Abstract
The pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) led to the death of millions of people worldwide and thousands more infected individuals developed sequelae due to the disease of the new coronavirus of 2019 (COVID-19). The development of several studies has contributed to the knowledge about the evolution of SARS-CoV2 infection and the disease to more severe forms. Despite this information being debated in the scientific literature, many mechanisms still need to be better understood in order to control the spread of the virus and treat clinical cases of COVID-19. In this article, we carried out an extensive literature review in order to bring together, in a single article, the biological, social, genetic, diagnostic, therapeutic, immunization, and even socioeconomic aspects that impact the SAR-CoV-2 pandemic. This information gathered in this article will enable a broad and consistent reading of the main aspects related to the current pandemic.
Keywords: SARS-CoV-2, COVID-19, coronavirus, pandemic, infection
Introduction
On December 8, 2019, a viral infection, previously unidentified, characterized by severe pneumonia, was reported in an individual who frequented a small local fish and wildlife market in the city of Wuhan, Hubei Province, China (Lu H. et al., 2020). The analysis based on nucleotide sequencing technology of the virus genome isolated from the blood of sick individuals led to the identification of a new coronavirus as the causative agent of the outbreak (Gorbalenya et al., 2020). Initially, the infected individuals were those who visited the seafood market or consumed foods of animal origin probably infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Later, a more in-depth analysis through contact tracing of patients positive for COVID-19 revealed that several individuals with no history of trips to the seafood market also tested positive for the disease, indicating the possibility of transmission from person to person (Chan J. F. et al., 2020; Chen J. et al., 2020; Ji et al., 2020).
During the second week of January 2020, due to the travel season of the Spring Festival, the new SARS-CoV-2 spread to other provinces of China and thus to other countries (Lu H. et al., 2020). The first case of SARS-CoV-2 infection confirmed outside of China was in Thailand on January 13, 2020. On January 16, 2020, the first case was confirmed in Japan. As of January 25, 2020, the number of confirmed cases had reached 2062, including in countries such as Hong Kong, Macao, Australia, Malaysia, Singapore, France, South Korea, Taiwan, United States, Vietnam, Nepal, and Sweden (Dhama et al., 2020). Due to the severity of this outbreak and its ability to spread internationally, the World Health Organization (WHO) declared a global health emergency on January 31, 2020. On March 11, 2020, a pandemic was declared (Cucinotta and Vanelli, 2020). Data published by the WHO showed that up to December 3, 2021, 22,105,872 people had been confirmed infected by the new coronavirus, 614,964 had died, and 312,827,402 doses of vaccine were administered (World Health Organization [WHO], 2021a).
This review aims to explore and summarize the available evidence on the main viral characteristics, immune response, diagnostic methods, therapeutic options and candidate/approved vaccines against SARS-CoV-2, so that this information can serve as a basis for a better understanding of future studies on SARS-CoV-2 and COVID-19.
SARS-CoV-2
Morphological, Genomic Structure and Replication of SARS-CoV-2
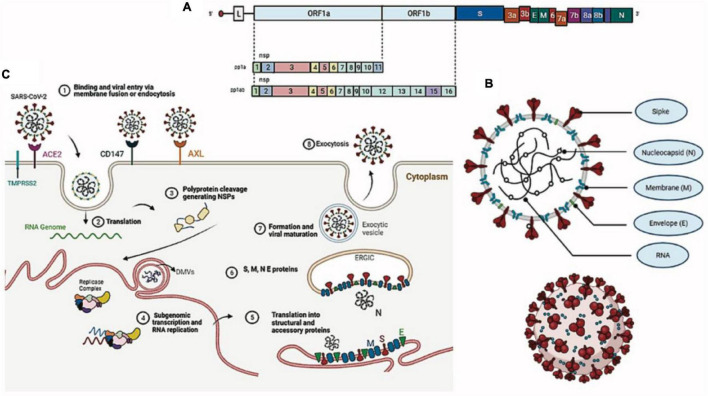
Coronaviruses are spherical, enveloped viruses of approximately 120 nm in diameter, containing a helical symmetry nucleocapsid, with a single-stranded RNA genome of positive polarity, non-segmented, 29.9 kb in size (NC_045512.2), and a GC content of 38% (Chan K. et al., 2020; Figure 1B). Its genome is composed of 13 open reading frames (ORFs) (Lu R. et al., 2020) encoding 7096 amino acids that constitute four structural proteins spike (S), envelope (E), membrane (M), and nucleocapsid (N) (Table 1) and 15 non-structural proteins (NSP1, NSP2, NSP3, NSP4, NSP5, NSP6, NSP7, NSP8, NSP9, NSP10, NSP12, NSP13, NSP14, NSP15, and NSP16), in addition to eight accessory proteins (3a, 3b, 6, 7a, 7b, 8b, 9b, and ORF14) that perform numerous functions in the processes of virus replication and assembly (Wu A. et al., 2020; Figure 1A). In comparison to SARS-CoV, SARS-CoV-2 lacks protein 8a but has a longer 8b protein, with approximately 121 amino acids, and a shorter 3b protein, containing 22 amino acids (Chan K. et al., 2020; Wu A. et al., 2020). More than 380 amino acid substitutions, located mainly in the NSP3, NSP2, and S proteins, have been identified between SARS-CoV-2 isolates and the consensus sequence, leading to divergence in the functional and pathogenic traits between it and other coronaviruses (Wu A. et al., 2020). We synthesized the role of structural proteins in the SARS-CoV-2 replication cycle (Table 1).
FIGURE 1.
Morphological structure, genome, and replication of SARS-CoV-2. (A) Viral genome. (B) Viral particle. (C) SARS-CoV-2 replication cycle.
TABLE 1.
SARS-CoV-2 structural proteins and their roles in the viral replication cycle.
| Protein | Function | References |
| Spike (S) | Divided into two subunits (S1 and S2) it is able to bind to the host cell through its receptor-binding domain. The S1 subunit is involved in binding the virus to the host cell membrane, while the S2 subunit acts in fusing the viral envelope with the cell membrane. | Hoffmann et al., 2020; Lan et al., 2020; Wrapp et al., 2020 |
| Nucleocapsid (N) | It binds and packages viral RNA into ribonucleoprotein complexes (RNP) located inside the viral envelope, forming a separate layer from the M, E, and S envelope proteins. This protein is recruited into the replication-transcription complex by NSP3 and, therefore, it is believed to be also involved in viral genome replication. | Chang et al., 2020; Cong et al., 2020; Yao et al., 2020 |
| Envelope (E) | Protein E has a transmembrane domain and is relatively small in size, with about 75 amino acids, which aid in the assembly and release of virions. |
Nieto-Torres et al., 2015; Venkatagopalan et al., 2015; Lu R. et al., 2020 |
| Membrane (M) | It has about 222 amino acids and is the most present protein in the viral particle, giving definitive shape to the virion envelope. This protein works simultaneously with proteins E, N, and S and plays an important role in ribonucleic acid (RNA) packaging and virus assembly. | Tang X. et al., 2020 |
The SARS-CoV-2 replication cycle begins with the binding of the S protein of the virus with the angiotensin-2 converting enzyme (ACE-2) of the host, considered the specific receptor of SARS-CoV-2 in human cells (Hoffmann et al., 2020). ACE2 is a type I membrane glycoprotein responsible for the conversion of angiotensin II into angiotensin 1–7 and is expressed in the lungs, nose, heart, intestine, and kidneys (Yan et al., 2020). The S protein of SARS-CoV-2 is a class I fusion protein that protects its fusion domain, keeping it hidden and inactive until the virus finds a host cell with its receptor, where it is then proteolytically cleaved into a hook-shaped structure that is necessary for its incorporation into the membrane of the target cell (Li and Petrovsky, 2016). The binding of the S protein to ACE-2 is dominated by polar contacts mediated by hydrophilic residues located in its C-terminal domain and promotes a cleavage event dependent on the endosomal cysteine proteases CatB and CatL or the transmembrane serine protease TMPRSS2, which exposes the protein S fusion peptide in order to favor viral entry (Hoffmann et al., 2020; Wang et al., 2020a; Yan et al., 2020). Two possible new SARS-CoV-2 receptors were recently identified: the tyrosine kinase AXL receptor and the transmembrane surface glycoprotein CD147 (inducer of extracellular matrix metalloproteinase or basigin), both capable of interacting with the S protein (Wang et al., 2020b; Wang S. et al., 2021). In addition, the cell receptor neuropilin-1 (NRP1), expressed in the olfactory epithelium, also seems to facilitate SARS-CoV-2 infection by interacting with the S protein (Daly et al., 2020).
After viral entry, the SARS-CoV-2 genomic RNA serves as a transcript that allows the translation of two polyproteins (pp1a and pp1ab), encoded in the 3′ two-thirds of the genome as ORF1a and ORF1b (Zhou P. et al., 2020). These polyproteins are cleaved by the action of two viral proteases (NSP3-PLpro and NSP5-Mpro), generating 16 non-structural proteins that are assembled into the replicase–transcriptase complex, which will later give rise to genomic and subgenomic RNAs (Fehr and Perlman, 2015). NSPs also induce cell membrane rearrangement to form double-membrane vesicles (DMVs), where the replication-transcription complex (RTC) is anchored (Ziebuhr et al., 2000; Angelini et al., 2013). The other third of the genome, at the 5′ end, encodes structural (S, E, M, and N) and accessory proteins (3a, 3b, 6, 7a, 7b, 8b, 9b, and ORF14) (Siu et al., 2008; Fehr and Perlman, 2015; Wu A. et al., 2020). In addition to complexes between viral proteins, different interaction complexes are formed between structural and non-structural proteins of the virus and host cell proteins (Srinivasan et al., 2020).
The replication of SARS-CoV-2 is a complex process that involves atypical RNA revision by NSP14, one of the non-structural proteins generated by the cleavage of the pp1ab polyprotein (Romano et al., 2020). Such a revision mechanism present in coronaviruses, unknown among RNA viruses before their discovery in SARS-CoV, results in replication error rates more than 10 times lower than those of other RNA viruses (approximately 10–6 mutations/nucleotide/cycle/nucleotide/cycle) (Rausch et al., 2020).
Structural proteins translated from subgenomic mRNAs are inserted into the endoplasmic reticulum (ER) and pass through the secretory pathway to the ER–Golgi intermediate compartment (ERGIC), where the S protein is cleaved into two subunits, S1 and S2 (Jaimes et al., 2020). The newly synthesized viral genome forms a complex with the N protein in the ERGIC for the assembly of new SARS-CoV-2 particles in an event mediated by the M protein with contributions from the E protein (Siu et al., 2008; Tseng et al., 2013). New viral particles emerge from ERGIC and are transported through vesicles to be released into the extracellular medium by exocytosis (Ulasli et al., 2010; Figure 1C).
Taxonomic Classification
Coronaviruses belong to the family Coronaviridae, which contains four genera (Alphacoronavirus, Betacoronavirus, Deltacoronavirus, and Gammacoronavirus) and includes species with a single-stranded RNA genome with positive polarity and 26–32 kb (International Committee on Taxonomy of Viruses [ICTV], 2019). SARS-CoV-2 belongs to the B strain of the genus Betacoronavirus, whose members infect only mammals (International Committee on Taxonomy of Viruses [ICTV], 2020; Mittal et al., 2020). Coronaviruses are zoonotic viruses with high mutation rates that infect a wide variety of wild and domestic animals and can also infect humans. Evolutionary analyses have shown that Alphacoronavirus and Betacoronavirus have bats and rodents as reservoirs, while birds are possible reservoirs of Deltacoronavirus and Gammacoronavirus (Chan et al., 2013, 2015).
The Origin
Until 2002, coronaviruses were not known to cause serious infections in humans. This scenario changed with the emergence of SARS-CoV in an animal market located in southern China, which later affected more than 8,000 people, with 774 deaths worldwide (Drosten et al., 2003; Lau et al., 2003). In 2012, a new coronavirus was identified as responsible for Middle East respiratory syndrome (MERS-CoV), infecting more than 2428 individuals and killing 838 (Zaki et al., 2012). SARS-CoV and MERS-CoV originated from bats and then jumped to another mammalian host, the civet of Himalayan palms (Paguma larvata) in the case of SARS-CoV and the dromedary camel (Camelus dromedarius) in the case of MERS-CoV, before infecting humans (Song et al., 2005; Azhar et al., 2014).
SARS-CoV-2 is the third beta-coronavirus to infect humans. Identified in late 2019, it probably originated from bats and, over time, accumulated mutations that gave it the capacity for zoonotic transmission (Zhou P. et al., 2020). The bat coronavirus RaTG13 seems to be the closest relative to SARS-CoV-2, since it shares 93.1% identity in the nucleotide sequence of the S gene and 98% identity in the amino acid sequence of the S protein (Wrapp et al., 2020; Zhou P. et al., 2020). The transmission route of SARS-CoV-2 (or its direct ancestor) from bats to humans, either directly or through an intermediate animal species, remains undefined (Banerjee et al., 2021). The complete genomic sequences of SARS-CoV-2 obtained from five patients at an early stage of the outbreak were almost identical, and they had 79.6% similarity with the SARS-CoV sequences (Li et al., 2020; Xu et al., 2020). Initially, the virus was named new coronavirus 2019 (2019-nCoV). On February 11, 2020, the Coronavirus Study Group of the International Committee of Virus Taxonomy officially named it SARS-CoV-2 based on phylogenetic analyses that showed similarity with SARS-CoV (Gorbalenya et al., 2020).
