Abstract
Previous studies have found that gene expression levels are associated with prognosis and some genes can be used to predict the survival risk of glioblastoma (GBM) patients. However, most of them just built the survival-related gene signature, and personal survival risk can be evaluated only in group. This study aimed to find the prognostic survival related genes of GBM, and construct survival risk prediction model, which can be used to evaluate survival risk by individual. We collected gene expression data and clinical information from the Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) databases. Cox regression analysis and LASSO-cox regression analysis were performed to get survival-related genes and establish the overall survival prediction model. The ROC curve and Kaplan Meier analysis were used to evaluate the prediction ability of the model in training set and two independent cohorts. We also analyzed the biological functions of survival-related genes by GO and KEGG enrichment analysis. We identified 99 genes associated with overall survival and selected 16 genes (IGFBP2, GPRASP1, C1R, CHRM3, CLSTN2, NELL1, SEZ6L2, NMB, ICAM5, HPCAL4, SNAP91, PCSK1N, PGBD5, INA, UCHL1 and LHX6) to establish the survival risk prediction model. Multivariate Cox regression analysis indicted that the risk score could predict overall survival independent of age and gender. ROC analyses showed that our model was more robust than four existing signatures. The sixteen genes can also be potential transcriptional biomarkers and the model can assist doctors on clinical decision-making and personalized treatment of GBM patients.
Keywords: GBM, prognostic risk model, overall survival, survival-related genes
1. Introduction
According to the 2016 WHO classification of tumors of the central nervous system, gliomas were classified into grade I–IV based on histopathology and genomics. Glioblastoma multiforme (GBM) is the highest malignant brain tumor as grade IV [1]. In the newest version of the classification of tumors of the central nervous system, numerous molecular changes with clinicopathologic utility that are important for the most accurate classification of CNS neoplasms were listed, such as IDH, TERT promoter, chromosomes 7/10, EGFR, which are important for glioblastoma [2]. Glioblastoma is the most common malignant primary brain tumor, accounting for 57.7% of all gliomas and 48.6% of all primary malignant central nervous system (CNS) tumors [3]. Although the treatment of glioblastoma has been greatly improved in diagnosis, surgery, traditional therapy such as chemoradiotherapy, and nursing, there are still efforts in immunotherapy, the overall prognosis is still discouraging, and the overall survival is still very poor due to the heterogeneity of glioblastoma [4]. For patients diagnosed with GBM, the median survival was about 12–15 months [5,6], and the 5-year survival rate was less than 10% [7].
With the development of high-throughput sequencing technology, many potential biomarkers related to the diagnosis and prognosis for treatment decision making of glioblastoma have been found, such as MGMT (O6 methylguanine DNA methyltransferase), H3F3A (H3 histone, family 3A), IDH (isocitrate dehydrogenase), EGFR (epidermal growth factor receptor), and PTEN (phosphatase and tensin homolog) [7,8,9,10,11]. The status of these biomarkers can provide a basis for individualized and targeted diagnosis and treatment management of patients.
Previous studies have shown that gene expression profiling can be used to classify glioma patients and identify patients with different overall survival and clinical characteristics [12,13,14,15,16,17]. GBM is divided into Proneural, Neural, Classical, and Mesenchymal subtypes by gene expression-based molecular classification, the result shows that different subtypes may require different therapeutic approaches [11,16]. Gene expression profiling can also be used to identify risk genes associated with survival and prognosis, and to assess the survival risk of patients. Zhang et al. identified 16 endoplasmic reticulum (ER) stress-related genes (CYP2E1, SLN, BRCA1, CISD2, LRRK2, BMP2, MYH7, HSPB1, DNM1L, SHISA5, RNF185, RCN1, SPP1, RPN2, PDIA3 and ATP2A2), and established an ER stress risk model based on The Cancer Genome Atlas (TCGA) glioma database to reflect the immune characteristics and predict the prognosis of glioma patients [18]. Yin et al. analyzed the gene expression profiles and identified five novel biomarkers (PTPRN, RGS14, G6PC3, IGFBP2 and TIMP4) that have potential in the prognosis prediction of GBM [19]. Wang et al. identified 14 autophagy-related genes (MTMR14, LENG9, P4HB, TCIRG1, HSPA5, DRAM1, CTSD, S100A8, CCL2, MSTN, UBQLN4, PHYHIP, RRAGB and ZKSCAN3) associated with the overall survival of patients with glioblastoma, and built a novel autophagy-related signature for the prediction of prognosis [20]. Cao et al. built a four-gene signature-derived (OSMR, HOXC10, SCARA3 and SLC39A10) risk score model to predict the survival and treatment response of GBM patients [21]. Pan et al. identified two genes (GRIA2 and RYR3) strongly associated with survival of GBM, and the two-gene signature was a robust prognostic model to predict GBM survival [22].
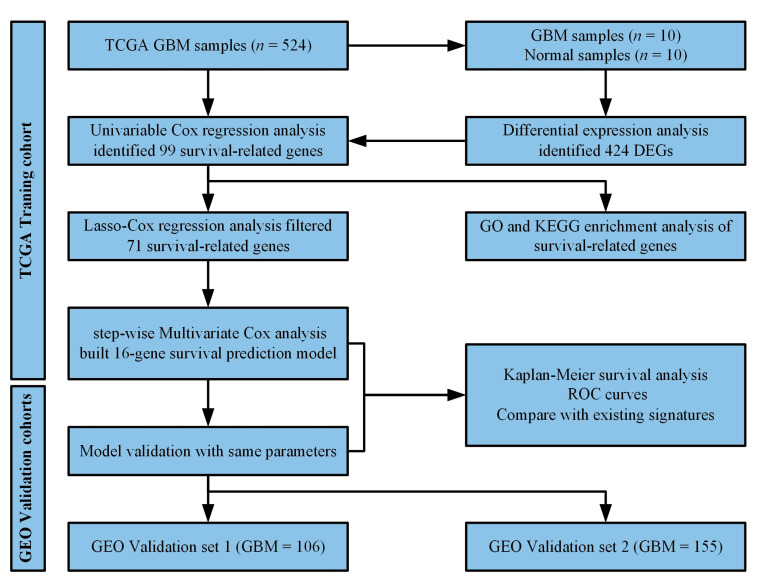
However, most of the studies just identified the survival-related genes and constructed a survival risk prediction model based on the prognostic signature, the model can generate a risk score for each patient, the risk score threshold divided patients into different risk groups. However, the risk score thresholds (usually the median risk score) are changed in different groups of patients collected from different institutions in these studies, which may be not convenient in clinical application. Based on public databases, we aim to explore novel prognostic biomarkers for survival prediction. By analyzing the genes expression profiles of GBM patients downloaded from Gene Expression Omnibus (GEO) and TCGA databases, we identified the prognostic genes and constructed a robust risk score model based on 16 genes, and we take the same risk score threshold in different groups. Therefore, our model can be used to predict survival risk for the individual patient with newly diagnosed glioblastoma. The workflow diagram of this study was shown in Figure 1.
Figure 1.
Schematic diagram of the study design and development of survival risk prediction model.
2. Materials and Methods
2.1. TCGA Dataset
We downloaded the GBM gene expression array dataset (Affymetrix HT Human Genome U133 Array Plate Set, level 1) and clinical information from TCGA [10]. After removing duplicated samples, ten normal control samples and 524 disease samples were obtained, the clinical information included age, gender, survival time, survival status, and so on. TCGA data set served as the training set.
2.2. The GEO Dataset
We downloaded three raw data sets of gene expression from GEO: GSE4412 [15] and GSE4271 [16] are all generated by Affymetrix human genome U133A array Plate Set. After removing duplicate samples, GSE4412 contained 50 glioblastoma samples and GSE4271 contained 56 glioblastoma samples. We also downloaded the gene expression profiles of GSE16011 [17], which contained 155 glioblastoma samples, gene expression profiles were generated by Affymetrix Gene Chip Human Genome U133 Plus 2.0 Array Plate Set. GSE4412 and GSE4271 patients’ data were combined as validation set 1 and GSE16011 patients’ data were used as validation set 2.
