Abstract
It has been shown that resistance to clarithromycin, a major cause of failure in Helicobacter pylori eradication therapy, is associated with point mutations in the 23S rRNA gene. We sought to apply the preferential homoduplex formation assay (PHFA), a novel technique for the efficient detection of point mutations, to detection of the mutations. PHFA was performed on streptavidin-coated microtiter plates with biotin- and dinitrophenyl-labeled amplicons to detect the wild-type gene or each mutant gene. DNA samples were extracted from gastric juice specimens of 412 patients with H. pylori infection and were applied to the assay. The detection threshold of PHFA was as few as 10 gene copies. The sensitivity of PHFA for the detection of H. pylori infection was higher than those of culture and the rapid urease test. A total of 337 (81.8%) samples had the wild-type gene, 38 (9.2%) had the A2144G mutation, and 37 (9.0%) contained both the wild type and a mutation (A2144G in 30 samples, A2143G in 5 samples, and A2143G plus A2144G in 2 samples). About half the strains isolated from patients with mixed infection were susceptible by the agar dilution method (MIC, <0.1 mg/liter). Therefore, PHFA can detect clarithromycin-resistant H. pylori strains, even in patients with mixed infections with the wild type, that are not detectable by the agar dilution method.
Helicobacter pylori is a gram-negative bacterium that infects the human gastric mucosa (28). The bacterium plays an important role in the pathogenesis of chronic gastritis, peptic ulcer diseases, gastric adenocarcinoma, and gastric mucosa-associated lymphoid tissue lymphoma (6, 17, 20, 30). A National Institutes of Health Consensus Development Conference has recommended the addition of antimicrobial agents to antisecretory drugs for the treatment of patients with H. pylori-associated peptic ulcer diseases (16). Amoxicillin, clarithromycin, and metronidazole are the antimicrobial agents that are most often used, and therapy with two of these antibiotics plus an antisecretory drug usually achieves an eradication rate above 80% (2, 4, 9, 10, 27). Virtually all H. pylori strains are susceptible to amoxicillin, while 5 to 10% of strains are resistant to clarithromycin and 5 to 50% of strains are resistant to metronidazole (1, 5, 14, 15, 24, 29).
Resistance to clarithromycin is associated with one of three known point mutations in domain V of the 23S rRNA gene of H. pylori: adenine to guanine at position 2143 (A2143G), adenine to guanine at position 2144 (A2144G), and adenine to cytosine at position 2143 (A2143C) (3, 7, 18, 25). An isolated culture of H. pylori is necessary for the detection of these mutations by conventional techniques since the 23S rRNA gene is well conserved among various bacterial species and bacterial contamination will interfere with the assay. By selecting PCR primers highly specific for H. pylori, however, we have established a PCR system that can amplify a segment of the H. pylori 23S rRNA gene containing the mutation sites without isolation of H. pylori (12). The results were well correlated with the clinical outcomes of clarithromycin-based antimicrobial therapy, with the eradication rate being 2 of 5 (40%) patients for patients infected with a mutant and 29 of 34 (85%) patients for patients infected with the wild type (12).
We previously used restriction fragment length polymorphism (RFLP) analysis to identify the mutations. That technique required special instruments and may not be suitable for clinical application. In contrast, the PCR-dependent preferential homoduplex formation assay (PHFA; PCR-PHFA), originally used for determination of HLA types (19), can be performed on ready-made microtiter plates with a conventional spectrophotometer for enzyme-linked immunosorbent assay. We conducted this study to establish a PCR-PHFA system for the detection of H. pylori 23S rRNA gene mutations associated with clarithromycin resistance.
MATERIALS AND METHODS
Patients.
Materials were obtained from 412 patients (males, n = 254; females, n = 158; mean age, 51.6 years; age range, 22 to 75 years) with H. pylori infection as determined by positivity by at least two of the following tests: culture, rapid urease test (RUT; Institute of Immunology, Co., Ltd., Tokyo, Japan), gastric juice-based ureA PCR (8, 31), 13C-urea breath test (13C-UBT), and serology (HM-CAP; Enteric Products, Inc., Westbury, N.Y.). Their endoscopic findings were as follows: gastric ulcer in 125 patients, duodenal ulcer in 96 patients, and nonulcer dyspepsia in 191 patients. None of them had previously received antimicrobial therapy against H. pylori.