Genetic Variants of SARS-CoV-2
Since the beginning of the pandemic, the genome plasticity of SARS-CoV-2 was evidenciated with the detection of multiple sites in the genome under positive selection (Velazquez-Salinas et al., 2020). Viruses belonging to the same strain, but containing different subsets of mutations, can be classified as different variants that are characterized by their transmissibility, disease severity, and ability to escape humoral immunity. Increased transmissibility is demonstrated by the ability of a variant to compete with other variants and to exhibit a higher effective reproduction rate and/or secondary attack rate compared to other circulating variants (Korber et al., 2020; Volz et al., 2021). Next, we summarize the molecular characteristics of the main variants of SARS-CoV-2 (Table 2).
TABLE 2.
Description/features of SARS-CoV-2 variants of concern.
| Variant/WHO label | Lineage | Description | Additional aminoacid/key mutations |
| Alpha (Davies et al., 2021; Galloway et al., 2021; Volz et al., 2021; Walensky et al., 2021) | B. 1. 1. 7 | UK lineage of concern, associated with the N501Y mutation United Kingdom, September-2020 +++ Transmissibility ++ Severity |
+S: 484K +S:452R N501Y P681H Deletions: H69-V70 Y144/145 |
| Beta (Tegally et al., 2021; Wibmer et al., 2021) | B. 1. 351 | South Africa, May-2020 +Transmissibility Severity: possible |
+S:L18F N501Y K417N E484K |
| Gamma (Candido et al., 2021; Wang P. et al., 2021) | P.1 | Brazilian lineage with a number of spike mutations with likely functional significance E484K, K417T, and N501Y. Brazil, Dez-2020 ++ Transmissibility Severity: possible |
+S:681H N501Y K417T E484K |
| Delta (European Centre for Disease Prevention and Control [ECDC], 2021; Motozono et al., 2021) | B. 1. 617. 2 | Predominantly India lineage with several spike mutations. India, October-2020 +++ Transmissibility +++ Severity |
+S:417N +S:484K L452R E484Q |
| Omicron (Callaway, 2021; Gao et al., 2021) | B. 1. 1. 529 | Several mutations that are found in other variants of concern and that are thought to make the virus more infectious. Multiple countries, November-2021 |
D614G N501Y K417N |
The mutations identified throughout the pandemic in the morphology of the virus clearly demonstrate, in addition to its adaptive capacity, its ability to develop evolutions in order to increase its ability to escape the host’s immune response, as well as make its entry into the cell easier, such mutations imply an increase in transmissibility or harmful alteration in the epidemiology of COVID-19; an increase in virulence or change in the clinical presentation of the disease; and/or diminished effectiveness of social and public health measures or available diagnostics, vaccines and therapies. Therefore, the wide vaccination coverage of the world population and the maintenance of measures to control the spread of the virus are the only efficient measures to contain these evolutions in the pathogenesis of the virus and in the effective control of the pandemic.
Genetics of the Human Host in SARS-CoV-2 Infection
The role of human host genetic variability in the evolution of SARS-CoV-2 infection has been extensively proposed due to the great heterogeneity in the clinical manifestations of COVID-19 and the variation in mortality rates between populations and ethnicities, which are strong indicators of the modulatory effect of host genetics on its pathogenesis (Chakravarty, 2021; Fricke-Galindo and Falfán-Valencia, 2021; Mohammadpour et al., 2021).
Studies on the genetic predisposition to COVID-19 have been of various kinds, such as meta-analyses (Benetti et al., 2020; Poulton et al., 2020; Rendeiro et al., 2020; Bernal et al., 2021), in silico approaches (Vique-Sanchez, 2020; Wang et al., 2020c; Calcagnile et al., 2021), in vitro (Hashizume et al., 2021), case–control studies (Amraei et al., 2020; Sakuraba et al., 2020), and case series (Asselta et al., 2020; Russo et al., 2020; Wang et al., 2020d).
In case–control studies, there are two main approaches: (i) those rationally based on functional evidence (Lambert et al., 2005; Heurich et al., 2014; Cao et al., 2020; Maucourant et al., 2020; Wang et al., 2020d; Secolin et al., 2021), which search for specific candidate genes and investigate allele frequencies or differences in gene expression levels; and (ii) those based on genomic searches, such as whole-genome sequencing (WGS) (Wang et al., 2020f), whole-exome sequencing (WES) Zhang et al., 2020b; Kosmicki et al., 2021), and genome-wide association studies (GWAS) (Ellinghaus et al., 2020; Hu J. et al., 2020; Pairo-Castineira et al., 2021). Supplementary File organizes the associations reported in the literature by cytogenetic location, discriminating the evidence of association by type of study (WGS, WES, GWAS, case-control, meta-analyses, and functional evidence). A total of 51 regions could be identified containing approximately 86 candidate genes.
In this context, altered immune responses, such as those caused by primary immunodeficiencies (PIDs), may be important in disease progression, where at least one case of death has already been reported during coronavirus infection (Szczawinska-Poplonyk et al., 2013). More severe viral infections have been associated with the presence of PID (Dropulic and Cohen, 2011), which constitute a group of more than 350 rare diseases that together have a considerable prevalence (McCusker et al., 2018). Genetically, PIDs are heterogeneous, most often monogenic. The registry of rare diseases ORPHANET1 recognizes at least 308 genes involved in PIDs, which makes their diagnosis complex and their prevalence underestimated.
Total exome sequencing of SARS-CoV-2 identified 24 functional variants in eight genes, TLR3, UNC93B1, TICAM1, TBK1, IRF7, IFNAR2, IRF3, and IFNAR1 (Zhang et al., 2020a), of which the first six are on the list of 308 genes known to be associated with PID. Additionally, the ADAM17 gene, which is also on the list, has been implicated in the ability of SARS-CoV-2 to infect cells and modulate the inflammatory response (Heurich et al., 2014; Palau et al., 2020; Zipeto et al., 2020).
Few studies have addressed the direct relationship between PID and COVID-19 directly. A meta-analysis suggests a correlation between the prevalence of selective IgA deficiency, the most common PID, and the prevalence of COVID-19 (Naito et al., 2020). A case series with two men from two families indicated that mutations in the TLR7 gene, one of those implicated in PID, were present in these patients with severe COVID-19 (Van der Made et al., 2020). An important prospective study suggests a 10-fold higher mortality rate from COVID-19 in children with PID (Delavari et al., 2020).
Together, these observations show that studies on the genetic modulation of COVID-19 by PID-related genes are consistent. Thus, Supplementary File shows that of the 51 chromosomal regions associated with COVID-19, 37 (72.5%) also contain PID-related genes, as described in ORPHANET. Among these 51 regions, those present on chromosomes 6, 19, and 21 stand out. On chromosome 6, more specifically in the MHC class I and II regions (6p21.32, 6p21.33, 6p22.1), the evidence of association with COVID-19 comes from WGS, GWAS, functional (expression and affinity) studies, meta-analyses, and case–control studies, and this region contains several PID-related genes. On chromosome 19, the most consistently associated region is the one containing the KIR gene complex (19q13.42), whose evidence comes from case–control, meta-analysis, and functional studies (Sakuraba et al., 2020; Bernal et al., 2021; Rendeiro et al., 2021) and which contains PID-related genes. Two regions of chromosome 21 (21q22.11, 21q22.3) also have multiple lines of evidence showing an association with COVID-19, the most strongly associated being the one that contains the TMPRSS2 and MX1 genes. TMPRSS2 is already well established as an important marker for the ease of viral entry (Andolfo et al., 2020; Asselta et al., 2020; Latini et al., 2020; Sajuthi et al., 2020; Senapati et al., 2020; Vastrad et al., 2020; Wang et al., 2020c; Mohammad et al., 2021; Schönfelder et al., 2021; Torre-Fuentes et al., 2021), while MX1 is a gene responsive to interferon and is closely directed to the activity of response to viral infections, including SARS-CoV-2 infection (Russo et al., 2020; Bizzotto et al., 2020).
The regulation of important genes in COVID-19 has also been addressed by epigenetic studies, which have shown that methylation patterns (Corley and Ndhlovu, 2020) and microRNA expression (Fulzele et al., 2020; Widiasta et al., 2020) are altered in the disease. The roles of microRNAs in the regulation of key genes in COVID-19 make them important candidate biomarkers. Another important aspect of their action is their binding to viral mRNAs to silence them and act as antivirals, as shown by in silico studies (Fulzele et al., 2020).
The genes involved in viral infection reported in the literature are ACE2, TMPRSS2, ADAM17, NRP1, and NRP2. One literature review (Fulzele et al., 2020) identified 12 relevant microRNAs in the regulation of ACE2 (miR-18, miR-125b, miR-132, miR-143, miR-181, miR-200, miR-145, miR-155, miR-212, miR-421, miR-482-3p, and miR-4262). In addition to these microRNAs experimentally deduced to target ACE2, an in silico search was performed in the miRDB database (Chen and Wang, 2020) for possible microRNAs targeting the ACE2, TMPRSS2, ADAM17, NRP1, and NRP2 genes. Only microRNAs with a score greater than 94 were considered. The search identified candidate microRNAs to be relevant biomarkers in the regulation of these genes. Additionally, seven microRNAs were described as candidates targeting the mRNAs of viral genes (Fulzele et al., 2020), making them an important group of markers.
Finally, a relevant group of microRNAs has been described as regulators of the inflammatory response (Tahamtan et al., 2018), of which some belong to the aforementioned list of microRNAs involved in regulating the expression of genes related to SARS-CoV-2 infection or targeting viral genes. Table 3 lists the microRNAs that may be relevant in COVID-19.
TABLE 3.
MicroRNAs possibly relevant in COVID-19, according to their type of evidence.
| MicroRNAs with in silico evidence of regulation of the expression of genes related to SARS-CoV-2 infection: |
| hsa-miR-124-3p, hsa-miR-1297, hsa-miR-153-5p, hsa-miR-26a-5p, hsa-miR-26b-5p, hsa-miR-3133, hsa-miR-3163, hsa-miR-331-3p, hsa-miR-33a-3p, hsa-miR-3646, hsa-miR-4465, hsa-miR-4500, hsa-miR-506-3p, hsa-miR-5094, hsa-miR-548ae-3p, hsa-miR-548ah-3p, hsa-miR-548aj-3p, hsa-miR-548am-3p, hsa-miR-548aq-3p, hsa-miR-548j-3p, hsa-miR-548x-3p, hsa-miR-578, hsa-miR-6844, hsa-miR-7977, hsa-miR-92a-1-5p |
| MicroRNAs with experimental evidence of regulation of ACE2 expression: |
| hsa-miR-125b, hsa-miR-145, hsa-miR-181, hsa-miR-200, hsa-miR-212, hsa-miR-421, hsa-miR-482-3p, hsa-miR-18, hsa-miR-132, hsa-miR-143, hsa-miR-155, hsa-miR-4262 |
| MicroRNAs with in silico evidence of regulation of viral gene expression: |
| hsa-miR-15b-5p, hsa-miR-15a-5p, hsa-miR-548c-5p, hsa-miR-548d-3p, hsa-miR-409-3p, hsa-miR-30b-5p, hsa-miR-505-3p |
| MicroRNAs with evidence of regulation of the inflammatory response: |
| hsa-miR-21, hsa-miR-24, hsa-miR-124, hsa-miR-145, hsa-miR-146, hsa-miR-149, hsa-miR-155, hsa-miR-181a, hsa-miR-181b, hsa-miR-181c, hsa-miR-181d |
Correlating the most recent findings, host genetics linked to the immune response are strongly suggested as a predictor of the prognosis of COVID-19. Initial evidence pointing to genes such as ACE2 as important for viral entry into the host cell is not as relevant as was expected. Rather, viral entry is now more clearly linked to genes such as TMPRSS2 and MX1, which are near each other on chromosome 21. TMPRSS2 is crucial for the initial phenomena of infection and is responsible for the viral response related to interferon.
In this scenario, evidence emerged for specific immune response genes, such as KIR and MHC genes, in addition to a potential association with multiple PID genes. Therefore, the current data suggest that the influence of human host genetics on COVID-19 seems to be polygenic and focused on the genetic modulation of the immune response, making it increasingly less likely that the broad spectrum of COVID-19 manifestations is determined by oligogenic models. Additionally, the relevant role of multiple immunorelevant genes also favors the role of epigenetic regulation in these genes, especially microRNAs.
There are several pieces of evidence on the role of host genetics influencing the dynamics of SARS-CoV-2 infection, the variability of genes involved in the immune response has a direct impact on the course of COVID-19, but the magnitude of genetic diversity makes this elucidation more complex. However, concentrating efforts in order to discover the key genes involved in viral pathogenesis, as well as in the escape of the immune response and, based on this, building a panel with the main findings, can strongly contribute to the targeting of cases, enabling a more accurate view on how the evolution of cases can occur, and thus outline a more effective preventive and therapeutic planning, contributing to a better dynamics of care services, as well as favoring the studies of vaccines used today and those that are still under research.