All the raw data was preprocessed by the R package AFFY and the background correction and normalization were performed using robust multi-array analysis (RMA). SVA package [23] was used to remove batch effects.
The clinical information of all patients is shown in Table S1.
2.3. Differential Expression Analysis
There are 10 normal samples served as the control group, and 10 tumor samples were randomly selected from TCGA training set served as the GBM group. The differentially expressed genes (DEGs) between the GBM group and the control group were analyzed by the limma software package [24] in R (version 4.0.5). Generally, the genes whose > 1 and adjusted p-value < 0.05 is considered to be statistically significant DEGs [25]. In this study, the genes were considered as DEGs by the standards of whose adjusted p-value < 0.01 and > 2.5. R package pheatmap [26] was used to draw heatmap while ggplot2 (https://cran.r-project.org/web/packages/ggplot2/, accessed on 20 August 2021) was used to draw volcano map.
2.4. Identify Genes Associated with Survival
Univariable Cox regression analysis was performed with differentially expressed genes in the survival R software package (https://cran.r-project.org/web/views/Survival.html, accessed on 20 August 2021) in order to analyze the correlation between each gene and overall survival. Genes with log-rank p < 0.05 were considered to be associated with overall survival [27]. The survival-related genes were further filtered by the Lasso-Cox regression model [28,29,30] in the glmnet R package (https://cran.r-project.org/web/packages/glmnet/index.html, accessed on 20 August 2021) to reduce the dimensionality of inputted variables.
2.5. Construction of Survival Risk Model
In order to optimize the model, step-wise Multivariate Cox analysis was used to further filter the survival risk related genes and construct the survival risk model by the “survival”, “survminer” packages:
| Risk score = h0 × exp(gene1 × coefficient1 + gene2 × coefficient2 + genei × coefficienti) | (1) |
Here, h0 is a constant, genei is the gene expression level. We assigned risk scores to each sample in the training cohort according to these gene expression levels. The patients were divided into low-risk group and high-risk group by the median risk score. The prognostic value of the model was evaluated by Kaplan-Meier survival analysis, Log-rank test, and time-dependent receiver operating characteristic (ROC) curve with “survival”, “survminer”, and “timeROC” packages in R.
2.6. Risk Model Validation
The validation set 1 and validation set 2 were employed to validate the robustness of diagnostic accuracy on overall survival by the prediction model. ROC curve and Kaplan Meier analysis were used to verify the prognostic value of GBM patients. p-value < 0.05 was considered statistically significant. Multivariate Cox analysis was used to validate whether the risk score was an independent risk factor for GBM survival.
2.7. Compare with Existing Signatures
As mentioned at first, lots of genes have been found related to overall survival and could be used to predict the prognosis of patients with GBM. We compared the prognostic capability of our model with signatures reported in other studies, including the five-gene signature (PTPRN, RGS14, G6PC3, IGFBP2 and TIMP4) screened by Yin et al. [19], the two-gene signature (GRIA2 and RYR3) screened by Pan et al. [22], the five-gene signature (DES, RANBP17, CLEC5A, HOXC11 and POSTN) screened by Wang et al. [25], the four-gene signature (LHX2, MEOX2, SNAI2 and ZNF22) derived by Cheng et al. [31].
For each signature, we first conducted Multivariate Cox regression analysis and built the prognostic model with its genes in TGCA cohort. Then according to the median risk score, the patients were divided into low- and high-risk groups in TGCA cohort and two validation cohorts. Kaplan-Meier analysis and ROC curves were performed to evaluate the prognostic power of each model.
2.8. GO and KEGG Enrichment Analysis of Survival-Related Genes
In order to identify potential molecular biomechanisms of genes related to survival, we conducted functional enrichment analyses including GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways analysis and visualized the results by “ggplot2” package.
3. Results
3.1. Differentially Expressed Genes Can Clearly Distinguish GBM Patients from Normal Samples
In total, 424 differentially expressed genes were identified by differential expression analysis of 10 normal brain tissue samples and 10 GBM tumor samples. Among these genes, 327 were down regulated and 97 were up regulated in GBM tumor tissue (Figure 2A). These differentially expressed genes can clearly distinguish GBM samples from normal samples, tumor samples were tightly clustered together and notably separated from normal samples (Figure 2B). The 424 differently expressed genes were listed in Table S2.
Figure 2.
Identification of DEGs between GBM and normal brain tissues. (A) Volcano plots of differentially expressed genes showing the log (Fold Change) of mRNA in GBM compared to normal brain tissues, and the corresponding–log10 (adjusted p-value). Genes with adjusted p-value below 0.01 and fold change above 2.5 (below −2.5) were marked with red (blue) dots; (B) Heatmap of differentially expressed genes. GBM samples can be clearly distinguished from normal samples according to differentially expressed genes that are from (A). The color bar means the expression level of differentially expressed genes.
3.2. 99 DEGs Were Significantly Related to Survival
Univariate Cox regression analysis was performed to identify the overall survival related genes in the training set. 99 genes were identified as potential survival-related biomarkers of GBM patients (p < 0.05) (Table S3). LASSO Cox regression analysis was performed to further analyze these 99 survival-related genes to avoid overfitting [32,33], genes were filtered by 10-fold cross-validation with a maximum of 1000 iterations, and 28 genes were screened for subsequent analysis (Figure 3).
Figure 3.
Lasso-cox gene screening process. (A) LASSO coefficient profiles of the 99 genes in TCGA GBM cohort; (B) Selection of the best parameter (lambda) in the LASSO-Cox model.
3.3. The Risk Score of Sixteen-Gene Model Were Strongly Associated with Overall Survival of GBM Patients
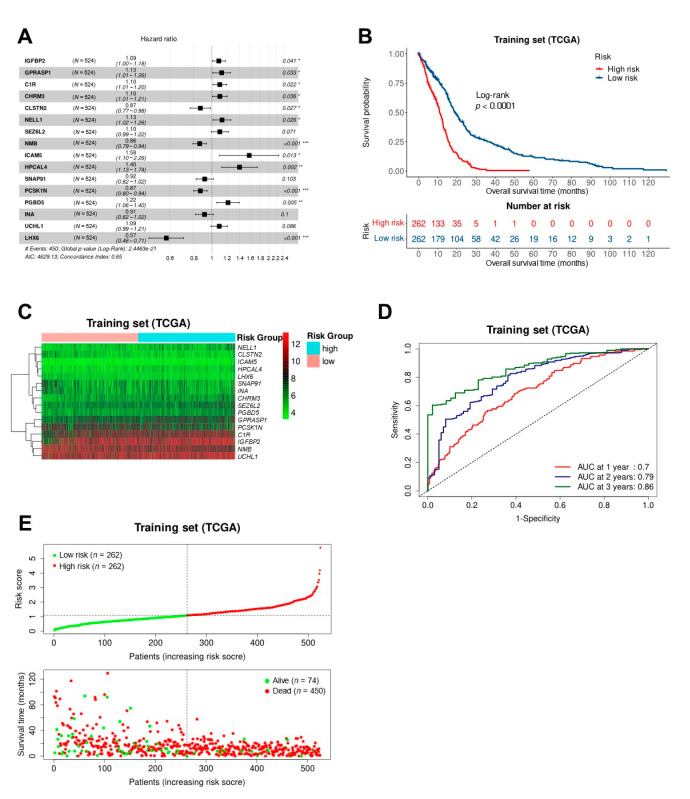
Furthermore, multivariate Cox analysis narrowed the list down further to 16 survival related genes and we constructed the survival risk model with these genes, which can be used to predict the overall survival risk of patients with GBM (Figure 4A). Of these genes, CLSTN2, NMB, SNAP91, PCSK1N, INA, and LHX6 were favorable prognostic factors for glioblastoma survival, whose risk ratio (HR) < 1, the regression coefficient < 0. IGFBP2, GPRASP1, C1R, CHRM3, NELL1, SEZ6L2, ICAM5, HPCAL4, PGBD5, and UCHL1 were risk factors, whose HR > 1, the regression coefficient > 0. The risk score of each patient was generated as follows: 0.04701 × exp(0.0842 × IGFBP2 + 0.1187 × GPRASP1 + 0.0962 × C1R + 0.0965 × CHRM3 − 0.1409 × CLSTN2 + 0.1231 × NELL1 + 0.0965 × SEZ6L2 − 0.149 × NMB + 0.4547 × ICAM5 + 0.3367 × HPCAL4 − 0.0876 × SNAP91 − 0.1406 × PCSK1N + 0.1966 × PGBD5 − 0.0901 × INA + 0.0897 × UCHL1 − 0.5555 × LHX6) (Table 1).