H. pylori strains.
H. pylori was isolated from biopsy specimens obtained from the antrum and cultured on Columbia agar with 5% (vol/vol) horse blood and Dent antibiotic supplement (Oxoid, Basingstoke, United Kingdom) at 37°C for 5 days under microaerobic conditions by using Campy-Pak Systems (BBL, Cockeysville, Md.). Identification of H. pylori was based on colony morphology, microscopy, and positive urease, catalase, and oxidase activities.
DNA samples.
DNA samples were extracted from gastric juice or biopsy specimens. Gastric juice was aspirated from the stomach through an endoscope or was collected with a capsuled absorbent nylon string (Entero-Test; HDC Corp., San Jose, Calif.). DNA was extracted by using SepaGene (Sanko Junyaku, Tokyo, Japan) (8, 12). Gastric biopsy specimens were obtained from the antrum during endoscopic examination, and DNA was extracted with the Qiagen Tissue Kit (Qiagen, Chatsworth, Calif.).
PCR amplicons.
PCR primers targeting the 23S rRNA gene were designed as described previously, with the sense primer (5′-TGTAGTGGAGGTGAAAATTCCTCC-3′; positions 2101 to 2125) labeled with dinitrophenyl (DNP) and the antisense primer (5′-GATATTCCCATTAGCAGTGCT-3′; positions 2172 to 2192) labeled with biotin. Double-labeled amplicons were prepared by PCR amplification with a pair of 5′ biotin- and 5′ DNP-labeled PCR primers and with the DNA of either the wild type or one of the mutants (A2143G, A2144G, or A2143C) as a template. Unlabeled amplicons were prepared by PCR with a pair of unlabeled PCR primers and template DNA extracted from the samples. The amplification cycle consisted of denaturation at 94°C for 5 min, followed by 50 cycles of denaturation (94°C, 30 s), annealing (55°C, 30 s), and extension (72°C, 30 s).
PHFA.
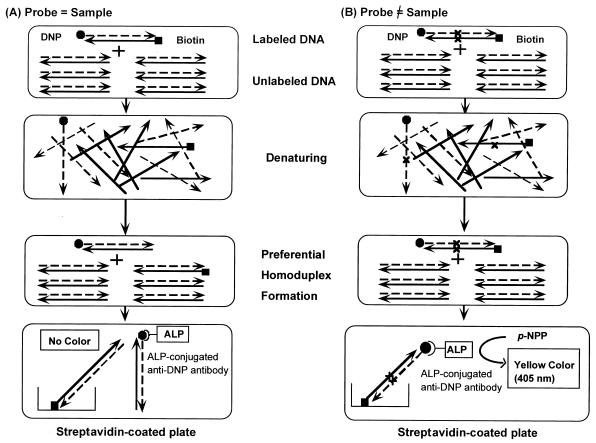
A mixture of unlabeled sample amplicon (15 μl) and one of the double-labeled amplicons (1 μl) was first denatured at 98°C for 10 min and was then slowly cooled to 70°C strictly at a rate of 1°C per 10 min; annealing occurs only when all nucleotide pairs are complementary. Only when the unlabeled amplicon was identical to the double-labeled amplicon was the number of double-labeled amplicons decreased due to hybridization. Those amplicons labeled with biotin were captured on a streptavidin-coated microtiter plate. After adding anti-DNP antibody-conjugated alkaline phosphatase and p-nitrophenyl phosphate, the absorbance at 405 nm was increased if the captured amplicons were also labeled with DNP. The absorbance was expressed as the percentage of the absorbance measured with no addition of unlabeled amplicons. The procedure is outlined in Fig. 1.
FIG. 1.
PCR-PHFA. Double-labeled PCR amplicons were prepared with biotin- and DNP-labeled primers (probes). Unlabeled amplicons were prepared with unlabeled primers (samples). Labeled and unlabeled amplicons were mixed and were denatured at 98°C for 10 min and were then cooled slowly to 70°C at a rate of 1°C/10 min. (A) When labeled and unlabeled amplicons were identical, the population of double-labeled double-stranded DNA decreased due to dilution with the unlabeled molecule. (B) When two amplicons differed by even a single base, preferential homoduplex formation occurred. The population of double-labeled double-stranded DNA did not decrease. Double-labeled double-stranded DNA was captured on a streptavidin-coated microtiter plate via biotin-streptavidin affinity and was quantified with alkaline phosphatase (ALP)-conjugated anti-DNP antibody and p-nitrophenyl phosphate (p-NPP) as the substrate.
Sensitivity and specificity of PCR-PHFA for detection of H. pylori DNA.
