Abstract
Specific and sensitive tests for the detection and typing of group A rotavirus strains are needed for a more comprehensive knowledge of the epidemiology of rotaviral infection. In this study 500 stool specimens taken from 1996 to 1998 from children with acute diarrhea in Buenos Aires were examined. Group A rotavirus was unequivocally demonstrated in 62% of the samples tested by enzyme-linked immunosorbent assay (ELISA) for detection of VP6 antigen, polyacrylamide gel electrophoresis of double-stranded RNA, and reverse transcription-PCR (RT-PCR) for amplification of the VP7:G (1,062 bp) and VP4:P (876 bp) genes. Only five positive specimens were found by RT-PCR but not by ELISA. G and P typing was carried out by nested amplification of variable sequences of the VP7 and the VP4 genes with six G- and five P-type-specific primers (multiplex PCR). Results obtained by this method showed the prevalence of the following G and P types: G1, 39%; G2, 43%; G4, 4%; P[8], 16%; P[4], 71%. Unexpectedly, the G-P type combination most frequently found was G2P[4] (43%) rather than G1P[8] (12%), which is the most commonly found worldwide. Unusual strains of the type G1P[4] accounted for 14% of the total, while mixed infections with more than one type were found in 10% of the samples. Detection of fecal rotavirus-specific immunoglobulin M (IgM) and IgA antibodies in consecutive samples of two patients taken at daily intervals demonstrated that high levels of IgM and IgA antibodies were detected on day 1 after the onset of disease and that the samples remained positive for about 10 days, after which virus shedding was no longer observed. Multiplex PCR offers a sensitive and specific alternative to determine the prevalence of group A rotavirus G and P types and to identify the emergence of uncommon strains, whereas detection of fecal IgM and IgA antibodies represents a useful supplement to virus detection for the diagnosis of current or recently acquired infections.
Rotaviruses have been recognized as the major etiologic agents of acute gastroenteritis in infants and young children worldwide (14, 20, 35). Rotavirus serotypes are specified by two outer capsid proteins, VP4 and VP7, encoded by different genome segments (13, 32). VP7 and VP4 proteins elicit, independently, neutralizing antibodies and specify the virus G (outer shell glycoprotein) and P (for protease-susceptible protein) serotypes, respectively. VP4, the product of gene 4, is the viral hemagglutinin and appears to be responsible for restriction of growth in tissue culture and virulence in experimental animals (50). Proteolytic cleavage of this protein enhances rotavirus infectivity (41). Rotavirus serotypes have been established on the basis of a 20-fold or higher difference in reciprocal neutralization titers with hyperimmune homologous and heterologous antisera (57–59). Because the genes encoding these proteins segregate independently of each other during reassortment, a dual-serotyping system to account for the specificities of both VP7 and VP4 has been adopted (32, 44).
On the basis of the VP7 protein, 14 different G types have been described so far; among these, 10 serotypes were associated with acute gastroenteritis in humans (31). Four of these rotavirus serotypes (G1 to G4) are the most common etiologic agents of childhood diarrhea worldwide for which vaccines have been developed (36, 37). Typing of human group A rotavirus by molecular and immunological methods has been reported (6, 15, 18, 26, 27, 40, 57). P serotypes have been defined by Gorziglia et al. (25) by using polyclonal antibodies to baculovirus-expressed VP4 protein. They showed that rotavirus serotype P[8] with G1, G3, and G4 specificities (prototype strains Wa, Ku, P, and VA70) was present in isolates from children with acute diarrhea whereas type P[4] combined with virulent G2 (DS-1-like strain) and P[6] with G1 to G4 specificities (prototype strains M37, 1076, McN13, and STE) were isolated from asymptomatic newborns excreting rotavirus. These strains were classified into three genetic and three antigenic types, designated P1A, P1B, and P2, respectively. Since VP4 is a minor outer protein with only 250 copies of the molecule per viral particle, monoclonal antibodies to this protein are rather difficult to obtain (5) for the average laboratory. In addition, preparation of the necessary reagents is laborious and time-consuming (21, 23). To overcome these problems, the typing of rotavirus P strains can be accomplished by identification of genetically different VP4 genes by reverse transcription-PCR (RT-PCR), as previously reported (9, 17, 33, 52, 53).
Analysis of prevalent VP7 and VP4 genes is important for evaluating candidate rotavirus vaccines. The prevalence of G types in Argentine children infected with group A rotavirus was previously assessed with monoclonal antibodies to types G1 to G4 by an enzyme-linked immunosorbent assay (ELISA) (24). Many studies using VP4 (P) genotyping methods demonstrated a worldwide combination of one P genotype, P[8], with G1, G3, and G4, whereas P[4] was frequently associated with G2 (16, 17, 52). Studies carried out in India revealed the prevalence of a different G-P combination: genotype P[6] frequently associated with an unusual G serotype G9 (49). Accordingly, similar studies performed in Brazil also demonstrated the prevalence of unusual human strains, bearing P[8] in combination with G5, among children with acute gastroenteritis (53).
The asymptomatic nature of neonatal rotavirus infection may be explained by the acquisition of maternal antibodies during early life. A recent study (48) demonstrated that the lack of maternal antibodies to P serotypes predisposes neonates to infections with unusual rotavirus strains. According to these authors it was demonstrated that rotavirus strains infecting newborns have unique neutralizing antigens (P serotypes) on their outer capsids that are different from those found on rotavirus strains causing gastroenteritis in older children.
Specific markers of rotavirus infection are rotavirus specific immunoglobulin A (IgA) and IgM antibodies in duodenal juice; however, both salivary and fecal antirotavirus antibodies can be taken as indicators of intestinal immune responses in young children (1, 6). Furthermore, the detection of such antibodies in stool samples from both symptomatic and asymptomatic children can be taken as a marker of recently acquired infections since IgM and IgA coproantibodies remain for long periods after the onset of clinical disease and in the absence of viral shedding.
This report describes the characterization of rotavirus strains isolated from infants and children at three different hospitals in Buenos Aires southern districts between 1996 and 1998 by G and P genotyping by RT-PCR, electropherotyping, and detection of group A rotavirus-specific IgM and IgA antibodies in stool samples. Individual samples taken from children with acute diarrhea and consecutive samples taken from two patients monitored at daily intervals after the onset of disease were evaluated by these methods.
MATERIALS AND METHODS
Viruses.
Human rotavirus strains Wa (G1P[8]), DS-1 (G2P[4]), P (G3P[8]), and VA 70 (G4P[8]), propagated in cultures of MA104 cells, were used in this study. These prototype strains were kindly provided by J. Gomez, Viral Gastroenteritis Unit, Argentine Reference Center. Two animal rotavirus strains were included as controls, simian rotavirus SA11 (serotype G3) and bovine UK (serotype G6).
A total of 500 stool samples collected in three consecutive winter seasons (April to July) from 1996 to 1998 were used in this study; all submitted samples were taken from children (age range: 6 months to 2 years; 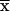 = 13 months) suffering acute diarrhea of unknown viral etiology since all patients were negative for known enteric bacteria and parasites. All samples were submitted to our laboratory for differential diagnosis of rotavirus diarrhea. Laboratory diagnosis of other enteric viruses was not performed for any of these samples. Requested patient data included age, sex, dates of disease onset and specimen collection, initial symptoms, duration of illness, degree of dehydration when observed, and diarrhea severity.
