Abstract
Standard treatment of hepatitis delta virus (HDV) infection remains pegylated‐interferon alfa (peg‐IFNα) in most centers, which is not only associated with rather low efficacy but several adverse events. Hepatitis B core‐related antigen (HBcrAg) is linked to intrahepatic covalently closed circular DNA levels and has previously been suggested as response predictor in IFN‐based treatment of hepatitis B virus (HBV) mono‐infection. This study aimed to investigate the value of HBcrAg in the management of patients with HBV/HDV co‐infection undergoing peg‐IFNα treatment. The Hep‐Net‐International‐Delta‐Hepatitis‐Intervention Trial‐2 study included 120 patients co‐infected with HBV/HDV. Patients were treated for 96 weeks with peg‐IFNα and either tenofovir or placebo. Ninety‐nine patients with HDV‐RNA results 24 weeks after end of treatment (FU24) were included in this analysis, of whom 32 patients (32.3%) had undetectable HDV RNA at FU24. HBcrAg was measured at baseline, week 12, 24, 48, 96, and FU24. HBcrAg levels showed no significant correlation with HDV RNA but were significantly linked to treatment outcome. HBcrAg levels < 4.5 log IU/mL at baseline, week 24, and week 48 had high negative predictive value (NPV) for achieving undetectable HDV RNA at FU24 (81.8%, 87.1% and 95.0%, respectively). Similarly, HBcrAg levels at week 96 were significantly higher in patients with viral relapse until FU24 (3.0 vs. 3.63 log IU/mL; P = 0.0089). Baseline, week 24, and week 48 HBcrAg levels were also associated with the likelihood of achieving HBsAg level < 100 IU/mL at FU24 (HBcrAg < 3.0 log IU/mL: NPV 91.7%, 90.4% and 92.3%, respectively). Test statistics improved when combining HBcrAg with additional viral and clinical parameters. Conclusion: HBcrAg is linked to treatment response to peg‐IFNα in patients with HBV/HDV co‐infection and could be a promising marker to determine treatment futility.
Abbreviations
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- BL
baseline
- cccDNA
covalently closed circular DNA
- EOT
end of treatment/week 96
- FU24
follow‐up 24 weeks after end of treatment
- HBcAg
hepatitis B core antigen
- HBcrAg
hepatitis B core‐related antigen
- HBeAg
hepatitis B e antigen
- HBsAg
hepatitis B surface antigen
- HBV
hepatitis B virus
- HDV
hepatitis D virus
- HIDIT‐II
Hep‐Net‐International‐Delta‐Hepatitis‐Intervention Trial‐2
- IQR
interquartile range
- NPV
negative predictive value
- Peg‐IFNα
pegylated‐interferon alfa
- PPV
positive predictive value
- ROC
receiving operator characteristic
- TDF
tenofovir
- w12
treatment week 12
- w24
treatment week 24
- w48
treatment week 48
- w96
treatment week 96/end of treatment
Chronic hepatitis D virus (HDV) infection, also known as hepatitis delta, is the most severe form of chronic viral hepatitis and frequently leads to liver cirrhosis, hepatic decompensation, and the development of hepatocellular carcinoma.( 1 ) HDV is a defective satellite virus, and the RNA genome only encodes the HDV antigen. Therefore, virus life cycle and the production of infectious viral particles depend on host polymerases and the presence of hepatitis B surface antigen (HBsAg).( 2 ) As the HDV RNA does not encode for a viral enzyme, development of antiviral treatment is challenging. Current standard treatment regimen consists of pegylated‐interferon alfa (peg‐IFNα), which induces HDV‐RNA response (undetectable HDV RNA at 24 weeks after treatment) in about one quarter of patients. Of note, even among the responders, HDV‐RNA negativity is not durable in many cases.( 3 , 4 , 5 ) New drugs such as the entry inhibitor bulevirtide and lonafarnib are currently tested in phase 3 studies, and bulevirtide has recently been approved based on results of phase 2 studies in Europe.( 6 , 7 , 8 ) However, broad access to bulevirtide remains uncertain at this point (such as due to treatment costs), and it may not be available in most centers for quite some time. Moreover, it is currently unclear whether combination treatment with peg‐IFNα will be required to achieve functional cure and avoid indefinite treatment duration.( 9 ) However, treatment with peg‐IFNα leads to a variety of severe side effects, whereas the number of patients achieving a maintained virological response remains rather limited, so far. Therefore, biomarkers to predict antiviral response and to identify patients with a reasonable chance of benefiting from antiviral therapy are urgently needed.( 10 )
Recently, hepatitis B core‐related antigen (HBcrAg) has been identified as an additional virological marker of hepatitis B virus (HBV) infection. HBcrAg simultaneously measures denatured hepatitis B e antigen (HBeAg), HBV core antigen (HBcAg) and an artificial core‐related protein.( 11 ) It has been shown that HBcrAg levels correlate with HBV DNA and, most importantly, with intrahepatic transcriptional activity of covalently closed circular DNA (cccDNA) in both HBeAg‐positive and HBeAg‐negative patients.( 12 ) Furthermore, levels of HBcrAg could be linked to different phases of chronic HBV infection and are suggested as an additional marker to distinguish between HBeAg‐negative patients infected with chronic HBV, with active and inactive disease.( 13 , 14 ) Importantly, HBcrAg has also been suggested as a useful marker to predict response to peg‐IFNα therapy in both HBeAg‐positive and HBeAg‐negative patients.( 15 )
Because presence of HBV is essential for HDV infection and persistence, the analyses of promising serological and molecular markers of HBV infection, in particular those reflecting the transcriptional activity of intrahepatic cccDNA, deserve further attention in HBV/HDV co‐infection, especially in the setting of antiviral treatment. So far, HBcrAg levels have not been analyzed in HBV/HDV co‐infected patients during antiviral treatment. Therefore, we aimed to investigate the clinical utility of HBcrAg levels before and during treatment with peg‐IFNα in patients with HDV treated in the international, multicenter Hep‐Net International Delta Hepatitis Intervention Trial‐2 (HIDIT‐II) trial.
Methods
Study Cohort
The HIDIT‐II was a prospective international, multicenter study including 120 HBV/HDV co‐infected patients who were randomly assigned to either receive peg‐IFNα plus tenofovir (TDF) or plus placebo for 96 weeks. Patients with chronic HDV infection and compensated liver disease without human immunodeficiency virus or hepatitis C virus infection were eligible for the study. Further information is available at www.clinicaltrials.gov (NCT00932971).( 4 )
A total of 99 patients with HDV‐RNA results available at posttreatment week 24 were included in our analysis. For 21 patients, HDV‐RNA results at the 24‐week follow‐up (FU24) were not available; those patients were excluded from this study. Baseline characteristics for the excluded patients are available in Supporting Table S1.
Study Endpoints
According to the primary endpoint of the HIDIT‐II trial and the recommendations from the 2019 European Association for the Study of the Liver–American Association for the Study of Liver Diseases HBV Treatment Endpoints Conference, three endpoints were chosen for the current study( 10 , 16 ):
Undetectable HDV RNA 24 weeks after end of treatment
Undetectable HDV RNA at the end of treatment
HBsAg decline to a level < 100 IU/mL 24 weeks after end of treatment (as a predictor of HBsAg loss during longer follow‐up)
Measurement of Serological and Virological Markers
HBcrAg was measured at baseline (BL), treatment week 12 (w12), 24 (w24), 48 (w48), 96/end of treatment (w96/EOT), and FU24 using the Lumipulse G (Fujirebio, Ghent, Belgium) according to the manufacturer’s instructions. The assay’s validated linear measurement range is from 3 log to 7 log U/mL. Samples with HBcrAg above 7 log U/mL were diluted and retested in order to calculate the quantitative HBcrAg level. HBV DNA, HBsAg, and HDV RNA were measured as part of the HIDIT‐II study at the central laboratory at BL and the respective study time points including w12, w24, w48, w96, and FU24. HDV‐RNA measurement was performed using the Cobas TaqMan system with an inhouse assay (Roche Diagnostics, Mannheim, Germany). The test shows linearity over a range from 3 × 102 to 107 copies/mL and a lower limit of detection of 15 copies/mL.( 4 , 17 )
Statistics
Data were expressed as frequencies (%) for categorical variables or as median (with interquartile range [IQR]) for quantitative variables. Categorical data were analyzed with the Fisher ´s exact test. Mann‐Whitney U test was used to analyze continuous data. HBcrAg cutoff values were previously defined as ≤ 3.0 log U/mL (lower limit of detection) and ≤ 4.5 log U/mL and compared with cutoffs selected by generating receiving operator characteristic (ROC) curves and Youden index. Univariate and multivariate analyses were performed by logistic regression analysis using backward variable selection for multivariate analysis. Variables that presented a P value < 0.05 in the univariate analysis were included in the multivariate analysis. However, total number of variables in the multivariate analysis was restricted to 1 per 10 events with regard to the primary endpoint.( 18 ) Statistical analyses were performed using Excel software (version 16.37 for Mac; Microsoft Cooperation, Redmond, WA), GraphPad Prism (version 7.05 for Windows; GraphPad Software, La Jolla, CA, www.graphpad.com), and SPSS software (version 26; SPSS Inc., Chicago, IL).