The cutoff value for the absorbance at 405 nm was determined by using a serially diluted plasmid containing wild-type or mutant nucleotide sequences. The sensitivity was evaluated by testing DNA samples (10 ng each) obtained from 100 clinically isolated H. pylori strains. The specificity was evaluated by testing for a cross-reaction with DNA samples extracted from 12 bacterial species other than H. pylori: Staphylococcus aureus, Citrobacter freundii, Escherichia coli, Pseudomonas aeruginosa, Salmonella enterica serotype Typhimurium, Enterobacter cloacae, Campylobacter jejuni, Morganella morganii, Providencia rettgeri, Acinetobacter calcoaceticus, Proteus mirabilis, and Klebsiella pneumoniae.
Detection of H. pylori infection by PCR-PHFA.
The results of the PCR-PHFAs with gastric juice samples were compared with those of culture and RUT with biopsy specimens for 254 patients who were positive for H. pylori by the definition described above.
Determination of clarithromycin resistance by PCR-PHFA.
Resistance to clarithromycin was determined by PCR-PHFA with the double-labeled wild-type, A2143G, A2144G, and A2143C amplicons by using gastric juice samples from 412 patients. The results were compared with the results of MIC assessments for strains isolated from the same patients. The MIC was determined by the agar dilution method with agar plates containing clarithromycin at concentrations ranging from 0.05 to 100 mg/liter. The MIC was defined as the minimum concentration of clarithromycin at which bacterial growth was inhibited. The breakpoint was chosen on the basis of a value described previously (11). H. pylori strains were classified as resistant to clarithromycin when the MIC was >1 mg/liter.
Comparison of gastric juice and biopsy specimens as PCR-PHFA samples.
DNA was extracted from gastric juice samples and biopsy specimens obtained from 60 patients during endoscopic examination. Both DNA samples were applied to the PCR-PHFA, and the results were compared.
Comparison between PCR-PHFA and PCR-RFLP for detection of clarithromycin resistance-associated mutation.
DNA samples extracted from 94 cultures of H. pylori were applied to both PCR-PHFA and PCR-RFLP analysis. PCR-RFLP analysis was performed as described previously (12). Briefly, after PCR amplification of a segment of the 23S rRNA gene containing the mutation sites associated with clarithromycin resistance, A2143G and A2144G point mutations were detected by digestion with restriction enzymes MboII and BsaI, respectively. Mutant genes would yield by agarose gel electrophoresis two bands instead of the one undigested band that appears in the case of the wild-type gene.
Changes in clarithromycin resistance because of antimicrobial therapy.
Changes in clarithromycin resistance because of antimicrobial therapy were determined for patients who received antimicrobial therapy against H. pylori by PCR-PHFA of gastric juice samples collected before and after therapy. Eradication was assessed by 13C-UBT performed more than 6 weeks after the completion of drug administration.
RESULTS
Sensitivity and specificity of PCR-PHFA.
As few as 10 molecules of plasmid containing either the wild-type, A2143G, A2144G, or A2143C sequence resulted in a positive PCR-PHFA result when the cutoff index was set at 30% (Table 1). DNAs from all 100 H. pylori strains isolated were detected by PCR-PHFA. The specificity of PCR-PHFA was confirmed by the absence of cross-reactions with DNA samples extracted from 12 bacterial species other than H. pylori.
TABLE 1.
PCR-PHFA with cloned templatesa
| Unlabeled templates
|
Index values for double-labeled probesb
|
|||
|---|---|---|---|---|
| Type | No. of molecules | AA probe | GA probe | AG probe |
| AA | 1,000 | 1.5 | 62.5 | 79.6 |
| AA | 100 | 8.8 | 82.7 | 85.3 |
| AA | 50 | 4.9 | 82.8 | 88.7 |
| AA | 10 | 14.9 | 85.6 | 87.6 |
| GA | 1,000 | 63.1 | 1.6 | 87.0 |
| GA | 100 | 58.5 | 2.5 | 92.2 |
| GA | 50 | 57.6 | 2.5 | 86.3 |
| GA | 10 | 80.2 | 26.1 | 83.8 |
| AG | 1,000 | 65.6 | 82.4 | 1.7 |
| AG | 100 | 64.6 | 82.2 | 2.7 |
| AG | 50 | 82.2 | 92.4 | 2.7 |
| AG | 10 | 72.6 | 92.4 | 9.2 |
Abbreviations: AA, wild type; GA, A2143G mutation; AG, A2144G mutation.
The index values are the absorbance at 405 nm obtained with the DNA template as a percentage of the absorbance obtained without the template. The A2143C templates and the A2143C probe had similar results (data not shown).