= 13 months) suffering acute diarrhea of unknown viral etiology since all patients were negative for known enteric bacteria and parasites. All samples were submitted to our laboratory for differential diagnosis of rotavirus diarrhea. Laboratory diagnosis of other enteric viruses was not performed for any of these samples. Requested patient data included age, sex, dates of disease onset and specimen collection, initial symptoms, duration of illness, degree of dehydration when observed, and diarrhea severity.
Stool suspensions of 10 to 20% were made in phosphate-buffered saline, pH 7.2. Samples prepared in this way were evaluated with a commercial ELISA kit (Pathfinder; Kallestad Diagnostics, Austin, Tex.), by an ELISA method specific for the VP6 rotavirus antigen, by electropherotyping of the double-stranded RNA (dsRNA) genome by polyacrylamide gel electrophoresis (PAGE), and finally by screening for the presence of rotavirus copro-IgM and -IgA antibodies by capture ELISA assays. G and P genotyping was assessed by RT-PCR with generic and type-specific primers for 100 randomly selected samples.
Ten of these samples were tested for the presence of rotavirus particles by electron microscopy (EM).
ELISA for antigen detection.
An ELISA consisting of a double-antibody sandwich assay using goat antirotavirus (VP6-specific) antibodies labelled with biotin (N-hydroxysuccinimidobiotin; H1757; Sigma Chemical Co., St. Louis, Mo.) and avidin-conjugated horseradish peroxidase (HRPO) (P0347; DAKO A/S, Glostrup, Denmark) as described previously (21, 23). Briefly, a 96-well polystyrene microtiter plate (Nunc, Roskilde, Denmark) was coated with 50 μl of affinity-purified (protein G-Sepharose 4B; Pharmacia, Uppsala, Sweden) goat antirotavirus VP6 (0.5 μg/well) in bicarbonate buffer, pH 9.6. The plate was incubated for 1 h at room temperature in a wet chamber. After this and the following steps the plates were washed three times with phosphate-buffered saline (0.5 M NaCl final concentration)-Triton X-100 (0.2% [vol/vol]) (22). Stool suspensions were diluted in ELISA dilution buffer, i.e., 1% (wt/vol) bovine serum albumin in washing solution, and were added to duplicate empty wells in 50-μl volumes. Serial dilutions of purified bovine rotavirus and noninfected MA104 cells were included in each plate (50 μl/well) as positive and negative antigen controls, respectively. Plates were incubated for 1 h at 37°C or overnight at 4°C. After the plates were washed, a 1/1,000 dilution of biotin-labelled antirotavirus IgG was added and the plates were incubated further for 1 h at 37°C, followed by a 30-min incubation with an appropriate dilution of avidin-conjugated HRPO. Substrate (o-phenylenediamine [OPD]) was added for color development according to standard procedures. The optical density (OD) was measured at a wavelength of 490 nm (ELISA reader Max Line; Molecular Devices, Sunnyvale, Calif.). The ELISA E value was calculated as the difference between the OD for rotavirus antigen and that for negative-control antigen. ELISA cutoff values corresponding to E ≤ 0.2 were calculated by testing 50 stool samples taken from healthy age- and sex-matched children without known diarrhea episodes during the last 8 months (control group).
All samples were additionally tested with a commercial ELISA kit from Kallestad.
ELISA for antibody detection.
Detection of rotavirus-specific IgM and IgA antibodies in stool samples was performed as μ and α capture ELISAs, respectively (46).
The following reagents included in the tests were obtained from DAKO A/S: rabbit anti-human IgM (A0425) and rabbit anti-human IgA (A0262). Bicarbonate buffer (pH 9.6), washing buffer, and dilution buffer were the same as described above for the rotavirus antigen detection assay.
(i) Rotavirus IgM (μ capture ELISA).
Microtiter test plates (Maxisorp; Nunc) were coated with 50 μl of rabbit anti-human IgM (μ chain specific; IgG fraction) diluted in bicarbonate buffer (pH 9.6; 50 μl/well). Plates were incubated for 1 h at room temperature in a wet chamber. After this and the following steps the plates were washed three times with washing solution as described before. Fifty microliters of stool samples serially diluted in dilution buffer was added to two duplicate wells (one for rotavirus antigen and one for noninfected cell control antigen), with two wells for each dilution. Following a 1-h incubation at 37°C and another wash, 50-μl volumes of rotavirus antigen (108 50% tissue culture infective doses/ml) and noninfected MA104 cell control antigen were added to duplicate wells. After overnight incubation at 4°C the plates were washed and subsequently incubated for 1 h at 37°C with 10 ng (calculated as IgG) of biotin-labelled goat antirotavirus IgG/well. After this step, the plates were treated with avidin-conjugated HRPO followed by OPD as described before for the antigen detection assay.
ELISA cutoff values were obtained by testing stool samples from the control group. A cutoff value was defined as three standard deviations above the arithmetic mean E value of the negative samples from the control group corresponding to E ≤ 0.2.
(ii) Rotavirus IgA (α capture ELISA).
The test was performed similarly to the rotavirus IgM antibody ELISA with the exception that rabbit anti-human IgA was used as the catching antibody instead of rabbit anti-human IgM. ELISA cutoff levels were obtained by testing stool samples from the control group as for the IgM assay; a sample was considered to be negative if the E value was ≤0.2.
Viral dsRNA and PAGE.
Viral RNA was extracted from fecal suspensions by acid-phenol-chloroform and alcohol precipitation according to methods published elsewhere (30). Duplicate extracted dsRNA samples were diluted in 10 μl of sterile distilled H2O for RT-PCR and in electrophoresis sample buffer for PAGE analysis.
In some samples an additional purification step with CF11 cellulose was required to remove substances inhibitory to the RT-PCR enzymes (56).
RT-PCR.
A 10-μl portion of each dsRNA-extracted sample was used as the template for RT to synthesize cDNA copies from both strands. The RNA was denatured at 95°C and quickly chilled on ice for 2 min. The reaction volume was brought to 25 μl by adding the RT reaction mixture containing 50 mM Tris-HCl, pH 8.3; 50 mM KCl; 10 mM MgCl2; 10 mM dithiothreitol and 0.5 mM spermidine; 500 μM (each) dATP, dCTP, dTTP, and dGTP (Promega, Madison, Wis.); 0.4 μM concentrations of primers beg and end (26) for the VP7 gene (1,062 bp) and primers 1 and 2 (16) for amplification of an 876-bp fragment of the VP4 gene; 7 U of avian myeloblastosis virus (M5101; Promega); and 20 U of RNasin RNase inhibitor (N2511; Promega). Oligonucleotide primers were purchased by DNAgency (Malvern, Pa.). Dimethyl sulfoxide (DMSO; 5% [vol/vol]) was added to the RT mixture, and cDNA synthesis was performed for 1 h 30 min in a water bath at 42°C.