Ethics
The study protocol of the HIDIT‐II study, including associated scientific analysis, was approved by the respective national competent authorities and by the ethics committees at each participating institution (EudraCT No. 2008‐005560‐13). Each patient provided written informed consent.
Results
Study Cohort and Treatment Outcome
In total, 99 patients with available HDV‐RNA results at week 24 at EOT (FU24) were included in the analysis. Median age of the patients was 47 years (IQR 42‐60), most were male (66.7%), and 37.4% had evidence of liver cirrhosis. Further detailed baseline characteristics of patients included in the analysis are found in Table 1.
TABLE 1.
Baseline Characteristics
| Patients, Total (n = 99) | Peg‐IFNα Plus TDF (n = 48) | Peg‐IFNα Plus Placebo (n = 51) | |
|---|---|---|---|
| Sex | |||
| Male | 66 (66.7%) | 34 (70.8%) | 32 (62.7%) |
| Female | 35 (33.3%) | 14 (29.2%) | 19 (37.3%) |
| Age | |||
| Median (IQR) | 47 (42‐60) | 46 (39.3‐58) | 52 (45‐64) |
| HDV RNA | |||
| <300 copies/mL | 5 (5.1%) | 3 (6.3%) | 2 (3.9%) |
| Median log10 copies/mL (IQR) | 5.17 (4.27‐5.76) | 5.15 (4.17‐5.93) | 5.07 (3.94‐5.58) |
| >105 copies/mL | 53 (53.5%) | 26 (54.2%) | 27 (52.9%) |
| HBV DNA | |||
| Negative | 8 (8.1%) | 2 (4.2%) | 6 (11.8%) |
| Median log10 IU/mL (IQR) | 1.93 (1.3‐3.22) | 1.56 (1.30‐3.18) | 2.13 (1.3‐3.42) |
| <100 IU/mL | 38 (38.4%) | 24 (50%) | 14 (27.5%) |
| >2,000 UI/mL | 20 (20.2%) | 8 (16.7%) | 12 (23.5%) |
| HBsAg | |||
| Median log10 IU/mL (IQR) | 3.92 (3.51‐4.2) | 3.98 (2.42‐4.2) | 3.89 (3.38‐4.21) |
| <1,000 IU/mL | 10 (10.1%) | 4 (8.3%) | 6 (11.8%) |
| HBeAg | |||
| Positive | 18 (18.2%) | 10 (20.8%) | 8 (15.7%) |
| Missing | 10 (10.1%) | 5 (10.4%) | 5 (9.8%) |
| HBcrAg | |||
| Median log U/mL (IQR) | 4.11 (3‐4.76) | 4.18 (3.31‐4.84) | 3.76 (3‐4.57) |
| ≤3 log U/mL | 27 (27.3%) | 7 (14.6%) | 20 (39.2%) |
| 3‐4.5 log U/mL | 39 (39.4%) | 23 (47.9%) | 16 (31.4%) |
| >4.5 log U/mL | 33 (33.3%) | 18 (37.5%) | 15 (29.4%) |
| ALT | |||
| Median IU/L (IQR) | 85 (58‐149) | 79 (54‐136) | 96.5 (60.5‐170.3) |
| AST | |||
| Median IU/L (IQR) | 58 (44‐99) | 56 (42.5‐96.5) | 60 (47‐99) |
| Bilirubin | |||
| Median µmol/L (IQR) | 12 (8‐17) | 13 (8‐17) | 12 (8‐17) |
| Albumin | |||
| Median g/L (IQR) | 41 (39‐45) | 42 (39.8‐45) | 41 (39‐44) |
| Thrombocytes | |||
| Median 1,000/µL (IQR) | 175 (128‐199) | 175 (132‐215.5) | 175 (123‐198) |
| INR | |||
| Median (IQR) | 1.07 (1.01‐1.15) | 1.07 (1.01‐1.16) | 1.07 (1‐1.14) |
| Cirrhosis | 37 (37.4%) | 19 (39.6%) | 18 (35.3%) |
Quantitative values are depicted as median with IQR; categorical values are depicted as numbers and percentages.
Abbreviation: INR, international normalized ratio.
At FU24, HDV RNA was undetectable in 32 patients (32.3%) (responders), and 67 patients (67.7%) showed detectable HDV RNA at FU24 (nonresponders). At w96 (EOT), 45 patients had undetectable HDV RNA, but 16 patients (35.6%) experienced viral relapse until FU24. Of note, 3 of the 54 patients with detectable HDV RNA at w96 spontaneously cleared the virus and showed undetectable HDV RNA at FU24 (5.6%).
Correlation of HBcrAg With HDV RNA, HBV DNA, and HBsAg at BL and During Treatment
Levels of HBcrAg were determined at BL, w12, w24, w48, w96, and FU24. At baseline, most patients showed HBcrAg levels < 5 log IU/mL (81% vs. 18%), and 27% even had a value ≤ 3 log IU/mL (Supporting Fig. S1). During treatment, levels of HBcrAg showed an increase from BL to w12, which was followed by a more prominent decline until w48 compared with the second half of treatment duration. Median HBcrAg levels did not differ significantly between end of treatment and FU24 (3.45 vs. 3.36 IU/mL; P = 0.996) (Fig. 1). When only including HBeAg‐negative patients, median levels of HBcrAg were overall lower, but kinetics during treatment remained unchanged including the previously described increase at w12 (Supporting Fig. S2). Median HBcrAg levels at baseline, w12, w24, w48, w96, and FU24 did not differ significantly between patients receiving either peg‐IFNα and placebo or peg‐IFNα and TDF (data not shown).
FIG. 1.
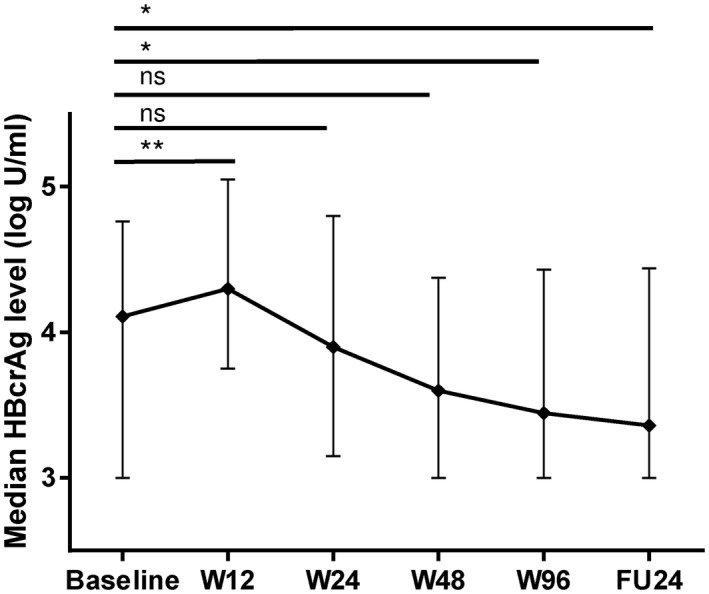
HBcrAg kinetics during treatment. Median HBcrAg levels (log U/mL) with IQR at different time points of treatment in the total cohort. *P ≤ 0.05; **P ≤ 0.01. Abbreviation: ns, not significant.
Correlations of HBcrAg and HDV RNA, HBV DNA, and HBsAg are depicted in Fig. 2. At the different time points there was no or only a weak positive correlation between HBcrAg and HDV RNA. The correlation slightly increased during treatment, and the highest correlation could be observed at EOT (w96) (r: 0.48; P < 0.0001). Moderate positive correlations were detected between HBcrAg and HBV DNA or HBsAg at baseline, w12, w24, w48, w96, and FU24 (Fig. 2). No or only very weak negative correlations were observed between HBcrAg and aspartate aminotransferase (AST), alanine aminotransferase (ALT), or AST‐to–platelet ratio index score (Supporting Fig. S3).
FIG. 2.