Figure 4.
Establishment of the survival risk model. (A) 16 genes were eventually selected to establish prognostic model. ***: p < 0.001, **: p < 0.01, *: p < 0.05; (B) Kaplan-Meier curve of overall survival in high- and low-risk group of the GBM patients in the training cohort; (C) Heatmap of 16 genes expression patterns; (D) ROC curves of the model for overall survival at 1-, 2-, and 3- years; (E) The distribution risk scores in the TCGA cohort.
Table 1.
The details of the 16 genes used in the prognostic prediction model.
| Gene Name | Coef. | HR | 95% CI | p-Value |
|---|---|---|---|---|
| IGFBP2 | 0.0842 | 1.0878 | 1.0036–1.1790 | 0.0405 |
| GPRASP1 | 0.1187 | 1.1260 | 1.0099–1.2555 | 0.0325 |
| C1R | 0.0962 | 1.1010 | 1.0142–1.1951 | 0.0216 |
| CHRM3 | 0.0965 | 1.1013 | 1.0063–1.2054 | 0.0361 |
| CLSTN2 | −0.1409 | 0.8686 | 0.7665–0.9842 | 0.0272 |
| NELL1 | 0.1231 | 1.1310 | 1.0151–1.2600 | 0.0256 |
| SEZ6L2 | 0.0965 | 1.1013 | 0.9918–1.2230 | 0.0710 |
| NMB | −0.1490 | 0.8616 | 0.7935–0.9354 | 0.0004 |
| ICAM5 | 0.4547 | 1.5757 | 1.0990–2.2592 | 0.0134 |
| HPCAL4 | 0.3367 | 1.4004 | 1.1277–1.7389 | 0.0023 |
| SNAP91 | −0.0876 | 0.9162 | 0.8245–1.0180 | 0.1034 |
| PCSK1N | −0.1406 | 0.8688 | 0.8007–0.9427 | 0.0007 |
| PGBD5 | 0.1966 | 1.2173 | 1.0611–1.3966 | 0.0050 |
| INA | −0.0901 | 0.9139 | 0.8208–1.0175 | 0.1003 |
| UCHL1 | 0.0897 | 1.0938 | 0.9874–1.2117 | 0.0859 |
| LHX6 | −0.5555 | 0.5738 | 0.4645–0.7087 | 0.0000 |
The patients were divided into high-risk group and low-risk group according to the median risk score. Kaplan-Meier survival analysis and log-rank test of high-risk and low-risk GBM patients showed that there was a significant difference in the overall survival between the two groups (p < 0.0001). Patients in the low-risk group had a better prognosis, that was to say, the patients had longer overall survival, the median overall survival of high and low risk groups respectively were 10.23 and 16.12 months (Figure 4B). The time-dependent ROC curves were generated to assess the ability to discriminate the prognostic risk of the model. The areas under the curve (AUC) of predicting 1-, 2-, and 3-years OS in TCGA dataset were 0.7, 0.79, and 0.86, respectively (Figure 4D). The expression patterns of 16 genes in high and low risk groups of TCGA cohort were shown in Figure 4C. With the increase of risk score, the expression level of CLSTN2, NMB, SNAP91, PCSK1N, INA, and LHX6 were lower and lower, and the expression level of the other genes were higher and higher. The distribution of risk scores and survival information for patients in high and low risk groups were shown in Figure 4E, higher risk score patients have a shorter survival time.
3.4. The Model Was Sufficient and Effective in Predicting Overall Survival in GBM External Verification
Two external validation sets were then used to evaluate the prediction efficiency of the model. Using the same model and parameters, patients in the validation sets were also divided into high-risk and low-risk groups. Similar to the training set, the overall survival of the high-risk group was significantly shorter than that of the low-risk group (p < 0.05), the median overall survival of high and low risk groups in validation set 1 respectively are 9.42 and 18.78 months, the median overall survival of high and low risk groups in validation set 2 respectively were 7.79 and 16.18 months (Figure 5A,B). The AUC values of 1-, 2-, 3-years overall survival prediction in validation sets, respectively, are 0.75, 0.74, 0.79 and 0.61, 0.71, 0.74 (Figure 5C,D). The expression patterns of 16 genes in verification set 1 and 2 were shown in Figure 5E,F, the expression patterns with the increase of risk score of these genes in validation cohorts have similar trends as in the training set. Figure 5G,H showed the distribution of risk scores and survival information of patients in the validation sets, it also shows that the higher the risk score, the shorter the patient’s survival. These results indicated the accuracy of the survival risk model constructed by 16 genes in predicting the prognosis of patients with glioblastoma.
Figure 5.
Validation and evaluation of the effectiveness of the survival risk model. (A,B) Kaplan-Meier survival analysis curves of validation set 1, 2; (C,D) Time-dependent ROC curves of validation set 1, 2 for the first, second, and third years; (E,F) Heatmap of 16 gene expression profiles in validation set 1, 2. Red parts indicate higher expression and green parts indicate lower expression; (G,H) Risk scores distribution and survival status scatter plots of patients in validation set 1, 2.
To further verify the independence of the prognostic model, Univariate and Multivariate Cox regression analyses were performed. The result showed that the prognosis risk score was significantly associated with overall survival, independent of clinical factors including age and gender (Table 2).
Table 2.
Univariate and multivariate analyses of prognostic factors and overall survival of GBM patients.
| Training Set | Univariate Cox Regression Analysis | Multivariate Cox Regression Analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p-Value | HR | 95% CI | p-Value | |
| Risk (high/low) | 2.719 | 2.218–3.334 | 6.352 × 10−22 | 2.471 | 2.006–3.044 | 1.839 × 10−17 |
| Age (≥60/<60) | 1.866 | 1.544–2.255 | 1.093 × 10−10 | 1.583 | 1.304–1.922 | 3.341 × 10−06 |
| Gender (male/female) | 0.852 | 0.703–1.033 | 0.103 | 0.957 | 0.789–1.162 | 0.657 |
| Validation Set 1 | ||||||
| Risk (high/low) | 2.476 | 1.584–3.872 | 6.99 × 10−05 | 2.286 | 1.429–3.657 | 5.605 × 10−04 |
| Age (≥60/<60) | 1.796 | 1.061–3.041 | 0.029 | 1.365 | 0.788–2.364 | 0.267 |
| Gender (male/female) | 1.233 | 0.798–1.905 | 0.346 | 1.099 | 0.708–1.707 | 0.673 |
| Validation Set 2 | ||||||
| Risk (high/low) | 2.293 | 1.573–3.343 | 1.592 × 10−05 | 2.17 | 1.481–3.179 | 7.026 × 10−05 |
| Age (≥60/<60) | 2.449 | 1.734–3.459 | 3.637 × 10−07 | 2.35 | 1.655–3.337 | 1.785 × 10−06 |
| Gender (male/female) | 0.902 | 0.637–1.277 | 0.56 | 0.823 | 0.579–1.168 | 0.275 |
3.5. The Sixteen-Gene Model Was More Robust and Effective Compaired with Four Existing-Survival-Related Gene Signatures
We also built the survival prediction model with four existing survival-related signatures We evaluated the ability of these models for predicting the survival risk of GBM patients in the three cohorts. The result showed that Yin’s signature also was robust in predicting the prognosis risk of patients with GBM (log-rank test p < 0.05 in all three cohorts; Figure 6A and Figure S1A,H). The smaller the p-value, the greater the difference in survival between the two groups. The results of Kaplan-Meier survival analysis, ROC curves, and AUC values of different models for predicting 1-, 2-, 3-years overall survival in training cohort were shown in Figure 6A–G. The results of Kaplan-Meier survival analysis, ROC curves, and AUC values of these models to predict 1-, 2-, 3-years overall survival in validation sets were shown in Figure S1. Although the models can divide GBM patients into low and high-risk groups, the difference in survival between the two groups was mostly insignificant. The performance of our model and the models of the existing four signatures was summarized in Table 3. As shown in the table, most of the AUC values of our model were bigger than the existing models in the three cohorts. The performance of these four models for survival risk prediction was very poor in the validation set 1, the p-values of Pan’s, Wang’s and Cheng’s signature are greater than 0.05, and the AUC values of the four models were only about 0.5–0.6 for predicting 1-, 2-, 3-years overall survival. And the AUC values of our model for predicting 1-, 2-, 3-years overall survival in three cohorts were more stable. These results demonstrated that our model was more robust and powerful to predict overall survival than the four existing signatures.