Conditions for the PCR were as follows. The reaction volume was brought to 10 μl by adding the PCR mixture, which contained 0.25 μM concentrations of primers beg and end (VP7) and 1 and 2 (VP4), 1 μl of the PCR buffer supplied with the enzyme, 0.75 U of Taq DNA polymerase B (Promega), 1 μg of bovine serum albumin (Sigma Chemical Co.), 100 μM concentrations of each of the deoxynucleoside triphosphates, and 2 μM MgCl2. Capillary tubes were loaded with PCR mixtures and then placed in a thermocycler (IT Idaho Technology). PCR consisted of 1 cycle at 92°C for 1 min; 30 cycles of 92°C for 2 s, 42°C for 10 s, and 72°C for 30 s; and 1 cycle at 72°C for 3 min. A 10-μl aliquot of the amplification product was electrophoresed through 1.5% agarose (Promega) in Tris-acetic acid-EDTA buffer (0.089 M Tris, 0.089 M acetic acid, 0.002 M EDTA [pH 7.5] containing 0.5 μg of ethidium bromide/ml) and visualized with an UV transilluminator.
Typing by multiplex PCR.
PCR products from the RT-PCR described above were used as templates for a second amplification round with a cocktail of specific primers which amplify variable regions of the VP7 gene, G types (26), and variable regions of the VP4 gene, P types (52) (DNAgency). This method is referred to as multiplex PCR. The 1,062-bp (VP7) amplified products and corresponding 876-bp (VP4) samples obtained after the first RT-PCR were either used directly (1 μl) or cut out and extracted from the agarose gel as purified DNA (1 μl) after electrophoresis. Conditions for the multiplex PCR were otherwise the same as those for the RT-PCR.
RESULTS
ELISA for detection of rotavirus antigens and antibodies.
The sensitivity of the ELISA performed with biotinylated antibodies for antigen detection was tested by using serial 10-fold dilutions of highly purified bovine rotavirus and a corresponding noninfected control. The minimal amount of antigen detected by this method was estimated to be about 0.1 ng/ml, equivalent to 4 × 106 viral particles/ml.
A total of 500 human stool samples and 50 samples from healthy children were evaluated in parallel in-house ELISA and ELISA with a commercial kit from Kallestad (K-ELISA). Of 500 samples evaluated by both methods, 62% of the samples from patients with acute gastroenteritis were positive in both assays; however, 4 samples were only positive by the in-house ELISA, as confirmed by a positive PAGE and RT-PCR analysis. On the other hand, three samples positive by the K-ELISA were negative by both PAGE and RT-PCR (results not shown).
None of the samples taken from healthy children showed values exceeding the estimated ELISA cutoff OD value. These samples were also negative in the RT-PCR.
Results obtained with the μ capture and α capture ELISA for determination of IgM and IgA antibodies in stool samples showed that among the total antigen-positive samples 32.92% were also positive for IgM antibodies whereas 7.93% of the IgM-positive samples had an undetectable amount of virus, regardless of the detection method employed. Accordingly, among the antigen-positive samples 39.02% showed detectable levels of IgA. These antibodies were found in 20.63% of the antigen-negative samples (Table 1). Neither IgM and IgA antibodies nor rotavirus antigens were found in samples taken from healthy children. These results suggested that for diagnostic purposes, detection of fecal rotavirus antibodies in diarrhea samples in the absence of virus can be used as supplement to antigen detection.
TABLE 1.
Relationship between group A rotavirus antigens and antibodies in stool samples
| Rotavirus antigen assay
|
No. (%) of samples:
|
||||
|---|---|---|---|---|---|
| Result | No. (%) of samples | Positive for rotavirus antibody:
|
Negative for rotavirus antibodies | ||
| IgM | IgA | IgM + IgA | |||
| Positive | 310 (62) | 34 (10.97) | 53 (17.07) | 68 (21.95) | 155 (50) |
| Negative | 190 (38) | 6 (3.17) | 30 (15.87) | 9 (4.76) | 145 (76.19) |
Detection of the rotavirus genome by PAGE and identification of rotavirus particles by EM.
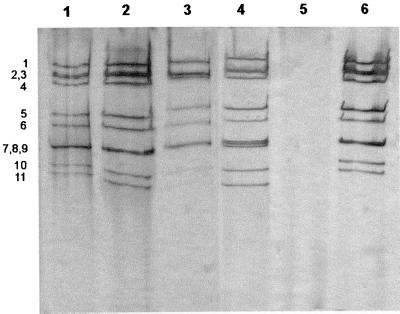
An electropherotype profile (4-2-3-2) characteristic of group A was demonstrated by PAGE in samples positive by ELISA. A total of 257 (51.4%) of 500 tested samples were positive in both assays; however, about 18% of the ELISA-positive samples were not confirmed to be positive for the rotavirus genome. Most of the rotavirus strains detected by PAGE showed the long electrophoretic pattern (Fig. 1) with the exception of three strains which exhibited short electropherotypes.
FIG. 1.
PAGE analysis of human rotavirus dsRNA. PAGE analysis of the dsRNA genome extracted from stool suspensions taken from children with acute diarrhea is shown. All samples (lanes 1, 2, 3, and 6) exhibited the typical 4-2-3-2 pattern of group A rotavirus. Lane 4, bovine rotavirus strain UK; lane 5, extraction procedure applied to a pool of stool suspensions taken from healthy children. Numbers denote positions of dsRNA segments.
A total of 10 selected samples positive in both assays were further analyzed by EM for the presence of rotavirus particles. Samples were selected according to the results obtained by PAGE and ELISA to ensure the presence of enough rotavirus particles to be visualized by EM since the sensitivities of PAGE and EM are similar (see below). A total of 257 samples fulfilled this criterion; however, only 10 of these were randomly selected for EM analysis. All samples showed the typical structure of the rotavirus double-shelled particles. Other enteric viruses were not found in the few analyzed samples.
Comparison ELISA, PAGE, and RT-PCR.
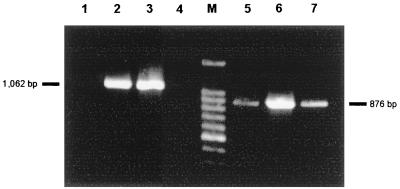
When the samples were evaluated by the three methods, no significant differences between ELISA and RT-PCR were observed; however only 82% of the samples positive by ELISA and RT-PCR were positive by PAGE. The positive results obtained by RT-PCR were not dependent on the different dsRNA extraction and purification methods used. Amplifications of the whole VP7 gene (1,062 bp) and a fragment of the VP4 gene (856 bp) by RT-PCR in stool samples from different patients, compared to molecular weight markers, are shown in Fig. 2. Only a few samples contained substances inhibitory to the RT reaction. To overcome this problem, a further purification of the dsRNA by CF11 cellulose was included after the acid-phenol-chloroform extraction step. After treatment with CF11 cellulose, purified material yielded dsRNA templates suitable for RT-PCR amplification.
FIG. 2.
Amplification of the VP7 and VP4 genes by RT-PCR. Amplification products corresponding to the VP7 (lanes 2, 3, and 4) and VP4 genes (lanes 5, 6, and 7) in three different patient samples are shown. Lane 1, negative control; lane M, molecular weight markers (100-bp ladder). The sizes of amplified bands are indicated.
