed to cancer.[30] These data suggest that chronic infection allow may not induce tumorigenesis in a host with a healthy innate and adaptive immune system that efficiently fights with the pathogens. However, if the hosts have defects in key signaling, for example, APC/β-catenin, or have accumulated mutations, the risks of tumorigenesis are vigilantly high.
Bacterial Virulence Factors
The bacterial type 3 secretion system (T3SS) directly injects effectors into the host cytoplasm using its filament like-needle, in a finely coordinated manner.[31,32] The Salmonella genome contains up to 23 pathogenicity islands (SPIs).[33,34] These SPIs encode a wide range of virulence factors and secretion system apparatuses.[35,36] T3SS is encoded by SPI1 and SPI2 as major virulence determinants of Salmonella infection, synthesizing over 60 effectors.[37] The primary role of SPI is in epithelial invasion and inflammation and the primary role of SPI2 in intracellular proliferation and survival.[35,38,39]
Bacterial proteins regulate epithelial and immunological cells to dampen the inflammatory response, which helps bacterial survive and maintains self-limited chronic inflammation in the host. AvrA represents the least characterized effector from the SPI1 which is originally thought to be non-virulent.[39] A common aspect of infection-related cancer is the induction of chronic inflammation through various mechanisms.[40,41] On the other hand, acute inflammation is a short-term response that either clears the pathogenic bacteria from the host through recruitment of immunological cells or results in fatal consequences (host death) if the host immune response is excessive. Thus, it does not lead to the development of cancer. Whereas most bacteria induce inflammation in the host, some pathogenic bacteria have evolved the ability to temper the inflammatory response to create a suitable niche for their survival and proliferation in the host, not killing the host or host cells they infected. This is an important characteristic common in many T3SS pathogens with numerous virulence factors that induce strong proinflammatory reactions, such as Salmonella.[42] AvrA is the major anti-inflammatory protein from SPI-1 of S. enterica, which plays a crucial role in establishing chronic infection for pathogen survival in the host.
AvrA is a close homolog to a family of acetyltransferases expressed in several enteric pathogens, including YopJ/P in Yersinia pseudotuberculosis and VopA in Vibrio parahaemolyticus.[43] AvrA exerts anti-inflammatory activities through inhibition of NF-κB and c-Jun N-terminal kinase (JNK) pathways, resulting in reduced secretion of inflammatory mediators.[44] The AvrA gene is present in most S. enterica isolates from humans and animals. This is particularly the case for non-typhoid Salmonella as its prevalence was reported to be 98%–100% (514/523 and 185/185),[45,46] whereas it was absent in typhoid strains, which has been linked to gallbladder cancer. On the contrary, AvrA protein expression is known to vary markedly with clinical presentation. AvrA protein is not often produced by clinical isolates from systemic disease, but it is often detectable in those from limited enteritis.[47] Interestingly, whereas expressions of most effectors from SPI1 are controlled at the transcriptional level.[48] It is now clear that expression of bacterial proteins, such as AvrA, are regulated in a post-transcriptional manner and that mRNA transcription takes place constitutively in all AvrA-positive bacterial strains,[49] as AvrA remains silent when cloned into E. coli.[50]
S. enterica is equipped with the carbon storage regulator (Csr) system, consisting of a small RNA-binding protein (CrsA) and non-coding RNAs (CsrB/CsrC).[51] CsrA binding to the 5′ untranslated and/or early coding regions of mRNAs alters mRNA translation, turnover, and/or transcript elongation, leading to either decay or stabilization of specific mRNA targets, whereas CsrB/CsrC containing multiple CsrA-binding sites bind and sequester CsrA, thereby antagonizing CsrA activities.[51,52] Overexpression of either CsrA or CsrB shuts down AvrA expression, but that constitutional CsrA expression was required for AvrA expression.[49]
An AOM-DSS animal model is to replicate the colitis-associated CRC in humans. In the AOM-DSS mice, we showed that colorectal tumor incidence indeed significantly increased (almost doubled) in the AvrA+ Salmonella Typhimurium-infected mice, compared with mice without bacterial infection or mice infected with AvrA−Salmonella.[7] In human tumor samples, AvrA protein was immunohistochemically detectable in colorectal mucosa from CRC patients more often than that from patients with no colorectal pathologies.[29] A higher risk of colon carcinoma formation after infection with wild-type vs. ΔprgH mutant Salmonella Typhimurium (lacking the T3SS) strains.[30] Other bacterial factors are also shown novel roles in the development of cancer, as reviewed in recently published papers.[14,53]
Microbial Post-Translational Modifications and Carcinogenesis