Transmission
The transmissibility of SARS-CoV-2 is not known with precision. It is believed that the ingestion of infected animals as a food source is the main cause of zoonotic transmission (Ji et al., 2020). As SARS-CoV-2 is highly similar to SARS-CoV, bats could be the host of the new coronavirus. In addition, Malaysian pangolins (Manis javanica) can harbor coronaviruses very similar to SARS-CoV-2 and are a potential natural reservoir of the virus (Zhou P. et al., 2020). Human-to-human transmission can occur through droplets containing infectious particles spread by speech, coughing, or sneezing, which can reach the mucous membranes of the eyes, nose, or mouth as portals, or by direct contact with contaminated surfaces, such as stainless steel, plastic, glass, and cardboard for at least several hours (Doremalen et al., 2020; Fan et al., 2020; Xiang Ong et al., 2020). As respiratory viruses have the highest transmission rate when the patient is symptomatic, because it is during this period that the viral load reaches a peak, it is believed that the same occurs with COVID-19 (Zhou F. et al., 2020). However, the possibility of viral transmission from an asymptomatic individual (Bai et al., 2020) is not excluded because there is evidence of asymptomatic or presymptomatic spread of SARS-CoV-2, highlighting its ability to colonize and replicate in the throat during the initial infection (Arons et al., 2020; Pan et al., 2020; Wölfel et al., 2020).
The genetic material of SARS-CoV-2 has been detected in the feces, whole blood, and urine of patients with COVID-19, but it has not been documented whether transmission by these means is possible (Young et al., 2020). The possibility of fecal aerosol transmission was described in a report based on circumstantial evidence, and this may have caused the community outbreak of COVID-19 in a high-rise building in Canton, China (Kang et al., 2020). Little is known about the vertical transmission of SARS-CoV-2, and further studies are needed to assess its transmissibility from pregnant woman to fetus (Chen H. et al., 2020; Hu X. et al., 2020). However, in March 2020, the first proven case of transplacental transmission of SARS-CoV-2 was described, involving a pregnant woman affected by COVID-19 during late pregnancy, with detection of the viral genome in the amniotic fluid collected before rupture of the placenta (Vivanti et al., 2020).
To date, there is no evidence of viral transmission from pets to humans. Ferrets and cats are highly susceptible to SARS-CoV-2, while dogs have low susceptibility; other animals, including pigs, chickens, and ducks, are not susceptible to the virus under experimental conditions (Shi et al., 2020). Interestingly, viral transmission between cats has been observed (Shi et al., 2020). Another study showed that 22 cats in France and two of 10 cats in China from patients with COVID-19 had SARS-CoV-2 infection with mild respiratory and digestive symptoms (Sailleau et al., 2020). This indicates that cats, being common pets, can theoretically transmit the virus to other animals and, possibly, humans. Still, there is no clear evidence that transmission of SARS-CoV-2 has occurred from cats to humans. In ferrets, SARS-CoV-2 is able to replicate in the upper respiratory tract without causing serious illness or death (Shi et al., 2020). Recently, an outbreak of SARS-CoV-2 in visions (Neovison mink) was reported on several farms in the Netherlands with transmission events to humans via respiratory droplets promoting secondary transmission of a mink SARS-CoV-2 variant back for humans (Oude Munnink et al., 2021; Shuai et al., 2021). Between April 26 and November 22, 2020, 14 outbreaks of COVID-19 occurred on commercial mink farms in Utah, one outbreak in a commercial mink farm in Wisconsin, and another in Oregon, USA (United States of America). Clinical signs included respiratory signs and sudden death from a total of 12,330 deaths among 145,757 susceptible animals (Eckstrand et al., 2021).
Repeat infections of SARS-CoV-2 between humans and animals (spillback) can lead to the emergence of new animal reservoirs, with risk of secondary infection (spillover) for humans through an animal reservoir, which can lead to the appearance of variants of SARS-CoV-2, as described above. Such events are of great concern, as the formation of wild virus reservoirs, the appearance of a mutant strain with increased transmissibility and severity of SARS-CoV-2 in humans puts efforts for the long-term control of COVID-19 at risk and they also threaten vulnerable animal populations that are particularly susceptible to lethal diseases.
Diagnosis of COVID-19
Faced with the pandemic caused by the new coronavirus, the early and safe diagnosis of the infection is extremely important to interrupt the transmission of the disease and assist in making decisions such as isolation and/or distancing of people, which will provide more time for public health implementation measures that may have positive impacts in reducing the problems associated with COVID-19.
According to the WHO, the diagnosis of COVID-19 can be clinical or epidemiological, using the International Code of Diseases (ICD) ICD-10 Z20.9 (contact with exposure to unspecified communicable disease) as a record. In the face of clinical manifestations suggestive of the disease and when laboratory confirmation is inconclusive or not available, the patient is considered infected and the ICD UO7.2 (unidentified virus - attributed to a clinical or epidemiological diagnosis of COVID) should be used for recording –19, when laboratory confirmation is inconclusive or unavailable. Includes diagnosis of a probable case or suspected case of COVID-19). On the other hand, those with a diagnosis confirmed by laboratory tests must be registered using the code ICD-UO7.1 (identified virus – attributed to a COVID-19 diagnosis confirmed by laboratory tests) (Brazil Ministério da Saúde, 2020).
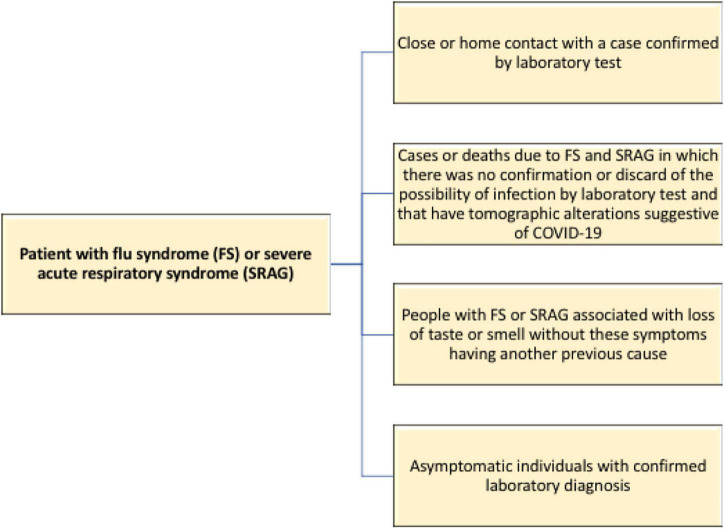
The Brazilian Ministry of Health included new criteria for the characterization of COVID-19 cases, going beyond the laboratory tests already adopted. Given the difficulties of testing, the agency allowed for the following individuals to be deemed infected by the new coronavirus (Brazil Ministério da Saúde, 2020; Figure 2).
FIGURE 2.
Flowchart for identifying individuals possibly infected with the new coronavirus in the absence of laboratory tests.
SARS-CoV-2 was initially characterized by sequencing its genome, which provided the necessary information to develop quantitative or real-time polymerase chain reaction (PCR) tests for viral detection (Zhou P. et al., 2020; Zhu et al., 2020). Different laboratory diagnostic tools are used in clinical practice to confirm cases of SARS-CoV-2 infection and differ in terms of sample type, collection time, specificity, and sensitivity (Table 4).
TABLE 4.
Diagnostic methods for SARS-CoV-2 infection.
| Test | Type of sample | Pros and cons | References |
| RT-PCR | Nasopharyngeal smear or saliva. Collection within 7 days of symptoms | Gold standard test for detecting SARS-CoV-2. The accuracy of the test depends on the stage of the disease and the degree of viral multiplication. Higher sensitivities are reported depending on which genetic targets are used in performing the test | Xiao et al., 2020; Zou et al., 2020; Erster et al., 2021 |
| Enzyme-Linked Immunosorbent Assay (ELISA) IgM, IgA, IgG |
Serum | Useful for diagnosing previous infections. Important for population serological surveillance and research activities. It is not useful for diagnosing acute infection. Descending titles over time (2–3 months) |
Huang A. T. et al., 2020; Huang M. et al., 2020; Krajewski et al., 2020; Li et al., 2020; Lucas et al., 2020; Bichara et al., 2021 |
| Lateral flow immunoassay (LFIA) (Antigen or antibody) |
Nasal or nazopharinzeal smear, serum or whole blood | Detects acute infection through the presence of viral antigens. Previous infection by detection of antibodies. Average time 15–20 min. Low cost. Low sensitivity and specificity of 56.2 and 99.5%, respectively |
Canetti et al., 2020; Dinnes et al., 2020; Prazuck et al., 2020 |
| Loop-mediated isothermal amplification (LAMP) | Nasal or nazopharinzeal smear | The accuracy of the test depends on the stage of the disease and the degree of viral multiplication. Highly effective, fast results, and limited cost | Broughton et al., 2020; Huang W. E. et al., 2020; Bektaş et al., 2021; Jones et al., 2021; Juscamayta-López et al., 2021 |
| CRISPR/Cas | Nasal or nazopharinzeal smear | High sensitivity and specificity rates and low analysis costs. With 100% sensitivity and specificity | Broughton et al., 2020; Hou et al., 2020; Lucia et al., 2020 |
| Biosensors | Nazopharinzeal smear | Tecnologia rápida e altamente sensível. Ausência de reatividade cruzada com outros coronavirus | Seo et al., 2020; Zhao et al., 2021 |
The COVID-19 pandemic highlighted the importance of laboratory diagnostic methods. Currently, nucleic acid amplification methods represent the gold standard for the diagnosis of COVID-19 infection with several RT-PCR-based tests approved by different national and international regulatory agencies. However, despite the high sensitivity of RT-PCR, the need for trained professionals and expensive instruments and reagents that limit its application, especially in low-income countries, drive the development of diagnostic methods to overcome the limitations of RT-PCR, among them, low-cost diagnostic strategies are promising and can be used for the effective diagnosis of COVID-19 infection in low- and middle-income countries. Rapid antigen and antibody tests and immunoenzymatic serological tests represent the most widely used techniques for monitoring the spread of SARS-CoV-2 infection. It is important to note that, despite the low cost of such techniques, the low sensitivity and specificity of LFIAs and ELISAs, the use of such point-of-care tests enabled the implementation of effective health surveillance systems that allowed for the effective management of the COVID pandemic -19, thus limiting the number of infections.
The scientific community in a short period developed several useful methods to correctly diagnose a suspected case of SARS-CoV-2 infection. Despite the limitations of some laboratory diagnostic methods, it is necessary to take into account not only the test to be used but also the patient’s medical history, the time of exposure to individuals infected with SARS-CoV-2, the type of sample to be used, be collected and analyzed, and how to interpret the result. The integration of all these elements will provide a solid foundation for correctly diagnosing COVID-19 infection and effectively managing the COVID-19 pandemic.
Treatment Approaches
According to the WHO, there are still no specific drugs for COVID-19. In the early days of the pandemic, many governments around the world implemented, to some degree, measures recommended by the WHO to limit the spread of the virus, such as self-isolation, social distancing, hand washing, closing schools and universities, and mask wearing in public places (Prather et al., 2020). Due to the lack of specific therapy, some drugs used in other infectious diseases have been applied against COVID-19 (Table 5) in clinical practice, although their effectiveness is controversial.
TABLE 5.
Medicines used in the treatment of COVID-19.
| Drug name | Class | Mechanism of action | Adverse effect | References |
| Chloroquine and Hydroxychloroquine | Antiparasitic | Inhibition of host cell receptor glycosylation to block viral entry, acidification of the endosomal and proteolytic process. | High doses can lead to respiratory arrest, cardiac arrest, and hypokalemia. | Savarino et al., 2003; Yamamoto et al., 2004; Eze et al., 2021 |
| Lopinavir/Ritonavir | Antiviral | Inhibition of 3CL protease. Inhibition of viral replication. |
Risks for pediatric patients | Chu et al., 2004; Kim et al., 2016 |
| Remdesivir | Antiviral | RNA-dependent RNA polymerase inhibitor. Block viral replication |
There is no information on whether overdosing can cause any adverse effects | Hoehl et al., 2020; Li and De Clercq, 2020; Wang et al., 2020e |
| Heparin | Anticoagulant and anti-inflammatory | Heparin binds to the RBD of the SARS-CoV-2 protein S, inhibiting viral infection | Platelet count usually decreases to between days 5 and 12 | Kawase et al., 2012; Zhou et al., 2015; Hoffmann et al., 2020; Mycroft-West et al., 2020; Tang N. et al., 2020; Yamamoto et al., 2020 |
| Tocilizumab | Monoclonal antibody | IL-6 inhibiting receptor. Cytokine storm reduction blockade. |
Overdose-neutropenia | Guaraldi et al., 2020; Rosas et al., 2020 |
| Anakinra | Immune Response Modulator | Monoclonal antibody that acts against the IL-1 receptor | Rheumatoid arthritis (incidence > 10%) | Conti et al., 2020 |
| Baricitinib | Immune Response Modulator | Antiviral activity Inhibitor of clathrin-mediated endocytosis Janus kinases 1 and 2 (JAK1/2 inhibitor) |
Multiple adverse reactions |
Cantini et al., 2020; Richardson et al., 2020 |
| Camostat Mesilate | Antiviral | TMPRSS2 inhibitor that prevents replication Viral Blocks viral mutation |
Rash, pruritus, nausea, abnormal values from laboratory tests and diarrhea | Uno, 2020 |
| Molnupiravir | Antiviral | It works by inducing mutagenesis in viral RNA, causing the newly formed RNA strand chain to terminate | Mild adverse effects | Kabinger et al., 2021; Zhou et al., 2021 |
| Paxlovid | Antiviral | It inhibits viral replication at a stage known as proteolysis, which occurs before viral RNA replication | Absent |
Wang et al., 2020g; Ahmad et al., 2021; Pfizer, 2021 https://www.pfizer.com/news/press-release/press-release-detail/pfizers-novel-covid-19-oral-antiviral-treatment-candidate |
The unfortunate SARS-CoV-2 pandemic in early 2020 posed a challenge to all researchers to find potential therapeutic agents for treatment. There were extensive efforts to reuse approved drugs during the COVID-19 pandemic. This strategy offers several advantages over developing an entirely new drug, with reduced risk of failure because safety has already been evaluated. Currently, there are antiviral therapies developed to induce a direct effect on SARS-CoV-2, either by blocking viral entry into host cells or by controlling viral enzymes with a significant contribution to genome replication. A big step forward in the control of severe cases and deaths related to COVID-19.