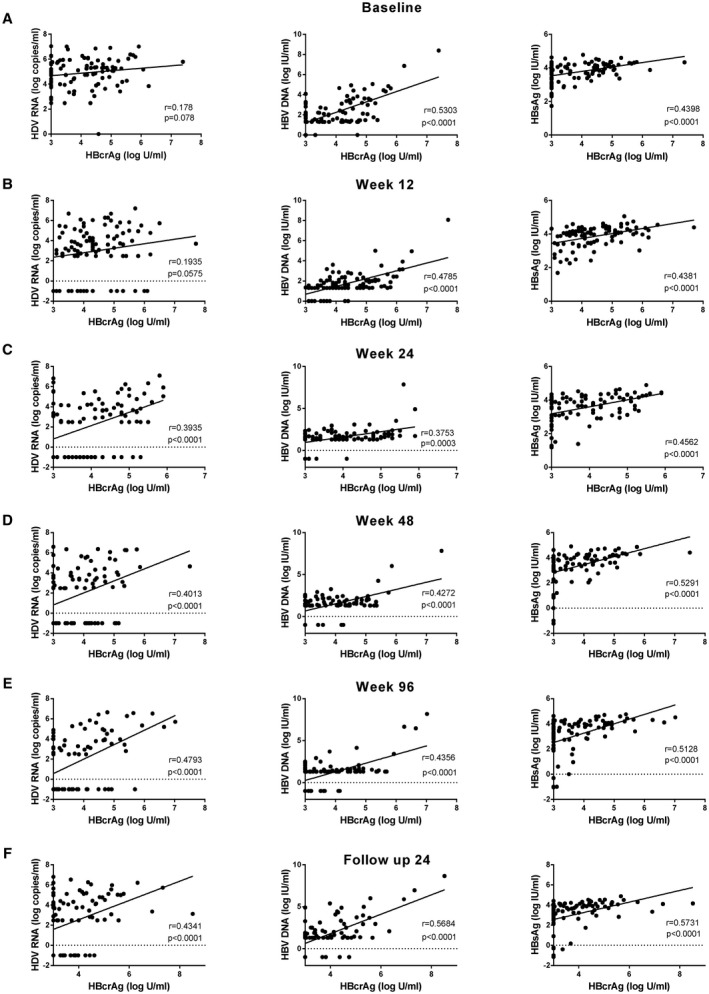
Correlation of HBcrAg levels with HDV RNA, HBV DNA, and HBsAg levels at baseline (A), w12 (B), w24 (C), w48 (D), w96 (E), and FU24 (F).
When separating according to treatment regimens, slightly stronger positive correlations between HBcrAg and HDV RNA at BL, w24, and w48 were observed in patients treated with peg‐IFNα plus placebo compared to treatment with peg‐IFNα plus TDF. The correlation between HBcrAg and HDV RNA was slightly stronger at w96 and FU24 than at w48 and similar between both treatment groups. At w48 and w96, there was no or only a weak correlation between HBcrAg and HBV DNA or HBsAg in the TDF group, whereas persistent moderate positive correlations were observed in the placebo group (Supporting Fig. S4).
Predictive Value of Baseline and On‐Treatment HBcrAg Levels for Undetectable HDV RNA at EOT
Median levels of HBcrAg at baseline, w24, and w48 differed significantly between patients with undetectable and those with detectable HDV RNA at the end of treatment (3.56 vs. 4.34 log U/mL [P = 0.0197], 3.7 vs. 4.1 [P = 0.0142], and 3.4 vs. 3.92 IU/mL [P = 0.0309], respectively) (Fig. 3A‐F). A similar trend was observed for HBeAg‐negative patients. For these patients, median levels of HBcrAg at baseline were significantly lower in EOT responders compared with nonresponders (3.4 vs. 4.15 log U/mL; P = 0.0398) and numerically lower at w24 and w48 (Supporting Fig. S5).
FIG. 3.
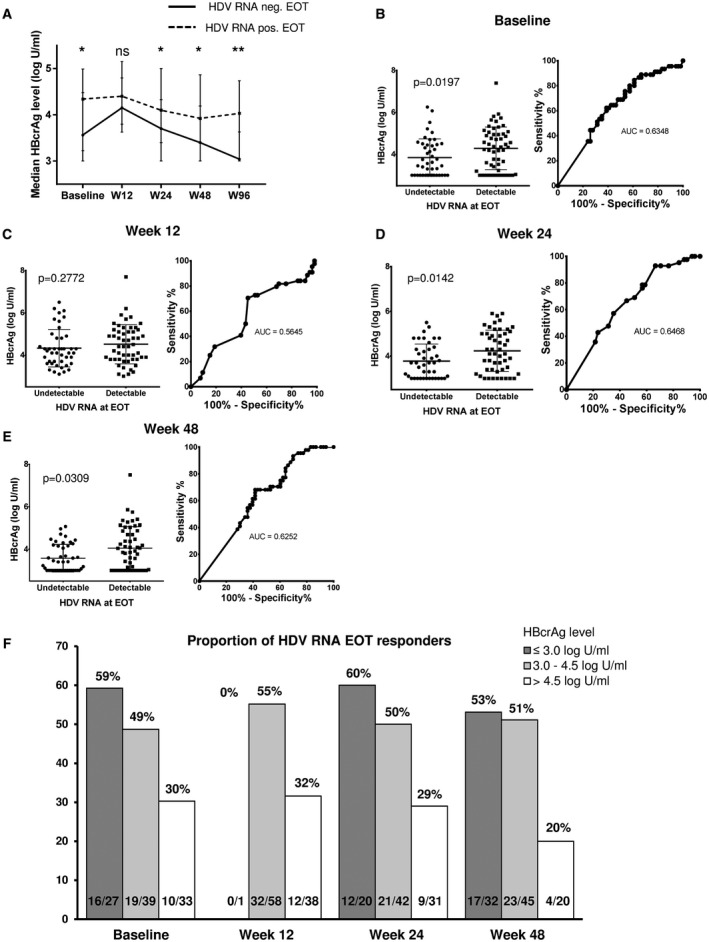
(A) Median HBcrAg levels (log U/mL) at different time points during treatment separated into responders at EOT (undetectable HDV RNA at w96; n = 45) and nonresponders at EOT (n = 54). Median HBcrAg levels and ROC curves at baseline (B), w12 (C), w24 (D), and w48 (E) for undetectable HDV RNA at EOT. (F) Proportions of EOT responders according to HBcrAg level at each time point. *P ≤ 0.05; **P ≤ 0.01. Abbreviations: AUC, area under the curve; ns, not significant.
Only 10 (30%) of the 33 patients with baseline HBcrAg levels > 4.5 log U/mL were HDV RNA–negative at EOT, whereas this was the case for 49% (19 of 39) and even 59% (16 of 27) of patients with baseline HBcrAg levels between 3.0 and 4.5 log U/mL and ≤ 3.0 log U/mL, respectively (Fig. 3F). Using the Youden index, 4.72 log U/mL was identified as optimal HBcrAg cutoff to distinguish EOT responders and nonresponders. Only 6 of 26 patients (23%) with a baseline HBcrAg level > 4.72 log U/mL were HDV RNA–negative at EOT, whereas this was the case in 53% of those with levels ≤ 4.72 log IU/mL (sensitivity 86.7%, specificity 37.0%, positive predictive value [PPV] 53.4%, negative predictive value [NPV] 76.9%, P = 0.0110) (Table 2).
TABLE 2.
