Abstract
The P (VP4) and G (VP7) genotypes of 167 group A rotavirus strains obtained during the period 1996 to 1998 from 149 children living in a suburban community in Guinea-Bissau, western Africa, were determined by the reverse transcription-PCR technique. A total of nine combinations including five different P types and five different G types were identified. The globally common genotype pairs P[8], G1; P[4], G2; P[8], G3 and P[8], G4 were underrepresented in this study area. We found a substantial year-to-year variation in the occurrence of the genotype combinations. In 1996 and 1997, P[6], G2 was the most frequent, whereas P[8], G1 was more common in 1998. The unusual type P[9], G3 and a few mixed infections were detected. Sixteen percent of the rotavirus-positive samples were nontypeable.
Rotavirus (RV) is a major cause of acute diarrhea in children worldwide and an important cause of child death in the developing world (20). RV, a member of the Reoviridae family, has a triple-layered capsid with two major outer capsid surface proteins, VP4 and VP7, which are involved in virus neutralization following natural infection and which are thought to be important in the development of protective immunity. The antigenic specificities of VP4 and VP7 are termed P (for protease-sensitive protein) and G (for glycoprotein), respectively (4, 10). A description of the most common P and G genotypes and their corresponding serotypes is important for designing relevant RV vaccine formulations. Whereas genotype distributions have been described in studies from Europe, the Americas, and Asia (1, 12, 16, 17, 20, 25, 28, 30, 31), only a few reports have emerged from Africa (3, 8, 22, 23, 29), and none of these are from western Africa. From this region only a single RV serotyping study, from The Gambia, has been reported (27). RV infections in Guinea-Bissau demonstrate a consistent temporal pattern characterized by a seasonal peak of infections during the cooler months from January to March, with very few infections in the intervening months (21). The objective of the present study was to describe P and G genotype combinations of RV from children in Bissau, the capital of Guinea-Bissau.
MATERIALS AND METHODS
Study population and identification of RV.
The study was undertaken in the four suburban districts of Bandim I, Bandim II, Belem, and Mindera of Bissau from 15 January 1996 to 24 March 1998. The surveillance population consisted of three groups. The first group (group I) included all children born between 31 June 1994 and 1 January 1997 and residing in or moving to 603 randomly selected houses included in a prospective diarrhea surveillance study with weekly morbidity visits. If the child had diarrhea on the day of the visit, a stool sample was collected. The second group (group II) included 200 children born into families living in these houses between 15 January 1996 and 15 January 1997; these children were monitored for 2 years from birth with weekly stool sampling irrespective of diarrheal status. The third group (group III) included all children living in the study area from January 1996 to January 1997 who were hospitalized at the central hospital of Simaõ Mendes and who had a stool sample analyzed upon admission, again irrespective of their symptomatology. Diagnosis of RV was carried out on stool samples by an enzyme-linked immunosorbent assay (DAKO IDEA, Copenhagen, Denmark), and positive samples were stored at −20°C before genotyping at the National Public Health Laboratory, Bissau, Guinea-Bissau, in the period from November 1997 to May 1998. If the same child was shedding RV twice within a period of 2 weeks, only the first RV isolate was included in the analyses.
Oligonucleotide primers.
The nucleotide positions and sequences of primers (5′ to 3′) used to identify the G genotype were as follows: 9con 1 (nucleotides [nt] 37 to 56), TAGCTCCTTTTAATGTATGG; 9con 2 (nt 922 to 941), GTATAAAATACTTGCCACCA; 9T1-1 (nt 176 to 195), TCTTGTCAAAGCAAATAATG; 9T1-2 (nt 262 to 281), GTTAGAAATGATTCTCCACT; 9T-3P (nt 484 to 503), GTCCAGTTGCAGTGTTAGC; 9T-4 (nt 423 to 440), GGGTCGATGGAAAATTCT; 9T-9B (nt 131 to 147), TATAAAGTCCATTGCAC. For P genotyping, the sequences of primers (5′ to 3′) were as follows: con 3 (nt 11 to 32), TGGCTTCGCTCATTTTATAGACA; con 2 (nt 868 to 887), ATTTCGGACCATTTATAACC; 1T-1 (nt 339 to 356), TCTACTTGGATAACGTGC; 2T-1 (nt 474 to 494), CTATTGTTAGAGGTTAGAGTC; 3T-1 (nt 259 to 278), TGTTGATTAGTTGGAT TCAA; 4T-1 (nt 385 to 482), TGAGACATGCAATTGGAC; 5T-1 (nt 575 to 594), ATCATAGTTAGTAGTCGG.