Figure 6.
Comparison of ability for survival prediction by the sixteen-gene signature and four published prognostic signatures in the training cohort. (A–D) Kaplan-Meier analysis of four published signatures for the overall survival of glioblastoma patients in training cohort; (E–G) The ROC curves of different models for predicting 1-, 2-, 3-year overall survival in the training cohort.
Table 3.
The performance of our model and the models of existing four signatures.
| Training Set | Our Model | The Model of Yin’s Signature | The Model of Pan’s Signature | The Model of Wang’s Signature | The Model of Cheng’s Signature |
|---|---|---|---|---|---|
| p-value (between low and high risk) | <0.0001 | 0.0012 | 0.0092 | <0.0001 | 0.00046 |
| AUC value of 1 year | 0.7 | 0.59 | 0.53 | 0.67 | 0.57 |
| AUC value of 2 years | 0.79 | 0.64 | 0.57 | 0.68 | 0.61 |
| AUC value of 3 years | 0.86 | 0.63 | 0.57 | 0.7 | 0.66 |
| Validation Set 1 | |||||
| p-value (between low and high risk) | <0.0001 | 0.015 | 0.1 | 0.092 | 0.19 |
| AUC value of 1 year | 0.75 | 0.68 | 0.62 | 0.42 | 0.57 |
| AUC value of 2 years | 0.74 | 0.7 | 0.6 | 0.55 | 0.68 |
| AUC value of 3 years | 0.79 | 0.65 | 0.64 | 0.49 | 0.63 |
| Validation Set 2 | |||||
| p-value (between low and high risk) | <0.0001 | 0.00039 | 0.0013 | 0.0047 | <0.0001 |
| AUC value of 1 year | 0.61 | 0.6 | 0.55 | 0.63 | 0.61 |
| AUC value of 2 years | 0.71 | 0.57 | 0.68 | 0.72 | 0.74 |
| AUC value of 3 years | 0.74 | 0.61 | 0.74 | 0.85 | 0.82 |
3.6. The Risk Scores Were Association with Some Critical Clinicopathological Parameters
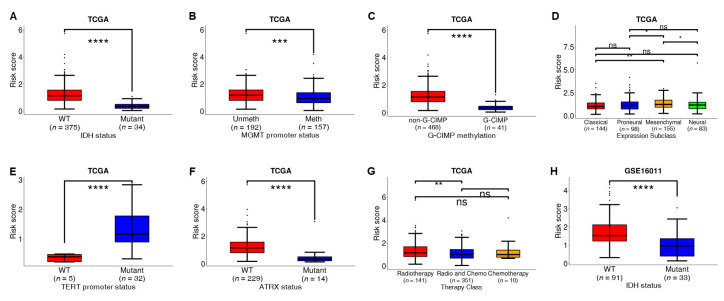
In training set and validation set 2, the correlation between risk score and some important clinicopathological features (therapeutic modalities, key molecular biomarkers, etc.) was analyzed. The results showed that there was a statistically significant difference in risk score between GBM patients with IDH wild type and IDH mutation type, GBM patients with IDH wild type have higher risk scores both in TCGA cohort and GSE16011 cohort (Figure 7A,H), which was also demonstrated in previous studies [20,21,34]. The higher risk score was associated with MGMT promoter unmethylation subtype, non-methylated subtype, Mesenchymal subtype, TERT mutation type, ATRX wild type, and radiotherapy alone (Figure 7B–G), and high-risk score means poor prognosis, they were all risk factors for survival. All these results indicate that a high-risk score was associated with short survival.
Figure 7.
Correlation between risk score and clinicopathology. (A,H) Comparison of risk scores in IDH-WT GBM and IDH-Mutant GBM in TCGA cohort and GSE16011 cohort; (B) Comparison of risk scores in MGMT-promoter- methylation, and MGMT-promoter- unmethylation GBM in the TCGA cohort; (C) Comparison of risk scores in G-CIMP and non-G-CIMP methylation GBM in the TCGA cohort; (D) Comparison of risk scores in Proneural, Neural, Classical, and Mesenchymal GBM in the TCGA cohort; (E) Comparison of risk scores in TERT-WT and TERT-Mutant GBM in TCGA cohort; (F) Comparison of risk scores in ATRX-WT and ATRX-Mutant GBM in the TCGA cohort; (G) Comparison of risk scores for therapeutic modalities in the TCGA cohort (only radiotherapy, radiotherapy and chemotherapy, only chemotherapy). ****: p ≤ 0.0001, ***: p ≤ 0.001, **: p ≤ 0.01, *: p ≤ 0.05, ns: p > 0.05.
To confirm and further convince the independent of the prognostic efficacy of the risk model, the clinical-pathological factors such as therapeutic modalities, key genes status, and expression subtypes, GBM patients were stratified by these factors, and Kaplan Meier survival analysis was performed in the training cohort and validation set 2 cohort. Kaplan Meier survival analysis was also performed for high-risk and low-risk groups of GBM patients combined with clinical parameters. As shown in Figures S2 and S3, there were generally and significantly differences in overall survival between high-risk and low-risk groups of patients among stratified GBM patients by clinical-pathological factors. These results suggest that the model can distinguish the high-risk subgroup and the low-risk subgroup in each clinical subtype, the risk score can effectively predict the survival risk of patients with glioblastoma. Univariate and Multivariate Cox regression analyses were also performed with patients who having the clinical-pathological and molecular characteristics information in TCGA training set (206 samples) and validation set 2 (92 samples). The results showed that the prognosis risk score of our model was significantly associated with overall survival, independent of clinical and molecular characteristics such as IDH status, MGMT promoter status (Table S4). The analysis also shows that some molecular markers and clinical information also have the ability of survival risk assessment, such as Karnofsky Performance Status (KPS) score.
3.7. The Biological Pathway of Survival-Related Genes Involved In
We constructed the model using 16 survival-related genes, due to the number of these 16 genes being too small to enrich for significant pathways, we used all 99 survival-related genes for pathway analysis. Functional enrichment analysis of 99 survival-related genes was conducted, and the results are shown in Figure 8. For molecular functions (MF), the major enriched GO terms were enzyme inhibitor activity, phospholipase inhibitor activity, ion channel binding, etc. (Figure 8A). For cellular components (CC), the genes were mainly enriched in the synaptic membrane, transport vesicle, synaptic vesicle, etc. (Figure 8B). For the biological processes (BP), the major enriched GO terms were the modulation of chemical synaptic transmission, regulation of trans-synaptic signaling, synapse organization, etc. (Figure 8C). According to KEGG analysis, these genes are mainly enriched in Pertussis, Complement and coagulation cascades, ECM receptor interaction, and many other pathways (Figure 8D).
Figure 8.
The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of genes associated with overall survival in GBM patients. (A) The top 5 statistically significant enriched terms in molecular function; (B) The top 5 statistically significant enriched terms in cellular components; (C) The top 5 statistically significant enriched terms in biological process; (D) The top 5 enriched KEGG pathways (Some are not statistically significant).