Performance of Proposed Cutoff Values
| Time Point | Cutoff HBcrAg (log U/mL) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | P Value | |
|---|---|---|---|---|---|---|---|
| HDV RNA neg. EOT | Baseline | ≤3.0 | 35.6 | 79.3 | 59.3 | 59.7 | 0.1144 |
| ≤4.5 | 77.8 | 42.6 | 53.0 | 69.7 | 0.0354 | ||
| ≤4.72* | 86.7 | 37.0 | 53.4 | 76.9 | 0.0110 | ||
| w12 | ≤3.0 | 0 | 98.1 | 0 | 54.2 | >0.999 | |
| ≤4.35* | 70.5 | 54.7 | 56.4 | 69.1 | 0.0146 | ||
| ≤4.5 | 72.7 | 49.1 | 54.2 | 68.4 | 0.0371 | ||
| w24 | ≤3.0 | 28.6 | 84.3 | 60.0 | 58.9 | 0.2042 | |
| ≤4.5 | 78.6 | 43.1 | 53.2 | 71.0 | 0.0299 | ||
| ≤4.85* | 92.9 | 33.3 | 53.4 | 85.0 | 0.0023 | ||
| w48 | ≤3.0 | 38.6 | 38.6 | 71.7 | 53.1 | 0.3858 | |
| ≤3.73* | 90.9 | 30.2 | 53.1 | 80.0 | 0.0122 | ||
| ≤4.5 | 68.2 | 58.5 | 57.7 | 68.9 | 0.0138 | ||
| HDV RNA neg.FU24 | Baseline | ≤3.0 | 40.6 | 79.1 | 48.2 | 73.6 | 0.0537 |
| ≤4.5 | 81.3 | 40.3 | 39.4 | 81.8 | 0.0411 | ||
| ≤4.70* | 90.6 | 37.3 | 40.9 | 89.3 | 0.0040 | ||
| w12 | ≤3.0 | 0 | 98.5 | 0 | 67.7 | >0.999 | |
| ≤4.35* | 74.2 | 51.5 | 41.8 | 81.0 | 0.0272 | ||
| ≤4.5 | 74.2 | 45.5 | 39.0 | 79.0 | 0.0770 | ||
| w24 | ≤3.0 | 34.5 | 84.4 | 50.0 | 74.0 | 0.0564 | |
| ≤4.05* | 79.3 | 59.4 | 46.9 | 86.4 | 0.0007 | ||
| ≤4.5 | 86.2 | 42.2 | 40.3 | 87.1 | 0.0086 | ||
| w48 | ≤3.0 | 53.1 | 76.9 | 53.1 | 76.9 | 0.0054 | |
| ≤3.63* | 75.0 | 60.0 | 48.0 | 83.0 | 0.0013 | ||
| ≤4.5 | 96.9 | 29.2 | 40.3 | 95.0 | 0.0026 | ||
| w96 † | ≤3.0 | 58.6 | 68.8 | 77.3 | 47.8 | 0.1205 | |
| ≤3.56* | 86.2 | 56.3 | 78.1 | 69.2 | 0.0051 | ||
| ≤4.5 | 100 | 25.0 | 70.7 | 100 | 0.0122 | ||
| HBsAg < 100 IU/mL FU24 | Baseline | ≤3.0* | 57.1 | 77.4 | 29.6 | 91.7 | 0.0193 |
| ≤4.5 | 78.6 | 36.9 | 17.2 | 91.2 | 0.3674 | ||
| w12 | ≤3.0 | 0 | 98.8 | 0 | 86.5 | >0.9999 | |
| ≤3.75* | 46.2 | 78.6 | 25.0 | 90.4 | 0.0809 | ||
| ≤4.5 | 76.9 | 40.5 | 16.7 | 91.9 | 0.3586 | ||
| w24 | ≤3.0 | 41.7 | 81.5 | 25.0 | 90.4 | 0.1238 | |
| ≤3.85* | 83.3 | 55.6 | 21.7 | 95.7 | 0.0142 | ||
| ≤4.5 | 83.3 | 34.6 | 15.9 | 93.3 | 0.3253 | ||
| w48 | ≤3.0 | 64.3 | 72.3 | 28.1 | 92.3 | 0.0122 | |
| ≤3.40* | 78.6 | 63.1 | 26.2 | 94.6 | 0.0068 | ||
| ≤4.5 | 100 | 24.1 | 18.2 | 100 | 0.0671 | ||
| w96 | ≤3.0 | 78.6 | 67.9 | 29.0 | 95.0 | 0.0020 | |
| ≤3.13* | 100 | 26.2 | 18.4 | 100 | 0.0349 | ||
| ≤4.5 | 85.7 | 66.7 | 30.0 | 96.6 | 0.0003 |
The P value was calculated by using the Fisher’s exact test to compare categorical data, in this case treatment response according to HBcrAg cutoff values. Bold values indicates P ≤ 0.05.
Identified by Youden index.
Only patients with negative HDV RNA at w96 were included (n = 45) to assess test performances for relapse at FU24.
Similarly, patients with HBcrAg levels ≤ 3.0 log U/mL, 3.0‐4.5 log U/mL, and > 4.5 log U/mL at w48 achieved undetectable HDV RNA later on at EOT in 53% (17 of 32), 51% (23 of 45) and 20% (4 of 20), respectively (Fig. 3F). Optimal w48 cutoff suggested using the Youden index was 3.73 U/mL (sensitivity 90.9%, specificity 30.2%, PPV 53.1%, NPV 80.0%, P = 0.0122) (Table 2). The highest NPV was detected for the cutoff of 4.85 U/mL at w24. Only 3 patients (3 of 20, 15%) with an HBcrAg level > 4.85 U/mL were HDV RNA–negative at EOT, whereas this was the case for 53% (39 of 73) of patients with HBcrAg levels ≤ 4.85 U/mL (sensitivity 92.9%, specificity 33.3%, PPV 53.4%, NPV 85.0%, P = 0.0023).
Predictive Value of HBcrAg Levels for Undetectable HDV RNA at Posttreatment Week 24
Patients with undetectable HDV RNA at FU24 showed significantly lower median HBcrAg levels at BL (3.43 vs. 4.33 log U/mL; P = 0.0072), w24 (3.3 vs. 4.25; P = 0.0005), w48 (3.0 vs. 4.04 log U/mL; P = 0.0003), and w96 (3.0 vs. 3.83 log U/mL; P = 0.0001) (Fig. 4A,C‐F). Comparable to the kinetics in EOT responders, HBcrAg levels of patients with negative HDV RNA at FU24 increased from BL to w12. The median change of HBcrAg levels from BL to w12 (delta HBcrAg) was significantly higher in treatment responders compared with nonresponders and explains the consecutive more pronounced HBcrAg decline from w12 to w24 in treatment responders (Fig. 4B). However, overall fraction of increase or decrease of HBcrAg from baseline to w12 did not differ significantly between responders and nonresponders (increase in responders vs. nonresponders: 84% [26 of 31] versus 68% [45 of 66]; P = 0.1412). If only including HBeAg‐negative patients, comparable results were obtained (Supporting Fig. S6).
FIG. 4.
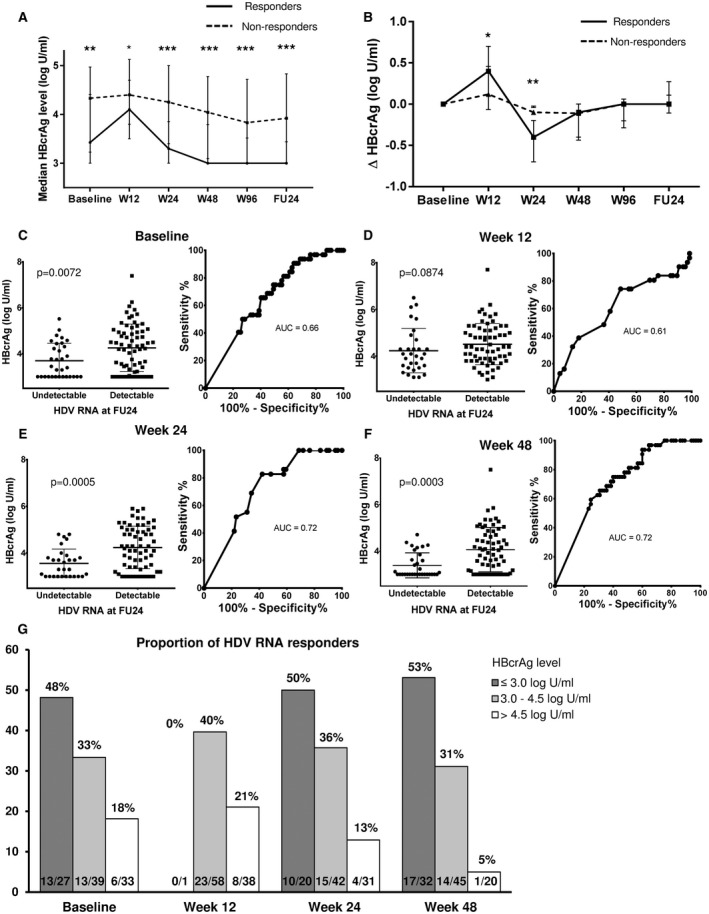
(A) Median HBcrAg levels (log U/mL) at different time points during treatment separated into responders at FU24 (undetectable HDV RNA at FU24; n = 32) and nonresponders at FU24 (n = 67). (B) Median change of HBcrAg (delta HBcrAg) at different time points during treatment. Median HBcrAg levels and ROC curves at baseline (C), w12 (D), w24 (E), and w48 (F) for undetectable HDV RNA at FU24. (G) Proportions of FU24 responders according to HBcrAg level at each time point. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
At baseline, 18% (6 of 33) of the patients with HBcrAg levels > 4.5 log U/mL were HDV RNA–negative at w24, whereas this was the case in 33% (13 of 39) and 48% (13 of 27) of the patients with levels between 3.0 and 4.5 log U/mL and ≤ 3.0 log U/mL, respectively (Fig. 4G). A baseline HBcrAg level ≤ 4.70 log U/mL was identified as optimal threshold for response prediction linked to a high NPV. Of note, only 3 of 28 patients (11%) with a baseline HBcrAg level > 4.70 log U/mL were HDV RNA–negative at FU24 (sensitivity 90.6%, specificity 37.3%, PPV 40.9%, NPV 89.3%, P = 0.004) (Table 2). At w24, 50% (10 of 20), 36% (15 of 42), and 13% (4 of 31) of patients with HBcrAg levels ≤ 3.0 log U/mL, 3.0‐4.5 log U/mL, or > 4.5 log U/mL became HDV RNA–negative at FU24, respectively (Fig. 4G). Optimal w24 cutoff according to the Youden index was 4.05 log U/mL (sensitivity 79.3, specificity 59.4, PPV 46.9%, NPV 86.4%, P = 0.0007) (Table 2). At w48, only 5% of patients (1 of 20) with HBcrAg levels > 4.5 log U/mL became HDV RNA–negative at FU24, whereas this was the case for 53% (17 of 32) of patients with HBcrAg levels ≤ 3 log U/mL (Fig. 4G). The optimal w48 cutoff according to the Youden index was 3.63 log IU/mL (sensitivity 75%, specificity 60.0%, PPV 48.0%, NPV 83.0%, P = 0.0013) (Table 2).