Vaccine Platforms
Multiple vaccine platforms have already been approved for emergency use against COVID-19. As of August 19, 2021, 20 vaccine candidates were within the WHO evaluation process for commercialization (World Health Organization [WHO], 2021b; Table 6).
TABLE 6.
Vaccines available and in development for protecting against SARS-CoV-2.
| Manufacturer | Vaccine name | Platform | Evaluation status | Status |
| BioNTech Manufacturing GmbH | BNT162b2/COMIRNATY Tozinameran (INN) | Nucleoside modified mRNA | Finished | Approved for use |
| AstraZeneca, AB | AZD1222 Vaxzevria | Recombinant ChAdOx1 adenoviral vector encoding the Spike protein antigen of the SARS-CoV-2. | Finished | Approved for use |
| Serum Institute of India Pvt. Ltd | Covishield (ChAdOx1_nCoV-19) | Recombinant ChAdOx1 adenoviral vector encoding the spike protein antigen of the SARS-CoV-2. | Main data finalized | Approved for use |
| Janssen–Cilag International NV | Recombinant, Ad26.COV2.S | Recombinant, replication-incompetent adenovirus type 26 (Ad26) vectored vaccine encoding the (SARS-CoV-2) spike protein | Finished | Approved for use |
| Moderna Biotech | mRNA-1273 | mRNA-based vaccine encapsulated in lipid nanoparticle (LNP) | Finished | Approved for use |
| Beijing Institute of Biological Products Co., Ltd. (BIBP) | SARS-CoV-2 Vaccine (Vero Cell), Inactivated (lnCoV) | Inactivated, produced in Vero cells | Finished | Approved for use |
| Sinovac Life Sciences Co., Ltd. | COVID-19 Vaccine (Vero Cell), Inactivated/CoronavacTM | Inactivated, produced in Vero cells | Finished | Approved for use |
| The Gamaleya National Center | Sputnik V | Human Adenovirus Vector-based COVID-19 vaccine | Waiting, waiting for submission | Approved for use |
| Bharat Biotech, India | SARS-CoV-2 Vaccine, Inactivated (Vero Cell)/COVAXIN | Whole-Virion Inactivated Vero cells | In progress | Approved for use |
| Sinopharm/WIBP | Inactivated SARS-CoV-2 Vaccine (Vero Cell) | Inactivated, produced in Vero cells | In progress | In progress. Not approved |
| CanSinoBio | Ad5-nCoV | Recombinant novel coronavirus vaccine (adenovirus type 5 vector) | - | In progress. Not approved |
| Nonavax | NVX-CoV2373/Covovax | Recombinant nanoparticle prefusion spike protein formulated with Matrix-M™ adjuvant | – | In progress. Not approved |
| Sanofi | CoV2 preS dTM-AS03 vaccine | Recombinant, adjuvanted | – | In progress. Not approved |
| Serum Institute of India Pvt. Ltd | NVX-CoV2373/Covovax | Recombinant nanoparticle prefusion spike protein formulated with Matrix-M™ adjuvant | – | In progress. Not approved. |
| Clover Biopharmaceuticals | SCB-2019 | Novel recombinant SARS-CoV-2 spike (S)-Trimer fusion protein | – | In progress. Not approved |
| Urevac | Zorecimeran (INN) concentrate and solvent for dispersion for injection; Company code: CVnCoV/CV07050101 | mRNA-based vaccine encapsulated in lipid nanoparticle (LNP) | – | In progress. Not approved |
| Vector State Research Center of Virology and Biotechnology | EpiVacCorona | Peptide antigen | – | In progress. Not approved |
| Zhifei Longcom | Recombinant Novel Coronavirus Vaccine (CHO Cell) | Recombinant protein subunit | – | In progress. Not approved |
| IMBCAMS | SARS-CoV-2 Vaccine, Inactivated (Vero Cell) | Inactivated | – | In progress. Not approved |
| BioCubaFarma | Soberana 01, Soberana 02 Soberana Plus Abdala | SARS-CoV-2 spike protein conjugated chemically to meningococcal B or tetanus toxoid or aluminum | – | In progress. Not approved |
Adapted from World Health Organization [WHO] (2021c). –, No information.
The efficacy of an effective vaccine depends on the long-term response of specific antibodies to viral antigens from plasma cells, as well as the development of persistent memory of T cells and B cells. In the case of SARS-CoV infection, adaptive humoral and cellular immune responses are crucial to the elimination of infection.
Due to the need to ensure the immunity of the population, clinical trials of SARS-CoV-2 vaccines have accelerated their development process. To date, few adverse events have been reported in clinical trials of such vaccines, and the safety of those that have been tested more extensively is promising.
Along with safety, the efficacy of the vaccine is important. The FDA recommended that vaccines show at least 50% efficacy compared to placebo, defined primarily by (i) reduction in COVID-19 cases, (ii) reduction in COVID-19 severity, or (iii) reduction in COVID-19 severity infections by SARS-CoV-2 (Food and Drug Administration [FDA], 2020).
Inactivated Virus
Inactivated virus vaccines are being developed by isolating SARS-CoV-2 from samples of patients hospitalized with COVID-19. Then the virus is used to infect a cell line in the laboratory, after which it is chemically inactivated with β-propiolactone (Gao et al., 2020). This type of platform exposes the vaccinated individual to several viral proteins instead of a single target, and depending on the inactivation process, the structural integrity of the viral antigens can be affected to favor a T cell response toward a Th2 profile (Tregoning et al., 2020). In 2020, a pilot study of a candidate inactivated SARS-CoV-2 vaccine (BBIBP-CorV) demonstrated high productivity and good genetic stability over the SARS-CoV-2 vaccine (Wang et al., 2020h).
Attenuated Virus
This type of platform simulates natural infection using deoptimized viral genomes that are not translated efficiently in hosts (Tregoning et al., 2020). With a low level of infection, these vaccines induce robust and long-lasting immune responses (Yong et al., 2019). However, the storage of these vaccines at low temperature and their contraindication in immunocompromised patients and the elderly may be considered disadvantages (Sarmiento et al., 2016).
Viral Vectors
Vaccines using recombinant virus vectors work similarly to an endogenous pathogen, expressing the target protein in the cytoplasm of the host cell. This location favors the response of cytotoxic T cells, establishing the cell-mediated immunity that is crucial for the elimination of virus-infected cells (Coughlan, 2020). Vaccines using adenoviral vectors can induce potent antibodies and T cell responses, with variations in intensity depending on the serotype (Tan et al., 2004). Currently, there are two vaccine candidates against SARS-CoV-2 in phase I/II clinical trials that use the measles virus (NCT04497298) or the vesicular stomatitis virus (NCT04569786) as a vector (Jon, 2020).
Nucleic Acid
In addition, DNA or RNA vaccination is also able to trigger humoral and cellular immune responses through the activation of CD4+ helper T cells and CD8+ cytotoxic T cells, respectively. Upon entering the cell, DNA vaccines are detected by a variety of innate immune receptors, such as interferon gene stimulator (STING)/TANK binding kinase 1 (TBK1). The IRF3 pathway, the inflammasome, and many other factors are involved in the mode of action of the DNA vaccine (Li and Petrovsky, 2016). Two studies have analyzed the immunogenicity of these vaccine platforms against SARS-CoV-2 in animal models (Smith et al., 2010; Patel et al., 2020).
Recombinant Protein Subunit
Subunit vaccines, particularly those based on the RBD of the SARS-CoV S protein, contain the main antigenic determinants that can induce neutralizing antibodies and CD8+ T cell responses (Bonavia et al., 2003; He et al., 2006). This characteristic provides useful information for designing safe and effective vaccines against SARS-CoV-2, since the RBD of the S protein of SARS-CoV-2 contains similar epitopes. However, they are vaccines that generally require adjuvants or nanoparticles to increase their immunogenicity. A subunit vaccine (StriFK-FH002C) prevented hamsters from transmitting the virus to other, unvaccinated hamsters in cohabitation, causing a lower viral load in the upper respiratory tract of hamsters vaccinated after the challenge, showing that the vaccine is a candidate for SARS-CoV-2 (Wu et al., 2021).
Despite this success, the goal of achieving global herd immunity through vaccination, which would allow for the abandonment of other non-pharmaceutical interventions and restrictions on our social, cultural, and recreational activities has not yet been achieved in all countries. This is due to a number of reasons and unexpected developments that challenged and delayed the progress of vaccination. One of the most striking developments has been the rapid local and global emergence of SARS-CoV-2 variants with different transmission and immune evasion properties. This not only started new waves of infections but also impacted the evaluation of the vaccine’s efficacy in clinical trials. Another major obstacle to global immunity is represented by a strong imbalance in the worldwide distribution of vaccines. A global pandemic strategy combined between prevention measures and vaccination will minimize the burden of infection for the poor, but also for rich countries, and depends on the equitable distribution of vaccines to establish large-scale immunity at the global level and contain the COVID-19 pandemic.
Socioeconomic Aspects of the Pandemic
Individuals affected by COVID-19 are potentially at risk of physiological and economic harm. The decline of the economy began with the decrease in activity in the travel, tourism, and export sectors (Organization for Economic Cooperation and Development [OECD], 2020; Khan et al., 2020), but soon its generalized effects hindered production and consumption, with layoffs and bankruptcies in all sectors.
In 2020, the pandemic scenario accentuated the decline in the economies of Latin American and Caribbean countries, which have low economic growth (Organization for Economic Cooperation and Development [OECD], 2020). This scenario, together with the economic slowdown faced in previous years and the drop in activity caused by the pandemic, has negatively affected living standards and well-being in the countries of the region (Organization for Economic Cooperation and Development [OECD], 2020). The most vulnerable population will be low-income people, approximately 74% of whom work informally (Organization for Economic Cooperation and Development [OECD], 2020). By itself, the pandemic attempts to affect the present and the future with regard to the aspect of life of the people, which includes material goods (income, quality of employment, and housing) and aspects such as education, ability to form skills, in addition to emotional well-being (Khan et al., 2020; Organization for Economic Cooperation and Development [OECD], 2020).
The global economic recovery is predicted to be slow; for 2021, it will increase almost 6% differently from the previous year, with an increase of 3.5%; however, this is far from sufficient to ensure the necessary impetus for growth of the gross domestic product worldwide (Organization for Economic Cooperation and Development [OECD], 2021). With the advancement of large-scale vaccination, the manufacturing sector is gradually growing as trade and the reopening of borders progress, promoting an increase in job creation (Organization for Economic Cooperation and Development [OECD], 2021). In contrast, there is caution about the arrival of enough vaccines for enterprises of low-income individuals, especially in underdeveloped or developing countries, enabling a further weakening of economic growth, with acute increase in poverty and potentially in financing problems because the global economic and social impact of keeping borders closed outweighs the costs of making vaccines, tests, and health supplies more widely available for these countries (Khan et al., 2020; Organization for Economic Cooperation and Development [OECD], 2021).
With the increase in the unemployment rate, governments and central banks should intervene with increased spending and lower interest rates to increase consumer demand and investment, respectively (International Labour Organization [ILO], 2020). Even so, estimating the economic costs of a global disease at this time is still uncertain, since the pandemic has spiraling effects on the national and global economy, which means that any economic shock in a country will quickly spread to other countries due to the commercial and financial links associated with globalization (International Labour Organization [ILO], 2020).
Low- and middle-income countries remain vulnerable to the pandemic, in addition to suffering dramatic social and economic consequences. In this scenario, ensuring the emergence and success of the adoption of new forms of economic development and governance models would not only help to reduce the socioeconomic discrepancies affected by the pandemic, but also the risk associated with vulnerable populations. These social changes must be the result of a reflection that enables the generation of new behaviors, and reflections and actions on the socioeconomic aspects of the pandemic. However, this issue raised by COVID-19’s control policies appears to have received little attention in the relevant economic literature. And with that, the outbreak aggravated existing vulnerabilities, injustices, and distrust in society.
Final Considerations
This study presents an overview of the current context of COVID-19, offering a summary of the effects on health and socioeconomic status, viral characteristics, transmission, therapeutic options, vaccine prospects, immune response, and available diagnostic tools. It is possible that only when the pandemic ends will we be able to more accurately assess the economic and social consequences of this catastrophic event and that only then will we be able to extract sufficient knowledge to fight future epidemics, especially in matters of global public health.