In 16 survival-related genes, GPRASP1, CHRM3, CLSTN2, NELL1, SEZ6L2, ICAM5, HPCAL4, SNAP91, PCSK1N, PGBD5, INA, UCHL1 and LHX6 were down regulated, IGFBP2, C1R and NMB were up regulated compared with normal tissue samples. GPRASP1 has been found overexpressed in brain, pancreatic, and breast cancers as compared to their respective normal tissues, and it may be a biomarker for early-stage cancer [35]. CHRM3 may play a vital role in the deficits of the thalamus–cortical connectivities and is associated with schizophrenia dysfunction [36]. Overexpression of CHRM3 or activation of CHRM3 by carbachol promoted cell proliferation, migration, and castration resistance in prostate cancer [37]. CLSTN2 is associated with episodic memory and late-onset Alzheimer’s disease [38,39]. NELL1 has been found to be associated with a variety of tumors, which may inhibit the progress of cancer. That makes it a promising candidate biomarker of tumor suppressor genes and a potential target for tumor therapy [40,41,42,43,44]. SEZ6L2 is highly expressed in colorectal cancer and high expression means a poor prognosis [45]. It also is an important regulator of drug-resistant cells and tumor spheroid cells in lung adenocarcinoma [46]. ICAM5 is an intercellular adhesion molecule and may play a role in tumorigenesis and perineural invasion, most likely through P13K/Akt signaling pathway [47]. ICAM-5 regulates T cell activity and participates in the immune privilege of the brain. It may be a useful anti-inflammatory agent for the treatment of various inflammatory brain diseases [48]. Some studies have found that HPCAL4 may be the core gene of GBM and a potential therapeutic target [49]. It also has been found can affect many cellular processes, such as homeostasis, learning and memory, cancer, and pain [50]. SNAP91 is associated with Alzheimer’s disease [51], schizophrenia [52], Parkinson’s disease [53] and colorectal cancer [54]. PCSK1N has an association with various neurodegenerative diseases, widely expressed in neurons throughout the brain [55,56]. PGBD5 which encodes an active DNA transposase was highly expressed in various childhood and adult solid tumors [57]. The PGBD5 DNA transposase confers a synthetic dependency on DNA damage repair and signaling, and expression of PGBD5 induces DNA damage, which requires both DNA damage repair and DNA damage signaling, resulting in apoptosis if impaired by their selective inhibitors [58]. It also has been found to be associated with frontotemporal dementia [59]. INA is a strong prognostic factor in gliomas [60]. UCHL1 may play an important role in maintaining axonal function after cerebral ischemia [61]. UCHL1 is also associated with many types of cancers including lung, colorectal, breast cancer, and pancreatic, which can promote tumor progression [62,63]. LHX6 is associated with many kinds of cancers and can inhibit the proliferation, invasion, and migration of breast cancer cells by modulating the PI3K/Akt/mTOR and Wnt/β-catenin signaling pathways [64,65,66]. LHX6 plays a tumor-suppressing role in MC-LR-induced liver cancer through the Wnt/β-catenin and P53 pathways [67]. IGFBP2 plays an essential role in cognitive development during early life, it is a potential target for learning and memory impairment therapies and neurodegenerative diseases [68]. IGFBP2 is overexpressed and promotes many key oncogenic processes, including epithelial-to-mesenchymal transition, cellular migration, invasion, angiogenesis, stemness, transcriptional activation, and epigenetic programming in many solid tumors [69]. IGFBP2 promotes tumor progression in pancreatic ductal adenocarcinoma through the STAT3 pathway [70]. C1R is up-regulated in cutaneous squamous cell carcinoma [71]. DNA-methylation of C1R is significantly correlated with overall survival in acute myeloid leukemia [72]. NMB is expressed in non-tumorigenic cells, but its expression is low to undetectable in malignant cells [73], and several studies have shown that NMB may act as a tumor suppressor [74,75,76,77].
4. Discussion
Glioblastoma is a kind of central nervous system tumor with a poor prognosis. Combined with clinical and overall survival information, we analyzed in-depth the gene expression data of GBM patients and constructed the survival risk prediction model. We found some survival-related genes and analyzed their molecular biomechanisms. These genes can be used as potential biomarkers in diagnosis and treatment. The results and our model can support clinical decision-making for the diagnosis and prognosis of the disease, so as to provide effective treatment and obtain better prognostic outcomes.
In this study, a total of 424 differentially expressed genes were obtained between GBM and normal brain tissue. Univariate Cox analysis showed that 99 genes were associated with overall survival. The survival-related genes were selected for further analysis. In the training set, 16 genes related to overall survival were screened by LASSO-Cox regression analysis and multiple Cox regression analysis. We found that the expression levels of CLSTN2, NMB, SNAP91, PCSK1N, INA and LHX6 were positively correlated with the overall survival of glioblastoma, while the expression levels of IGFBP2, GPRASP1, C1R, CHRM3, NELL1, SEZ6L2, ICAM5, HPCAL4, PGBD5 and UCHL1 were negatively correlated with overall survival. All these genes have been found to be associated with diseases or cancers. It reflects from the side the biological significance of the survival-related genes we selected and the effectiveness of our model.
Based on these 16 genes, we built a prognostic survival risk model. The patients were divided into low-risk group and high-risk group by the median risk score. Two external datasets verified the effectiveness of the model, the results show that the model was more robust and can effectively predict the survival risk of patients. Because we took the same risk score threshold in different groups, we didn’t have to adjust the risk cutoff value of high and low risk groups in different GBM patients’ datasets, so our model can be used to predict survival risk for a new individual patient of glioblastoma.
We also found that GBM patients with IDH wild type, MGMT promoter unmethylation, Mesenchymal subtype, TERT mutation have higher risk scores, which means these are all negative factors. The model can distinguish the high-risk subgroup and the low-risk subgroup in each clinical subtype.
Our model was more robust and effective compared with the existing four gene-related signatures, maybe because we used three steps to selected survival-related genes and constructed the model, trying to search for survival-related gene combinations as best as possible and reduce the redundancy in genes to construct the optimal model.
There are some limitations to our work. First of all, our differential expression analysis only contained a very limited number of normal samples. The number of genes included in the study is only more than 12,000, which may overlook some potential mRNAs. Secondly, this study did not explore the protein expression of these 16 genes and their prognostic effects. Thirdly, the reliability of the scoring model needs further clinical and experimental verification. Fourthly, the genes expression data we used were only generated by Affymetrix human genome U133A array Plate Set and Affymetrix Gene Chip Human Genome U133 Plus 2.0 Array Plate Set, our model may not be suitable for data generated by other chips, because the data generated by different types of chips may be very different.
Acknowledgments
The authors sincerely acknowledge the patients who provided research samples, their families, and the public databases: TCGA, GEO.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biomedicines10020317/s1, Table S1: Demographic and clinical characteristics of patients, Table S2: Differentially expressed genes, Table S3: Survival related genes, Table S4: Univariate and Multivariate analyses of clinical and molecular factors for overall survival of GBM patients in TCGA cohort and GSE16011 cohort, Figure S1: Comparisons of Kaplan-Meier analysis, ROC curves of different models in validation cohorts, Figure S2: Prognostic significance of clinical indicators and risk scores in the TCGA cohort, Figure S3: Prognostic significance of clinical indicators and risk scores of the GSE16011 cohort.
Author Contributions
Z.Y. and L.L. designed the study. Z.Y. implemented the algorithm. Z.Y., M.D. and L.L. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (61772375, 61936013, 71921002), The National Social Science Fund of China (18ZDA325), National Key R&D Program of China (2019YFC0120003), Natural Science Foundation of Hubei Province of China (2019CFA025); and Independent Research Project of School of Information Management Wuhan University (413100032).
Institutional Review Board Statement
Ethical review and approval were waived for this study, due to the data of the article is obtained from the public database, and ethical approval has been applied.
Informed Consent Statement
Patient consent was waived due to the data of the article being obtained from the public database, and informed consent statements had been applied.