The process of tumorigenesis is initiated when a replication-competent cell (stem cell or partially differentiated descendent of a stem cell) acquires a mutation in a “gatekeeping” pathway that endows it with a selective growth advantage. In some cancers, the gatekeeper has been identified (eg, APC, RB1, and NF1 in tumors of the colon, eye, and nervous system, respectively).[12] Mutation of the apc gene appears to be a “gate keeper” event.[54] APC negatively regulates the expression of ß-catenin, a transcriptional regulator of crypt cell proliferation. With inactivating mutations in APC, ß-catenin is upregulated and translocates to the nucleus to bind Tcf-4. Loss of APC and increased ß-catenin induce several pro-inflammatory and tumor-promoting genes, including c-Myc, cyclin D1, and cyclooxygenase-2 (Cox-2).[55,56] Cyclin D1 is a key cell cycle regulator required for G1-S transition. Cox-2 is the rate-limiting biosynthetic enzyme for prostaglandins that participate in a wide range of inflammatory and neoplastic processes. Genetic polymorphisms that control susceptibility to key mutations and that regulate inflammatory responses to these mutations are believed to contribute to an individual's risk of CRC.[57,58]
Microbiota promote tumorigenesis in the intestine through crosstalk with the host. Several proteobacteria have evolved global regulation systems of post-transcriptional gene expression for their various virulence factors to be responsive to changes in the environment and to flourish in specialized host niches. In addition to Salmonella, infection with the other bacteria, such as bacterium C. rodentium, increases epithelial cell proliferation and promotes chemically initiated tumors in the colon of mice through the ß-catenin pathway.[59] The Min mouse, which harbors a germline mutation in one allele of APC, the tumor suppressor gene, is a model for the early steps of human CRC. APC Min mice infected with C. rodentium were found to have a 4-fold increase in the number of colonic adenomas, compared with uninfected Min mice. Most of the colonic adenomas were in the distal colon, where C. rodentium-induced hyperplasia occurs. These data demonstrate that bacterial infection promotes colon tumor formation in genetically susceptible mice.[59] In human colonic epithelial cells, it is reported that B. fragilis toxin, a bacterial toxin, triggers ß-catenin nuclear localization. Subsequently, c-myc transcription and translation are induced and persistent cellular proliferation ensues.[60] We have demonstrated that Salmonella-epithelial interactions also influence ß-catenin signaling and its upstream regulator Wnt in infection and development of CRC.[53,61–65]Fusobacterium nucleatum is among the significantly enriched bacteria in stools and tissue biopsies of patients with CRC.[66,67]F. nucleatum promotes intestinal tumorigenesis through stimulating the β-catenin signaling via its FadA adhesin. Our observation and that of others indicated that bacteria may render the host susceptible to oncogenic transformation in the colon through the Wnt/ß-catenin pathway.[64]
Post-Translational Modifications (PTMs) of proteins provide highly versatile tools and tricks used by both prokaryotic and eukaryotic cells to regulate the activity of key proteins. PTMs include the addition of simple chemical groups, such as phosphate, acetyl, methyl, or hydroxyl groups; more complex groups, such as adenosine monophosphate, adenosine diphosphate-ribose, sugars, or lipids; and small polypeptides, such as ubiquitin or ubiquitin-like proteins, as well as modifications of specific amino acid side chains. PTMs are increasingly recognized as key strategies used by many bacterial and viral pathogens to modulate host proteins critical for their infection.[68] Human oncoviruses which persist in host cells and express latent gene products, such as human papillomavirus (HPV), Kaposi's sarcoma-associated herpesvirus, Epstein-Barr virus, human T-lymphotropic virus type-1, hepatitis B virus (HBV), and adenovirus, commonly exploit PTMs (specifically ubiquitination/deubiquitination) on host key signaling pathways.[69,70] The best-known example is p53 and Rb inactivation by HPV E6/E7.
Bacterial effectors (particularly from T3SS) have been described to catalyze host PTMs directly, target host PTMs indirectly, or exert PTMs on their own effectors.[71,72] Whereas YopJ of Yersinia, a homolog to AvrA, acts as deubiquitinase and acetyltransferase, inhibiting host NF-κB and mitogen-activated protein kinase (MAPK) pathways,[73,74] we have discovered that Salmonella AvrA possesses both deubiquitination and acetyltransferase properties.[42,51,63] Specific targets for deubiquitination include inhibitor α of NF-κB (IκBα) and β-catenin, thus inhibiting NF-κB pathways and activating the β-catenin pathway, the latter of which is expected to have a profound effect on colorectal carcinogenesis.
Furthermore, deubiquitinating enzymes regulate myriad ubiquitin-mediated signaling pathways closely linked to inflammation and cell deaths, such as Wnt, NF-κB, and nucleotide oligomerization domain (NOD)-like receptors 1 and 2 (NOD1/2) pathways, besides rescuing target proteins from degradation.[75–78] Host targets of AvrA O (threonine)- and Nε(lysine)-acetylation include mitogen-activated protein kinase kinase (MAPKK)[79] and p53.[42,80] Acetylation of critical amino acid residues in MAPKK blocks MAPKK phosphorylation activities, thus inhibiting downstream c-JNK and NF-κB signaling pathways.[42] This may in turn compensate by activating signal transducer and activator of transcription 3 signaling and inflammation as seen in the AvrA-induced colorectal carcinogenesis model.[8]
TP53 is known to be muted in the majority of cancer, acetylation of mutated or wild-type TP53 may have potential consequences in cancer progression.[81] Acetylation of p53 has been shown to increase its stability and transcriptional activities.[82] Bacterial regulation of p53 was first discovered in the context of Salmonella infection (first published on March 11, 2010).[80] We have shown that bacterial AvrA can target acetylation of p53 for bacterial survival in the intestinal epithelial cells.[80] Bacterial degradation of p53 was also reported in the H. pylori-infected gastric epithelial cells (Epub on June 12, 2010).[83] Later, it is shown that H. pylori CagA protein induces degradation of p53 protein in a p14ARF-dependent manner.[84]