Moreover, HBcrAg levels at EOT were significantly linked to viral relapse until FU24. A total of 45 patients had undetectable HDV‐RNA levels at EOT, and viral relapse until FU24 was detected in 16 of these patients (35.6%). HBcrAg levels at w96 were significantly lower in sustained responders compared with relapsers (3.0 vs. 3.63 log U/mL; P = 0.0089). Of note, all patients with an HBcrAg level > 4.5 log U/mL at EOT experienced viral relapse (sensitivity 100%, specificity 25.0%, PPV 70.7%, NPV 100%, P = 0.0122) (Table 2 and Supporting Fig. S7).
Predictive Value of HBcrAg Levels for HBsAg Decline
Apart from undetectable HDV RNA, HBsAg loss is defined as an endpoint of finite antiviral treatment of HBV/HDV co‐infection.( 16 ) Because HBsAg loss is rare, HBsAg decline to a low level of < 100 IU/mL was selected in the current study, which is frequently proposed as a predictor of HBsAg loss during long‐time follow‐up.( 19 )
Data analysis showed that HBcrAg levels at baseline, w24, and w48 differed significantly between patients with HBsAg levels < 100 IU/mL and ≥ 100 IU/mL at FU24 (3.0 vs. 4.83 log U/mL [P = 0.0451]; 3.1 vs. 4.0 log U/mL [P = 0.0148]; and 3.0 vs. 3.77 log U/mL [P = 0.0062], respectively) (Fig. 5).
FIG. 5.
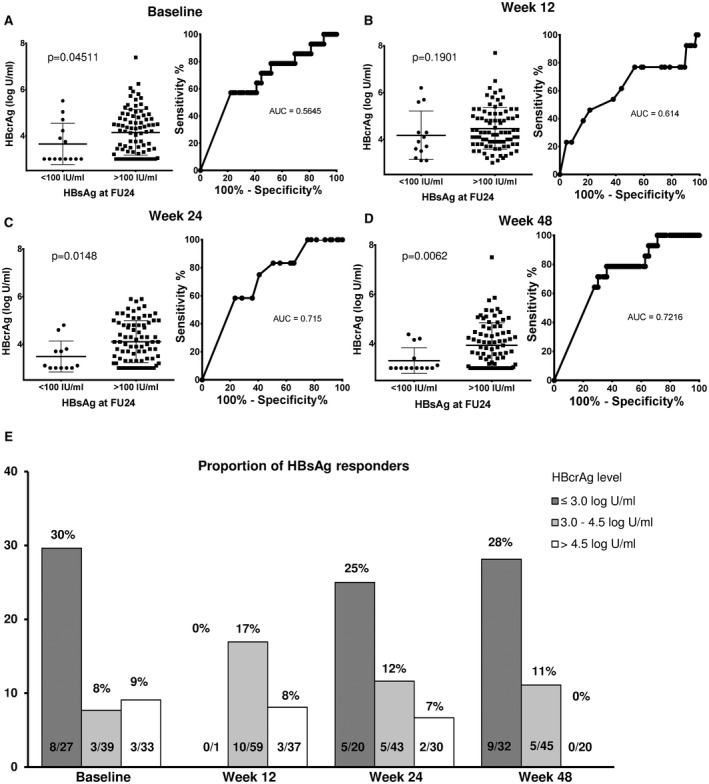
(A‐D) Median HBcrAg levels (log U/mL) at different time points during treatment according to HBsAg response (HBsAg < 100 IU/mL) at FU24. Median HBcrAg levels and ROC curves at baseline (A), w12 (B), w24 (C), and w48 (D) for HBsAg < 100 IU/mL at FU24. (E) Proportions of HBsAg responders according to HBcrAg level at each time point.
A baseline HBcrAg threshold of 3.0 log U/mL had a high NPV for achieving HBsAg levels < 100 IU/mL at w24 (NPV 91.7%). Only 8% (6 of 72) of patients with baseline HBcrAg levels > 3.0 U/mL showed HBsAg levels < 100 IU/mL at FU24, whereas this was documented in 30% (8 of 27) of the patients with baseline HBcrAg levels ≤ 3.0 log U/mL (sensitivity 57.1%, specificity 77.7%, PPV 29.6%, NPV 91.7%, P = 0.0190). At w24 and w48, NPVs of the cutoff ≤ 3.0 log U/mL were comparable to that at baseline (90.4% and 92.3%, respectively). Using the Youden index, 3.85 log U/mL was identified as the optimal cutoff to distinguish between HBsAg responders and nonresponders at w24. Only 2 of 47 patients (4%) with HBcrAg levels > 3.85 log U/mL showed HBsAg levels < 100 IU/mL at FU24 (sensitivity 83.3%, specificity 55.6%, PPV 21.7%, NPV 95.7%, P = 0.0142). Comparable test statistics were achieved for the cutoff of 3.4 log U/mL at w48 identified by Youden index. All patients with HBcrAg levels > 4.5 log U/mL at week 48 (n = 20) showed HBsAg levels > 100 IU/mL at FU24 (Fig. 5 and Table 2).
Combination of Virological Parameters for the Prediction of Treatment Response
To further assess the potential additional value of HBcrAg, we decided to analyze other predictors of treatment response. Kinetics of HBsAg levels were comparable to HBcrAg kinetics during treatment, and median levels of HBsAg differed significantly between treatment responders at EOT and FU24 (Supporting Fig. S8). Patients with negative HDV RNA at FU24 showed a more prominent median HBsAg decline from BL to week 24 than nonresponders (delta HBsAg −0.2598 log IU/mL vs. 0 log IU/mL; P = 0.0044). In contrast to HBcrAg kinetics, no significant increase from BL to week 12 was detectable for HBsAg. However, concordant or discordant change in HBcrAg and HBsAg levels from baseline to week 12 or 24 was not associated with treatment response. The Youden index cutoffs to predict treatment response at FU24 were calculated and are found in Supporting Table S2. Overall, NPVs for treatment response at different time points during treatment were comparable but slightly lower than those calculated for HBcrAg (HBcrAg vs. HBsAg: baseline 89.3% vs. 82.1%, w12 81.0% vs. 78.7%, w24 86.4% vs. 79.7%, and w48 83% vs. 85.3%).
When combining proposed cutoffs for HBcrAg and HBsAg, test statistics to predict treatment response at FU24 were further improved (Table 3). Only 2 of 21 patients (9.5%) with baseline HBsAg levels > 3.745 log IU/mL and HBcrAg levels > 4.7 log U/mL became HDV RNA–negative at FU24 (sensitivity 93.1%, specificity 32.2%, PPV 40.3%, NPV 90.5%, P = 0.0084). At w24, 88% (30 of 34) of patients with HBsAg levels > 3.166 log IU/mL and HBcrAg levels > 4.05 log U/mL remained HDV RNA–positive at FU24, while only 4 patients (12%) were HDV RNA–negative (sensitivity 85.7%, specificity 51.7%, PPV 46.2%, NPV 88.2%, P = 0.0009). Comparable results were achieved for combined cutoffs at w48.