RNA extraction.
The RV double-stranded RNA (dsRNA) was isolated from the stool specimens with the RNaid kit, (Bio 101, Inc., La Jolla, Calif.) by a modification of previously published procedures (11). One milliliter of a 20% stool suspension was prepared by mixing the sample with 50 mM Tris-HCl, pH 7.5. For reference strains, a similar suspension was prepared by mixing 500 μl of RV-containing cell culture supernatant with 30 μl of 1 M Tris-Cl, pH 7.5. The suspension was vigorously vortexed, followed by centrifugation for 1 min at 5,400 × g. Four hundred microliters of the supernatant was mixed gently with 800 μl of 6 M guanidine thiocyanate (GITC) (Boehringer GmbH, Mannheim, Germany) dissolved in a 50 mM Tris-Cl buffer, pH 7.5. After incubation at 56°C for 10 min with intermittent gentle mixing by inversion, 10 μl of RNaid matrix was added, and the tube was shaken gently for 5 min at room temperature.
The RNaid matrix with bound RNA was pelleted by centrifugation at 1,300 × g for 1 min, and the supernatant was discarded. Four consecutive wash steps followed, the first step with a washing solution of 4 M GITC dissolved in 50 mM Tris-Cl, pH 7.5, and the last three steps with a wash solution consisting of 50 mM Tris-Cl, pH 7.5, 0.5 mM EDTA, and 50% ethanol. For each washing step, the pellet was completely resuspended by gentle vortexing in 1,200 μl of wash solution, followed by centrifugation at 1,300 × g for 1 min, and then the supernatant was discarded. After the last wash, the tube was centrifuged at 10,000 × g for 2 min, and the remaining supernatant was aspirated with a pipette. The samples were then left to dry overnight at room temperature or desiccated under vacuum. The RNA was extracted twice from the matrix by each time adding 30 μl of double-distilled H2O (ddH2O), followed by incubation for 20 min at 65°C with intermittent vigorous vortexing. After centrifugation for 2 min at 10,000 × g, the two RNA-containing supernatants were pooled and kept at −20°C until use.
RT reaction.
In two separate PCR tubes, 3 μl of RNA extract was mixed with 1 μl of 20 μM 9Con 1 or con 3, for the G and P typing, respectively. After being heated to 97°C for 5 min, the tubes were immediately put on ice for 1 min, centrifuged to collect the liquid, and incubated for 5 min further at 65°C to complete primer annealing. At 42°C, a solution containing 3.0 μl of 10× PCR buffer (100 mM Tris-Cl [pH 8.3], 500 mM KCl), 3.0 μl of 2 mM deoxynucleoside triphosphates (dNTP) (a 2 mM concentration each of dATP, dGTP, dCTP, and dTTP), 6.0 μl of 25 mM MgCl2, 7 U of avian myeloblastosis virus reverse transcriptase (Promega Corp., Madison, Wis., or Molecular Genetics Resources), and ddH2O to bring the volume to 26 μl was added to each tube, followed by incubation at 42°C for 1 h.
One-step PCR.
A 45-μl solution containing 34.8 μl of ddH2O, 4.5 μl of 10× PCR buffer, 4.5 μl of 2 mM dNTP, and 0.2 μl of 5-U/μl Taq DNA polymerase was added to each completed RT reaction mixture. Finally, 1 μl of 20 μM G mixture (20 μM concentrations of each of the 9T-1, 9T1-2, 9T-3P, 9T-4, and 9T-9B primers, corresponding to G1, G2, G3, G4, and G9) and 1 μl of 20 μM P mixture (20 μM concentrations of each of the 1T-1, 2T-1, 3T-1, 4T-1, and 5T-1 primers, corresponding to P[8], P[4], P[6], P[9], and P[10]) were added to the tubes for G and P genotyping, respectively. The tubes were subsequently subjected to 30 cycles of 1 min at 94°C, 2 min at 50°C, and 2 min at 72°C. To confirm the specificity of this RT-PCR method, tissue culture supernatants of the reference strains Wa (G1), DS-1 (P[4], G2), P (G3), ST3 (G4), M37 (P[6]), WI61 (P[8], G9), K8 (P[9]), and 69M (P[10]) were analyzed.