To compare the sensitivities of ELISA and RT-PCR, 10-fold serial dilutions of three pooled samples were made and tested by each method. It was found that all three samples remained positive by RT-PCR when diluted 100-fold; in contrast, this dilution was not detected by ELISA. With respect to the number of positive samples detected by each test, only five additional positives were found by RT-PCR. The overall sensitivity of the RT-PCR was dependent on the introduction of 5% DMSO and Mg2+ ions in RT reaction mixtures. The addition of DMSO decreased the amount of dsRNA template needed for amplification of both VP7 and VP4 genes from microgram amounts in the absence of DMSO to nanogram amounts when DMSO was present. On the other hand, the presence of this reagent in PCR mixtures impaired substantially the sensitivity of the assay.
The estimated numbers of rotavirus particles detected per milliliter by each method were as follows: 106 (ELISA), 104 (RT-PCR), and 1011 (PAGE).
Culture methods have generally been regarded as the ultimate standard for the diagnosis of viral infections, but this presupposes the presence of viable virus particles in the samples. This is not a requirement for immunochemical methods such as ELISA or dsRNA detection methods such as PAGE and RT-PCR. Taking into account the high level of sensitivity extensively reported for most PCR methods, we have considered RT-PCR the “gold standard” against which other tests may be judged.
To establish the diagnostic sensitivity and specificity of PAGE, in-house ELISA, and the commercial ELISA kit, each method was compared with RT-PCR, giving the following results: for in-house ELISA, sensitivity was 98.4%, specificity was 100%, positive predictive value was 100%, negative predictive value was 97.3%, and correspondence estimated from these values was 99%; for the commercial ELISA kit, sensitivity was 97.1%, specificity was 98.3%, positive predictive value was 99%, negative predictive value was 95.1%, and correspondence was 97.6%; for PAGE, sensitivity was 81.8%, specificity was 100%, positive predictive value was 100%, negative predictive value was 95.1%, and correspondence was 97.6%.
Typing.
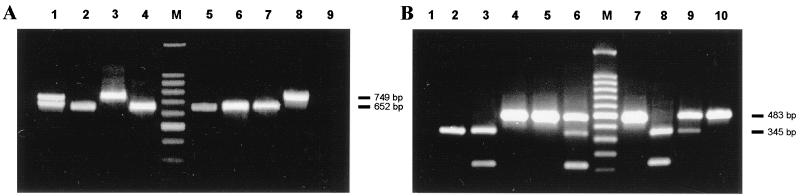
G and P typing by multiplex PCR of 100 randomly selected samples showed the prevalence of the following G and P serotypes: G1, 39%; G2, 43%; G4, 4%; coinfections with both G1 and G2, 7%; P[8], 16%; P[4], 71%; P[8] and P[4] simultaneously, 3%. No other G or P types were found in the samples evaluated. In Fig. 3 are shown the patterns of the amplified G and P types obtained in agarose gels with different clinical samples, compared to molecular weight markers. The presence of two different G types with a single P type and the presence of two P types with only one G serotype were found in 10% of the samples tested, suggesting either coinfections with two rotaviruses or the presence of nontypeable strains (Fig. 3A, lane 1; Fig. 3B, lanes 6 and 9). Of note, an additional amplification product of 100 bp was detected in a few patient samples when the samples were subjected to P typing by multiplex PCR (Fig. 3B, lanes 3, 6, and 8). The size of this band cannot be attributed to any of the P types detected by this method.
FIG. 3.
Typing of human group A rotavirus VP7 and VP4 genes by multiplex PCR. (A) Amplification products of the VP7 genes in stool samples taken from eight different patients with acute diarrhea. Lanes 3 and 8, type G1; lanes 2, 4, 5, 6, and 7, type G2; lane 1, coinfection with both G1 and G2 types. (B) Amplification products of the VP4 genes in stool samples taken from nine different patients with acute diarrhea. Lanes 2, 3, and 8, type P[8]; lanes 4, 5, 7, and 10, type P[4]; lanes 6 and 9, coinfection with both P[8] and P[4] types. Negative controls consisted of multiplex PCR applied to the VP7 gene (panel A, lane 9) and to the VP4 gene (panel B, lane 1) of bovine group A rotavirus strain UK. Lane M, molecular weight markers (100-bp ladder). Sizes of amplified bands are indicated.
The combinations of G and P types most frequently found were G1 with P[8] and G2 with P[4]; however, strains bearing the unusual combination of G1 with P[4] were detected in 14% of the samples (Table 2). Two approaches were used for G and P typing of samples by multiplex PCR: the second round of amplification was performed with either 1 μl of purified DNA fragments, 1,062 bp (G) and 876 bp (P), extracted from agarose gel or with 1 μl of the first PCR mixture. Results with purified DNA fragments were considerably better in terms of sensitivity and the clear-cut definition of resolved bands since a positive reaction was still seen when the dilution of purified cDNA template was increased twofold. The minimal amount of dsRNA needed for a positive RT-PCR was in the range of 20 to 80 pg of RNA template, corresponding to 2 to 8 ng of RNA in the original sample.
TABLE 2.
VP4 (P) and VP7 (G) typing of human group A rotavirus by multiplex PCR
| G type | Prevalence (%) ofa:
|
||||
|---|---|---|---|---|---|
| P[8] | P[4] | P[8] + P[4] | Nontypeable | Total | |
| G1 | 12 | 14 | 3b | 10c | 39 |
| G2 | 43 | 43 | |||
| G4 | 4 | 4 | |||
| G1 + G2 | 7b | 7 | |||
| Nontypeable | 7c | 7 | |||
| Total | 16 | 71 | 3 | 10 | 100 |
Percentage of total evaluated samples.
Mixed infections included three G1P[8] plus P[4] samples and seven G1 plus G2P[4] samples.
Nontypeable included 10 samples of type G1 combined with untypeable P and 7 samples of untypeable G combined with P[4].
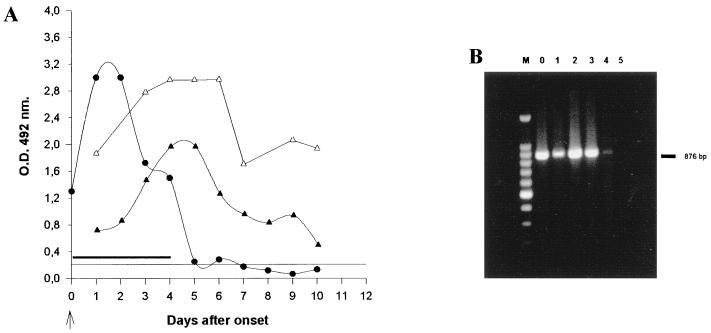
The presence of rotavirus demonstrated by ELISA, RT-PCR, and IgM and IgA antibodies in consecutive samples from a patient with acute diarrhea monitored at daily intervals indicated that antibodies in stools can be taken as markers of recently acquired infection since IgA and IgM were still present when no viral shedding (10 days after onset) was seen (Fig. 4).
FIG. 4.