Author Contributions
AV conceptualized the idea of the article for review. MT performed the literature search, analyzed the cited articles, and wrote the manuscript. CB, MV, MQ, IV, ES, CC, and MA carefully reviewed the manuscript and made changes and additions to its intellectual content. All authors approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Funding
The authors thank the National Council for Scientific and Technological Development (CNPq, # 301869/2017-0 and # 401235/2020-3), the Coordination for the Improvement of Higher Education Personnel (CAPES), and the Pro-Rectory of Research and Graduate Program of the Federal University of Pará (PROPESP/UFPA) for financial support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.789882/full#supplementary-material
References
- Ahmad B., Batool M., Ain Q. U., Kim M. S., Choi S. (2021). Exploring the binding mechanism of PF-07321332 SARS-CoV-2 protease inhibitor through molecular dynamics and binding free energy simulations. Int. J. Mol. Sci. 22:9124. 10.3390/ijms22179124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoroso A., Magistroni P., Vespasiano F., Bella A., Bellino S., Puoti F., et al. (2021). HLA and AB0 polymorphisms may influence SARS-CoV-2 infection and COVID-19 severity. Transplantation 105 193–200. 10.1097/TP.0000000000003507 [DOI] [PubMed] [Google Scholar]
- Amraei R., Yin W., Napoleon M. A., Suder E. L., Berrigan J., Zhao Q., et al. (2020). CD209L and CD209 are receptors for SARS-CoV-2. BioRxiv [Preprint]. 10.1101/2020.06.22.165803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfo I., Russo R., Lasorsa A. V., Cantalupo S., Rosato B. E., Bonfiglio F., et al. (2020). Common variants at 21q22.3 locus influence MX1 gene expression and susceptibility to severe COVID-19. SSRN Electron. J. 24:102322. 10.1101/2020.12.18.20248470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini M. M., Akhlaghpour M., Neuman B. W., Buchmeier M. J. (2013). Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio 4:e00524–13. 10.1128/mBio.00524-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzurez A., Naka I., Miki S. (2021). Association of HLA-DRB1*09:01 with severe COVID-19. HLA 98 37–42. 10.1111/tan.14256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arons M. M., Hatfield K. M., Reddy S. C., Kimball A., James A., Jacobs J. R., et al. (2020). Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N. Engl. J. Med. 382 2081–2090. 10.1056/NEJMoa2008457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselta R., Paraboschi E. M., Mantovani A., Duga S. (2020). ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID-19 severity in Italy. Aging 12 10087–10098. 10.1101/2020.03.30.20047878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar E. I., El-Kafrawy S. A., Farraj S. A., Hassan A. M., Al-Saeed M. S., Hashem A. M., et al. (2014). Evidence for Camel-to-Human Transmission of MERS Coronavirus. N. Engl. J. Med. 370 2499–2505. 10.1056/NEJMoa1401505 [DOI] [PubMed] [Google Scholar]
- Bai Y., Yao L., Wei T., Tian F., Jin D., Chen L., et al. (2020). Presumed asymptomatic carrier transmission of COVID-19. J. Am. Med. Assoc. 323 1406–1407. 10.1001/jama.2020.2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Doxey A. C., Mossman K., Irving A. T. (2021). Unraveling the zoonotic origin and transmission of SARS-CoV-2. Ecol. Evol. 36 180–184. 10.1016/j.tree.2020.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barquera R., Collen E., Di D., Buhler S., Teixeira J., Llamas B., et al. (2020). Binding affinities of 438 HLA proteins to complete proteomes of seven pandemic viruses and distributions of strongest and weakest HLA peptide binders in populations worldwide. HLA 96 277–298. 10.1111/tan.13956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bektaş A., Covington M. F., Aidelberg G., Arce A., Matute T., Núñez I., et al. (2021). Accessible LAMP-enabled rapid test (ALERT) for detecting SARS-CoV-2. Viruses 13:742. 10.3390/v13050742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetti E., Tita R., Spiga O., Ciolfi A., Birolo G., Bruselles A., et al. (2020). ACE2 gene variants may underlie interindividual variability and susceptibility to COVID-19 in the Italian population. Eur. J. Hum. Genet. 28 1602–1614. 10.1038/s41431-020-0691-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlyamani I., Venet F., Coudereau R., Gossez M., Monneret G. (2020). Monocyte HLA-DR measurement by flow cytometry in COVID-19 patients: an interim review. Cytom. Part A 97 1217–1221. 10.1002/cyto.a.24249 [DOI] [PubMed] [Google Scholar]
- Bernal E., Gimeno L., Alcaraz M. J., Quadeer A. A., Moreno M., Martínez-Sánchez M. V., et al. (2021). Activating Killer-cell Immunoglobulin-like receptors are associated with the severity of COVID-19. J. Infecs. Dis. 224 229–240. 10.1093/infdis/jiab228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bichara C. D. A., Amoras E. S. G., Vaz G. L., Torres M. K. S., Queiroz M. A. F., Amaral I. P. C., et al. (2021). Dynamics of anti-SARS-CoV-2 IgG antibodies post-COVID-19 in a Brazilian Amazon population. BMC Infect. Dis. 21:443. 10.1186/s12879-021-06156-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birol R. L. W., Birol I. (2020). Retrospective in silico HLA predictions from COVID-19 patients reveal alleles associated with disease prognosis. medRxiv [Preprint]. 10.1101/2020.10.27.20220863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizzotto J., Sanchis P., Abbate M., Lage-Vickers S., Lavignolle R., Toro A., et al. (2020). SARS-CoV-2 infection boosts MX1 antiviral effector in COVID-19 patients. iScience 23:101585. 10.1016/j.isci.2020.101585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorsi I., Carrega P., Venanzi Rullo E., Ducatelli R., Falco M., Freni J., et al. (2021). HLA-C*17 in COVID-19 patients: hints for associations with severe clinical outcome and cardiovascular risk. Immunol. Lett. 234 44–46. 10.1016/j.imlet.2021.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonavia A., Zelus B. D., Wentworth D. E., Talbot P. J., Holmes K. V. (2003). Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J. Virol. 77 2530–2538. 10.1128/jvi.77.4.2530-2538.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosso M., Thanaraj T. A., Abu-Farha M., Alanbaei M., Abubaker J., Al-Mulla F. (2020). The two faces of ACE2: the role of ACE2 receptor and its polymorphisms in hypertension and COVID-19. Mol. Ther. Methods Clin. Dev. 18 321–327. 10.1016/j.omtm.2020.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazil Ministério da Saúde (2020). Diretrizes Para Diagnóstico e Tratamento da COVID-19. Brasília: Brazil Ministério da Saúde. [Google Scholar]
- Broughton J. P., Deng X., Yu G., Fasching C. L., Servellita V. (2020). CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 38 870–874. 10.1038/s41587-020-0513-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcagnile M., Forgez P., Iannelli A., Bucci C., Alifano M., Alifano P. (2021). Molecular docking simulation reveals ACE2 polymorphisms that may increase the affinity of ACE2 with the SARS-CoV-2 Spike protein. Biochimie 180 143–148. 10.1016/j.biochi.2020.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. (2021). Heavily mutated Omicron variant puts scientists on alert. Nature 600:21. 10.1038/d41586-021-03552-w [DOI] [PubMed] [Google Scholar]
- Candido S., Mishra S., Crispim M. A. E., Sales F. C., De Jesus J. G., Andrade P. S., et al. (2021). Genomics and epidemiology of a novel SARS-CoV-2 lineage in Manaus, Brazil. MedRxiv [Preprint]. 10.1101/2021.02.26.21252554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canetti D., Dell’Acqua R., Riccardi N., Della Torre L., Bigoloni A., Muccini C., et al. (2020). SARS-CoV-2 IgG/IgM rapid test as a diagnostic tool in hospitalized patients and healthcare workers, at a large teaching hospital in northern italy, during the 2020 COVID-19 pandemic. New Microbiol. 43 161–165. Epub 2020 Oct 31. [PubMed] [Google Scholar]
- Cantini F., Niccoli L., Matarrese D., Nicastri E., Stobbione P., Goletti D., et al. (2020). Bariticinib therpay in COVID-19: a pilot study on safety and clinical impact. J. Infect. 81 318–356. 10.1016/j.jinf.2020.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Li L., Feng Z., Wan S., Huang P., Sun X., et al. (2020). Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 6 4–7. 10.1038/s41421-020-0147-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty S. (2021). COVID-19: the effect of host genetic variations on host-virus interactions. J. Proteome Res. 20 139–153. 10.1021/acs.jproteome.0c00637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. F., Yuan S., Kok K. H., To K. K., Chu H., Yang J., et al. (2020). A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395 514–523. 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. F. W., Kai-Wang K., Tse H., Jin D. Y., Yuen K. Y. (2013). Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trend Microbiol. 21 544–555. 10.1016/j.tim.2013.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. F. W., Lau S. K. P., To K. K. W., Cheng V. C. C., Woo P. C. Y., Yuen K. Y., et al. (2015). Middle east respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2 465–522. 10.1128/CMR.00102-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K., Dorosky D., Sharma P., Abbasi S. A., Dye J. M., Kranz D. M., et al. (2020). Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Sience 369 1261–1265. 10.1126/science.abc0870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Hou M., Chang C., Hsiao C., Huang T. (2020). The SARS coronavirus nucleocapsid protein – Forms and functions. Antiviral Res. 103 39–50. 10.1016/j.antiviral.2013.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Guo J., Wang C., Luo F., Yu X., Zhang W., et al. (2020). Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. Lancet 395 809–815. 10.1016/S0140-6736(20)30360-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Huang C., Zhang Y., Zhang S., Jin M. (2020). Severe acute respiratory syndrome coronavirus 2-specific antibodies in pets in Wuhan, China. J. Infect. 81 e68–e69. 10.1016/j.jinf.2020.06.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Wang X. (2020). MiRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 48 D127–D131. 10.1093/nar/gkz757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C. M., Cheng V. C. C., Hung I. F. N., Wong M. M. L., Chan K. H., Chan K. S., et al. (2004). Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax 59 252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y., Ulasli M., Schepers H., Mauthe M., V’kovski P., Kriegenburg F., et al. (2020). Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J. Virol. 94:e01925–19. 10.1128/JVI.01925-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti P., Ronconi G., Caraffa A., Gallenga C. E., Ross R., Frydas I., et al. (2020). Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by coronavirus-19 (COVID-19 or SARS-CoV-2): anti-inflammatory strategies. J. Biol. Regul. Homeost. Agents 34 327–331. 10.23812/CONTI-E [DOI] [PubMed] [Google Scholar]
- Corley M., Ndhlovu L. (2020). DNA methylation analysis of the COVID-19 host cell receptor, angiotensin i converting enzyme 2 gene (ACE2) in the respiratory system reveal age and gender differences. Med. Pharmacol. 11:7107. 10.20944/preprints202003.0295.v1 32283112 [DOI] [Google Scholar]
- Correale M., Croella F., Leopizzi A., Mazzeo P., Tricarico L., Mallardi A., et al. (2021). The evolving phenotypes of cardiovascular disease during COVID - 19 pandemic. Cardiovasc. Drugs Ther. 10.1007/s10557-021-07217-8 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlan L. (2020). Factors which contribute to the immunogenicity of non-replicating adenoviral vectored vaccines. Front. Immunol. 11:909. 10.3389/fimmu.2020.00909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta D., Vanelli M. (2020). WHO declares COVID-19 a pandemic. Acta Biomed. 91 157–160. 10.23750/abm.v91i1.9397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly J. L., Simonetti B., Klein K., Chen K., Williamson M. K., Antón-Plágaro C., et al. (2020). Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 370 861–865. 10.1126/science.abd3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N. G., Abbott S., Barnard R. C., Jarvis C. I., Kucharski A. J., Munday J. D., et al. (2021). Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 372:eabg3055. 10.1126/science.abg3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delavari S., Abolhassani H., Abolnezhadian F., Babaha F., Iranparast S., Ahanchian H., et al. (2020). Impact of SARS-CoV-2 pandemic on patients with primary immunodeficiency. J. Clin. Immunol. 41 345–355. 10.1007/s10875-020-00928-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhama K., Patel S. K., Pathak M., Yatoo M. I., Tiwari R., Malik Y. S., et al. (2020). An update on SARS-CoV-2/COVID-19 with particular reference to its clinical pathology, pathogenesis, immunopathology and mitigation strategies. Travel. Med. Infect. Dis. 37:101755. 10.1016/j.tmaid.2020.101755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinnes J., Deeks J. J., Adriano A., Berhane S., Davenport C., Dittrich S., et al. (2020). Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 8:CD013705. 10.1002/14651858.CD013705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doremalen N., Bushmaker T., Morris D., Holbrook M., Gamble A., Williamson B., et al. (2020). Aerosol and surface stability of HCoV-19 (SARS-CoV-2) compared to SARS-CoV-1. Engl. J. Med. 382 1564–1567. 10.1056/NEJMc2004973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dropulic L. K., Cohen J. I. (2011). Severe viral infections and primary immunodeficiencies. Clin. Infect. Dis. 53 897–909. 10.1093/cid/cir610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Günther S., Preiser W., Werf S., Brodt H. R., Becker S., et al. (2003). Identification of a novel coronavirus in patients with severe acute respiratory syndrome. Engl. J. Med. 348 1967–1976. 10.1056/NEJMoa030747 [DOI] [PubMed] [Google Scholar]