Data Availability Statement
The data sets used in this study can be downloaded from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE4412(GSE4412), accessed on 20 August 2021, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE4271(GSE4271), accessed on 20 August 2021, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE16011(GSE16011), accessed on 20 August 2021, https://portal.gdc.cancer.gov/(TCGA), accessed on 20 August 2021, their published articles including additional files.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Louis D.N., Perry A., Wesseling P., Brat D.J., Cree I.A., Figarella-Branger D., Hawkins C., Ng H.K., Pfister S.M., Reifenberger G., et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neurooncology. 2021;23:1231–1251. doi: 10.1093/neuonc/noab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ostrom Q.T., Patil N., Cioffi G., Waite K., Kruchko C., Barnholtz-Sloan J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2013–2017. Neurooncology. 2020;22:iv1–iv96. doi: 10.1093/neuonc/noaa200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan A.C., Ashley D.M., Lopez G.Y., Malinzak M., Friedman H.S., Khasraw M. Management of glioblastoma: State of the art and future directions. CA Cancer J. Clin. 2020;70:299–312. doi: 10.3322/caac.21613. [DOI] [PubMed] [Google Scholar]
- 5.Xu Y.Y., Gao P., Sun Y., Duan Y.R. Development of targeted therapies in treatment of glioblastoma. Cancer Biol. Med. 2015;12:223–237. doi: 10.7497/j.issn.2095-3941.2015.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stupp R., Mason W.P., van den Bent M.J., Weller M., Fisher B., Taphoorn M.J., Belanger K., Brandes A.A., Marosi C., Bogdahn U., et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 7.Sturm D., Witt H., Hovestadt V., Khuong-Quang D.A., Jones D.T., Konermann C., Pfaff E., Tonjes M., Sill M., Bender S., et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 8.Ceccarelli M., Barthel F.P., Malta T.M., Sabedot T.S., Salama S.R., Murray B.A., Morozova O., Newton Y., Radenbaugh A., Pagnotta S.M., et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noushmehr H., Weisenberger D.J., Diefes K., Phillips H.S., Pujara K., Berman B.P., Pan F., Pelloski C.E., Sulman E.P., Bhat K.P., et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brennan C.W., Verhaak R.G., McKenna A., Campos B., Noushmehr H., Salama S.R., Zheng S., Chakravarty D., Sanborn J.Z., Berman S.H., et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P., et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim Y.W., Koul D., Kim S.H., Lucio-Eterovic A.K., Freire P.R., Yao J., Wang J., Almeida J.S., Aldape K., Yung W.K. Identification of prognostic gene signatures of glioblastoma: A study based on TCGA data analysis. Neurooncology. 2013;15:829–839. doi: 10.1093/neuonc/not024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckel-Passow J.E., Lachance D.H., Molinaro A.M., Walsh K.M., Decker P.A., Sicotte H., Pekmezci M., Rice T., Kosel M.L., Smirnov I.V., et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N. Engl. J. Med. 2015;372:2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan W., Zhang W., You G., Zhang J., Han L., Bao Z., Wang Y., Liu Y., Jiang C., Kang C., et al. Molecular classification of gliomas based on whole genome gene expression: A systematic report of 225 samples from the Chinese Glioma Cooperative Group. Neurooncology. 2012;14:1432–1440. doi: 10.1093/neuonc/nos263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.William A.F., Edmundo C.-V., Zixing F., Steve H., Timothy C., Linda M.L., Paul S.M., Stanley F.N. Gene Expression Profiling of Gliomas Strongly Predicts Survival. Cancer Res. 2004;64:6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 16.Phillips H.S., Kharbanda S., Chen R., Forrest W.F., Soriano R.H., Wu T.D., Misra A., Nigro J.M., Colman H., Soroceanu L., et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 17.Gravendeel L.A.M., Kouwenhoven M.C.M., Gevaert O., de Rooi J.J., Stubbs A.P., Duijm J.E., Daemen A., Bleeker F.E., Bralten L.B.C., Kloosterhof N.K., et al. Intrinsic Gene Expression Profiles of Gliomas Are a Better Predictor of Survival than Histology. Cancer Res. 2009;69:9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Q., Guan G., Cheng P., Cheng W., Yang L., Wu A. Characterization of an endoplasmic reticulum stress-related signature to evaluate immune features and predict prognosis in glioma. J. Cell Mol. Med. 2021;25:3870–3884. doi: 10.1111/jcmm.16321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin W., Tang G., Zhou Q., Cao Y., Li H., Fu X., Wu Z., Jiang X. Expression Profile Analysis Identifies a Novel Five-Gene Signature to Improve Prognosis Prediction of Glioblastoma. Front. Genet. 2019;10:419. doi: 10.3389/fgene.2019.00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q.W., Liu H.J., Zhao Z., Zhang Y., Wang Z., Jiang T., Bao Z.S. Prognostic Correlation of Autophagy-Related Gene Expression-Based Risk Signature in Patients with Glioblastoma. Onco. Targets Ther. 2020;13:95–107. doi: 10.2147/OTT.S238332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao M., Cai J., Yuan Y., Shi Y., Wu H., Liu Q., Yao Y., Chen L., Dang W., Zhang X., et al. A four-gene signature-derived risk score for glioblastoma: Prospects for prognostic and response predictive analyses. Cancer Biol. Med. 2019;16:595–605. doi: 10.20892/j.issn.2095-3941.2018.0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan Y., Zhang J.H., Zhao L., Guo J.C., Wang S., Zhao Y., Tao S., Wang H., Zhu Y.B. A robust two-gene signature for glioblastoma survival prediction. J. Cell Biochem. 2020;121:3593–3605. doi: 10.1002/jcb.29653. [DOI] [PubMed] [Google Scholar]
- 23.Jeffrey T., Leek W.E.J., Hilary S., Parker Elana J., Fertig Andrew E., Jaffe Y.Z., John D. Storey and Leonardo Collado Torres. sva: Surrogate Variable Analysis. Bioconductor; 2020. R Package Version 3.38.0. [Google Scholar]
- 24.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y., Liu X., Guan G., Zhao W., Zhuang M. A Risk Classification System with Five-Gene for Survival Prediction of Glioblastoma Patients. Front. Neurol. 2019;10:745. doi: 10.3389/fneur.2019.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dailey A.L. Metabolomic Bioinformatic Analysis. Methods Mol. Biol. 2017;1606:341–352. doi: 10.1007/978-1-4939-6990-6_22. [DOI] [PubMed] [Google Scholar]
- 27.George B., Seals S., Aban I. Survival analysis and regression models. J. Nucl. Cardiol. 2014;21:686–694. doi: 10.1007/s12350-014-9908-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang E., Hou X., Hou B., Zhang M., Song Y. A risk prediction model of DNA methylation improves prognosis evaluation and indicates gene targets in prostate cancer. Epigenomics. 2020;12:333–352. doi: 10.2217/epi-2019-0349. [DOI] [PubMed] [Google Scholar]
- 29.Zhang C., Zhang Z., Zhang G., Zhang Z., Luo Y., Wang F., Wang S., Che Y., Zeng Q., Sun N., et al. Clinical significance and inflammatory landscapes of a novel recurrence-associated immune signature in early-stage lung adenocarcinoma. Cancer Lett. 2020;479:31–41. doi: 10.1016/j.canlet.2020.03.016. [DOI] [PubMed] [Google Scholar]