TABLE 3.
Performance of Proposed Cutoff Values
| Time Point | Cutoff HBsAg (log IU/mL) and HBcrAg (log U/mL) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | P Value | |
|---|---|---|---|---|---|---|---|
| HDV RNA neg. FU24 | Baseline | HBsAg ≤ 3.745 and/or HBcrAg ≤ 4.7 | 93.1 | 32.2 | 40.3 | 90.5 | 0.0084 |
| w12 | HBsAg ≤ 3.873 and/or HBcrAg ≤ 4.35 | 77.4 | 40.6 | 38.7 | 78.8 | 0.1088 | |
| w24 | HBsAg ≤ 3.166 and/or HBcrAg ≤ 4.05 | 85.7 | 51.7 | 46.2 | 88.2 | 0.0009 | |
| w48 | HBsAg ≤ 3.305 and/or HBcrAg ≤ 3.63 | 86.2 | 56.7 | 49.0 | 89.5 | 0.0001 | |
| w96/relapse* | HBsAg ≤ 2.803 and/or HBcrAg ≤ 3.56 | 93.1 | 50.0 | 77.1 | 80.0 | 0.0017 |
The P value was calculated by using the Fisher’s exact test to compare categorical data, in this case treatment response according to cutoff values. Bold values indicates P ≤ 0.05.
Only patients with negative HDV RNA at w96 were included to assess test performances for relapse at FU24.
HBcrAg, HBsAg, HDV RNA, and Absence of Cirrhosis as Risk Factors for Treatment Failure
To develop stopping rules before or during treatment, univariate and multivariate binary logistic regression analyses were performed to assess additional parameters associated with treatment failure, defined as HDV‐RNA persistence at FU24. Clinical and virological parameters at baseline, w12, and w24 were included in the univariate analysis. HBV‐DNA levels were excluded due to the small sample size when ruling out patients receiving TDF. Levels of HBcrAg, HBsAg, and HDV RNA at baseline, w12 (except for HBcrAg), and w24 were significantly associated with HDV‐RNA persistence at FU24 in the univariate analysis. Interestingly, presence of cirrhosis was identified as a protective parameter against treatment failure (Supporting Table S3). In the multivariate‐analysis baseline HBcrAg, baseline HDV RNA and absence of cirrhosis remained significantly associated with treatment failure. When including HBsAg in the model, baseline HBsAg and HDV RNA remained independently associated with HDV‐RNA persistence (Supporting Table S4A). In our cohort, 69% of patients (46 of 67) with HDV‐RNA persistence at FU24 would have been correctly identified as future nonresponders by applying the HBsAg cutoff at baseline (HBsAg > 3.745 log IU/mL). By additionally applying the identified cutoff for HBcrAg at baseline (HBcrAg > 4.7 log U/mL), 6 additional patients who remained HDV RNA–positive at FU24 would have been identified correctly (78%, n = 52 of 67). At treatment week 24, high levels of HBcrAg were associated with HDV‐RNA persistence. However, values did not reach statistical significance in the multivariate analysis (Supporting Table S4B). When applying the selected cutoff of HBsAg to the cohort, 76% of patients (51 of 67) would have been correctly identified as future nonresponders at w24. By additional application of the HBcrAg cutoff, 10 additional patients (n = 61 of 67 [91%]) would have been identified as future nonresponders at this on‐treatment time point. At w12, neither HBsAg nor HBcrAg remained independently associated with treatment failure (Supporting Table S4C). At the end of treatment, absence of cirrhosis, high levels of HBcrAg, and high levels of HBsAg were independently associated with viral relapse at FU24 (Supporting Table S4D). By additionally applying the HBcrAg cutoff of > 3.56 log U/mL to the HBsAg cutoff of > 2.803 log IU/mL to our cohort, 1 additional patient would have been identified to show viral relapse at FU24 (n = 15 of 16; 94%).
Discussion
In this analysis we showed that HBcrAg could serve as a promising baseline or on‐treatment marker to predict the outcome of peg‐IFNα‐based antiviral treatment in patients with HDV. At baseline and during antiviral treatment, levels of HBcrAg positively correlated with HBV DNA and HBsAg but to a lesser extend with HDV RNA. Interestingly, levels of HBcrAg differed significantly between treatment responders and non‐responders throughout the entire study period. By combining levels of HBcrAg with additional predictive parameters at baseline, treatment week 24 or end of treatment, the ratio of correctly identifying future non‐responders at FU24 increased notably.
To our knowledge, so far, only a single study analyzed the correlation of HBcrAg in HDV positive patients.( 20 ) In this study the authors analyzed virological parameters of patients with HDV to characterize the interplay among HBV, HDV, and their correlation to the severity of liver disease. In line with previous publications, high anti‐HDV immunoglobulin M levels were identified as a marker of advanced stage of chronic HDV.( 21 ) HBcrAg was associated with higher ALT levels in the univariate but not in the multivariate analysis. There was a weak positive correlation among HBcrAg and HBV DNA, HBsAg and HDV RNA, and patients with negative HDV RNA showed significantly lower levels of HBcrAg than those with detectable HDV RNA. Importantly, patients undergoing antiviral treatment had been excluded from the study, and only off‐treatment patients had been analyzed.( 20 ) Therefore, our study, which includes patients during antiviral treatment, complements the results by Ricco et al. We showed that overall the positive correlation between HBcrAg and HBV and HBsAg persists during treatment, although being more prominent in the peg‐IFNα TDF‐free treatment group. This is understandable, as TDF inhibits HBV‐DNA replication; therefore, levels of HBV DNA are reduced. In our cohort there was no correlation between HBcrAg and HDV RNA at baseline, but developed during treatment and persisted until 24 weeks after treatment, regardless of treatment regimen. One possible explanation might be that by reduced HBV replication during treatment, the cccDNA use, reflected by HBcrAg levels, is dominated by HDV. Nevertheless, the broad antiviral effect of peg‐IFNα is not included in this hypothesis, and its effect on HDV RNA remains elusive. Therefore, it remains debatable whether the changes in correlations really reflect the HBV/HDV interplay, as the complex interaction between the two viruses might hardly be completely described by these bimodal correlations. Furthermore, it is possible that the stronger positive correlation is simply due to mathematical reasons in calculating the correlation coefficient. Patients with lower HBcrAg at baseline experience a higher chance of becoming HDV RNA–negative during treatment. Therefore, the correlation of remaining (detectable) HDV‐RNA and HBcrAg levels becomes more pronounced.
Current focus of research in HBV mono‐infection is functional cure, which is closely related to the reduction of cccDNA copy numbers or transcriptional activity.( 22 ) Treatment endpoints in HDV remain a matter of debate. However, when aiming for functional and permanent cure in HDV, the cccDNA is certainly also the most important target, as it is most likely the main source of HBsAg required for HDV replication.( 10 ) Interestingly, a study by Pollicino et al. showed slightly lower amounts of cccDNA in HDV infected patients than HBV mono‐infected patients.( 23 ) However, the ratio of cccDNA to intracellular HBV DNA showed higher proportions of cccDNA in HDV‐positive patients, and HBsAg amounts per cccDNA molecule were significantly higher in the HDV‐positive patients. Furthermore, a significant correlation between levels of HDV RNA and HBV cccDNA was detected.