Detection of virus and coproantibodies in consecutive samples. (A) Consecutive samples from two patients taken at daily intervals and tested for rotavirus antigens (●) and rotavirus IgM (▵) and IgA (▴) antibodies. —, RT-PCR-positive samples. The arrow indicates illness onset. (B) Amplification products obtained by RT-PCR with VP4-specific primers (876 bp) in consecutive samples. Lane 0, disease onset; lanes 1 to 5, 1 to 5 days after onset, respectively; lane M, molecular weight markers (100-bp ladder).
The presence of these antibodies was detected after 10 days, with elevated concentrations on day 6, and a marked decline 10 days after onset. The patients tested on consecutive days were 6 and 7 months old and had no previous history of severe diarrhea.
DISCUSSION
The methods of choice for detection of rotavirus in stool samples should have high degrees of sensitivity, specificity, and reproducibility, which ensure consistency of performance in the laboratory. ELISA for the detection of viral antigens is the method commonly employed in many laboratories in combination with either electropherotype determination by PAGE or detection of viral particles by EM. The overall sensitivities of these methods are in the range of 108 to 109 viral particles/ml for PAGE; the most sensitive ELISA described detects as few as 105 to 106 viral particles/ml, whereas for a positive EM reaction 108 viral particles/ml are required (2, 12, 56).
For rotavirus infection, where levels of virus shedding are usually very high, all of these methods are suitable for diagnostic purposes (2, 8, 19, 28).
Nevertheless, when samples are taken in a late phase of the infection or when samples different from stools, such as throat swabs, cerebrospinal fluid, and respiratory secretions, where the amount of rotavirus is expected to be very low, are used, more-sensitive techniques such as RT-PCR amplification methods are required (3, 33, 60).
Taking into account that cell culture methods for human rotavirus stool samples are reported to be 75% as efficient as antigen detection methods (35), culture procedures are not considered the gold standard against which other tests may be judged. Recently reported data (10, 29, 42) indicated that the main drawbacks associated with the use of the latex agglutination assays for diagnostic purposes are the low sensitivity and specificity of these assays compared to most ELISA methods. Recently reported data indicated that RT-PCR for direct detection of rotavirus in stool samples may be considered the gold standard method (47). In the present study, a comparison of diagnostic sensitivities and specificities of the in-house ELISA, commercial ELISA kit, and PAGE for direct detection of group A rotavirus in stool samples, with RT-PCR as the standard method, was made. The data presented here indicate that for the rapid screening of a large number of samples the in-house ELISA method was able to detect rotavirus in stool samples with sensitivity (98.4%) and specificity (100%) similar to those of RT-PCR at a considerably lower cost and without previous treatment of the sample. In laboratories with a restricted budget, handling a large number of specimens, the ELISA method should in the long run offer worthwhile savings compared to RT-PCR. ELISA was considerably more sensitive than electropherotyping by PAGE. Nevertheless, for typing purposes the method of choice is the multiplex PCR since this method allows the simultaneous typing and identification of uncommon emergent rotavirus strains. The use of PCR enabled the genotyping of rotavirus-positive specimens that could not be typed by ELISA with type-specific monoclonal antibodies. Furthermore, it may be necessary to use several monoclonal antibodies directed to different epitopes of the same serotype because of epitope polymorphism within a serotype (7). On the other hand, removal of inhibitory substances present in stool samples is occasionally needed for a successful PCR (56). In our experience extraction procedures including or not including the CF11 purification step could be used for obtaining dsRNA free of substances inhibitory of the RT-PCR enzymes. Nevertheless, in a very small number of samples further purification of extracted RNA was necessary for the removal of inhibitors that hampered RT reactions (3, 33, 56). Multiplex PCR with a mixture of type-specific primers allows the typing of the rotavirus present in the samples with sensitivity and accuracy. The test is rather easy to perform since all type-specific primers are added in one step. This design allows simultaneous detection of coinfections with different viruses and identification of new nontypeable strains by only one amplification run.
The results presented here showed the prevalence of serotypes G2 and G1, which are the most commonly found in other parts of the world and which are the types included in available vaccines.
Consistent with the findings of previous published studies (9, 17, 26, 52, 53), G1P[8] and G2P[4] were the G-P type combinations frequently found among the tested samples; however, G2P[4] showed a greater prevalence (43%) than G1P[8] (12%). These results showed a different distribution of G-P combinations with respect to the G-P types previously found among children in the United States (17, 26, 47) or among children from New Delhi (39, 49). According to these reports, G1P[8] was the most commonly found in the United States and was distributed equally with G2P[4] strains among Indian children.
Unexpectedly, unusual combinations of G1P[4] were found in 14% of the samples. These rare combinations were usually present in single-rotavirus infections. Coinfections with only one G type and two P types and two different G types with a single P type were observed in 3 and 7% of tested samples, respectively. These results need further confirmation since they perhaps represent nontypeable G or P types coinfecting the same patient; specific primers for the unusual G5 type, which is, however, commonly found in developing countries (47, 53), were not included in the multiplex PCR. Another possible explanation for these results is the presence of coinfections with two rotavirus strains sharing identical G or P types. Similar findings were recently reported (47) in an extensive survey conducted in 10 U.S. cities, where unusual types were found in 1.4% (G1P[4]) and 0.3% (G2P[8]) of 348 rotavirus strains examined by immunoassays and molecular methods including RT-PCR and hybridization. Furthermore, additional amplification products of small size (100 bp) were seen in a few patient samples (Fig. 3B, lanes 3, 6, and 8); these products cannot be related to any of the known P types (52). One possible explanation for these results is the presence in these samples of coinfections with P[8] and a new recombinant strain. The epidemiological implications of these uncommon strains remain to be elucidated. Patient and control stool samples were taken from young children (6 to 24 months of age; mean age, 13 months) whose ages corresponded to the peak acquisition age of the rotaviral infection reported for most developing countries, including the population under study (A. Castello, M. Argüelles, G. Villegas, and G. Glikmann, unpublished data). Of note, no difference in age distribution was evident among the children suffering acute diarrhea caused by different genotypes. Accordingly, neither age distribution nor appearance of unusual genotypes was related to diarrheal severity.
The intestinal immune response to the infecting rotavirus strain(s) was evaluated by detection of copro-IgA and -IgM antibodies in single and consecutive samples from children with moderate to severe diarrhea episodes.
Determination of IgM and IgA antibodies in individual stool samples from children suffering from acute diarrhea demonstrated that high levels of IgM were present in 32.92% of patient samples positive for viral antigens whereas 39.02% of patients with detectable amounts of virus were positive for IgA. Nevertheless, 7.93% of patients without detectable amounts of virus showed high levels of IgM whereas IgA was present in 20.63% of these samples. Furthermore, 155 of 310 antigen-positive samples did not show either IgM or IgA antibodies. These results suggest that detection of rotavirus-specific IgM and IgA in the absence of virus is probably more likely for a recently acquired infection and can be used as a supplement to virus detection for diagnostic purposes. In the present survey, determination of copro-IgM and -IgA antibodies was performed in order to evaluate markers of the intestinal immune response during acute diarrhea episodes regardless of their protective effect on either primary or secondary rotavirus infections.
Bishop et al. (1) demonstrated that IgA coproconversion is a valuable alternative method for detection of symptomatic and asymptomatic rotavirus infections in young children.