PCR-PHFA compared with culture and RUT.
The sensitivity of PCR-PHFA for the detection of H. pylori infection in 254 patients with H. pylori infection was compared with those of culture and RUT. PCR-PHFA demonstrated the highest sensitivity (243 of 254; 95.7%), followed by culture (89.0%) and RUT (84.3%) (Table 2). All 20 subjects negative by culture and RUT but positive by PCR-PHFA were positive by both serology and ureA PCR. Two subjects were negative by PCR-PHFA, culture, and RUT but were still positive by 13C-UBT and serology.
TABLE 2.
Comparison of PHFA, culture, and RUTa
| PCR PHFA result | No. of samples with the following resulta:
|
||||
|---|---|---|---|---|---|
| CX+, RUT+ | CX+, RUT− | CX−, RUT+ | CX−, RUT− | Total | |
| Positive | 205 | 15 | 3 | 20b | 243 |
| Negative | 3 | 3 | 3 | 2 | 11 |
| Total | 208 | 18 | 6 | 22 | 254 |
All patients were positive for H. pylori by at least two of the following methods: culture, RUT, ureA PCR, 13C-UBT, and serology. CX, culture; +, positive result; −, negative result.
The 20 patients were positive by both ureA PCR and serology.
Prevalence of mutant infection as determined by PCR-PHFA.
A total of 412 gastric juice samples were positive for H. pylori DNA by PCR-PHFA. Three hundred thirty-seven (81.8%) of the DNAs were determined to be wild-type DNA and 38 (9.2%) were found to have the A2144G mutation. The remaining 37 samples (9.0%) had both wild-type and mutant DNAs (Table 3). They were designated mixed infections, in contrast to pure infections, in which only one type of the 23S rRNA gene was detected. The prevalence of mutant infection, including mixed infection, was 18.2% among the patients in the present study. No pure A2143G mutant infection was found in this study. No A2143C mutant was found as a pure or mixed infection.
TABLE 3.
Prevalence of 23S rRNA mutant infection as determined by PCR-PHFAa
| Type | No. of samples | Prevalence (%) |
|---|---|---|
| Pure wild type | 337 | 81.8 |
| Pure mutant | ||
| A2143G | 0 | 0.0 |
| A2144G | 38 | 9.2 |
| Mixed | ||
| Wild type + A2143G | 5 | 1.2 |
| Wild type + A2144G | 30 | 7.3 |
| Wild type + A2144G + A2144G | 2 | 0.5 |
| Total | 412 | 100 |
No A2143C mutation was found in either pure or mixed infections.
Comparison of PCR-PHFA and MIC testing.
The PCR-PHFA result with gastric juice was compared with the result of MIC testing by the conventional agar dilution method with isolated strains (n = 186). PCR-PHFA indicated pure wild-type infection in 149 patients, pure mutant infection in 18 patients, and mixed infection in 19 patients. For the majority (147 of 149) of isolates from patients with pure wild-type infection, as determined by PCR-PHFA, the MIC was below 0.2 mg/liter, and for all isolates from patients with pure mutant infection, the MIC was above 3.13 mg/liter. For the isolates obtained from patients with mixed infection, the MIC was below 0.1 mg/liter for 10 of 19 isolates and above 3.13 mg/liter for 9 of 19 isolates.
Comparison of gastric juice and biopsy specimens as PCR-PHFA samples.
By using DNA samples extracted from gastric juice, 12 of 60 patients were shown to have mixed infections. However, only 6 of 12 DNA samples obtained from their biopsy specimens revealed mixed infections and the remaining 6 samples were determined to be infected with only the wild type. In contrast, only one mixed infection was found by using biopsy specimens, with which gastric juice-based PCR-PHFA indicated pure wild-type infection (Table 4).
TABLE 4.
Comparison of gastric juice and biopsy specimens as PCR-PHFA samplesa
| Biopsy result | No. of gastric juice specimens infected with the following:
|
|||
|---|---|---|---|---|
| Pure wild type | Mixed | Pure mutant | Total | |
| Pure wild type | 38 | 6 | 0 | 44 |
| Mixed | 1 | 6 | 0 | 7 |
| Pure mutant | 0 | 0 | 9 | 9 |
| Total | 39 | 12 | 9 | 60 |
Mutant indicates either the A2143G or the A2144G mutant.
Comparison of PCR-PHFA and PCR-RFLP analysis.