Treatment with peg‐IFNα has been shown to be able to induce degradation of cccDNA in HBV‐infected patients.( 24 ) However, the likelihood for durable response appears to be linked to the amount of transcriptionally active cccDNA at baseline. In chronic HBV infected patients, it has been shown that HBcrAg correlates well with the amount and transcriptional activity of HBV cccDNA.( 25 , 26 ) Moreover, HBcrAg levels correlate with HBV RNA, which also reflects levels of HBV cccDNA.( 27 ) Of note, both HBcrAg and HBV RNA have been suggested as valuable response predictors when using peg‐IFNα treatment in HBeAg‐positive and HBeAg‐negative patients mono‐infected with HBV among plenty of studies.( 27 , 28 , 29 , 30 , 31 , 32 ) However, so far no data were available with regard to HBV/HDV co‐infection. Importantly, in our study we showed that HBcrAg does not only decrease during antiviral treatment but could serve as a predictive marker of peg‐IFNα treatment response in patients with HDV. At baseline and during antiviral treatment, levels of HBcrAg showed high NPVs for achieving undetectable HDV RNA at FU24. Therefore, HBcrAg levels at baseline could serve as a helpful marker to better select patients benefiting from antiviral treatment. Importantly, patients with low chances of achieving positive treatment outcome were identified with high accuracy by combining HBcrAg with other viral and clinical markers. By combining HBcrAg with HBsAg, test statistics to predict treatment response were further improved. Not only at baseline but also during treatment, the evaluation of HBcrAg served as a marker to identify patients not benefiting from continuing antiviral therapy. At w24, HBcrAg levels showed high NPVs for achieving treatment endpoints. Therefore, HBcrAg‐based stopping rules that identify patients with high risk of treatment failure could be derived. By applying these stopping rules, patients would be spared the continuation of a probable ineffective but side effect–rich antiviral treatment. Similar to baseline, test performances were improved by combining HBcrAg and HBsAg as compared with each marker as a single parameter. This underlines the importance of virological markers reflecting HBV cccDNA transcriptional activity in the context of maintained virological response in chronic HDV infection. For the establishment of stopping rules, early on‐treatment time points are preferable. Interestingly, at w12, levels of HBcrAg and, to a lesser extent, HBsAg differed less significantly between treatment responders and nonresponders. This is due to a more pronounced increase from baseline to w12 in treatment responders, especially for HBcrAg. A recently published study showed an early on‐treatment increase in HBV DNA to be associated with suppression of HDV RNA at the end of treatment.( 33 ) The authors demonstrated that HBV DNA increase paralleled an increase of hepatic inflammation and markers of cell death (M30) in several patients. However, early on‐treatment HBcrAg levels and HDV RNA outcome after follow‐up were not assessed during this analysis. It might be possible that the observed increase of HBcrAg levels in treatment responders in our analysis might be due to apoptosis of hepatocytes.
In addition to baseline or early on‐treatment time points, the level of HBcrAg at the end of treatment could also help to predict relapse, and therefore identify patients in need of further therapeutic strategies. The documented predictive value of the HBcrAg level at the end of treatment appears to further support an important role of HBV cccDNA as indicator for a permanent cure. Moreover, it fits very well with the predictive value of HBcrAg for off‐treatment response after nucleotide/nucleoside cessation in HBV mono‐infected patients documented in some studies, which is also believed to be linked to a lower amount of transcriptional active cccDNA.( 34 , 35 , 36 )
However, the implementation of HBcrAg into clinical practice has to be taken with caution, and further studies to validate this marker and to establish and confirm specific cutoffs are needed. The HIDIT‐II trial was not designed to evaluate stopping rules during antiviral treatment. Therefore, confirmation in a validation cohort would be necessary, especially with regard to the proposed treatment algorithm. Another obvious limitation of our study is the limited follow‐up time of 24 weeks. Late relapses after FU24 are known. Unfortunately, no data extending FU24 is available in our cohort, so far. Although endpoints for HDV treatment have not yet been defined definitely, negative HDV RNA 24 weeks after the end of antiviral therapy is the preferred endpoint for finite antiviral treatment according to current recommendations.( 10 , 16 ) However, analyses of viral parameters, including HBcrAg in patients with late relapse, are of special interest and should be included in future studies. In addition, our analysis only included patients receiving peg‐IFNα as antiviral treatment. Currently, new antiviral substances like lonafarnib are being tested, and results so far are promising.( 8 , 37 ) Moreover, the entry‐inhibitor bulevirtide will certainly improve response rates, but it is possible that access to this treatment might be an important limitation.( 6 , 7 ) However, the analysis of HBcrAg in patients during antiviral treatment with these new substances would be interesting and could further evaluate the role of HBcrAg in HDV‐positive individuals.
In summary, we analyzed levels of HBcrAg in patients with HDV undergoing antiviral treatment. Significant differences were observed between treatment responders and nonresponders, and cutoff values were calculated to distinguish between these groups. Test statistics and prediction models were further improved when combining HBcrAg with additional virological and clinical markers. Further analyses are needed to validate HBcrAg as a marker for on‐treatment monitoring and to verify the identified cutoffs.
Supporting information
Supplementary Material
Supported by Fujirebio Europe, Gilead Sciences, F. Hoffmann‐La Roche and HepNet Study House.
Potential conflict of interest: Dr. Cornberg advises and is on the speakers’ bureau for Abbvie, Gilead, and MSD. He advises Novartis, Roche, and Spring Bank. He is on the speakers’ bureau for Falk. Dr. Deterding is on the speakers’ bureau for Gilead, Alnylam, MSD, Merck Falk, and AbbVie. Dr. Yurdaydin is on the speakers’ bureau and received grants from Eiger Biopharma. He consults for, advises, is on the speakers’ bureau for, and received grants from Gilead Biopharma. He also received grants from AbbVie. Dr. Maasoumy is on the speakers’ bureau for and received grants from Fujirebio. Dr. Wedemeyer consults for, advises, is on the speakers’ bureau for, is an investigator for, and received grants from AbbVie. He received grants, consults for, advises, and is on the speakers’ bureau for BioTest, He is an investigator for, received grants from, consults, and advises Bristol‐Myers Squibb, Gilead, and Novartis. He is an investigator, consults, and advises for Altimmune and MYR GmbH. He is an investigator for, consults for, advises, and is on the speakers’ bureau for Janssen. He received grants from, consults, and advises for Roche. He is an investigator for, received grants from, and is on the speakers’ bureau for Merck/MSD. He consults and advises BTG, Dicerna, Enanta, Siemens, and Vir Biotechnology and is an investigator for Transgene. Dr. Manns consults for, is on the speakers’ bureau for, and received grants from Roche, Bristol‐Myers Squibb, and Gilead. He consults for Enyo Pharma and Curevac. Dr. Sandmann received honoraria from Dr. Falk Pharma.
References
Author names in bold designate shared co‐first authorship.
- 1. Hughes SA, Wedemeyer H, Harrison PM. Hepatitis delta virus. Lancet 2011;378:73‐85. [DOI] [PubMed] [Google Scholar]
- 2. Lempp FA, Ni Y, Urban S. Hepatitis delta virus: insights into a peculiar pathogen and novel treatment options. Nat Rev Gastroenterol Hepatol 2016;13:580‐589. [DOI] [PubMed] [Google Scholar]
- 3. Wedemeyer H, Yurdaydìn C, Dalekos GN, Erhardt A, Çakaloğlu Y, Değertekin H, et al. Peginterferon plus adefovir versus either drug alone for hepatitis delta. N Engl J Med 2011;364:322‐331. [DOI] [PubMed] [Google Scholar]
- 4. Wedemeyer H, Yurdaydin C, Hardtke S, Caruntu FA, Curescu MG, Yalcin K, et al. Peginterferon alfa‐2a plus tenofovir disoproxil fumarate for hepatitis D (HIDIT‐II): a randomised, placebo controlled, phase 2 trial. Lancet Infect Dis 2019;19:275‐286. [DOI] [PubMed] [Google Scholar]