Consistent with these findings, it was previously shown that fluctuations in levels of rotavirus IgA coproantibodies are sensitive indicators of rotavirus reinfections (6) since after an acute episode of diarrhea a greater-than-threefold increase of copro-IgA was detected in stool samples from young children taken at weekly intervals. Studies of mice have shown that a single inoculation of live virus in antibody-negative animals elicited a long-lasting protective immunity and that protection correlates with the presence of IgA intestinal antibodies but that high levels of serum neutralizing antibodies of the IgG type were not related to protection (12). Protection against diarrhea after adoptive transfer of CD8 spleen cells from immunized mice into syngeneic pups before rotavirus inoculation was reported previously (43). The importance of IgA intestinal antibodies and cellular immunity markers such as CD8 lymphocytes in protection against rotavirus disease was confirmed by other groups (4, 44, 54, 55). Protection against rotavirus disease has been correlated with titers of serum (45, 51) or stool (5, 39) rotavirus antibodies following natural infection of young children. Furthermore, detection of IgM and IgA coproantibodies to confirm recently acquired infections with other enteric viruses such as hepatitis A virus (38) or animal coronaviruses (11) has been reported. A recent study (1) of serum, fecal, and breast milk rotavirus antibodies determined in 68 mother-infant pairs demonstrated that IgA coproconversion was the most sensitive method for detection of symptomatic and asymptomatic rotavirus infection in children, compared to the direct detection of the virus in stools. The same study clearly demonstrated that after a primary rotavirus infection with rotavirus serotype G2P[4], followed by a reinfection with a rotavirus of a different serotype, G4P[8], 12 months later, a large increase in copro-IgA antibodies in the stool samples occurred at the onset of each infection; however, copro-IgA antibodies did not persist for >2 weeks after primary infection, whereas coproantibody increases persisted for >10 weeks after reinfection, resulting in a long-lasting copro-IgA response (IgA plateau). Accordingly, the results of the present study for consecutive samples from two children suggest a primary infection with the rotavirus serotype G4P[8] since both IgM and IgA antibodies were detected in high levels 6 to 7 days after onset, with a marked decline after 10 days when rotavirus shedding was no longer demonstrated by ELISA or RT-PCR and with complete recovery of clinical symptoms. Furthermore, since both children were only 6 to 7 month old and therefore probably lacked protection by maternal antibodies, it can be assumed that they probably suffered from a primary infection with the G4P[8] genotype detected by multiplex PCR.
In conclusion, the present study has shown that for diagnosis of a large number of samples, the use of a double-antibody sandwich ELISA with biotinylated antibodies provides a rapid, sensitive, and inexpensive procedure for the direct detection of rotavirus antigens in clinical specimens, with performance equal to that of RT-PCR.
Typing of rotavirus strains is a main application of PCR, since this method represents a very convenient alternative when type-specific monoclonal antibodies are not available (6, 24). An additional advantage of this method is the potential for identification of new reassortant or recombinant strains that are unable to be typed with primers directed to the known human genotypes.
Furthermore, detection of specific IgM and IgA antibodies represents a useful supplement to rotavirus detection methods in the diagnosis of current or recently acquired infections.
ACKNOWLEDGMENTS
Clinical samples were kindly provided by Ana Borsa from Children Hospital Sor María Ludovica, La Plata, and by Luciana Irczick from Hospital Materno Infantil de San Francisco Solano, Solano.
Marcelo H. Argüelles is a research fellow of the Comisión de Investigaciones Científicas (grant number 2482).
REFERENCES
- 1.Bishop R F, Bugg H C, Mazendycz P J, Lund J S, Gorell R J, Barnes G L. Serum, fecal and breast milk rotavirus antibodies as indices of infection in mother-infant pairs. J Infect Dis. 1996;174(Suppl. 1):S22–S29. doi: 10.1093/infdis/174.supplement_1.s22. [DOI] [PubMed] [Google Scholar]
- 2.Brandt C D, Kim H W, Rodriguez W J, Thomas L, Yolken R-H, Arrobio J O, Kappikian A Z, Parrot R H, Chanock R M. Comparison of direct electron microscopy, immune electron microscopy, and rotavirus enzyme-linked immunosorbent assay for detection of gastroenteritis viruses in children. J Clin Microbiol. 1981;13:976–981. doi: 10.1128/jcm.13.5.976-981.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buessa J, Colomina J, Raga J, Villanueva A, Prat J. Evaluation of reverse transcription and polymerase chain reaction (RT/PCR) for the detection of rotaviruses: application of the assay. Res Virol. 1996;147:353–361. doi: 10.1016/S0923-2516(97)85127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burns W B, Siadath-Pajouh M, Krishnaney A A, Greenberg H B. Novel protective effect of rotavirus VP6 specific IgA monoclonal antibodies that lack conventional neutralizing activity. Science. 1996;272:104–107. doi: 10.1126/science.272.5258.104. [DOI] [PubMed] [Google Scholar]
- 5.Coulson B. Typing of human rotavirus VP4 by an enzyme immunoassay using monoclonal antibodies. J Clin Microbiol. 1993;31:1–8. doi: 10.1128/jcm.31.1.1-8.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coulson B S, Grimwood K, Hudson I L, Barnes G L, Bishop R F. Role of coproantibody in clinical protection of children during reinfection with rotavirus. J Clin Microbiol. 1992;30:1678–1684. doi: 10.1128/jcm.30.7.1678-1684.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coulson B S, Kirkwood C. Relation of VP7 amino acid sequence to monoclonal antibody neutralization of rotavirus and rotavirus monotype. J Virol. 1991;65:5968–5974. doi: 10.1128/jvi.65.11.5968-5974.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cukor G, Blacklow N R. Human viral gastroenteritis. Microbiol Rev. 1984;48:157–179. doi: 10.1128/mr.48.2.157-179.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das B K, Gentsch J R, Cicirello E G, Woods P A, Gupta A, Ramachandran M, Kumar R, Bhan M K, Glass R I. Characterization of rotavirus strains from newborns in New Delhi, India. J Clin Microbiol. 1994;32:1820–1822. doi: 10.1128/jcm.32.7.1820-1822.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Beer M, Peenze I, da Costa Mendez V M, Steele A D. Comparison of electron microscopy, enzyme linked immunosorbent assay and latex agglutination for the detection of bovine rotavirus in faeces. J S Afr Vet Assoc. 1997;68:93–96. doi: 10.4102/jsava.v68i3.883. [DOI] [PubMed] [Google Scholar]