DNAs from 94 cultured samples of H. pylori were applied to both PCR-RFLP analysis and PCR-PHFA. PCR-RFLP analysis revealed pure wild-type infection in 80 samples and pure mutant infection in 14 samples; no mixed infections, as indicated by mixed RFLP patterns, were found. However, PCR-PHFA revealed 11 mixed infections. Six of the infections were determined to be pure wild-type infection by PCR-RFLP analysis and the other five were determined to be pure mutant infection. The results of the two assays agreed for the remaining nine infections (pure mutant infection). The frequency distribution of typing was significantly different between the two tests.
Changes in clarithromycin resistance because of antimicrobial therapy.
Seven patients with a mixed infection received antimicrobial therapy; four patients received the regimen lansoprazole at 30 mg twice daily (b.i.d.), amoxicillin at 500 mg four times daily (q.i.d.), and clarithromycin at 200 mg b.i.d. (LAC), and three patients received a regimen that included lansoprazole and amoxicillin but not clarithromycin. Eradication was achieved in none of the patients in the former group and one of the patients in the latter group. While the two patients who received the clarithromycin-free regimen still harbored mixed infection after unsuccessful eradication, only the mutant type remained in all four patients who had mixed infections before unsuccessful clarithromycin-based therapy (Table 5). Forty other patients with pure wild-type infection before therapy received the LAC regimen. A mutant strain appeared after unsuccessful therapy in one of them. Wild-type infection persisted in 3 patients, and the pathogen was eradicated from the remaining 36 patients.
TABLE 5.
Description of treatment failure with mutant strains
| Treatment and patient no. | Diseasea | Age (yr) | Sexb | Strain type before treatment | Strain type after treatment |
|---|---|---|---|---|---|
| Treatment without clarithromycinc | |||||
| 1 | DU | 47 | M | Wildd + A2144G | Wild + A2144G |
| 2 | GU | 38 | M | A2144G | A2144G |
| 3 | CG | 22 | F | Wild + A2144G | Wild + A2144G |
| 4 | DU | 47 | F | A2144G | A2144G |
| Treatment including clarithromycine | |||||
| 5 | CG | 54 | F | Wild + A2144G | A2144G |
| 6 | DU | 40 | M | Wild + A2144G | A2144G |
| 7 | GU | 38 | M | Wild + A2144G | A2144G |
| 8 | GU | 48 | M | Wild + A2144G | A2144G |
| 9 | GU | 60 | F | A2144G | A2144G |
| 10 | GU | 55 | F | A2144G | A2144G |
| 11 | CG | 55 | M | A2144G | A2144G |
| 12 | CG | 58 | M | Wild | A2144G |
DU, duodenal ulcer; GU, gastric ulcer; CG, chronic gastritis.
M, male; F, female.
Lansoprazole at 30 mg b.i.d. plus amoxicillin at 500 mg q.i.d. for 1 to 2 weeks.
Wild, wild type.
Lansoprazole at 30 mg b.i.d. plus amoxicillin at 500 mg q.i.d. and clarithromycin at 200 mg b.i.d. for 1 week.
DISCUSSION
We have previously shown that the PCR-RFLP assay with H. pylori-specific primers was clinically useful both for the detection of H. pylori infection and for the evaluation of clarithromycin resistance (12). Compared with that assay, the novel PCR-PHFA has several additional benefits. A number of samples can be processed simultaneously on a microtiter plate. The sensitivity has been improved, with the assay detecting as little as 10 molecules of DNA. Most importantly, PCR-PHFA, by using distinct probes for the wild type and each mutant, can detect each genotype separately, allowing the detection of mixed infections. In contrast, PCR-RFLP analysis failed to detect most mixed infections, as shown above, since PCR-RFLP analysis is prone to detect only the dominant genotype when several genotypes coexist.
The present study also demonstrated that the detection of mutant strains when coinfection with the wild type is present depends not only on the methodology but also on the sample, and gastric juice is preferable to biopsy specimens. The mutant and wild-type strains may inhabit distinct areas of the stomach, and the conventional assay for the detection of resistance, which uses cultured bacteria obtained from a biopsy specimen, may fail to detect one or the other strain. If the examination before treatment happened to detect the wild type and only the mutant survived the eradication therapy, the results may falsely indicate an induction of resistance. In comparison, the use of PCR-PHFA avoids the sampling error because it can use gastric juice, which contains bacterial bodies dropped from the entire area of the stomach.