Two-step PCR.
All nontypeable (NT) samples were reextracted, and the dsRNA was reverse transcribed and amplified by the one-step PCR method with 1 μl of 20 μM 9con 2 and 1 μl of 20 μM con 2 used instead of the corresponding G mixture and P mixture. If bands were visible after electrophoresis, the 9con 2 or the con 2 amplification products were subjected to a new PCR, where 5 μl of the PCR products was mixed with 0.5 μl of 20 μM 9con 1 or 0.5 μl of 20 μM con 3 primer, respectively. Then 0.5 μl of 20 μM concentrations of each of the G or P genotype-specific primers, 10 μl of 5 mM dNTP, 12 μl of 25 mM MgCl2, 10 μl of 10× PCR buffer, 0.2 μl of Taq DNA polymerase, and 59.8 μl of ddH2O were added to each tube. The tubes were subjected to 30 cycles of PCR with the same steps and cycles as those described for one-step PCR.
Agarose gel analysis.
The PCR products were analyzed on a 2% agarose gel (Sigma Chemical Co., St. Louis, Mo.), together with a 123-bp DNA ladder (Life Technologies Inc., Rockville, Md.), and visualized by ethidium bromide staining.
RESULTS
A total of 203 samples were RV positive. Eleven children shed RV twice within a 2-week period and, accordingly, only the first RV isolate from each of these children was included. Of the remaining 192 samples, 167 had sufficient sample material for further examination with RT-PCR. These 167 samples were collected from 149 children of whom 42 had participated in the prospective diarrhea surveillance study (group I), 97 were from the birth cohort (group II), and 10 were children admitted to the Simaõ Mendes hospital (group III). All samples were found in the months December to April in 1996 (58 samples), 1997 (101 samples), and 1998 (44 samples); no RV-positive samples were identified in the intervening months.
Specimens that yielded either P or G bands were reexamined with a repeated dsRNA extraction and the one-step G or P genotyping RT-PCR, respectively. Sixty specimens were successfully typed with the one-step RT-PCR. Specimens that yielded neither of the two bands (i.e., NT strains) were reexamined with a repeated dsRNA extraction and the two-step RT-PCR procedure for both genotypes. Eight specimens (i.e., four P types and four G types) were typed by this procedure. In total, 140 samples yielded P and/or G bands while, despite reexamination with the two-step amplification procedure, 27 samples remained NT.
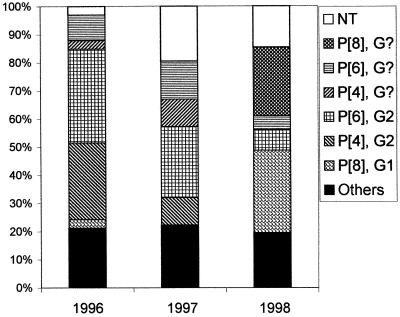
Table 1 shows that in the first 2 years of our survey, P[6], G2 was the most frequent genotype, while P[8], G1 was the most commonly isolated genotype in 1998 (Fig. 1). In 1996, 11 (33%) of the strains were P[6], G2, followed by 9 P[4], G2 (27%) and 3 P[6], G? (the RT-PCR for VP7 yielded no identifiable band) (9%) types. In 7 (21%) of the samples, only the P or the G type could be established. In 1997, 26 (28%) of the strains were P[6], G2, followed by 14 (15%) P[6], G? and 10 (11%) P[4], G? strains. In 31 cases (33%), only the P or the G type could be determined, and a total of 20 isolates (22%) were found to be NT. In 1998, only 3 (7%) were P[6], G2 and 12 (29%) were P[8], G1, while more than a third of the samples (36%) could only be classified with regard to either the G or the P type. Over the 3-year study, six cases of mixed infections were detected, with strains carrying double P bands identified as P[4] and P[6] or P[10]. In 1997, two cases of P[4]/P[6], G2 were observed, and in one case, the G genotype was NT. In 1998, two cases of P[4]/P[6], G1 and one case of P[4]/P[6] with no identifiable G type were found. The proportions of NT strains in 1996, 1997, and 1998 were 3, 22, and 15%, respectively. Figure 1 shows the relative distribution of the major genotypes during the 3 years.