- 11.El-Kanawati Z R, Tsunemitsu H, Smith D R, Saif L J. Infection and cross-protection studies of winter dysentery and calf diarrhea bovine coronavirus strains in colostrum-deprived and gnotobiotic calves. Am J Vet Res. 1996;57:48–53. [PubMed] [Google Scholar]
- 12.Estes M K. Advances in molecular biology: impact on rotavirus vaccine development. J Infect Dis. 1996;174(Suppl. 1):S37–S46. doi: 10.1093/infdis/174.supplement_1.s37. [DOI] [PubMed] [Google Scholar]
- 13.Estes M K, Cohen J. Rotavirus gene structure and function. Microbiol Rev. 1989;53:410–419. doi: 10.1128/mr.53.4.410-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Estes M K. Rotavirus and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1625–1655. [Google Scholar]
- 15.Flores J, Sears J, Perez-Schael I, White L, García D, Lanata C, Kapikian A Z. Identification of human rotavirus serotype by hybridization to polymerase chain reaction-generated probes derived from a hyperdivergent region of the gene encoding outer capsid protein VP7. J Virol. 1990;64:4021–4024. doi: 10.1128/jvi.64.8.4021-4024.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gentsch J R, Glass R I, Woods P A, Gouvea V, Gorziglia M, Flores J, Das B K, Bahn M K. Identification of group A rotavirus gene 4 by polymerase chain reaction. J Clin Microbiol. 1992;30:1365–1373. doi: 10.1128/jcm.30.6.1365-1373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gentsch J R, Woods P A, Ramachandran M, Das B K, Leite J P, Alfieri A, Kumar R, Bahn M K, Glass R I. Review of G and P typing results from a global collection of rotavirus strains: implications for vaccine development. J Infect Dis. 1996;174(Suppl. 1):S30–S36. doi: 10.1093/infdis/174.supplement_1.s30. [DOI] [PubMed] [Google Scholar]
- 18.Gerna G, Sarasini A, Arista S, Di Mateo A, Giovanelli L, Perea M, Halonen P. Prevalence of human rotavirus serotypes in some European countries. Scand J Infect Dis. 1990;22:5–10. doi: 10.3109/00365549009023112. [DOI] [PubMed] [Google Scholar]
- 19.Gilchrist M J R, Bretl T S, Moultney K, Knowlton D R, Ward R L. Comparison of seven kits for detection of rotavirus in fecal specimens with a sensitive, specific enzyme immunoassay. Diagn Microbiol Infect Dis. 1987;8:221–228. doi: 10.1016/0732-8893(87)90053-8. [DOI] [PubMed] [Google Scholar]
- 20.Glass R I, Kilgore P E, Holman R C, Shaoxiong J, Smith J C, Woods P A, Clarke M J, Ho M S, Gentsch J R. The epidemiology of rotavirus diarrhea in the United States: surveillance and estimates of disease burden. J Infect Dis. 1996;174(Suppl. 1):S5–S11. doi: 10.1093/infdis/174.supplement_1.s5. [DOI] [PubMed] [Google Scholar]
- 21.Glikmann G, Mordhorst C H, Koch C. Monoclonal antibodies for rapid diagnosis of influenza-A virus infections. Clin Diagn Virol. 1995;3:361–369. doi: 10.1016/0928-0197(94)00053-w. [DOI] [PubMed] [Google Scholar]
- 22.Glikmann G, Mordhorst C H. Secretory and serum immunoglobulin class-specific antibodies to mumps virus after a natural mumps infection. Serodiagn Immunother. 1987;1:275–285. [Google Scholar]
- 23.Glikmann G, Chen S, Mordhorst C H, Koch C. Monoclonal antibodies for rapid diagnostic of influenza-B virus in samples of patients with unknown respiratory infection. Clin Diagn Virol. 1995;4:27–42. doi: 10.1016/0928-0197(94)00053-w. [DOI] [PubMed] [Google Scholar]
- 24.Gomez J, Estes M K, Matson D, Bellinzoni R, Alvarez A, Grinstein S. Serotyping of human rotavirus in Argentina by ELISA with monoclonal antibodies. Arch Virol. 1990;112:249–259. doi: 10.1007/BF01323169. [DOI] [PubMed] [Google Scholar]
- 25.Gorziglia M, Larralde G, Kapikian A Z, Chanock R M. Antigenic relationship among human rotaviruses as determined by outer capsid protein VP4. Proc Natl Acad Sci USA. 1990;87:7155–7159. doi: 10.1073/pnas.87.18.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gouvea V, Glass R I, Woods P A, Taniguchi K, Clark H F, Forrester B, Fang Z Y. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol. 1990;28:276–282. doi: 10.1128/jcm.28.2.276-282.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green K Y, Hoshino Y, Ikegami N. Sequence analysis of the gene encoding the serotype specific glycoprotein (VP7) of two new human rotavirus serotypes. Virology. 1989;168:429–433. doi: 10.1016/0042-6822(89)90289-4. [DOI] [PubMed] [Google Scholar]
- 28.Hammond G W, Ahluwalia G S, Barker F G, Horsman G, Hazelton P R. Comparison of direct and indirect enzyme immunoassays with direct ultracentrifugation before electron microscopy for detection of rotaviruses. J Clin Microbiol. 1982;16:53–59. doi: 10.1128/jcm.16.1.53-59.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hendricks M K, Cuevas L E, Hart C A. Rotavirus diarrhea in Thai infants and children. Ann Trop Paediatr. 1995;15:147–152. doi: 10.1080/02724936.1995.11747763. [DOI] [PubMed] [Google Scholar]
- 30.Herring A J, Inglis N F, Ojeh C K, Snodgrass D R, Menzies J D. Rapid diagnosis of rotaviral infection by direct detection of viral nucleic acid in silver-stained polyacrylamide gels. J Clin Microbiol. 1982;16:473–477. doi: 10.1128/jcm.16.3.473-477.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoshino Y, Kapikian A Z. Rotavirus antigens. Curr Top Microbiol Immunol. 1994;185:179–227. doi: 10.1007/978-3-642-78256-5_7. [DOI] [PubMed] [Google Scholar]
- 32.Hoshino Y, Saif L J, Sereno M M, Midthun K, Flores J, Kapikian A Z, Chanock R M. Independent segregation of two antigenic specificities (VP3 and VP7) involved in neutralization of rotavirus infectivity. Proc Natl Acad Sci USA. 1985;82:8701–8704. doi: 10.1073/pnas.82.24.8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hussain M, Seth P, Broor S. Detection of group A rotavirus by reverse transcriptase and polymerase chain reaction in faeces from children with acute gastroenteritis. Arch Virol. 1995;140:1225–1233. doi: 10.1007/BF01322748. [DOI] [PubMed] [Google Scholar]
- 34.Hussain M, Seth P, Dar L, Broor S. Classification of rotavirus into G and P types with specimens from children with acute diarrhea in New Delhi, India. J Clin Microbiol. 1996;34:1592–1594. doi: 10.1128/jcm.34.6.1592-1594.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kapikian A Z, Chanock R M. Rotaviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1657–1708. [Google Scholar]
- 36.Kapikian A Z, Hoshino Y, Chanock R M, Perez-Schael I. Efficacy of a quadrivalent rhesus rotavirus-based human rotavirus vaccine aimed at preventing severe rotavirus diarrhea in infants and young adults. J Infect Dis. 1996;174(Suppl. 1):S65–S72. doi: 10.1093/infdis/174.supplement_1.s65. [DOI] [PubMed] [Google Scholar]