- Eckstrand C. D., Baldwin T. J., Rood K. A., Clayton M. J., Lott J. K., Wolking R. M., et al. (2021). An outbreak of SARS-CoV-2 with high mortality in mink (Neovison vison) on multiple Utah farms. PLoS Pathog. 17:e1009952. 10.1371/journal.ppat.1009952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein H., Diabetol C., Edelstein M. H., Guetta T., Barnea A., Waldman M., et al. (2021). Expression of the SARS - CoV - 2 receptorACE2 in human heart is associated with uncontrolled diabetes, obesity, and activation of the renin angiotensin system. Cardiovasc. Diabetol. 20:216. 10.1186/s12933-021-01275-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A., Invernizzi P., et al. (2020). Genomewide association study of severe Covid-19 with respiratory failure. N. Engl. J. Med. 383 1522–1534. 10.1056/nejmoa2020283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erster O., Shkedi O., Benedek G., Zilber E., Varkovitzky I., Shirazi R., et al. (2021). Improved sensitivity, safety, and rapidity of COVID-19 tests by replacing viral storage solution with lysis buffer. PLoS One 16:e0249149. 10.1371/journal.pone.0249149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control [ECDC] (2021). Emergence of SARS-CoV-2 B.1.617 Variants in India and Situation in the EU/EEA– 11 May 2021. Stockholm: ECDC. [Google Scholar]
- Eze P., Mezue K. N., Nduka C. U., Obianyo I., Egbuche O. (2021). Efficacy and safety of chloroquine and hydroxychloroquine for treatment of COVID-19 patients-a systematic review and meta-analysis of randomized controlled trials. Am. J. Cardiovasc. Dis. 11 93–107. [PMC free article] [PubMed] [Google Scholar]
- Fan J., Liu X., Pan W., Douglas M. W., Bao S. (2020). Epidemiology of coronavirus disease in gansu province, China, 2020. Emerg. Infect. Dis. 26 1257–1265. 10.3201/eid2606.20025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A. R., Perlman S. (2015). Coronaviruses: an overview of their replication and pathogenesis. Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 1282 1–23. 10.1007/978-1-4939-2438-7_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Food and Drug Administration [FDA] (2020). Research, Development and Licensure of Vaccines to Prevent COVID-19 Guidance for Industry. Available online at: https://www.fda.gov/media/139638/download (Acess 23 outubro de 2020). [Google Scholar]
- Fricke-Galindo I., Falfán-Valencia R. (2021). Genetics insight for COVID-19 susceptibility and severity: a review. Front. Immunol. 12:622176. 10.3389/fimmu.2021.622176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulzele S., Sahay B., Yusufu I., Lee T. J., Sharma A., Kolhe R., et al. (2020). COVID-19 virulence in aged patients might be impacted by the host cellular MicroRNAs abundance/profile. Aging Dis. 11 509–522. 10.14336/AD.2020.0428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway S., Paul P., MacCannell D. R., Johansson M. A., Brooks J. T., MacNeil A., et al. (2021). Emergence of SARS-CoV-2 B.1.1.7 Lineage. MMWR Morb. Mortal Wkly. Rep. 70 95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., et al. (2020). Development of an inactivated vaccine candidate for SARS-CoV-2. Science 369 77–81. 10.1126/science.abc1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S. J., Guo H., Luo G. (2021). Omicron variant (B.1.1.529) of SARS-CoV-2, a global urgent public health alert!. J. Med. Virol. 10.1002/jmv.27491 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmati D., Bramanti B., Serino M. L., Secchiero P., Zauli G., Tisato V. (2020). COVID-19 and individual genetic susceptibility/receptivity: role of ACE1/ACE2 genes, immunity, inflammation and coagulation. might the double x-chromosome in females be protective against SARS-COV-2 compared to the single x-chromosome in males? Int. J. Mol. Sci. 21 1–23. 10.3390/ijms21103474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez J., Albaiceta G. M., García-clemente M., López-larrea C. (2020). Angiotensin-converting enzymes (ACE, ACE2) gene variants and COVID-19 outcome. Gene 762:145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Baker S. C., de Groot R. J., Drosten C., Gulyaeva A. A. (2020). The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 5 536–544. 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guaraldi G., Meschiari M. M., Cozzi-Lepri A., Milic J., Tonelli R., Menozzi M., et al. (2020). Tocilizumab in patients with severe COVID-19: a retrospective cohort study. Lan. Rheumatol. 2 e474–e484. 10.1016/S2665-9913(20)30173-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashizume M., Gonzalez G., Ono C., Takashima A., Iwasaki M. (2021). Population-specific ACE2 single-nucleotide polymorphisms have limited impact on SARS-CoV-2 infectivity in vitro. Viruses 13 1–10. 10.3390/v13010067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Li J., Jiang S. (2006). A single amino acid substitution (R441A) in the receptor-binding domain of SARS coronavirus spike protein disrupts the antigenic structure and binding activity. Biochem. Biophys. Res. Commun. 344 106–113. 10.1016/j.bbrc.2006.03.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurich A., Hofmann-Winkler H., Gierer S., Liepold T., Jahn O., Pohlmann S. (2014). TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 88 1293–1307. 10.1128/jvi.02202-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehl S., Rabenau H., Berger A., Kortenbusch M., Cinatl J., Bojkova D., et al. (2020). Evidence of SARS-CoV-2 infection in returning travelers from Wuhan, China. Engl. J. Med. 382 1278–1280. 10.1056/NEJMc2001899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Krüger N., Müller M., Drosten C., Pöhlmann S. (2020). The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. Cell 181 271.e8–280.e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou T., Zeng W., Yang M., Chen W., Ren L. (2020). Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathog. 16:e1008705. 10.1371/journal.ppat.1008705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Li C., Wang S., Li T., Zhang H. (2020). Genetic variants are identified to increase risk of COVID-19 related mortality from UK Biobank data. medRxiv [Preprint]. 10.1101/2020.11.05.20226761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Gao J., Luo X., Feng L., Liu W., Chen J., et al. (2020). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vertical transmission in neonates born to mothers with coronavirus disease 2019 (COVID-19) pneumonia. Ostet. Gynecol. 136 1–3. 10.1097/AOG.0000000000003926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A. T., Garcia-Carreras B., Hitchings M. D. T., Yang B., Katzelnick L. C., Rattigan S. M., et al. (2020). A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat. Commun. 11:4704. 10.1038/s41467-020-18450-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Lu Q., Zhao H., Zhang Y., Sui Z., Fang L., et al. (2020). Temporal antibody responses to SARS-CoV-2 in patients of coronavirus disease 2019. Cell. Discov. 6:64. 10.1038/s41421-020-00209-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W. E., Lim B., Hsu C. C., Xiong D., Wu W., Yu Y., et al. (2020). RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 13 950–961. 10.1111/1751-7915.13586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubacek J. A., Dusek L., Majek O., Adamek V. (2020). ACE I/D polymorphism in Czech first-wave SARS-CoV-2-positive survivors. Clin. Chim. Acta J. 519 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Committee on Taxonomy of Viruses [ICTV] (2019). Available online at: https://talk.ictvonline.org/taxonomy/p/taxonomy-history?taxnode_id=202001847 (Acess 31 de agosto de 2021). [Google Scholar]
- International Committee on Taxonomy of Viruses [ICTV] (2020). Available online at: https://talk.ictvonline.org/ictv-reports/ictv_9th_report/positive-sense-rna-viruses-2011/w/posrna_viruses/222/coronaviridae (Acess 21 de julho de 2020). [Google Scholar]
- International Labour Organization [ILO] (2020). Available online at: https://www.ilo.org/global/topics/coronavirus/lang–en/index.htm (Acess 25 de outubro de 2020). [Google Scholar]
- Ishii T. (2020). Human leukocyte antigen (HLA) class I susceptible alleles against COVID-19 increase both infection and severity rate. Cureus 12 1–12. 10.7759/cureus.12239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturrieta-Zuazo I., Rita C. G., García-Soidán A., de Malet Pintos-Fonseca A., Alonso-Alarcón N., Pariente-Rodríguez R., et al. (2020). Possible role of HLA class-I genotype in SARS-CoV-2 infection and progression: a pilot study in a cohort of Covid-19 Spanish patients. Clin. Immunol. 219:108572. 10.1016/j.clim.2020.108572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes J., Millet J., Whittaker G. (2020). Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1/S2. iScience 23:101212. 10.1016/j.isci.2020.101212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W., Wang W., Zhao X., Zai J., Li X. (2020). Cross species transmission of the newly identified coronavirus 2019-nCoV. J. Med. Virol. 92 433–440. 10.1002/jmv.25682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jon C. (2020). Merck One of big pharma’s biggest players, reveals its COVID-19 vaccine and therapy plans. Science 10.1126/science.abd0121 [Epub ahead of print]. [DOI] [Google Scholar]
- Jones L., Bakre A., Naikare H., Kolhe R., Sanchez S., Mosley Y. Y. C., et al. (2021). Isothermal amplification and fluorescent detection of SARS-CoV-2 and SARS-CoV-2 variant virus in nasopharyngeal swabs. PLoS One 16:e0257563. 10.1371/journal.pone.0257563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juscamayta-López E., Valdivia F., Horna H., Tarazona D., Linares L., Rojas N., et al. (2021). A multiplex and colorimetric reverse transcription loop-mediated isothermal amplification assay for sensitive and rapid detection of novel SARS-CoV-2. Front. Cell infect. Microb. 11:653616. 10.3389/fcimb.2021.653616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabinger F., Stiller C., Schmitzová J., Dienemann C., Kokic G., Hillen H. S., et al. (2021). Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nature 28 740–746. 10.1038/s41594-021-00651-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M., Wei J., Yuan J., Guo J., Zhang Y., Hang J., et al. (2020). Probable evidence of fecal aerosol transmission of SARS-CoV-2 in a high-rise building. Ann. Intern. Med. 173 974–980. 10.7326/M20-0928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase M., Shirato K., van der Hoek L., Taguchi F., Matsuyama S. (2012). Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol. 86 6537–6545. 10.1128/JVI.00094-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Adil S. F., Alkhathlan H. Z., Tahir M. N., Saif S., Khan M., et al. (2020). COVID-19: a global challenge with old history, epidemiology and progress so far. Molecules 26:39. 10.3390/molecules26010039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor S. S., Omae Y., Nishida N., Sugiyama M., Kinoshita N., Suzuki T., et al. (2021). HLA-A*11:01:01:01, HLA-C*12:02:02:01-HLA-B*52:01:02:02, age and sex are associated with severity of Japanese COVID-19 with respiratory failure. Front. Immunol. 12:658570. 10.3389/fimmu.2021.658570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim U., Won E., Kee S., Jung S., Jang H. (2016). Combination therapy with lopinavir/ritonavir, ribavirin and interferon-α for middle east respiratory syndrome. Antivir. Ther. 21 455–459. 10.3851/IMP3002 [DOI] [PubMed] [Google Scholar]
- Korber B., Fischer W. M., Gnanakaran S., Yoon H., Theiler J., Abfalterer T., et al. (2020). Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182 812–827. 10.1016/j.cell.2020.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmicki J. A., Horowitz J. E., Banerjee N., Lanche R., Marcketta A., Maxwell E., et al. (2021). Pan-ancestry exome-wide association analyses of COVID-19 outcomes in 586,157 individuals. Am. J. Hum. Genet. 108 1350–1355. 10.1016/j.ajhg.2021.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewski R., Gołębiowska J., Makuch S., Mazur G., Agrawal S. (2020). Update on serologic testing in COVID-19. Clin. Chim Acta 510 746–750. 10.1016/j.cca.2020.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert D. W., Yarski M., Warner F. J., Thornhill P., Parkin E. T., Smith A. I., et al. (2005). Tumor necrosis factor-α convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2). J. Biol. Chem. 280 30113–30119. 10.1074/jbc.M505111200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge G., Yu J., Shan S., Zhou H., Fan S., et al. (2020). Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 81 215–220. 10.1038/s41586-020-2180-5 [DOI] [PubMed] [Google Scholar]
- Langton D. J., Bourke S. C., Lie B. A., Reiff G., Natu S., Darlay R., et al. (2021). The influence of HLA genotype on the severity of COVID-19 infection. HLA 98 14–22. 10.1111/tan.14284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latini A., Agolini E., Novelli A., Borgiani P., Giannini R., Gravina P., et al. (2020). COVID-19 and genetic variants of protein involved in the SARS-CoV-2 entry into the host cells. Genes 11 1–8. 10.3390/genes11091010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L. T., Fung Y. W. W., Wong F. P. F., Lin S. S. W., Wang C. R., Li H. L., et al. (2003). A real-time PCR for SARS-coronavirus incorporating target gene pre-amplification. Biochem. Biophys. Res. Commun. 312 1290–1296. 10.1016/j.bbrc.2003.11.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrer S., Rheinstein P. H. (2020). ABO blood groups, COVID-19 infection and mortality. Blood Cells, Mol. Dis. 89 1–5. 10.1016/j.bcmd.2021.102571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite M. D. M., Gonzalez-Galarza F. F., Silva B. C. C., da, Middleton D., Santos E. J. M. (2021). Predictive immunogenetic markers in COVID-19. Hum. Immunol. 82 247–254. 10.1016/j.humimm.2021.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., De Clercq E. (2020). Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat. Rev. Drug. Discov. 19 149–150. 10.1038/d41573-020-00016-0 [DOI] [PubMed] [Google Scholar]