- 30.Tibshirani R. The lasso method for variable selection in the Cox model. Stat. Med. 1997;16:385–395. doi: 10.1002/(SICI)1097-0258(19970228)16:4<385::AID-SIM380>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 31.Cheng Q., Huang C., Cao H., Lin J., Gong X., Li J., Chen Y., Tian Z., Fang Z., Huang J. A Novel Prognostic Signature of Transcription Factors for the Prediction in Patients With GBM. Front. Genet. 2019;10:906. doi: 10.3389/fgene.2019.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engebretsen S., Bohlin J. Statistical predictions with glmnet. Clin. Epigenetics. 2019;11:123. doi: 10.1186/s13148-019-0730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen R., Liu B., Li X., Yu T., Xu K., Ma J. Development and validation of an immune gene-set based prognostic signature for soft tissue sarcoma. BMC Cancer. 2021;21:144. doi: 10.1186/s12885-021-07852-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan H., Parsons D.W., Jin G., McLendon R., Rasheed B.A., Yuan W., Kos I., Batinic-Haberle I., Jones S., Riggins G.J., et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng X., Chang F., Zhang X., Rothman V.L., Tuszynski G.P. G-protein coupled receptor-associated sorting protein 1 (GASP-1), a ubiquitous tumor marker. Exp. Mol. Pathol. 2012;93:111–115. doi: 10.1016/j.yexmp.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 36.Wang Q., Cheng W., Li M., Ren H., Hu X., Deng W., Li M., Ma X., Zhao L., Wang Y., et al. The CHRM3 gene is implicated in abnormal thalamo-orbital frontal cortex functional connectivity in first-episode treatment-naive patients with schizophrenia. Psychol. Med. 2016;46:1523–1534. doi: 10.1017/S0033291716000167. [DOI] [PubMed] [Google Scholar]
- 37.Wang N., Yao M., Xu J., Quan Y., Zhang K., Yang R., Gao W.Q. Autocrine Activation of CHRM3 Promotes Prostate Cancer Growth and Castration Resistance via CaM/CaMKK-Mediated Phosphorylation of Akt. Clin. Cancer Res. 2015;21:4676–4685. doi: 10.1158/1078-0432.CCR-14-3163. [DOI] [PubMed] [Google Scholar]
- 38.Liu F., Arias-Vasquez A., Sleegers K., Aulchenko Y.S., Kayser M., Sanchez-Juan P., Feng B.J., Bertoli-Avella A.M., van Swieten J., Axenovich T.I., et al. A genomewide screen for late-onset Alzheimer disease in a genetically isolated Dutch population. Am. J. Hum. Genet. 2007;81:17–31. doi: 10.1086/518720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Preuschhof C., Heekeren H.R., Li S.C., Sander T., Lindenberger U., Backman L. KIBRA and CLSTN2 polymorphisms exert interactive effects on human episodic memory. Neuropsychologia. 2010;48:402–408. doi: 10.1016/j.neuropsychologia.2009.09.031. [DOI] [PubMed] [Google Scholar]
- 40.Peters I., Dubrowinskaja N., Hennenlotter J., Antonopoulos W.I., Von Klot C.A.J., Tezval H., Stenzl A., Kuczyk M.A., Serth J. DNA methylation of neural EGFL like 1 (NELL1) is associated with advanced disease and the metastatic state of renal cell cancer patients. Oncol. Rep. 2018;40:3861–3868. doi: 10.3892/or.2018.6732. [DOI] [PubMed] [Google Scholar]
- 41.Nakamura R., Oyama T., Tajiri R., Mizokami A., Namiki M., Nakamoto M., Ooi A. Expression and regulatory effects on cancer cell behavior of NELL1 and NELL2 in human renal cell carcinoma. Cancer Sci. 2015;106:656–664. doi: 10.1111/cas.12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao C.L., Zhang Q., Kong D.Y., Wu D., Su C.L., Tong J.X., Chen F., Zhang Q.F. MALDI-TOF Mass Array Analysis of Nell-1 Promoter Methylation Patterns in Human Gastric Cancer. Biomed Res. Int. 2015;2015:136941. doi: 10.1155/2015/136941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slovak M.L., Bedell V., Hsu Y.H., Estrine D.B., Nowak N.J., Delioukina M.L., Weiss L.M., Smith D.D., Forman S.J. Molecular Karyotypes of Hodgkin and Reed-Sternberg Cells at Disease Onset Reveal Distinct Copy Number Alterations in Chemosensitive versus Refractory Hodgkin Lymphoma. Clin. Cancer Res. 2011;17:3443–3454. doi: 10.1158/1078-0432.CCR-10-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhai Y., Wei R., Sha S., Lin C., Wang H., Jiang X., Liu G. Effect of NELL1 on lung cancer stemlike cell differentiation. Oncol. Rep. 2019;41:1817–1826. doi: 10.3892/or.2019.6954. [DOI] [PubMed] [Google Scholar]
- 45.An N., Zhao Y.Q., Lan H.T., Zhang M., Yin Y., Yi C. SEZ6L2 knockdown impairs tumour growth by promoting caspase-dependent apoptosis in colorectal cancer. J. Cell. Mol. Med. 2020;24:4223–4232. doi: 10.1111/jcmm.15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J.S., Kim H.Y., Won B., Kang S.W., Kim Y.N., Jang H. SEZ6L2 Is an Important Regulator of Drug-Resistant Cells and Tumor Spheroid Cells in Lung Adenocarcinoma. Biomedicines. 2020;8:500. doi: 10.3390/biomedicines8110500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maruya S.I., Myers J.N., Weber R.S., Rosenthal D.I., Lotan R., El-Naggar A.K. ICAM-5 (telencephalin) gene expression in head and neck squamous carcinoma tumorigenesis and perineural invasion! Oral. Oncol. 2005;41:580–588. doi: 10.1016/j.oraloncology.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 48.Tian L., Lappalainen J., Autero M., Hanninen S., Rauvala H., Gahmberg C.G. Shedded neuronal ICAM-5 suppresses T-cell activation. Blood. 2008;111:3615–3625. doi: 10.1182/blood-2007-09-111179. [DOI] [PubMed] [Google Scholar]
- 49.Yang J., Yang Q. Identification of Core Genes and Screening of Potential Targets in Glioblastoma Multiforme by Integrated Bioinformatic Analysis. Front. Oncol. 2020;10:615976. doi: 10.3389/fonc.2020.615976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alvaro C.G., Braz J.M., Bernstein M., Hamel K.A., Craik V., Yamanaka H., Basbaum A.I. Hippocalcin-like 4, a neural calcium sensor, has a limited contribution to pain and itch processing. PLoS ONE. 2020;15:e0226289. doi: 10.1371/journal.pone.0226289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu R.T., Yu Q., Zhou S.D., Yin Y.X., Hu R.G., Lu H.P., Hu B.L. Co-expression Network Analysis Reveals Novel Genes Underlying Alzheimer’s Disease Pathogenesis. Front. Aging Neurosci. 2020;12:605961. doi: 10.3389/fnagi.2020.605961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fromer M., Roussos P., Sieberts S.K., Johnson J.S., Kavanagh D.H., Perumal T.M., Ruderfer D.M., Oh E.C., Topol A., Shah H.R., et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yemni E.A., Monies D., Alkhairallah T., Bohlega S., Abouelhoda M., Magrashi A., Mustafa A., AlAbdulaziz B., Alhamed M., Baz B., et al. Integrated Analysis of Whole Exome Sequencing and Copy Number Evaluation in Parkinson’s Disease. Sci. Rep. 2019;9:3344. doi: 10.1038/s41598-019-40102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rademakers G., Massen M., Koch A., Draht M.X., Buekers N., Wouters K.A.D., Vaes N., De Meyer T., Carvalho B., Meijer G.A., et al. Identification of DNA methylation markers for early detection of CRC indicates a role for nervous system-related genes in CRC. Clin. Epigenet. 2021;13:80. doi: 10.1186/s13148-021-01067-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jarvela T.S., Lam H.A., Helwig M., Lorenzen N., Otzen D.E., McLean P.J., Maidment N.T., Lindberg I. The neural chaperone proSAAS blocks alpha-synuclein fibrillation and neurotoxicity. Proc. Natl. Acad. Sci. USA. 2016;113:E4708–E4715. doi: 10.1073/pnas.1601091113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Steenoven I., Koel-Simmelink M.J.A., Vergouw L.J.M., Tijms B.M., Piersma S.R., Pham T.V., Bridel C., Ferri G.L., Cocco C., Noli B., et al. Identification of novel cerebrospinal fluid biomarker candidates for dementia with Lewy bodies: A proteomic approach. Mol. Neurodegener. 