- 37.Kapikian A Z, Flores J, Vesikari T, Ruuska T, Madore H P, Green K I, Gorziglia M, Hoshino Y, Chanock R M, Midthun K, Perez-Schael I. Recent advances in development of a rotavirus vaccine for prevention of severe diarrheal illness of infants and young children. In: Mestecky J, Blair C, Ogra P L, editors. Immunology of milk and the neonate. New York, N.Y: Plenum Press; 1991. pp. 255–264. [DOI] [PubMed] [Google Scholar]
- 38.Locarnini S A, Coulepis A G, Kaldor J, Gust I D. Coproantibodies in hepatitis A: detection by enzyme-linked immunosorbent assay and immune electron microscopy. J Clin Microbiol. 1980;11:710–716. doi: 10.1128/jcm.11.6.710-716.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matson D O, O'Ryan M L, Herrera I, Pickering L K, Estes M K. Faecal antibody responses to symptomatic and asymptomatic rotavirus infections. J Infect Dis. 1993;167:577–583. doi: 10.1093/infdis/167.3.577. [DOI] [PubMed] [Google Scholar]
- 40.Matson D O, Estes M K, Burns J W, Greenberg H B, Taniguchi K, Urasawa Y. Serotype variation of human group A rotaviruses in two regions of the USA. J Infect Dis. 1990;162:605–614. doi: 10.1093/infdis/162.3.605. [DOI] [PubMed] [Google Scholar]
- 41.Mattion N M, Cohen J, Estes M K. The rotavirus proteins. In: Kapikian A Z, editor. Viral infections of the gastrointestinal tract. New York, N.Y: Marcel Dekker Inc.; 1994. pp. 169–250. [Google Scholar]
- 42.Nakata S, Adachi N, Ukae S, Kagawa K, Numata K, Urasawa S, Chiba S. Outbreaks of nosocomial rotavirus gastroenteritis in a pediatric ward. Eur J Pediatr. 1996;155:954–958. doi: 10.1007/BF02282886. [DOI] [PubMed] [Google Scholar]
- 43.Offit P A, Dudzik K I. Rotavirus-specific cytotoxic T lymphocytes passively protect against gastroenteritis in suckling mice. J Virol. 1990;64:6325–6328. doi: 10.1128/jvi.64.12.6325-6328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Offit P A, Clark H F, Blavat G, Greenberg H B. Reassortant rotaviruses containing structural proteins VP3 and VP7 from different parents induce antibodies protective against each parental serotype. J Virol. 1986;60:491–496. doi: 10.1128/jvi.60.2.491-496.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Ryan M L, Matson M D, Estes M K, Pickering I K. Anti-rotavirus G type specific and isotype specific antibodies in children with natural rotavirus infections. J Infect Dis. 1994;169:504–511. doi: 10.1093/infdis/169.3.504. [DOI] [PubMed] [Google Scholar]
- 46.Panum I, Thisthed E, Glikmann G, Obel N, Kofoed M, Sambo N H, Valerius E, Mordhorst C H. Respiratory syncytial virus: detection of secretory specific IgM and IgA antibodies by ELISA in nasopharyngeal aspirates from children with acute respiratory disease, a useful supplement to antigen detection. Clin Diagn Virol. 1997;8:219–226. doi: 10.1016/s0928-0197(97)10002-2. [DOI] [PubMed] [Google Scholar]
- 47.Ramachandran M, Gentsch J R, Parashar V D, Jin S, Woods P A, Holmes J L, Kirkwood C D, Bishop R F, Greenberg H B, Urasawa S, Gerna G, Coulson B S, Taniguchi K, Breese J S, Glass R I The National Rotavirus Strain Surveillance System Collaborating Laboratories. Detection and characterization of novel rotavirus strains in the United States. J Clin Microbiol. 1998;36:3223–3229. doi: 10.1128/jcm.36.11.3223-3229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramachandran M, Vij A, Kumar R, Das B K, Gentsch J R, Bahn M K, Glass R I. Lack of maternal antibodies to P serotypes may predispose neonates to infections with unusual rotavirus strains. Clin Diagn Lab Immunol. 1998;5:527–530. doi: 10.1128/cdli.5.4.527-530.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramachandran M, Das B K, Vij A, Kumar R, Bhambal N, Kesari N, Rawat H, Bahl L, Thakur S, Woods P A, Glass R I, Bahn M K, Glass R I. Unusual diversity of human rotavirus G and P genotypes in India. J Clin Microbiol. 1996;34:436–439. doi: 10.1128/jcm.34.2.436-439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruggeri F M, Greenberg H B. Antibodies to the trypsin cleavage peptide VP8* neutralize rotavirus by inhibiting binding of virions to target cells in culture. J Virol. 1991;65:2211–2219. doi: 10.1128/jvi.65.5.2211-2219.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryder R W, Singh N, Reeves W C, Kapikian A Z, Greenberg H B, Sack R B. Evidence of immunity induced by naturally acquired rotavirus and Norwalk virus infections on two remote Panamanian islands. J Infect Dis. 1985;151:99–105. doi: 10.1093/infdis/151.1.99. [DOI] [PubMed] [Google Scholar]
- 52.Santos N, Riepenhoff-Talty M, Clark H F, Offit P, Gouvea V. VP4 genotyping of human rotavirus in the United States. J Clin Microbiol. 1994;32:205–208. doi: 10.1128/jcm.32.1.205-208.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Timenetsky M D S T, Santos N, Gouvea V. Survey of rotavirus G and P types associated with human gastroenteritis in São Paulo, Brazil. J Clin Microbiol. 1994;32:2622–2624. doi: 10.1128/jcm.32.10.2622-2624.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ward R L, Bernstein D I. Lack of correlation between serum antibody titers and protection following vaccination with reassortant RRV vaccines. US Rotavirus Vaccine Efficacy Group Vaccine. 1995;13:1226–1232. doi: 10.1016/0264-410x(95)00060-e. [DOI] [PubMed] [Google Scholar]
- 55.Ward R L. Mechanisms of protection against rotavirus in humans and mice. J Infect Dis. 1996;174(Suppl. 1):S51–S58. doi: 10.1093/infdis/174.supplement_1.s51. [DOI] [PubMed] [Google Scholar]
- 56.Wilde J, Eiden J, Yolken R. Removal of inhibitory substances from human fecal specimens for detection of group A rotaviruses by reverse transcriptase and polymerase chain reactions. J Clin Microbiol. 1990;28:1300–1307. doi: 10.1128/jcm.28.6.1300-1307.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woods P A, Gentsch J, Gouvea V, Mata L, Simhon A, Santosham M, Bai Z S, Urasawa Y, Glass R I. Distribution of serotypes of human rotavirus in different populations. J Clin Microbiol. 1992;30:781–785. doi: 10.1128/jcm.30.4.781-785.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wyatt R G, James H D, Jr, Pittmann A L. Direct isolation in cell culture of human rotaviruses and their characterization into four serotypes. J Clin Microbiol. 1983;18:310–317. doi: 10.1128/jcm.18.2.310-317.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wyatt R G, Kapikian A Z, Mebus C A. Induction of cross-reactive serum neutralizing antibodies to human rotavirus in calves after in utero administration of bovine rotavirus. J Clin Microbiol. 1983;18:505–508. doi: 10.1128/jcm.18.3.505-508.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu L, Harbour D, McCrae M A. The application of polymerase chain reaction to the detection of rotavirus in faeces. J Virol Methods. 1990;27:29–38. doi: 10.1016/0166-0934(90)90143-4. [DOI] [PubMed] [Google Scholar]