The present study confirmed that induction of clarithromycin resistance through unsuccessful clarithromycin-based antimicrobial therapy was a rare event, if any. In most cases, the resistant mutant had already existed before treatment, frequently as a mixed infection with wild-type strains. We demonstrated that as many as 9% of patients had a mixed infection with a susceptible wild-type strain and resistant mutant strains, and half of the patients with the mixed infection might be falsely diagnosed as having infections only with susceptible strains when the results are judged by the agar dilution method with isolated strains.
Recently, Marais and colleagues (13) reported on a DNA enzyme immunoassay that can detect the three types of 23S rRNA gene mutations associated with clarithromycin resistance. By using biopsy specimens, they found mixed infection with wild-type and mutant H. pylori strains in 6 of 47 samples, pure wild-type infection in 14 of 47 samples, and pure mutant infection in 27 of 47 samples. The ratio of mixed to pure mutant infection reported by them, 6 to 27, was significantly lower than the ratio from our current data, 21 to 28 (P = 0.0197 by the chi-square test). Furthermore, for all strains involved with mixed infections in their study, MICs were higher than 4 mg/liter, indicating resistance, while for 10 of our 19 strains involved in mixed infection, as detected by PCR-PHFA, MICs were less than 0.1 mg/liter, which indicates susceptibility. Eradication rates were nevertheless low among the hosts of such strains when clarithromycin-based treatment was used. We have shown that gastric juice is better than biopsy specimen as a sample for the detection of a localized H. pylori infection (31), and this may also be the case when detecting localized colonization of resistant strains amid wild-type infection. PCR-PHFA with gastric juice as the sample is the first assay that can detect mixed infections caused by strains that are clinically resistant to clarithromycin but that may appear to be susceptible by MIC determinations.
The complete H. pylori genome has been sequenced, and it contains two 23S rRNA genes (23). When one of the genes is mutated and the other remains normal, the PCR-PHFA may yield a result similar to that obtained for a mixed infection. However, we think that this is unlikely, although it is theoretically possible, since PCR-RFLP analysis with cultured isolates never revealed a mixed pattern of digested and undigested PCR products. Taylor et al. (22) reported that the two 23S rRNA genes of resistant strains always had mutations at the same position, although the underlying mechanism for this phenomenon is not known. We confirmed these data for three clarithromycin-resistant strains in Japan by long-distance PCR of the nucleotides surrounding the 23S rRNA gene (data not shown).
Several studies in Western countries reported that the A2143G mutant and the A2144G mutant had similar prevalences. For example, Versalovic et al. (26) reported that 31 of 59 (53%) resistant strains were A2143G mutants and that 23 of 59 (39%) resistant strains were A2144G mutants. However, the current study showed that 70 of 75 (93%) mutant strains in Japan had the A2144G mutation.
Another mutation, A2143C, is also known to be associated with clarithromycin resistance and has been reported to account for 7% of the resistant strains in Western countries (21). The absence of the A2143C mutation in Japan is confirmed in the present study and is another characteristic of clarithromycin-resistant H. pylori strains in Japan. This geographical difference would be difficult to explain if random mutations at these sites happened frequently enough to induce clarithromycin resistance during clarithromycin-based therapy, supporting the idea that clarithromycin-resistant mutant strains coexist with the wild type.
In conclusion, we could detect H. pylori 23S rRNA gene mutations associated with clarithromycin resistance with a high sensitivity using PCR-PHFA. The assay can detect mixed infections with mutant and wild-type strains. We showed that the mixed infection, which may be falsely determined as a wild-type infection by conventional methods, is resistant to clarithromycin-based treatment, similar to pure mutant infection. Thus, the assay may be useful and may be required for the selection of appropriate regimens for eradication therapy.