- 5. Heidrich B, Yurdaydın C, Kabaçam G, Ratsch BA, Zachou K, Bremer B, et al. Late HDV RNA relapse after peginterferon alpha‐based therapy of chronic hepatitis delta. Hepatology 2014;60:87‐97. [DOI] [PubMed] [Google Scholar]
- 6. Bogomolov P, Alexandrov A, Voronkova N, Macievich M, Kokina K, Petrachenkova M, et al. Treatment of chronic hepatitis D with the entry inhibitor myrcludex B: first results of a phase Ib/IIa study. J Hepatol 2016;65:490‐498. [DOI] [PubMed] [Google Scholar]
- 7. Loglio A, Ferenci P, Uceda Renteria SC, Tham CYL, van Bömmel F, Borghi M, et al. Excellent safety and effectiveness of high‐dose myrcludex‐B monotherapy administered for 48 weeks in HDV‐related compensated cirrhosis: a case report of 3 patients. J Hepatol 2019;71:834‐839. [DOI] [PubMed] [Google Scholar]
- 8. Koh C, Canini L, Dahari H, Zhao X, Uprichard SL, Haynes‐Williams V, et al. Oral prenylation inhibition with lonafarnib in chronic hepatitis D infection: a proof‐of‐concept randomised, double‐blind, placebo‐controlled phase 2A trial. Lancet Infect Dis 2015;15:1167‐1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wedemeyer H, Schöneweis K, Bogomolov PO, Voronkova N, Chulanov V, Stepanova T, et al. GS‐13‐Final results of a multicenter, open‐label phase 2 clinical trial (MYR203) to assess safety and efficacy of myrcludex B in cwith PEG‐interferon alpha 2a in patients with chronic HBV/HDV co‐infection. J Hepatol 2019;70:e81. [Google Scholar]
- 10. Yurdaydin C, Abbas Z, Buti M, Cornberg M, Esteban R, Etzion O, et al. Treating chronic hepatitis delta: the need for surrogate markers of treatment efficacy. J Hepatol 2019;70:1008‐1015. [DOI] [PubMed] [Google Scholar]
- 11. Kimura T, Ohno N, Terada N, Rokuhara A, Matsumoto A, Yagi S, et al. Hepatitis B virus DNA‐negative dane particles lack core protein but contain a 22‐kDa precore protein without C‐terminal arginine‐rich domain. J Biol Chem 2005;280:21713‐21719. [DOI] [PubMed] [Google Scholar]
- 12. Suzuki F, Miyakoshi H, Kobayashi M, Kumada H. Correlation between serum hepatitis B virus core‐related antigen and intrahepatic covalently closed circular DNA in chronic hepatitis B patients. J Med Virol 2009;81:27‐33. [DOI] [PubMed] [Google Scholar]
- 13. Seto W‐K, Wong D K‐H, Fung J, Huang F‐Y, Liu K S‐H, Lai C‐L, et al. Linearized hepatitis B surface antigen and hepatitis B core‐related antigen in the natural history of chronic hepatitis B. Clin Microbiol Infect 2014;20:1173‐1180. [DOI] [PubMed] [Google Scholar]
- 14. Maasoumy B, Wiegand SB, Jaroszewicz J, Bremer B, Lehmann P, Deterding K, et al. Hepatitis B core‐related antigen (HBcrAg) levels in the natural history of hepatitis B virus infection in a large European cohort predominantly infected with genotypes A and D. Clin Microbiol Infect 2015;21:606.e1‐606.e10. [DOI] [PubMed] [Google Scholar]
- 15. van Campenhout MJH, Brouwer WP, van Oord GW, Xie Q, Zhang Q, Zhang N, et al. Hepatitis B core‐related antigen levels are associated with response to entecavir and peginterferon add‐on therapy in hepatitis B e antigen‐positive chronic hepatitis B patients. Clin Microbiol Infect 2016;22:571.e5‐e9. [DOI] [PubMed] [Google Scholar]
- 16. Cornberg M, Lok A‐F, Terrault NA, Zoulim F, Berg T, Brunetto MR, et al. Guidance for design and endpoints of clinical trials in chronic hepatitis B—report from the 2019 EASL‐AASLD HBV Treatment Endpoints Conference. J Hepatol 2020;72:539‐557. [DOI] [PubMed] [Google Scholar]
- 17. Mederacke I, Bremer B, Heidrich B, Kirschner J, Deterding K, Bock T, et al. Establishment of a novel quantitative hepatitis D virus (HDV) RNA assay using the Cobas TaqMan platform to study HDV RNA kinetics. J Clin Microbiol 2010;48:2022‐2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harrell FEJ, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med 1996;15:361‐387. [DOI] [PubMed] [Google Scholar]
- 19. Cornberg M, Wong VW‐S, Locarnini S, Brunetto M, Janssen HLA, Chan HL‐Y. The role of quantitative hepatitis B surface antigen revisited. J Hepatol 2017;66:398‐411. [DOI] [PubMed] [Google Scholar]
- 20. Ricco G, Popa DC, Cavallone D, Iacob S, Salvati A, Tabacelia D, et al. Quantification of serum markers of hepatitis B (HBV) and delta virus (HDV) infections in patients with chronic HDV infection. J Viral Hepat 2018;25:911‐919. [DOI] [PubMed] [Google Scholar]
- 21. Wranke A, Heidrich B, Ernst S, Calle Serrano B, Caruntu FA, Curescu MG, et al. Anti‐HDV IgM as a marker of disease activity in hepatitis delta. PLoS ONE 2014;9:e101002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lucifora J, Protzer U. Attacking hepatitis B virus cccDNA—the holy grail to hepatitis B cure. J Hepatol 2016;64(Suppl. 1):S41‐S48. [DOI] [PubMed] [Google Scholar]
- 23. Pollicino T, Raffa G, Santantonio T, Gaeta GB, Iannello G, Alibrandi A, et al. Replicative and transcriptional activities of hepatitis B virus in patients coinfected with hepatitis B and hepatitis delta viruses. J Virol 2011;85:432‐439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, et al. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 2014;343:1221‐1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wong D‐H, Seto W‐K, Cheung K‐S, Chong C‐K, Huang F‐Y, Fung J, et al. Hepatitis B virus core‐related antigen as a surrogate marker for covalently closed circular DNA. Liver Int 2017;37:995‐1001. [DOI] [PubMed] [Google Scholar]
- 26. Testoni B, Lebossé F, Scholtes C, Berby F, Miaglia C, Subic M, et al. Serum hepatitis B core‐related antigen (HBcrAg) correlates with covalently closed circular DNA transcriptional activity in chronic hepatitis B patients. J Hepatol 2019;70:615‐625. [DOI] [PubMed] [Google Scholar]
- 27. Farag MS, van Campenhout MJH, Pfefferkorn M, Fischer J, Deichsel D, Boonstra A, et al. Hepatitis B virus RNA as early predictor for response to pegylated interferon alpha in HBeAg‐negative chronic hepatitis B. Clin Infect Dis 2020;72:202‐211. [DOI] [PubMed] [Google Scholar]
- 28. Jansen L, Kootstra NA, van Dort KA, Takkenberg RB, Reesink HW, Zaaijer HL. Hepatitis B virus pregenomic RNA is present in virions in plasma and is associated with a response to pegylated interferon alfa‐2a and nucleos(t)ide analogues. J Infect Dis 2016;213:224‐232. [DOI] [PubMed] [Google Scholar]
- 29. van Bömmel F, van Bömmel A, Krauel A, Wat C, Pavlovic V, Yang L, et al. Serum HBV RNA as a predictor of peginterferon alfa‐2a response in patients with HBeAg‐positive chronic hepatitis B. J Infect Dis 2018;218:1066‐1074. [DOI] [PubMed] [Google Scholar]
- 30. Ma G, Lou B, Lv F, Zhao D, Zhang Z, Chen Y. HBcrAg and pg RNA and the therapeutic effect in HBeAg‐positive patients receiving anti‐viral therapy, baseline serum HBV‐RNA is a powerful predictor of response. J Viral Hepat 2020;27:837‐846. [DOI] [PubMed] [Google Scholar]
- 31. Chuaypen N, Posuwan N, Payungporn S, Tanaka Y, Shinkai N, Poovorawan Y, et al. Serum hepatitis B core‐related antigen as a treatment predictor of pegylated interferon in patients with HBeAg‐positive chronic hepatitis B. Liver Int 2016;36:827‐836. [DOI] [PubMed] [Google Scholar]
- 32. Zhang M, Li G, Shang J, Pan C, Zhang M, Yin Z, et al. Rapidly decreased HBV RNA predicts responses of pegylated interferons in HBeAg‐positive patients: a longitudinal cohort study. Hepatol Int 2020;14:212‐224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Anastasiou OE, Yurdaydin C, Maasoumy B, Hardtke S, Caruntu FA, Curescu MG, et al. A transient early HBV‐DNA increase during PEG‐IFNα therapy of hepatitis D indicates loss of infected cells and is associated with HDV‐RNA and HBsAg reduction. J Viral Hepat 2021;28:410‐419. [DOI] [PubMed] [Google Scholar]
- 34. Sonneveld MJ, van Oord GW, van Campenhout MJ, De Man RA, Janssen HLA, de Knegt RJ, et al. Relationship between hepatitis B core‐related antigen levels and sustained HBeAg seroconversion in patients treated with nucleo(s)tide analogues. J Viral Hepat 2019;26:828‐834. [DOI] [PubMed] [Google Scholar]
- 35. Hsu Y‐C, Nguyen MH, Mo L‐R, Wu M‐S, Yang T‐H, Chen C‐C, et al. Combining hepatitis B core‐related and surface antigens at end of nucleos(t)ide analogue treatment to predict off‐therapy relapse risk. Aliment Pharmacol Ther 2019;49:107‐115. [DOI] [PubMed] [Google Scholar]
- 36. Carey I, Gersch J, Wang B, Moigboi C, Kuhns M, Cloherty G, et al. Pre‐genomic HBV RNA and HBcrAg predict outcomes in HBeAg negative chronic hepatitis B patients suppressed on nucleos(t)ide analogue therapy. Hepatology 2020;72:42‐57. [DOI] [PubMed] [Google Scholar]
- 37. Yurdaydin C, Keskin O, Kalkan Ç, Karakaya F, Çalişkan A, Karatayli E, et al. Optimizing lonafarnib treatment for the management of chronic delta hepatitis: the LOWR HDV‐1 study. Hepatology 2018;67:1224‐1236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