- Li L., Petrovsky N. (2016). Molecular mechanisms for enhanced DNA vaccine immunogenicity. Expert Rev. Vaccines 15 313–329. 10.1586/14760584.2016.1124762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., et al. (2020). Early transmission dynamics in wuhan, china, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 382 1199–1207. 10.1056/NEJMoa2001316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littera R., Campagna M., Deidda S., Angioni G., Cipri S., Melis M., et al. (2020). Human leukocyte antigen complex and other immunogenetic and clinical factors influence susceptibility or protection to SARS-CoV-2 infection and severity of the disease course. The sardinian experience. Front. Immunol. 11:605688. 10.3389/fimmu.2020.605688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorent D., Nowak R., Roxo C., Lenartowicz E., Makarewicz A., Zaremba B., et al. (2021). Prevalence of Anti-SARS-CoV-2 Antibodies in Pozna n, after the First Wave of the COVID-19 Pandemic. Vaccines 9 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Stratton C. W., Tang Y. W. (2020). Outbreak of pneumonia of unknown etiology in Wuhan, China: the mystery and the miracle. J. Med. Virol. 92 401–402. 10.1002/jmv.25678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li D., Niu P., Yang B., Wu H., et al. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395 565–574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C., Wong P., Klein J., Castro T. B. R., Silva J., Sundaram M., et al. (2020). Yale IMPACT Team Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 584 463–469. 10.1038/s41586-020-2588-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucia C., Federico P. B., Alejandra G. C. (2020). An ultrasensitive, rapid, and portable coronavirus SARS-CoV-2 sequence detection method based on CRISPR-Cas12. bioRxiv [Preprint]. 10.1101/2020.02.29.971127 [DOI] [Google Scholar]
- Martínez-Sanz J., Jiménez D., Martínez-Campelo L., Cruz R., Vizcarra P., Sánchez-Conde M., et al. (2021). Role of ACE2 genetic polymorphisms in susceptibility to SARS-CoV-2 among highly exposed but non infected healthcare workers. Emerg. Microbes Infect. 10 493–496. 10.1080/22221751.2021.1902755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maucourant C., Filipovic I., Ponzetta A., Aleman S., Cornillet M., Hertwig L., et al. (2020). Natural killer cell immunotypes related to COVID-19 disease severity. Sci. Immunol. 5 1–14. 10.1126/SCIIMMUNOL.ABD6832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker C., Upton J., Warrington R. (2018). Primary immunodeficiency disorders. Allergy Asthma Clin. Immunol. 14 142–152. 10.1016/S0095-4543(05)70085-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal A., Manjunath K., Ranjan R. K., Kaushik S., Kumar S., Verma V. (2020). COVID-19 pandemic: insights into structure, function, and hACE2 receptor recognition by SARS-CoV-2. PLoS Pathog. 16:e1008762. 10.1371/journal.ppat.1008762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad A., Mara S. K., Alshawaf E., Abu-farha M. (2020). Structural analysis of ACE2 variant N720D demonstrates a higher binding affinity to TMPRSS2. Life Sci. 259 1–7. 10.1016/j.lfs.2020.118219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad S., Hashemi A., Thijssen M., Younes S., Alijan H. (2021). Human gene polymorphisms and their possible impact on the clinical outcome of SARS - CoV - 2 infection. Arch. Virol. 166 2089–2108. 10.1007/s00705-021-05070-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadpour S., Torshizi Esfahani A., Halaji M., Lak M., Ranjbar R. (2021). An updated review of the association of host genetic factors with susceptibility and resistance to COVID-19. J. Cell. Physiol. 236 49–54. 10.1002/jcp.29868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möhlendick B., Schönfelder K., Breuckmann K., Elsner C., Babel N., Balfanz P., et al. (2021). ACE2 polymorphism and susceptibility for SARS-CoV-2 infection and severity of COVID-19. Pharmacogenet. Genomics 31 165–171. 10.1097/FPC.0000000000000436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motozono C., Zahradnik M., Ikeda T., Saito A., Tan T., Ngare I., et al. (2021). An emerging SARS-CoV-2 mutant evading cellular immunity and increasing viral infectivity. bioRxiv [Preprint]. 10.1101/2021.04.02.438288 [DOI] [Google Scholar]
- Mycroft-West C. J., Su D., Pagani I., Rudd T. R., Elli S., Gandhi N. S., et al. (2020). Heparin inhibits cellular invasion by SARS-CoV-2: structural dependence of the interaction of the spike S1 receptor-binding domain with heparin. Thromb. Haemost. 120 1700–1715. 10.1055/s-0040-1721319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naemi F. M. A., Al-Adwani S., Al-Khatabi H., Al-Nazawi A. (2021). Association between the HLA genotype and the severity of COVID-19 infection among South Asians. J. Med. Virol. 93 4430–4437. 10.1002/jmv.27003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y., Takagi T., Yamamoto T., Watanabe S. (2020). Association between selective IgA deficiency and COVID-19. J. Clin. Biochem. Nutr. 67 122–125. 10.3164/jcbn.20-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres J. L., Verdiá-Báguena C., Jimenez-Guardeño J. M., Regla-Nava J. A., Castaño-Rodriguez C., Fernandez-Delgado R., et al. (2015). Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology 485 330–339. 10.1016/j.virol.2015.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novelli A., Andreani M., Biancolella M., Liberatoscioli L., Passarelli C., Colona V. L., et al. (2020). HLA allele frequencies and susceptibility to COVID-19 in a group of 99 Italian patients. HLA 96 610–614. 10.1111/tan.14047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organization for Economic Cooperation and Development [OECD] (2020). Available online at: https://www.oecd.org/coronavirus/policy-responses/covid-19-na-regiao-da-america-latina-e-caribe-implicacoes-sociais-e-economicas-e-politicas-prioritarias-433b9d11/ (accessed January 12, 2021). [Google Scholar]
- Organization for Economic Cooperation and Development [OECD] (2021). Available online at: https://www.oecd-ilibrary.org/sites/edfbca02-en/index.html?itemId=/content/publication/edfbca02-en&_ga=2.227241181.1503655903.1623254151-1665687391.1623254151 (accessed June 9, 2021). [Google Scholar]
- Oude Munnink B. B., Sikkema R. S., Nieuwenhuijse D. F., Molenaar R. J., Munger E., Molenkamp R., et al. (2021). Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 371 172–177. 10.1126/science.abe5901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A., Rawlik K., Pasko D., et al. (2021). Genetic mechanisms of critical illness in Covid-19. Nature 591 92–98. 10.1038/s41586-020-03065-y [DOI] [PubMed] [Google Scholar]
- Palau V., Riera M., Soler M. J. (2020). ADAM17 inhibition may exert a protective effect on COVID-19. Nephrol. Dial. Transplant. 35 1071–1072. 10.1093/ndt/gfaa093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L. L. M., Wang Q. (2020). Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 20 411–412. 10.1016/S1473-3099(20)30113-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A., Walters J., Reuschel E. L., Schultheis K., Parzych E., Gary E. N., et al. (2020). Intradermal-delivered DNA vaccine provides anamnestic protection in a rhesus macaque SARS-CoV-2 challenge model. bioRxiv [Preprint]. 10.1101/2020.07.28.225649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfizer (2021). Pfizer and BioNTech Conclude Phase 3 Study of COVID-19 Vaccine Candidate, Meeting All Primary Efficacy Endpoints. Available online at: https://www.pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-conclude-phase-3-study-covid-19-vaccine (accessed February 23, 2021). [Google Scholar]
- Poulton K., Wright P., Hughes P., Savic S., Welberry Smith M., Guiver M., et al. (2020). A role for human leucocyte antigens in the susceptibility to SARS-Cov-2 infection observed in transplant patients. Int. J. Immunogenet. 47 324–328. 10.1111/iji.12505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather K. A., Wang C. C., Schooley R. T. (2020). Reducing transmission of SARS-CoV-2. Science 368 1422–1424. 10.1126/science.abc6197 [DOI] [PubMed] [Google Scholar]
- Prazuck T., Colin M., Giachè S., Gubavu C., Seve A., Rzepecki V., et al. (2020). Evaluation of performance of two SARS-CoV-2 Rapid IgM-IgG combined antibody tests on capillary whole blood samples from the fingertip. PLoS One 15:e0237694. 10.1371/journal.pone.0237694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch J. W., Capoferri A. A., Katusiime M. G., Patro S. C., Kearney M. F. (2020). Low genetic diversity may be an Achilles heel of SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 117 24614–24616. 10.1073/pnas.2017726117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendeiro A. F., Casano J., Vorkas C. K., Singh H., Morales A., Desimone R. A., et al. (2020). Longitudinal immune profiling of mild and severe COVID-19 reveals innate and adaptive immune dysfunction and provides an early prediction tool for clinical progression. medRxiv [Preprint]. 10.1101/2020.09.08.20189092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendeiro A. F., Casano J., Vorkas C. K., Singh H., Morales A., DeSimone R. A., et al. (2021). Profiling of immune dysfunction in COVID-19 patients allows early prediction of disease progression. Life Sci. Alliance 4:e202000955. 10.26508/lsa.202000955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P., Griffin I., Tucker C. (2020). Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet 395 e30–e31. 10.1016/S0140-6736(20)30304-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R. (2020). A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells 9:1267. 10.3390/cells9051267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-López J. P., Carnalla-Cortés M., Pacheco-Olvera D. L., Ocampo-Godínez J. M., Oliva-Ramírez J., Moreno-Manjón J., et al. (2021). A bioinformatic prediction of antigen presentation from SARS-CoV-2 spike protein revealed a theoretical correlation of HLA-DRB1*01 with COVID-19 fatality in Mexican population: an ecological approach. J. Med. Virol. 93 2029–2038. 10.1002/jmv.26561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas I. O., Bräu N., Waters M., Go R., Hunter B. D., Bhagani S., et al. (2020). Tocilizumab in hospitalized patients with COVID-19 pneumonia. MedRxiv [Preprint]. 10.1101/2020.08.27.20183442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum J. T., Hamilton H., Weisman M. H., Reveille J. D., Winthrop K. L., Choi D. (2021). The effect of HLA-B27 on susceptibility and severity of COVID-19. J. Rheumatol. 48 621–622. 10.3899/JRHEUM.200939 [DOI] [PubMed] [Google Scholar]
- Russo R., Andolfo I., Lasorsa V. A., Iolascon A., Capasso M. (2020). Genetic analysis of the coronavirus SARS-CoV-2 host protease TMPRSS2 in different populations. Front. Genet. 11:872. 10.3389/fgene.2020.00872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailleau C., Dumarest M., Vanhomwegen J., Delaplace M., Caro V., Kwasiborski A. (2020). First detection and genome sequencing of SARS-CoV-2 in an infected cat in France. Transbound Emerg. Dis. 67 2324–2328. 10.1111/tbed.13659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajuthi S. P., DeFord P., Li Y., Jackson N. D., Montgomery M. T., Everman J. L., et al. (2020). Type 2 and interferon inflammation regulate SARS-CoV-2 entry factor expression in the airway epithelium. Nat. Commun. 11:5139. 10.1038/s41467-020-18781-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba A., Haider H., Sato T. (2020). Population difference in allele frequency of hla-c*05 and its correlation with covid-19 mortality. Viruses 12:1333. 10.3390/v12111333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmiento J. D., Villada F., Orrego J. C., Franco J. L., Trujillo-Vargas C. M. (2016). Adverse events following immunization in patients with primary immunodeficiencies. Vaccine 34 1611–1616. 10.1016/j.vaccine.2016.01.047 [DOI] [PubMed] [Google Scholar]
- Savarino A., Boelaert J. R., Cassone A., Majori G., Cauda R. (2003). Effects of chloroquine on viral infections: an old drug against today’s diseases. Lancet Infet. Dis. 3 722–727. 10.1016/s1473-3099(03)00806-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler E., Dribus M., Duffy B. F., Gragert L., Hock K., Farnsworth C. W. (2021). HLA genetic polymorphism in patients with coronavirus disease 2019 in Midwestern United States. HLA 98 370–379. 10.1111/tan.14387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönfelder K., Breuckmann K., Elsner C., Dittmer U., Fistera D., Herbstreit F., et al. (2021). Transmembrane serine protease 2 polymorphisms and susceptibility to severe acute respiratory syndrome coronavirus type 2 infection: a German Case-Control Study. Front. Genet. 12:667231. 10.3389/fgene.2021.667231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secolin R., de Araujo T. K., Gonsales M. C., Rocha C. S., Naslavsky M., De Marco L., et al. (2021). Genetic variability in COVID-19-related genes in the Brazilian population. Hum. Genome Var. 8:15. 10.1038/s41439-021-00146-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapati S., Kumar S., Singh A. K., Banerjee P., Bhagavatula S. (2020). Assessment of risk conferred by coding and regulatory variations of TMPRSS2 and CD26 in susceptibility to SARS-CoV-2 infection in human. J. Genet. 99:53. 10.1007/s12041-020-01217-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo G., Lee G., Kim M. J., Baek S. H., Choi M., Ku K. B., et al. (2020). Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano 14 5135–5142. 10.1021/acsnano.0c02823 [DOI] [PubMed] [Google Scholar]
- Shaath H., Vishnubalaji R., Elkord E., Alajez N. M. (2020). Single-cell transcriptome analysis highlights a role for neutrophils and inflammatory macrophages in the pathogenesis of severe COVID-19. Cells 9 1–19. 10.3390/cells9112374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., We Z., Zhong G., Yang H., Wang C., Huang B., et al. (2020). Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science 368 1016–1020. 10.1126/science.abb7015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkurnikov M., Nersisyan S., Jankevic T., Galatenko A., Gordeev I., Vechorko V., et al. (2021). Association of HLA class I genotypes with severity of coronavirus disease-19. Front. Immunol. 12:641900. 10.3389/fimmu.2021.641900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai L., Zhong G., Yuan Q., Wen Z., Wang C., He X., et al. (2021). Replication, pathogenicity, and transmission of SARS-CoV-2 in minks. Nat. Sci. Rev. 8:nwaa291. 10.1093/nsr/nwaa291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu L. Y., Lo K. J., Chan C. M., Kien F., Escriou N., Tsao S. W., et al. (2008). The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 82 11318–11330. 10.1128/JVI.01052-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L. R., Wloch M. K., Ye M., Reyes L. R., Boutsaboualoy S., Dunne C. E., et al. (2010). Phase 1 clinical trials of the safety and immunogenicity of adjuvanted plasmid DNA vaccines encoding influenza a virus H5 hemagglutinin. Vaccine 28 2565–2572. 10.1016/j.vaccine.2010.01.029 [DOI] [PubMed] [Google Scholar]