A recent study reported the impact of Salmonella infection on the early chromatin modifications occurring in the human gut microenvironment, which influence downstream immune responses in epithelial cells and immune cells.[85]S. Typhi-induced PTMs in histone methylation and acetylation associated with epithelial cells, natural killer T, mucosal-associated invariant T, T cell receptor-γδ, monocytes, and CD8+ T-cells that are related to both gene activation and silencing. It found that arginine methylation might regulate the early differentiation of effector-memory CD4+ T-cells following exposure to S. Typhi. Thus, bacterial infection involves various cell types in the intestine for pathogens to overcome the host responses.[85]
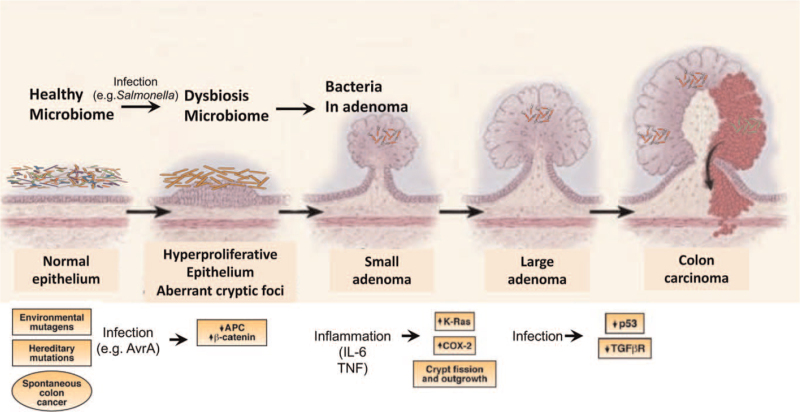
CRC has its own dysfunctions of several signaling pathways at the progression of CRC. A genetic model for CRC has been proposed in which the sequential accumulation of mutations in specific genes, including APC, Kirsten-ras, and p53, drives the transition from healthy colonic epithelia through increasingly dysplastic adenoma to CRC.[86] Inflammation is likely to be involved with sporadic as well as heritable CRC. The molecular mechanisms by which inflammation promotes cancer development are still being uncovered and could differ between colitis-associated and other forms of CRC.[87] It is known that distinct immune cells, cytokines, and other immune mediators are involved virtually in all steps of colon tumorigenesis, including initiation, promotion, progression, and metastasis. Now we know that bacterial factors manipulate the host signaling pathways, including DAN stability, Wnt/β-catenin, p53, transforming growth factor-β, and other inflammatory responses, thus contributing to the development of CRC [Figure 1].
Figure 1.
Microbial contribution to the progression of CRC. Bacterial factors manipulate the host signaling pathways, including APC/beta-catenin, p53, and other inflammatory responses, thus contributing to all steps of colon tumorigenesis, including initiation, promotion, progression, and metastasis. APC: Adenomatous polyposis coil; Cox-2: Cyclooxygenase-2; CRC: Colorectal cancer; IL-6: Interleukin 6; TNF: Tumor necrosis factor; TGFβR: Transforming growth factor-β receptor.
Bacterial Infection and Altered Intestinal Microbiome
Gut bacteria, fungi, virus, and archaea reportedly play important roles in the gut. These functions, among others, include food digestion, production of essential vitamins, absorption of nutrients, immune modulation, and resistance of colonization by pathogenic microbes.[88] Microbiome (including specific microbes, signaling pathways, and microbiota-related metabolites) contributes to the risk of CRC.[67] Changes in the intestinal microbiome can mediate or modify the effects of environmental factors on the risk of CRC (summarized in recent reviews[88,89]). Briefly, compositional alterations of stool and mucosal microbiomes across stages of colorectal tumorigenesis are characterized by ectopic overgrowth of oral pathogens and depletion of multiple commensal species.[66,67,90] Subsequent functional studies of CRC-enriched species have further led to the identification of pivotal mutagenic or tumor-promoting bacteria (eg, pks-positive E. coli, F. nucleatum, and Peptostreptococcus anaerobius) that may play direct causative roles in CRC.[91]
S. Typhimurium causes acute gut inflammation by using its virulence factors to invade the intestinal epithelium and survive in mucosal macrophages. It is known that acute infection changes the microbial community temporally. Reactive oxygen species generated during inflammation react with endogenous, luminal sulfur compounds (thiosulphate) to form a new respiratory electron acceptor, tetrathionate.[92] The genes conferring the ability to use tetrathionate as an electron acceptor produce a growth advantage for S. Typhimurium over the competing microbiota in the lumen of the inflamed intestine.[92] Thus, the ability to trigger intestinal inflammation is crucial for the biology of pathogens. S. Typhimurium could harvest energy by H2/fumarate respiration in the microbiota-colonized intestine.[93] Moreover, chronic infection may permanently change the profile and function of the microbiome in the intestine, thus altering the microenvironment for risks of tumor growth. Our published data have also shown that pathogenic bacteria, B. fragilis, accumulated in the tumors, based on the evidence from animal models and human samples.[11]
Butyrate-producing microbes decreased in adenoma and CRC. Interestingly, bacterial metabolite butyrate is known to downregulate the expression of 19 genes common to both serovars by a factor of twofold or more, and 17 of these genes localized to the SPI1. These included the SPI1 regulatory genes hilD and invF. Of the remaining two genes, ampH has 91% homology to an E. coli penicillin-binding protein and sopE2 encodes a type III-secreted effector protein associated with invasion but located at a separate site on the chromosome from SPI1.[94] Factors that affect the risk of CRC also affect the intestinal microbiome, including obesity; physical activity; and dietary intake of fiber, whole grains, and red and processed meat.[89,95] These factors, including Salmonella infection,[96] alter microbiome structure and function, along with the metabolic and immune pathways that mediate CRC development.