TABLE 1.
P and G genotypes of 167 rotavirus strains collected from 149 children in Bissau, Guinea-Bissau, during the period from January 1996 to April 1998
| P, G typesa | No. of strains in:
|
|||
|---|---|---|---|---|
| 1996 | 1997 | 1998 | 1996–1998 | |
| P[2], G8 | 0 | 0 | 2 | 2 |
| P[4], G2 | 9 | 10 | 0 | 19 |
| P[4], G3 | 1 | 0 | 0 | 1 |
| P[6], G1 | 2 | 1 | 0 | 3 |
| P[6], G2 | 11 | 26 | 3 | 40 |
| P[6], G3 | 1 | 0 | 0 | 1 |
| P[8], G1 | 1 | 0 | 12 | 13 |
| P[9], G3 | 0 | 1 | 0 | 1 |
| P[9], G4 | 0 | 0 | 1 | 1 |
| P[4], G? | 1 | 10 | 0 | 11 |
| P[6], G? | 3 | 14 | 2 | 19 |
| P[8], G? | 0 | 0 | 10 | 10 |
| P[9], G? | 0 | 0 | 1 | 1 |
| P[?], G1 | 1 | 1 | 1 | 3 |
| P[?], G2 | 2 | 5 | 1 | 8 |
| P[?], G4 | 0 | 1 | 0 | 1 |
| Mixed | 0 | 4 | 2 | 6 |
| NT | 1 | 20 | 6 | 27 |
| Total | 33 | 93 | 41 | 167 |
?, RT-PCR produced no identifiable band.
FIG. 1.
Relative distribution of the major genotype combinations, the NT strains, and the remaining combinations (others) during the period from January 1996 to April 1998 in suburban Bissau, Guinea-Bissau (n = 33 for 1996, 93 for 1997, and 41 for 1998).
DISCUSSION
The 167 isolates in this study were of nine distinct genotype combinations, with additional variation within the group of mixed strains and mixed infections. Studies of the distribution of RV genotypes from different geographic locations have suggested that the most prevalent strains causing childhood diarrhea worldwide have genotype P[8], G1 followed by strains having P[8], G4; P[4], G2 and P[8], G3 (2, 12, 16, 31). Genotypes P[8], G4 and P[8], G3, which are common worldwide, were not observed in our study, whereas P[8], G1; P[4], G2 and P[6], G2 were frequent. Combinations which included G3 or P[9] were observed in only five samples. The relative frequency of the P genotypes in South Africa (29) corresponded to the general worldwide distribution, with a high frequency of P[8] (64%) and a low frequency of P[6] (8%). Two studies of RV G serotypes from The Gambia and Kenya (14, 23, 31) showed that the most commonly identified serotype in both countries was G1, whereas in a recent genotype study from Malawi (8), a high proportion of G8 strains was detected. However, in other developing countries, the genotypes common worldwide have been found to be underrepresented, as in Indian and Brazilian children (7, 13, 19, 26). In India, strains of P[6] (P[6], G1; P[6], G2; P[6], G3 and P[6], G9), which primarily infect newborns, were common in children with diarrhea, and in Brazil, a number of uncommon serotypes (P[6], G1; P[6], G3; P[6], G4; P[3], G1 and P[3], G3) represented one-third of all RV-related diarrhea cases.
In addition, a novel G5 serotype was detected in Brazilian children with diarrhea (13), so some developing countries clearly have a high prevalence of strains not included in the recently licensed, but now suspended (6), rhesus-human reassortant rotavirus tetravalent (RRV-TV) vaccine.