2020;15:36. doi: 10.1186/s13024-020-00388-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Henssen A.G., Koche R., Zhuang J., Jiang E., Reed C., Eisenberg A., Still E., MacArthur I.C., Rodriguez-Fos E., Gonzalez S., et al. PGBD5 promotes site-specific oncogenic mutations in human tumors. Nat. Genet. 2017;49:1005–1014. doi: 10.1038/ng.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henssen A.G., Reed C., Jiang E., Garcia H.D., von Stebut J., MacArthur I.C., Hundsdoerfer P., Kim J.H., de Stanchina E., Kuwahara Y., et al. Therapeutic targeting of PGBD5-induced DNA repair dependency in pediatric solid tumors. Sci. Transl. Med. 2017;9:eaam9078. doi: 10.1126/scitranslmed.aam9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Broce I., Karch C.M., Wen N., Fan C.C., Wang Y., Tan C.H., Kouri N., Ross O.A., Hoglinger G.U., Muller U., et al. Immune-related genetic enrichment in frontotemporal dementia: An analysis of genome-wide association studies. PLoS Med. 2018;15:e1002487. doi: 10.1371/journal.pmed.1002487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Desestret V., Ciccarino P., Ducray F., Criniere E., Boisselier B., Labussiere M., Polivka M., Idbaih A., Kaloshi G., von Deimling A., et al. Prognostic stratification of gliomatosis cerebri by IDH1(R132H) and INA expression. J. Neuro-Oncol. 2011;105:219–224. doi: 10.1007/s11060-011-0587-4. [DOI] [PubMed] [Google Scholar]
- 61.Liu H., Povysheva N., Rose M.E., Mi Z., Banton J.S., Li W., Chen F., Reay D.P., Barrionuevo G., Zhang F., et al. Role of UCHL1 in axonal injury and functional recovery after cerebral ischemia. Proc. Natl. Acad. Sci. USA. 2019;116:4643–4650. doi: 10.1073/pnas.1821282116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pfoh R., Lacdao I.K., Saridakis V. Deubiquitinases and the new therapeutic opportunities offered to cancer. Endocr.-Relat. Cancer. 2015;22:T35–T54. doi: 10.1530/ERC-14-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu S.J., Gonzalez-Prieto R., Zhang M.D., Geurink P.P., Kooij R., Iyengar P.V., van Dinther M., Bos E., Zhang X.B., Le Devedec S.E., et al. Deubiquitinase Activity Profiling Identifies UCHL1 as a Candidate Oncoprotein That Promotes TGF beta-Induced Breast Cancer Metastasis. Clin. Cancer Res. 2020;26:1460–1473. doi: 10.1158/1078-0432.CCR-19-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Q., Liao J., He Z., Su Y., Lin D., Xu L., Xu H., Lin J. LHX6 Affects Erlotinib Resistance and Migration of EGFR-Mutant Non-Small-Cell Lung Cancer HCC827 Cells Through Suppressing Wnt/beta-Catenin Signaling. Oncotargets Ther. 2020;13:10983–10994. doi: 10.2147/OTT.S258896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bi Q.J., Men X.J., Han R., Li G.L. LHX6 inhibits the proliferation, invasion and migration of breast cancer cells by modulating the PI3K/Akt/mTOR signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2018;22:3067–3073. doi: 10.26355/eurrev_201805_15066. [DOI] [PubMed] [Google Scholar]
- 66.Hu Z., Xie L. LHX6 inhibits breast cancer cell proliferation and invasion via repression of the Wnt/beta-catenin signaling pathway. Mol. Med. Rep. 2015;12:4634–4639. doi: 10.3892/mmr.2015.3997. [DOI] [PubMed] [Google Scholar]
- 67.Chen H.Q., Zhao J., Li Y., Huang Y.J., Chen D.J., He L.X., Wang L.Q., Zheng C.F., Wang J., Cao J., et al. Epigenetic inactivation of LHX6 mediated microcystin-LR induced hepatocarcinogenesis via the Wnt/beta-catenin and P53 signaling pathways. Environ. Pollut. 2019;252:216–226. doi: 10.1016/j.envpol.2019.05.049. [DOI] [PubMed] [Google Scholar]
- 68.Khan S., Lu X., Huang Q., Tang J., Weng J., Yang Z., Lv M., Xu X., Xia F., Zhang M., et al. IGFBP2 Plays an Essential Role in Cognitive Development during Early Life. Adv. Sci. 2019;6:1901152. doi: 10.1002/advs.201901152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li T., Forbes M.E., Fuller G.N., Li J., Yang X., Zhang W. IGFBP2: Integrative hub of developmental and oncogenic signaling network. Oncogene. 2020;39:2243–2257. doi: 10.1038/s41388-020-1154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun L.H., Zhang X.B., Song Q.Q., Liu L., Forbes E., Tian W.J., Zhang Z.X., Kang Y.A., Wang H.M., Fleming J.B., et al. IGFBP2 promotes tumor progression by inducing alternative polarization of macrophages in pancreatic ductal adenocarcinoma through the STAT3 pathway. Cancer Lett. 2021;500:132–146. doi: 10.1016/j.canlet.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Riihila P., Viiklepp K., Nissinen L., Farshchian M., Kallajoki M., Kivisaari A., Meri S., Peltonen J., Peltonen S., Kahari V.M. Tumour-cell-derived complement components C1r and C1s promote growth of cutaneous squamous cell carcinoma. Br. J. Dermatol. 2020;182:658–670. doi: 10.1111/bjd.18095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bozic T., Lin Q., Frobel J., Wilop S., Hoffmann M., Muller-Tidow C., Brummendorf T.H., Jost E., Wagner W. DNA-methylation in C1R is a prognostic biomarker for acute myeloid leukemia. Clin. Epigenet. 2015;7:116. doi: 10.1186/s13148-015-0153-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Metz R.L., Patel P.S., Hameed M., Bryan M., Rameshwar P. Role of human HGFIN/nmb in breast cancer. Breast Cancer Res. 2007;9:R58. doi: 10.1186/bcr1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schmidt-Kittler O., Ragg T., Daskalakis A., Granzow M., Ahr A., Blankenstein T.J.F., Kaufmann M., Diebold J., Arnholdt H., Muller P., et al. From latent disseminated cells to overt metastasis: Genetic analysis of systemic breast cancer progression. Proc. Natl. Acad. Sci. USA. 2003;100:7737–7742. doi: 10.1073/pnas.1331931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rich J.N., Shi Q., Hjelmeland M., Cummings T.J., Kuan C.T., Bigner D.D., Counter C.M., Wang X.F. Bone-related genes expressed in advanced malignancies induce invasion and metastasis in a genetically defined human cancer model. J. Biol. Chem. 2003;278:15951–15957. doi: 10.1074/jbc.M211498200. [DOI] [PubMed] [Google Scholar]
- 76.Metz R.L., Yehia G., Fernandes H., Donnelly R.J., Rameshwar P. Cloning and characterization of the 5 flanking region of the HGFIN gene indicate a cooperative role among p53 and cytokine-mediated transcription factors-Relevance to cell cycle regulation. Cell Cycle. 2005;4:315–322. doi: 10.4161/cc.4.2.1468. [DOI] [PubMed] [Google Scholar]
- 77.Weterman M.A.J., Ajubi N., Vandinter I.M.R., Degen W.G.J., Vanmuijen G.N.P., Ruiter D.J., Bloemers H.P.J. Nmb, a Novel Gene, Is Expressed in Low-Metastatic Human-Melanoma Cell-Lines and Xenografts. Int. J. Cancer. 1995;60:73–81. doi: 10.1002/ijc.2910600111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets used in this study can be downloaded from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE4412(GSE4412), accessed on 20 August 2021, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE4271(GSE4271), accessed on 20 August 2021, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE16011(GSE16011), accessed on 20 August 2021, https://portal.gdc.cancer.gov/(TCGA), accessed on 20 August 2021, their published articles including additional files.