Summary and Future Direction
Emerging evidence has shown the single pathogen and its roles in promoting CRC, in the context of the microecology in the pathophysiology of CRC. The accumulated data suggest that the risk associated with this infection disproportionally affects individuals. IBD is a known risk factor for both CRC and salmonellosis, as this chronic condition is associated with recurrent episodes of intestinal inflammation and increased susceptibility to infection.[97,98] We have made progress in understanding the molecular mechanisms in bacterial infection and cancer, for example, bacterial proteins directly manipulate the key signaling pathways in the progression of CRC. In addition, typhoid toxin belongs to CDT toxins that induce DNA damage and cell cycle alternations[99] and bile enhances virulence of typhoidal Salmonella, but not that of non-typhoidal Salmonella,[100]F. nucleatum, and Hungatella hathewayi upregulated DNA methyltransferase in colonic epithelial cells to drive intestinal tumorigenesis.[91] Based on the current research, features of the enteric pathogens and intestinal microbiome might be used for CRC screening and modified for chemoprevention and treatment. Integrated prospective studies are urgently needed to investigate these strategies.[89]
The cutting-edge methods and tools, such as organoid cultures and “Omics,” have helped us to understand the molecular mechanisms and identify biomarkers for diagnosis.[89,101] and targets for treatment. Researchers have started to use organoids for modeling infection with pathogens, such as S. enterica[102–104] and gut–microbiota interactions.[11,105] Organoids have been used to address novel questions in host–microbe interactions, infectious diseases, and the resulting inflammatory conditions. Higher abundances of putrefying bacteria in the carcinoma stage of CRC. Acoustic reporter genes were used for non-invasive imaging of microorganisms in mammalian hosts, suggesting possible microbiome-based therapies to minimize the adverse effects of the gut microbiome in enteric diseases.[106]
In the future, more human data are needed to confirm the direct roles of infection in CRC development. Multiple disciplinary efforts are needed to further understand the role of bacteria in infection and cancer. The new insights will allow us to elucidate how enteric bacterial communities modulate tumorigenesis and identify novel approaches to target specific pathways that contribute to intestinal tumorigenesis.
Funding
This study was supported by grants from UIC Cancer Center, the NIDDK/National Institutes of Health (No. R01 DK105118, R01DK114126). The study sponsors play no role in the study design, data collection, analysis, and interpretation of data.
Conflicts of interest
None.
Footnotes
How to cite this article: Sun J. Impact of bacterial infection and intestinal microbiome on colorectal cancer development. Chin Med J 2022;135:400–408. doi: 10.1097/CM9.0000000000001979
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021; 71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Hannigan GD, Duhaime MB, Ruffin MT, Koumpouras CC, Schloss PD. Diagnostic potential and interactive dynamics of the colorectal cancer virome. mBio 2018; 9:e02248–e02318. doi: 10.1128/mBio.02248-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, et al. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut 2011; 60:631–637. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 4.Marchesi JR, Dutilh BE, Hall N, Peters WHM, Roelofs R, Boleij A, et al. Towards the human colorectal cancer microbiome. PLoS One 2011; 6:e20447.doi: 10.1371/journal.pone.0020447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer 2006; 118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 6.Kato I, Boleij A, Kortman GA, Roelofs R, Djuric Z, Severson RK, et al. Partial associations of dietary iron, smoking and intestinal bacteria with colorectal cancer risk. Nutr Cancer 2013; 65:169–177. doi: 10.1080/01635581.2013.748922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu R, Wu S, Zhang YG, Xia Y, Liu X, Zheng Y, et al. Enteric bacterial protein AvrA promotes colonic tumorigenesis and activates colonic beta-catenin signaling pathway. Oncogenesis 2014; 3:e105.doi: 10.1038/oncsis.2014.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu R, Wu S, Zhang Y-G, Xia Y, Zhou Z, Kato I, et al. Salmonella protein AvrA activates the STAT3 signaling pathway in colon cancer. Neoplasia 2016; 18:307–316. doi: 10.1016/j.neo.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000; 100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Zhang YG, Lu R, Wu S, Chatterjee I, Zhou D, Xia Y, et al. Vitamin D receptor protects against dysbiosis and tumorigenesis via the JAK/STAT pathway in intestine. Cell Mol Gastroenterol Hepatol 2020; 10:729–746. doi: 10.1016/j.jcmgh.2020.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med 2004; 10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 13.Sun J. Inflammation, Infection, and Microbiome in Cancers: Evidence, Mechanisms, and Implications. Switzerland: Springer; 2021. [Google Scholar]
- 14.Bleich RM, Arthur JC. Sun J. Microbiome and the Hallmarks of Cancer. Inflammation, Infection, and Microbiome in Cancers. Physiology in Health and Disease. Cham, Switzerland: Springer; 2021; Available from: https://link.springer.com/chapter/10.1007/978-3-030-67951-4_1#citeas. doi: 10.1007/978-3-030-67951-4_1. [Google Scholar]