A Brazilian study from 1996 and 1997 did not identify the G2 serotype among the 46 RV isolates tested (28), whereas in our study from the same years, the corresponding G2 genotype was the most frequently identified, with 24% of the strains having genotype P[6], G2. Thus, 69 G2 strains were identified, of which 40 were from children with diarrhea (data not shown). RV of genotype P[6] has previously been associated with asymptomatic infections in neonatal wards, but recent studies have documented several cases of diarrhea in children associated with RV strains of this type (7–9, 26). A year-to-year variation of the regionally circulating RV serotypes and genotypes has been observed in studies from different parts of the world (2, 12, 15, 18, 23, 30), and such changes were also observed in our study. Genotype P[6], G2 was the most prevalent during the first two epidemic seasons, i.e., in 1996 and 1997, while during the winter of 1998 this was superseded by P[8], G1. The importance of these year-to-year variations is still to be deciphered and is supposedly closely linked to the immunology of RV infections (5). In our study, strains from six children yielded double P bands identified as P[4] and P[6] or P[4] and P[10]. As reported earlier (22), mixed infections between members of different RV genetic subgroups represent a potential for natural reassortment between the genogroups. So far, the P[4]/P[6], G2 and P[4]/P[6], G1 genotypes have not been described in other studies and could represent either a coinfection with two different RVs or a regional natural reassortment between two wild-type RVs.
Despite several attempts to reextract the dsRNA and perform the two-step amplification RT-PCR, 27 (16%) of all samples remained NT. Fifteen of these samples did not yield visible bands after two-step amplification and gel electrophoresis and could represent potential new human RV genotypes. The inability to type these strains was not related to the time of specimen storage prior to analysis. A substantial year-to-year variation in the isolation frequency of NT strains was observed. Thus, in 1997, 21% of all RV strains were found to be NT, in contrast to 3% in 1996 and 15% in 1998. Whether this represents an epidemic of one or a few hitherto-undescribed genotypes awaits in-depth studies of these specimens.
Because most deaths due to RV diarrhea occur in Asia, Africa, and less-developed areas of Latin America, these are the regions which have the greatest potential for benefiting from an effective RV vaccine. However, the present study as well as similar studies from other low-income societies (26, 30) suggest that the diversity of genotypes may be greater than that represented in the RRV-TV vaccine. The finding of a high frequency of P[6] supports the current efforts to develop vaccines against diverse human RV P types (24).
The substantial genotypic diversity and the year-to-year variation in the frequency with which the various genotypes occur underscore the importance of a global surveillance over several years for further defining candidate RV vaccine antigens. In addition, detailed baseline data are needed to determine a shift in genotype distribution following attempts to vaccinate large child populations against RV.
ACKNOWLEDGMENTS
This study was supported by the EU-STD3 (contract no. TC*-CT94-0311), the WHO-V27/181/115, the Norwegian Research Council through grant 120779/730, L. Meltzers Høyskolefond, and the University of Bergen through grants to T.K.F. and H.S.
We thank Fransisco Dias for providing laboratory facilities at the Laboratorio National de Saude Publica, Bissau, and Peter Gerner-Smidt for laboratory support at Statens Serum Institute, Copenhagen. We are grateful to Bimal K. Das, All Indian Institute of Medical Sciences, New Delhi, for useful suggestions in connection with establishing the RT-PCR technique.