REFERENCES
- 1.Admek R J, Suerbaum S, Pfaffenbach B, Opferkuch W. Primary and acquired Helicobacter pylori resistance to clarithromycin, metronidazole, and amoxicillin—influence on treatment outcome. Am J Gastroenterol. 1998;93:386–389. doi: 10.1111/j.1572-0241.1998.00386.x. [DOI] [PubMed] [Google Scholar]
- 2.Bazzoli F, Zagari R M, Fossi S, Pozzato P, Alampi G, Siponi P. Short term low dose triple therapy for the eradication of Helicobacter pylori. Eur J Gastroenterol Hepatol. 1994;6:773–777. [Google Scholar]
- 3.Debets-Ossenkopp Y J, Sparrius M, Kusters J G, Kolkman J I, Vandenbroucke-Grauls C M. Mechanism of clarithromycin resistance in clinical isolates of Helicobacter pylori. FEMS Microbiol Lett. 1996;142:37–42. doi: 10.1111/j.1574-6968.1996.tb08404.x. [DOI] [PubMed] [Google Scholar]
- 4.The European Helicobacter pylori Study Group. Current European concepts in the management of Helicobacter pylori infection. Gut . 1997;41:8–13. doi: 10.1136/gut.41.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gotoh A, Kawakami Y, Akahane T, Akamatsu T, Shimizu T, Kiyosawa K, Katsuyama T. Susceptibility of Helicobacter pylori isolates against agents commonly administered for eradication therapy and the efficacy of chemotherapy. Microbiol Immunol. 1997;41:7–12. doi: 10.1111/j.1348-0421.1997.tb01166.x. [DOI] [PubMed] [Google Scholar]
- 6.Graham D Y, Lew G M, Klein P D, Evans D G, Evans D J, Saeed Z A, Malaty H M. Effect of treatment of Helicobacter pylori infection on the long-term recurrence of gastric or duodenal ulcer: a randomized, controlled study. Ann Intern Med. 1992;116:705–708. doi: 10.7326/0003-4819-116-9-705. [DOI] [PubMed] [Google Scholar]
- 7.Hulten K, Gibreel A, Skold O, Engstrand L. Macrolide resistance in Helicobacter pylori: mechanism and stability in strains from clarithromycin-treated patients. Antimicrob Agents Chemother. 1997;41:2550–2553. doi: 10.1128/aac.41.11.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawamata O, Yoshida H, Hirota K, Yoshida A, Kawaguchi R, Shiratori Y, Omata M. Nested-polymerase chain reaction for the detection of Helicobacter pylori infection with novel primers designed by sequence analysis of urease A gene in clinically isolated bacterial strains. Biochem Biophys Res Commun. 1996;219:266–272. doi: 10.1006/bbrc.1996.0216. [DOI] [PubMed] [Google Scholar]
- 9.Labenz J, Stolte M, Ruhl G H, Becker T, Tillenburg B, Borsch G. One-week low-dose triple therapy for the eradication of Helicobacter pylori infection. Eur J Gastroenterol Hepatol. 1995;7:9–11. [PubMed] [Google Scholar]
- 10.Lind T, Veldhuyzen S, Unge P, Spiller R, Bayerdorffer E, O'Morain C, Bardhan K D, Bradette M, Chiba N. Eradication of Helicobacter pylori using one-week triple therapies combining omeprazole with two antimicrobials: the MACH I study. Helicobacter. 1996;1:138–144. doi: 10.1111/j.1523-5378.1996.tb00027.x. [DOI] [PubMed] [Google Scholar]
- 11.Lind T, Megraud F, Unge P, Bayerdorffer E, O'Morain C, Spiller R, Veldhuyzen S, Bardhan K D, Hellblom M, Wrangstadh M, Zeijlon L, Cederberg C. The MACH2 study: role of omeprazole in eradication of Helicobacter pylori with 1-week triple therapies. Gastroenterology. 1999;116:248–253. doi: 10.1016/s0016-5085(99)70119-8. [DOI] [PubMed] [Google Scholar]
- 12.Maeda S, Yoshida H, Ogura K, Kanai F, Shiratori Y, Omata M. Helicobacter pylori-specific nested-PCR assay for the detection of 23S rRNA mutation associated with clarithromycin resistance. Gut. 1998;43:317–321. doi: 10.1136/gut.43.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marais A, Monteiro L, Occhialini A, Pina M, Lamouliatte H, Megraud F. Direct detection of Helicobacter pylori resistance to macrolides by a polymerase chain reaction/DNA enzyme immunoassay in gastric biopsy specimens. Gut. 1999;44:463–467. doi: 10.1136/gut.44.4.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Midolo P D, Lambert J R, Turnidge J. Metronidazole resistance: a predictor failure of Helicobacter pylori eradication by triple therapy. J Gastroenterol Hepatol. 1996;11:290–292. doi: 10.1111/j.1440-1746.1996.tb00078.x. [DOI] [PubMed] [Google Scholar]