- 15.Cianflone NFC. Salmonellosis and the GI tract: more than just peanut butter. Curr Gastroenterol Rep 2008; 10:424–431. doi: 10.1007/s11894-008-0079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleming-Dutra KE, Hersh AL, Shapiro DJS, Bartoces M, Enns EA, File TM, et al. Prevalence of inappropriate antibiotic prescriptions among us ambulatory care visits, 2010-2011. JAMA 2016; 315:1864–1873. doi: 10.1001/jama.2016.4151. [DOI] [PubMed] [Google Scholar]
- 17.Yang WH, Heithoff DM, Aziz PV, Sperandio M, Nizet V, Mahan MJ, et al. Recurrent infection progressively disables host protection against intestinal inflammation. Science 2017; 358:eaao5610.doi: 10.1126/science.aao5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhn KG, Falkenhorst G, Ceper TH, Dalby T, Ethelberg S, Molbak K, et al. Detecting non-typhoid Salmonella in humans by ELISAs: a literature review. J Med Microbiol 2012; 61:1–7. doi: 10.1099/jmm.0.034447-0. [DOI] [PubMed] [Google Scholar]
- 19.Falkenhorst G, Simonsen J, Ceper TH, van Pelt W, de Valk H, Sadkowska-Todys M, et al. Serological cross-sectional studies on Salmonella incidence in eight European countries: no correlation with incidence of reported cases. BMC Public Health 2012; 12:523.doi: 10.1186/1471-2458-12-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simonsen J, Strid MA, Mølbak K, Krogfelt KA, Linneberg A, Teunis P. Sero-epidemiology as a tool to study the incidence of Salmonella infections in humans. Epidemiol Infect 2008; 136:895–902. doi: 10.1017/S0950268807009314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dewulf EM, Cani PD, Claus SP, Fuentes S, Puylaert PG, Neyrinck AM, et al. Insight into the prebiotic concept: lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut 2013; 62:1112–1121. doi: 10.1136/gutjnl-2012-303304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keithlin J, Sargeant JM, Thomas MK, Fazil A. Systematic review and meta-analysis of the proportion of non-typhoidal Salmonella cases that develop chronic sequelae. Epidemiol Infect 2015; 143:1333–1351. doi: 10.1017/S0950268814002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ternhag A, Törner A, Svensson Å, Ekdahl K, Giesecke J. Short- and long-term effects of bacterial gastrointestinal infections. Emerg Infect Dis 2008; 14:143–148. doi: 10.3201/eid1401.070524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jess T, Simonsen J, Nielsen NM, Jørgensen KT, Bager P, Ethelberg S, et al. Enteric Salmonella or Campylobacter infections and the risk of inflammatory bowel disease. Gut 2011; 60:318–324. doi: 10.1136/gut.2010.223396. [DOI] [PubMed] [Google Scholar]
- 25.Koshiol J, Wozniak A, Cook P, Adaniel C, Acevedo J, Azocar L, et al. Salmonella enterica serovar Typhi and gallbladder cancer: a case-control study and meta-analysis. Cancer Med 2016; 5:3310–3235. doi: 10.1002/cam4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mughini-Gras L, Schaapveld M, Kramers J, Mooij S, Neefjes-Borst EA, van Pelt W, et al. Increased colon cancer risk after severe Salmonella infection. PLoS One 2018; 13:e0189721.doi: 10.1371/journal.pone.0189721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duijster JW, Hansen JV, Franz E, Neefjes JJC, Frisch M, Mughini-Gras L, et al. Association between Salmonella infection and colon cancer: a nationwide registry-based cohort study. Epidemiol Infect 2021; 149:e56.doi: 10.1017/S0950268821000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu R, Wu S, Liu X, Xia Y, Zhang YG, Sun J. Chronic effects of a Salmonella type III secretion effector protein AvrA in vivo. PLoS One 2010; 5:e10505.doi: 10.1371/journal.pone.0010505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu R, Bosland M, Xia Y, Zhang Y-G, Kato I, Sun J. Presence of Salmonella AvrA in colorectal tumor and its precursor lesions in mouse intestine and human specimens. Oncotarget 2017; 8:55104–55115. doi: 10.18632/oncotarget.19052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scanu T, Spaapen RM, Bakker JM, Pratap CB, Wu LE, Hofland I, et al. Salmonella manipulation of host signaling pathways provokes cellular transformation associated with gallbladder carcinoma. Cell Host Microbe 2015; 17:763–774. doi: 10.1016/j.chom.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 31.Costa TRD, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, et al. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Micro 2015; 13:343–359. doi: 10.1038/nrmicro3456. [DOI] [PubMed] [Google Scholar]
- 32.Gaytan MO, Martinez-Santos VI, Soto E, Gonzalez-Pedrajo B. Type three secretion system in attaching and effacing pathogens. Front Cell Infect Microbiol 2016; 6:129.doi: 10.3389/fcimb.2016.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayward MR, AbuOun M, La Ragione RM, Tchórzewska MA, Cooley WA, Everest DJ, et al. SPI-23 of S. Derby: role in adherence and invasion of porcine tissues. PLoS One 2014; 9:e107857.doi: 10.1371/journal.pone.0107857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabbagh SC, Forest CG, Lepage C, Leclerc J-M, Daigle F. So similar, yet so different: uncovering distinctive features in the genomes of Salmonella enterica serovars Typhimurium and Typhi. FEMS Microbiol Lett 2010; 305:1–13. doi: 10.1111/j.1574-6968.2010.01904.x. [DOI] [PubMed] [Google Scholar]