REFERENCES
- 1.Arista S, Vizzi E, Ferraro D, Cascio A, Di Stefano R. Distribution of VP7 serotypes and VP4 genotypes among rotavirus strains recovered from Italian children with diarrhea. Arch Virol. 1997;142:2065–2071. doi: 10.1007/s007050050224. [DOI] [PubMed] [Google Scholar]
- 2.Beards G, Desselberger U, Flewett T H. Temporal and geographical distributions of human rotavirus serotypes, 1983 to 1988. J Clin Microbiol. 1989;27:2827–2833. doi: 10.1128/jcm.27.12.2827-2833.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biritwum R B, Isomura S, Yamaguchi H, Toba M, Mingle J A. Seroepidemiological study of rotavirus infection in rural Ghana. Ann Trop Paediatr. 1984;4:237–240. doi: 10.1080/02724936.1984.11748343. [DOI] [PubMed] [Google Scholar]
- 4.Bishop R F. Natural history of human rotavirus infections. In: Kapikian A Z, editor. Viral infections of the gastrointestinal tract. 2nd ed. New York, N.Y: Marcel Dekker, Inc.; 1984. pp. 131–167. [Google Scholar]
- 5.Bishop R F, Barnes G L, Cipriani E, Lund J S. Clinical immunity after neonatal rotavirus infection: a prospective longitudinal study in young children. N Engl J Med. 1983;309:72–76. doi: 10.1056/NEJM198307143090203. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Intussusception among recipients of rotavirus vaccine—United States, 1998–1999. Morbid Mortal Weekly Rep. 1999;48:577–581. [PubMed] [Google Scholar]
- 7.Chatterjee B, Hussain M, Seth P, Broor S. Diversity of rotavirus strains infecting pediatric patients in New Delhi, India. J Trop Pediatr. 1996;42:207–210. doi: 10.1093/tropej/42.4.207. [DOI] [PubMed] [Google Scholar]
- 8.Cunliffe N A, Gondwe J, Broadhead R L, Molyneux M E, Woods P A, Bresee J S, Glass R I, Gentsch J R, Hart C A. Rotavirus G and P types in children with acute diarrhea in Blantyre, Malawi, from 1997 to 1998. J Med Virol. 1999;57:308–312. [PubMed] [Google Scholar]
- 9.Das B K, Gentsch J R, Cicirello H, Woods P, Gupta A, Ramachandran M, Kumar R, Glass R I. Characterization of rotavirus strains from newborns in New Delhi, India. J Clin Microbiol. 1994;32:1820–1822. doi: 10.1128/jcm.32.7.1820-1822.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Estes M K, Cohen J. Rotavirus gene structure and function. Microbiol Rev. 1989;53:410–449. doi: 10.1128/mr.53.4.410-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gentsch J R, Glass R I, Woods P, Gouvea V, Gorziglia M, Flores J, Das B K, Bhan M K. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992;30:1365–1373. doi: 10.1128/jcm.30.6.1365-1373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gentsch J R, Woods P, Ramachandran M, Das B K, Leite J P, Alfieri A, Kumar R, Bhan M K, Glass R I. Review of G and P typing results from a global collection of rotavirus strains: implications for vaccine development. J Infect Dis. 1996;174(Suppl. 1):30–36. doi: 10.1093/infdis/174.supplement_1.s30. [DOI] [PubMed] [Google Scholar]
- 13.Gouvea V, de Castro L, Timenetsky M C, Greenberg H, Santos N. Rotavirus serotype G5 associated with diarrhea in Brazilian children. J Clin Microbiol. 1994;32:1408–1409. doi: 10.1128/jcm.32.5.1408-1409.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanlon P, Hanlon L, Marsh V. Epidemiology of rotavirus in a periurban Gambian community. Ann Trop Paediatr. 1987;7:238–243. doi: 10.1080/02724936.1987.11748515. [DOI] [PubMed] [Google Scholar]
- 15.Hoshino Y, Kapikian A Z. Rotavirus antigens. Curr Top Microbiol Immunol. 1994;185:179–227. doi: 10.1007/978-3-642-78256-5_7. [DOI] [PubMed] [Google Scholar]
- 16.Huixia W, Taniguchi K, Urasawa T, Urasawa S. Serological and genomic characterization of human rotaviruses detected in China. J Med Virol. 1998;55:168–176. doi: 10.1002/(sici)1096-9071(199806)55:2<168::aid-jmv14>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 17.Kaga E, Iizuka M, Nakagomi T, Nakagomi O. The distribution of G (VP7) and P (VP4) serotypes among human rotaviruses recovered from Japanese children with diarrhea. Microbiol Immunol. 1994;38:317–329. doi: 10.1111/j.1348-0421.1994.tb01784.x. [DOI] [PubMed] [Google Scholar]