- 15.Miyaji H, Azuma T, Ito S, Yamazaki Y, Sato F, Hirai M, Kuriyama Y, Kato T, Kohri Y. Susceptibility of Helicobacter pylori isolates to metronidazole, clarithromycin and amoxycillin in vitro and in clinical treatment in Japan. Aliment Pharmcol Ther. 1997;11:1131–1136. doi: 10.1046/j.1365-2036.1997.00258.x. [DOI] [PubMed] [Google Scholar]
- 16.National Institutes of Health. Helicobacter pylori in peptic ulcer disease. NIH Consensus Development Panel on Helicobacter pylori in Peptic Ulcer Disease. JAMA. 1994;272:65–69. [PubMed] [Google Scholar]
- 17.Nomura A, Stemmermann G N, Chyou P H, Kato I, Perez-Perez G I, Blaser M J. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N Engl J Med. 1991;325:1132–1136. doi: 10.1056/NEJM199110173251604. [DOI] [PubMed] [Google Scholar]
- 18.Occhialini A, Urdaci M, Doucet-Populaire F, Bebear C M, Lamouliatte H, Megraud F. Macrolide resistance in Helicobacter pylori: rapid detection of point mutations and assays of macrolide binding to ribosomes. Antimicrob Agents Chemother. 1997;41:2724–2728. doi: 10.1128/aac.41.12.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oka T, Matsunaga H, Tokunaga K, Mitsunaga S, Juji T, Yamane A. A simple method for detecting single base substitutions and its application to HLA-DPB1 typing. Nucleic Acids Res. 1994;22:1541–1547. doi: 10.1093/nar/22.9.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parsonnet J, Friedman G D, Vandersteen D P, Chang Y, Vogelman J H. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 21.Stone G G, Shortridge D, Versalovic J, Beyer J, Flamn R K, Graham D Y, Ghoneim A T, Tanaka S K. PCR oligonucleotide ligation assay to determine the prevalence of 23S rRNA gene mutations in clarithromycin resistant Helicobacter pylori. Antimicrob Agents Chemother. 1997;41:712–714. doi: 10.1128/aac.41.3.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor D E, Ge Z, Purych D, Lo T, Hiratsuka K. Cloning and sequence analysis of two copies of a 23S rRNA gene from Helicobacter pylori and association of clarithromycin resistance with 23S rRNA mutations. Antimicrob Agents Chemother. 1997;41:2621–2628. doi: 10.1128/aac.41.12.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weldman J M, Fujii C, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 24.van Zwet A A, de Boer W A, Schneeberger P M, Weel J, Jansz A R, Thijs J C. Prevalence of primary Helicobacter pylori resistance to metronidazole and clarithromycin in The Netherlands. Eur J Clin Microbiol Infect Dis. 1996;15:861–864. doi: 10.1007/BF01691216. [DOI] [PubMed] [Google Scholar]
- 25.Versalovic J, Shortridge D, Kibler K, Griffy M V, Beyer J, Flamm R K, Tanaka S K, Graham D Y, Go M F. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40:477–480. doi: 10.1128/aac.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Versalovic J, Osato M S, Spakovsky K, Dore M P, Reddy R, Stone G G, Shortridge D, Flamn R K, Tanaka K, Graham D Y. Point mutation in the 23S rRNA gene of Helicobacter pylori associated with different levels of clarithromycin resistance. J Antimicrob Chemother. 1997;40:283–286. doi: 10.1093/jac/40.2.283. [DOI] [PubMed] [Google Scholar]
- 27.Walsh J H, Peterson W L. The treatment of Helicobacter pylori infection in the management of peptic ulcer disease. N Engl J Med. 1995;333:984–991. doi: 10.1056/NEJM199510123331508. [DOI] [PubMed] [Google Scholar]
- 28.Warren, J. R., and B. J. Marshall. 1983. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet 1 i: 1273–1275. [PubMed]
- 29.Weldon M J, Broadbent A, Chambers S, Nistry R, Ranganath L, Gould S R. A seven-day Helicobacter pylori treatment regimen using clarithromycin, omeprazole and tripotassium dicitrato bismuthate. Aliment Pharmcol Ther. 1996;10:279–283. doi: 10.1111/j.0953-0673.1996.00279.x. [DOI] [PubMed] [Google Scholar]
- 30.Wotherspoon A C, Doglioni C, Diss T C, Pan L, Moschini A, de Boni M, Isaacson P G. Regression of primary low-grade B-cell gastric lymphoma of mucosa-associated lymphoid tissue type after eradication of Helicobacter pylori. Lancet. 1993;342:575–577. doi: 10.1016/0140-6736(93)91409-f. [DOI] [PubMed] [Google Scholar]
- 31.Yoshida H, Hirota K, Shiratori Y, Nihei T, Amano S, Yoshida A, Kawamata O, Omata M. Use of a gastric juice-based PCR assay to detect Helicobacter pylori infection in culture-negative patients. J Clin Microbiol. 1998;36:317–320. doi: 10.1128/jcm.36.1.317-320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]