- 35.Hensel M. Evolution of pathogenicity islands of Salmonella enterica. Int J Med Microbiol 2004; 294:95–102. doi: 10.1016/j.ijmm.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 36.Kaur J, Jain SK. Role of antigens and virulence factors of Salmonella enterica serovar Typhi in its pathogenesis. Microbiol Res 2012; 167:199–210. doi: 10.1016/j.micres.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 37.Van Engelenburg SB, Palmer AE. Imaging type-III secretion reveals dynamics and spatial segregation of Salmonella effectors. Nat Methods 2010; 7:325–330. doi: 10.1038/nmeth.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuhle V, Hensel M. Cellular microbiology of intracellular Salmonella enterica: functions of the type III secretion system encoded by Salmonella pathogenicity island 2. Cell Mol Life Sci 2004; 61:2812–2826. doi: 10.1007/s00018-004-4248-z. [DOI] [PubMed] [Google Scholar]
- 39.Phoebe Lostroh C, Lee CA. The Salmonella pathogenicity island-1 type III secretion system. Microb Infect 2001; 3:1281–1291. doi: 10.1016/S1286-4579(01)01488-5. [DOI] [PubMed] [Google Scholar]
- 40.Kuper H, Adami HO, Trichopoulos D. Infections as a major preventable cause of human cancer. J Inter Med 2000; 248:171–183. doi: 10.1046/j.1365-2796.2000.00742.x. [DOI] [PubMed] [Google Scholar]
- 41.Merchant JL. Inflammation, atrophy, gastric cancer: connecting the molecular dots. Gastroenterology 2005; 129:1079–1082. doi: 10.1053/j.gastro.2005.07.038. [DOI] [PubMed] [Google Scholar]
- 42.Sun J. Pathogenic bacterial proteins and their anti-inflammatory effects in the eukaryotic host. Antiinflamm Antiallergy Agents Med Chem 2009; 8:214–227. doi: 10.2174/187152309789151986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu H, Jones RM, Neish AS. The Salmonella effector AvrA mediates bacterial intracellular survival during infection in vivo. Cell Microbiol 2012; 14:28–39. doi: 10.1111/j.1462-5822.2011.01694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X, Lu R, Xia Y, Wu S, Sun J. Eukaryotic signaling pathways targeted by Salmonella effector protein AvrA in intestinal infection in vivo. BMC Microbiol 2010; 10:326.doi: 10.1186/1471-2180-10-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huehn S, La Ragione RM, Anjum M, Saunders M, Woodward MJ, Bunge C, et al. Virulotyping and antimicrobial resistance typing of Salmonella enterica serovars relevant to human health in Europe. Foodborne Pathog Dis 2009; 7:523–535. doi: 10.1089/fpd.2009.0447. [DOI] [PubMed] [Google Scholar]
- 46.Prager R, Mirold S, Tietze E, Strutz U, Knüppel B, Rabsch W, et al. Prevalence and polymorphism of genes encoding translocated effector proteins among clinical isolates of Salmonella enterica. Int J Med Microbiol 2000; 290:605–617. doi: 10.1016/S1438-4221(00)80009-0. [DOI] [PubMed] [Google Scholar]
- 47.Streckel W, Wolff A-C, Prager R, Tietze E, Tschäpe H. Expression profiles of effector proteins SopB, SopD1, SopE1, and AvrA differ with systemic, enteric, and epidemic strains of Salmonella enterica. Mol Nutr Food Res 2004; 48:496–503. doi: 10.1002/mnfr.200400035. [DOI] [PubMed] [Google Scholar]
- 48.Ellermeier JR, Slauch JM. Adaptation to the host environment: Regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr Opin Microbiol 2007; 10:24–29. doi: 10.1016/j.mib.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 49.Kerrinnes T, Zelas ZB-B, Streckel W, Faber F, Tietze E, Tschäpe H, et al. CsrA and CsrB are required for the post-transcriptional control of the virulence-associated effector protein AvrA of Salmonella enterica. Int J Med Microbiol 2009; 299:333–341. doi: 10.1016/j.ijmm.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 50.Ben-Barak Z, Streckel W, Yaron S, Cohen S, Prager R, Tschäpe H. The expression of the virulence-associated effector protein gene avrA is dependent on a Salmonella enterica-specific regulatory function. Int J Med Microbiol 2006; 296:25–38. doi: 10.1016/j.ijmm.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 51.Timmermans J, Van Melderen L. Post-transcriptional global regulation by CsrA in bacteria. Cell Mol Life Sci 2010; 67:2897–2908. doi: 10.1007/s00018-010-0381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vakulskas CA, Potts AH, Babitzke P, Ahmer BMM, Romeo T. Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol Mol Biol Rev 2015; 79:193–224. doi: 10.1128/mmbr.00052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silbergleit M, Vasquez AA, Miller CJ, Sun J, Kato I. Oral and intestinal bacterial exotoxins: potential linked to carcinogenesis. Prog Mol Biol Transl Sci 2020; 171:131–193. doi: 10.1016/bs.pmbts.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jen J, Powell SM, Papadopoulos N, Smith KJ, Hamilton SR, Vogelstein B, et al. Molecular determinants of dysplasia in colorectal lesions. Cancer Res 1994; 54:5523–5526. [PubMed] [Google Scholar]
- 55.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 1999; 398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 56.Eisinger AL, Nadauld LD, Shelton DN, Peterson PW, Phelps RA, Chidester S, et al. The adenomatous polyposis coli tumor suppressor gene regulates expression of cyclooxygenase-2 by a mechanism that involves retinoic acid. J Biol Chem 2006; 281:20474–20482. doi: 10.1074/jbc.M602859200. [DOI] [PubMed] [Google Scholar]
- 57.Gryfe R, Di Nicola N, Gallinger S, Redston M. Somatic instability of the APC I1307K allele in colorectal neoplasia. Cancer Res 1998; 58:4040–4043. [PubMed] [Google Scholar]
- 58.Viet HT, Wagsater D, Hugander A, Dimberg J. Interleukin-1 receptor antagonist gene polymorphism in human colorectal cancer. Oncol Rep 2005; 14:915–918. [PubMed] [Google Scholar]