- 18.Kapikian A Z, Chanock R M. Rotaviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1657–1708. [Google Scholar]
- 19.Leite J P, Alfieri A A, Woods P, Glass R I, Gentsch J R. Rotavirus G and P types circulating in Brazil: characterization by RT-PCR, probe hybridization, and sequence analysis. Arch Virol. 1996;141:2365–2374. doi: 10.1007/BF01718637. [DOI] [PubMed] [Google Scholar]
- 20.Maunula L, van Bonsdorff C-H. Rotavirus serotypes and electropherotypes in Finland from 1986 to 1989. Arch Virol. 1986;140:877–890. doi: 10.1007/BF01314964. [DOI] [PubMed] [Google Scholar]
- 21.Mølbak K, Wested N, Højlyng N, Scheutz F, Gottschau A, Aaby P, da Silva A P. The etiology of early childhood diarrhea: a community study from Guinea-Bissau. J Infect Dis. 1994;169:581–587. doi: 10.1093/infdis/169.3.581. [DOI] [PubMed] [Google Scholar]
- 22.Mphahlele M, Steele A. Relative frequency of human rotavirus VP4 (P) genotypes recovered over a ten-year period from South African children with diarrhea. J Med Virol. 1995;47:1–5. doi: 10.1002/jmv.1890470102. [DOI] [PubMed] [Google Scholar]
- 23.Nakata S, Gatheru Z, Ukae S, Kobayashi N, Honma S, Muli J, Ogaja P, Nyangao J, Kiplagat E, Tukei P M, Steele M. Epidemiological study of the G serotype distribution of group A rotaviruses in Kenya from 1991 to 1994. J Med Virol. 1999;58:296–303. doi: 10.1002/(sici)1096-9071(199907)58:3<296::aid-jmv17>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 24.Offit P A, Clark H F. The rotavirus vaccine. Curr Opin Pediatr. 1999;11:9–13. doi: 10.1097/00008480-199902000-00003. [DOI] [PubMed] [Google Scholar]
- 25.Padilla-Noriega L, Mendez-Toss M, Menchaca G, Contreras J F, Romero-Guido P, Puerto F I, Guiscafre H, Mota F, Herrera I, Cedillo R, Munoz O, Calva J, Guerrero M L, Coulson B S, Greenberg H B, Lopez S, Arias C F. Antigenic and genomic diversity of human rotavirus VP4 in two consecutive epidemic seasons in Mexico. J Clin Microbiol. 1988;36:1688–1692. doi: 10.1128/jcm.36.6.1688-1692.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramachndran M, Das B K, Vij A, Kumar R, Bhambal S S, Kesari N, Rawat H, Bahl L, Thakur S, Woods P, Glass R I, Bhan M K, Gentsch J R. Unusual diversity of human rotavirus G and P genotypes in India. J Clin Microbiol. 1996;34:436–439. doi: 10.1128/jcm.34.2.436-439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rowland M, Goh S, Williams K, Campbell A, Beards G, Sanders R, Flewett T. Epidemiological aspects of rotavirus infection in young Gambian children. Ann Trop Paediatr. 1985;5:23–28. doi: 10.1080/02724936.1985.11748354. [DOI] [PubMed] [Google Scholar]
- 28.Santos N, Lima R C, Pereira C F, Gouvea V. Detection of rotavirus types G8 and G10 among Brazilian children with diarrhea. J Clin Microbiol. 1998;36:2727–2729. doi: 10.1128/jcm.36.9.2727-2729.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steele A, van Nekerk M, Mphahlele J. Geographic distribution of human rotavirus VP4 genotypes and VP7 serotypes in five South African regions. J Clin Microbiol. 1995;33:1516–1519. doi: 10.1128/jcm.33.6.1516-1519.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Timenetsky M, Santos N, Gouvea V. Survey of rotavirus G and P types associated with human gastroenteritis in Sao Paolo, Brazil, from 1986 to 1992. J Clin Microbiol. 1994;32:2622–2624. doi: 10.1128/jcm.32.10.2622-2624.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woods P, Gentsch J, Gouvea V, Mata L, Simhon A, Santosham M, Bai Z, Urasawa S, Glass R I. Distribution of serotypes of human rotavirus in different populations. J Clin Microbiol. 1992;30:781–785. doi: 10.1128/jcm.30.4.781-785.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]