- 59.Newman JV, Kosaka T, Sheppard BJ, Fox JG, Schauer DB. Bacterial infection promotes colon tumorigenesis in Apc(Min/+) mice. J Infect Dis 2001; 184:227–230. doi: 10.1086/321998. [DOI] [PubMed] [Google Scholar]
- 60.Wu S, Morin PJ, Maouyo D, Sears CL. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology 2003; 124:392–400. doi: 10.1053/gast.2003.50047. [DOI] [PubMed] [Google Scholar]
- 61.Sun J, Hobert ME, Duan Y, Rao AS, He TC, Chang EB, et al. Crosstalk between NF-kappaB and beta-catenin pathways in bacterial-colonized intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol 2005; 289:G129–G137. doi: 10.1152/ajpgi.00515.2004. [DOI] [PubMed] [Google Scholar]
- 62.Sun J, Hobert ME, Rao AS, Neish AS, Madara JL. Bacterial activation of beta-catenin signaling in human epithelia. Am J Physiol Gastrointest Liver Physiol 2004; 287:G220–G227. doi: 10.1152/ajpgi.00498.2003. [DOI] [PubMed] [Google Scholar]
- 63.Ye Z, Petrof EO, Boone D, Claud EC, Sun J. Salmonella effector AvrA regulation of colonic epithelial cell inflammation by deubiquitination. Am J Pathol 2007; 171:882–892. doi: 10.2353/ajpath.2007.070220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zha L, Garrett S, Sun J. Salmonella infection in chronic inflammation and gastrointestinal cancer. Diseases 2019; 7:28.doi: 10.3390/diseases7010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang YG, Wu S, Xia Y, Chen D, Petrof EO, Claud EC, et al. Axin1 prevents Salmonella invasiveness and inflammatory response in intestinal epithelial cells. PLoS One 2012; 7:e34942.doi: 10.1371/journal.pone.0034942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liang Q, Chiu J, Chen Y, Huang Y, Higashimori A, Fang J, et al. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin Cancer Res 2017; 23:2061–2070. doi: 10.1158/1078-0432.CCR-16-1599. [DOI] [PubMed] [Google Scholar]
- 67.Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med 2019; 25:679–689. doi: 10.1038/s41591-019-0406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ribet D, Cossart P. Pathogen-mediated posttranslational modifications: a re-emerging field. Cell 2010; 143:694–702. doi: 10.1016/j.cell.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shackelford J, Pagano JS. Tumor viruses and cell signaling pathways: deubiquitination versus ubiquitination. Mol Cell Biol 2004; 24:5089–5093. doi: 10.1128/MCB.24.12.5089-5093.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gustin JK, Moses AV, Fruh K, Douglas JL. Viral takeover of the host ubiquitin system. Front Microbiol 2011; 2:161.doi: 10.3389/fmicb.2011.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ribet D, Cossart P. Post-translational modifications in host cells during bacterial infection. FEBS Lett 2010; 584:2748–2758. doi: 10.1016/j.febslet.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 72.Dean P. Functional domains and motifs of bacterial type III effector proteins and their roles in infection. FEMS Microbiol Rev 2011; 35:1100–1125. doi: 10.1111/j.1574-6976.2011.00271.x. [DOI] [PubMed] [Google Scholar]
- 73.Zhou H, Monack DM, Kayagaki N, Wertz I, Yin J, Wolf B, et al. Yersinia virulence factor YopJ acts as a deubiquitinase to inhibit NF-(B activation. J Exp Med 2005; 202:1327–1332. doi: 10.1084/jem.20051194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mukherjee S, Hao Y-H, Orth K. A newly discovered post-translational modification - The acetylation of serine and threonine residues. Trends Biochem Sci 2007; 32:210–216. doi: 10.1016/j.tibs.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 75.Lork M, Verhelst K, Beyaert R. CYLD, A20 and OTULIN deubiquitinases in NF-(B signaling and cell death: So similar, yet so different. Cell Death Differ 2017; 24:1172.doi: 10.1038/cdd.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Witt A, Vucic D. Diverse ubiquitin linkages regulate RIP kinases-mediated inflammatory and cell death signaling. Cell Death Differ 2017; 24:1160.doi: 10.1038/cdd.2017.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Clague MJ, Coulson JM, Urbé S. Cellular functions of the DUBs. J Cell Sci 2012; 125:277–286. doi: 10.1242/jcs.090985. [DOI] [PubMed] [Google Scholar]
- 78.Citterio E. Fine-tuning the ubiquitin code at DNA double-strand breaks: deubiquitinating enzymes at work. Fronti Genet 2015; 6:282.doi: 10.3389/fgene.2015.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jones RM, Wu H, Wentworth C, Luo L, Collier-Hyams L, Neish AS. Salmonella AvrA coordinates suppression of host immune and apoptotic defenses via JNK pathway blockade. Cell Host Microbe 2008; 3:233–244. doi: 10.1016/j.chom.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 80.Wu S, Ye Z, Liu X, Zhao Y, Xia Y, Steiner A, et al. Salmonella typhimurium infection increases p53 acetylation in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol 2010; 298:G784–G794. doi: 10.1152/ajpgi.00526.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nguyen T-A, Menendez D, Resnick MA, Anderson CW. Mutant TP53 posttranslational modifications: challenges and opportunities. Hum Mutat 2014; 35:738–755. doi: 10.1002/humu.22506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reed SM, Quelle DE. p53 acetylation: regulation and consequences. Cancers (Basel) 2015; 7:30–69. doi: 10.3390/cancers7010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wei J, Nagy TA, Vilgelm A, Zaika E, Ogden SR, Romero-Gallo J, et al. Regulation of p53 tumor suppressor by Helicobacter pylori in gastric epithelial cells. Gastroenterology 2010; 139:1333–1343. doi: 10.1053/j.gastro.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wei J, Noto JM, Zaika E, Romero-Gallo J, Piazuelo MB, Schneider B, et al. Bacterial CagA protein induces degradation of p53 protein in a p14ARF-dependent manner. Gut 2015; 64:1040–1048. doi: 10.1136/gutjnl-2014-307295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sztein MB, Bafford AC, Salerno-Goncalves R. Salmonella enterica serovar Typhi exposure elicits ex vivo cell-type-specific epigenetic changes in human gut cells. Sci Rep 2020; 10:13581.doi: 10.1038/s41598-020-70492-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smith G, Carey FA, Beattie J, Wilkie MJ, Lightfoot TJ, Coxhead J, et al. Mutations in APC, Kirsten-ras, and p53 — Alternative genetic pathways to colorectal cancer. Proc Natl Acad Sci U S A 2002; 99:9433–9438. doi: 10.1073/pnas.122612899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Terzic J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology 2010; 138:2101–2114.e5.doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 88.Coker OO, Yu J. Sun J. Microbiome in Human Gastrointestinal Cancers. Springer, Inflammation, Infection, and Microbiome in Cancers. Physiology in Health and Disease. Cham:2021. [Google Scholar]
- 89.Song M, Chan AT, Sun J. Influence of the gut microbiome, diet, and environment on risk of colorectal cancer. Gastroenterology 2020; 158:322–340. doi: 10.1053/j.gastro.2019.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WK, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun 2015; 6:8727.doi: 10.1038/ncomms9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xia X, Wu WKK, Wong SH, Liu D, Kwong TNY, Nakatsu G, et al. Bacteria pathogens drive host colonic epithelial cell promoter hypermethylation of tumor suppressor genes in colorectal cancer. Microbiome 2020; 8:108.doi: 10.1186/s40168-020-00847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 2010; 467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nguyen BD, Cuenca VM, Hartl J, Gul E, Bauer R, Meile S, et al. Import of aspartate and malate by DcuABC drives H2/fumarate respiration to promote initial Salmonella gut-lumen colonization in mice. Cell Host Microbe 2020; 27:922–936.e6. doi: 10.1016/j.chom.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Hautefort I, Thompson A, et al. Butyrate specifically down-regulates Salmonella pathogenicity island 1 gene expression. Appl Environ Microbiol 2006; 72:946–949. doi: 10.1128/AEM.72.1.946-949.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sun J, Kato I. Gut microbiota, inflammation and colorectal cancer. Genes Dis 2016; 3:130–143. doi: 10.1016/j.gendis.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stecher B. The roles of inflammation, nutrient availability and the commensal microbiota in enteric pathogen infection. Microbiol Spectr 2015; 3: doi: 10.1128/microbiolspec.MBP-0008-2014. [DOI] [PubMed] [Google Scholar]
- 97.Dulai PS, Sandborn WJ, Gupta S. Colorectal cancer and dysplasia in inflammatory bowel disease: a review of disease epidemiology, pathophysiology, and management. Cancer Prev Res (Phila) 2016; 9:887–894. doi: 10.1158/1940-6207.CAPR-16-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jess T, Simonsen J, Nielsen NM, Jorgensen KT, Bager P, Ethelberg S, et al. Enteric Salmonella or Campylobacter infections and the risk of inflammatory bowel disease. Gut 2011; 60:318–324. doi: 10.1136/gut.2010.223396. [DOI] [PubMed] [Google Scholar]
- 99.Di Domenico EG, Cavallo I, Pontone M, Toma L, Ensoli F. Biofilm producing Salmonella typhi: chronic colonization and development of gallbladder cancer. Int J Mol Sci 2017; 18:1887.doi: 10.3390/ijms18091887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Johnson R, Ravenhall M, Pickard D, Dougan G, Byrne A, Frankel G. Comparison of Salmonella enterica serovars Typhi and Typhimurium reveals typhoidal serovar-specific responses to bile. Infect Immun 2018; 86:e00490-17.doi: 10.1128/iai.00490-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang YG, Xia Y, Sun J. A simple and sensitive method to detect vitamin D receptor expression in various disease models using stool samples. Genes Dis 2021; 8:939–945. doi: 10.1016/j.gendis.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang YG, Zhu X, Lu R, Messer JS, Xia Y, Chang EB, et al. Intestinal epithelial HMGB1 inhibits bacterial infection via STAT3 regulation of autophagy. Autophagy 2019; 15:1935–1953. doi: 10.1080/15548627.2019.1596485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang YG, Wu S, Xia Y, Sun J. Salmonella-infected crypt-derived intestinal organoid culture system for host-bacterial interactions. Physiol Rep 2014; 2:e12147.doi: 10.14814/phy2.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang YG, Sun J. Study bacteria-host interactions using intestinal organoids. Methods Mol Biol 2019; 1576:249–254. doi: 10.1007/7651_2016_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lau HCH, Kranenburg O, Xiao H, Yu J. Organoid models of gastrointestinal cancers in basic and translational research. Nat Rev Gastroenterol Hepatol 2020; 17:203–222. doi: 10.1038/s41575-019-0255-2. [DOI] [PubMed] [Google Scholar]
- 106.Bourdeau RW, Lee-Gosselin A, Lakshmanan A, Farhadi A, Kumar SR, Nety SP, et al. Acoustic reporter genes for noninvasive imaging of microorganisms in mammalian hosts. Nature 2018; 553:86–90. doi: 10.1038/nature25021. [DOI] [PMC free article] [PubMed] [Google Scholar]