- Song H. D., Tu C. C., Zhang G. W., Wang S. Y., Zheng K., Lei L. C., et al. (2005). Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. U.S.A. 102 2430–2435. 10.1073/pnas.0409608102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinetti T., Hirzel C., Fux M., Walti L. N., Schober P., Stueber F., et al. (2020). Reduced monocytic human leukocyte antigen-dr expression indicates immunosuppression in critically Ill COVID-19 patients. Anesth. Analg. 131 993–999. 10.1213/ANE.0000000000005044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S., Cui H., Gao Z., Liu M., Lu S., Mkandawire W., et al. (2020). Structural genomics of SARS-CoV-2 indicates evolutionary conserved functional regions of viral proteins. Viruses 12:360. 10.3390/v12040360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strafella C., Caputo V., Termine A., Barati S., Gambardella S., Borgiani P., et al. (2020). Analysis of ace2 genetic variability among populations highlights a possible link with covid-19-related neurological complications. Genes 11 1–10. 10.3390/genes11070741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczawinska-Poplonyk A., Jonczyk-Potoczna K., Breborowicz A., Bartkowska-Sniatkowska A., Figlerowicz M. (2013). Fatal respiratory distress syndrome due to coronavirus infection in a child with severe combined immunodeficiency. Influenza Other Respi. Viruses 7 634–636. 10.1111/irv.12059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahamtan A., Teymoori-Rad M., Nakstad B., Salimi V. (2018). Anti-inflammatory MicroRNAs and their potential for inflammatory diseases treatment. Front. Immunol. 9:1377. 10.3389/fimmu.2018.01377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi A., Matsui M. (2021). Identification of HLA-A*02:01-restricted candidate epitopes derived from the nonstructural polyprotein 1a of SARS-CoV-2 that may be natural targets of CD8 + T cell recognition in vivo. J. Virol. 95:e01837–20. 10.1128/jvi.01837-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Goh P., Fielding B., Shen S., Chou C., Fu J., et al. (2004). Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin. Diagn. Lab. Immunol. 11 362–371. 10.1128/cdli.11.2.362-371.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N., Bai H., Chen X., Gong J., Li D., Sun Z. (2020). Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. J. Thromb. Haemost. 18 1094–1099. 10.1111/jth.14817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Wu C., Li X., Song Y., Yao X., Wu X., et al. (2020). On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 7 1012–1023. 10.1093/nsr/nwaa036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., et al. (2021). Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 592 438–443. 10.1038/s41586-021-03402-9 [DOI] [PubMed] [Google Scholar]
- Tomita Y., Ikeda T., Sato R., Sakagami T. (2020). Association between HLA gene polymorphisms and mortality of COVID-19: an in silico analysis. Immun. Inflamm. Dis. 8 684–694. 10.1002/iid3.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre-Fuentes L., Matías-Guiu J., Hernández-Lorenzo L., Montero-Escribano P., Pytel V., Porta-Etessam J., et al. (2021). ACE2, TMPRSS2, and furin variants and SARS-CoV-2 infection in Madrid, Spain. J. Med. Virol. 93 863–869. 10.1002/jmv.26319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima Y., Nemoto K., Matsumoto S., Nakamura Y., Kiyotani K. (2020). SARS-CoV-2 genomic variations associated with mortality rate of COVID-19. J. Hum. Genet. 65 1075–1082. 10.1038/s10038-020-0808-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tregoning J. S., Brown E. S., Cheeseman H. M., Flight K. E., Higham S. L., Lemm N. M., et al. (2020). Vaccines for COVID-19. Clin. Exp. Immunol. 202 162–192. 10.1111/cei.13517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Y., Chang C., Wang S., Huang K., Wang C. (2013). Identifying SARS-CoV membrane protein amino acid residues linked to virus-like particle assembly. PLoS One 8:e64013. 10.1371/journal.pone.0064013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulasli M., Verheije M. H., de Haan C. A. M., Reggiori F. (2010). Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell Microb. 12 844–861. 10.1111/j.1462-5822.2010.01437.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y. (2020). Camostat mesilate therapy for COVID-19. Intern. Emerg. Med. 15 1577–1578. 10.1007/s11739-020-02345-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Made C. I., Simons A., Schuurs-Hoeijmakers J., Van Den Heuvel G., Mantere T., Kersten S., et al. (2020). Presence of genetic variants among young men with severe COVID-19. JAMA J. Am. Med. Assoc. 324 663–673. 10.1001/jama.2020.13719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastrad B., Vastrad C., Tengli A. (2020). Identification of potential mRNA panels for severe acute respiratory syndrome coronavirus 2 (COVID-19) diagnosis and treatment using microarray dataset and bioinformatics methods. 3 Biotech 10:422. 10.1007/s13205-020-02406-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Salinas L., Zarate S., Eberl S., Gladue D. P., Novella I., Borca M. V. (2020). Positive selection of ORF1ab, ORF3a, and ORF8 genes drives the early evolutionary trends of SARS-CoV-2 during the 2020 COVID-19 pandemic. Front. Microbiol. 11:550674. 10.3389/fmicb.2020.550674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatagopalan P., Daskalova S. M., Lopez L. A., Dolezal K. A., Hoguea B. G. (2015). Coronavirus envelope (E) protein remains at the site of assembly. Virology 478 75–85. 10.1016/j.virol.2015.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietzen H., Zoufaly A., Traugott M., Aberle J. (2020). NK cell receptor NKG2C deletion and HLA-E variants are risk factors for severe COVID-19. Res. Square 10.21203/rs.3.rs-34505/v1 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vique-Sanchez J. L. (2020). Potential inhibitors interacting in Neuropilin-1 to develop an adjuvant drug against COVID-19, by molecular docking. Bioorgan. Med. Chem. 33 1–8. 10.1016/j.bmc.2021.116040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivanti A. J., Vauloup-Fellous C., Prevot S., Zupan V., Suffee C., Do Cao J., et al. (2020). Transplacental transmission of SARS-CoV-2 infection. Nat. Comm. 11:3572. 10.1038/s41467-020-17436-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E., Mishra S., Chand M., Barrett J., Johnson R., Hopkins S., et al. (2021). Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: insights from linking epidemiological and genetic data. medRxiv [Preprint]. 10.1038/s41586-021-03470-x [DOI] [PubMed] [Google Scholar]
- Walensky R. P., Walke H. T., Fauci A. S. (2021). SARS-CoV-2 variants of concern in the United States-challenges and opportunities. JAMA J. Am. Med. Assoc. 325 1037–1038. 10.1001/jama.2021.2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., et al. (2020a). Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 181 894–904.e9. 10.1016/j.cell.2020.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Chen W., Zhang Z., Deng Y., Lian J., Du P., et al. (2020b). CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Sinal Transduto Alvo Ther. 5:283. 10.1038/s41392-020-00426-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Xu X., Zhou X., Chen P., Liang H., Li X., et al. (2020c). Molecular simulation of SARS-CoV- 2 spike protein binding to pangolin ACE2 or human ACE2 natural variants reveals altered susceptibility to infection. J. Gen. Virol. 101 921–924. 10.1099/jgv.0.001452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Zhang W., Zhang J., He J., Zhu F. (2020d). Distribution of HLA allele frequencies in 82 Chinese individuals with coronavirus disease-2019 (COVID-19). HLA 96 194–196. 10.1111/tan.13941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., et al. (2020e). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 30 269–271. 10.1038/s41422-020-0282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Huang S., Gao R., Zhou Y., Lai C., Li Z., et al. (2020f). Initial whole-genome sequencing and analysis of the host genetic contribution to COVID-19 severity and susceptibility. Cell Discov. 6:83. 10.1038/s41421-020-00231-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Hou H., Yao Y., Wu S., Huang M., Ran X., et al. (2020g). Systemically comparing host immunity between survived and deceased COVID-19 patients. Cell. Mol. Immunol. 17 875–877. 10.1038/s41423-020-0483-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhang Y., Huang B., Deng W., Quan Y., Wang W., et al. (2020h). Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell 182 713.e4–721.e4. 10.1016/j.cell.2020.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Casner R. G., Nair M. S., Wang M., Yu J., Cerutti G., et al. (2021). Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe 29 747.e4–751.e4. 10.1016/j.chom.2021.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Qiu Z., Hou Y., Deng X., Xu W., Zheng T., et al. (2021). AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 31 126–140. 10.1038/s41422-020-00460-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C. K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., et al. (2021). SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 27 622–625. 10.1038/s41591-021-01285-x [DOI] [PubMed] [Google Scholar]
- Widiasta A., Sribudiani Y., Nugrahapraja H., Hilmanto D., Sekarwana N., Rachmadi D. (2020). Potential role of ACE2-related microRNAs in COVID-19-associated nephropathy. Noncoding RNA Res. 5 153–166. 10.1016/j.ncrna.2020.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V. M., Guggemos W., Seilmaier M., Zange S., Müller M. A., et al. (2020). Virological assessment of hospitalized patients with COVID-2019. Nature 581 465–469. 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
- Wool G. D., Miller J. L. (2021). The impact of COVID-19 disease on platelets and coagulation. Pathobiology 88 15–27. 10.1159/000512007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization [WHO] (2021a). Disponível em: https://covid19.who.int/region/amro/country/br (Acess 3 de dezembro de 2021). [Google Scholar]
- World Health Organization [WHO] (2021b). Disponível em: https://www.who.int/pt/emergencies/diseases/novel-coronavirus-2019/covid-19-vaccines (Acess 31 de agosto de 2021). [Google Scholar]
- World Health Organization [WHO] (2021c). Disponível em: https://extranet.who.int/pqweb/sites/default/files/documents/Status_COVID_VAX_19August2021.pdf (Acess 31 de agosto de 2021). [Google Scholar]
- Wrapp D., Wang N., Corbett K. S., Goldsmith J. A., Hsieh C., Abiona O., et al. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367 1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., et al. (2020). Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 27 325–328. 10.1016/j.chom.2020.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Feng Z., Li P., Yu Q. (2020). Relationship between ABO blood group distribution and clinical characteristics in patients with COVID-19. Clin. Chim. Acta J. 509 1–5. 10.1016/j.cca.2020.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Huang X., Yuan L., Wang S., Zang Y., Xiong H. (2021). A recombinant spike protein subunit vaccine confers protective immunity against SARS-CoV-2 infection and transmission in hamsters. Sci. Transl. Med. 13:eabg1143. 10.1126/scitranslmed.abg1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Ong S. W., Tan Y. K., Chia P. W., Lee T. H., Ng O. K., Wong M. S. Y., et al. (2020). Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. JAMA 323 1610–1612. 10.1001/jama.2020.3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A. T., Tong Y. X., Gao C., Zhu L., Zhang Y. J., Zhang S. (2020). Dynamic profile of RT-PCR findings from 301 COVID-19 patients in Wuhan, China: a descriptive study. J. Clin. Virol. 127:104346. 10.1016/j.jcv.2020.104346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., et al. (2020). Evolution of the novel coronavirus from the ongoing wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 63 457–460. 10.1007/s11427-020-1637-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Kiso M., Sakai-Tagawa Y., Iwatsuki-Horimoto K., Imai M., Takeda M., et al. (2020). The anticoagulant nafamostat potently inhibits SARS-CoV-2 S protein-mediated fusion in a cell fusion assay system and viral infection in vitro in a cell-type-dependent manner. Viruses 12:629. 10.3390/v12060629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N., Yang R., Yoshinaka Y., Amari S., Nakano T., Cinatl J., et al. (2004). HIV protease inhibitor nelfinavir inhibits replication of SARS-associated coronavirus. Biochem. Biophys. Res. Commun. 318 719–725. 10.1016/j.bbrc.2004.04.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Gou Y., Zhou Q. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 360 1444–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H., Song Y., Chen Y., Wu N., Xu J., Sun C., et al. (2020). Molecular architecture of the SARS-CoV-2 virus. Cell 183 730.e13–738.e13. 10.1016/j.cell.2020.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong C. Y., Ong H. K., Yeap S. K., Ho K. L., Tan W. S. (2019). Recent advances in the vaccine development against Middle East respiratory syndrome-coronavirus. Front. Microbiol. 10:1781. 10.3389/fmicb.2019.01781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young B. E., Ong S. W. X., Kalimuddin S., Low J. G., Tan S. Y., Loh J., et al. (2020). Singapore 2019 novel coronavirus outbreak research team. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA 323 1488–1494. 10.1001/jama.2020.3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung Y. L., Cheng C. K., Chan H. Y., Xia J. T., Lau K. M., Wong R. S. M., et al. (2021). Association of HLA-B22 serotype with SARS-CoV-2 susceptibility in Hong Kong Chinese patients. HLA 97 127–132. 10.1111/tan.14135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A. M., Van Boheemen S., Bestebroer T. M., Osterhaus A. D., Fouchier R. A. (2012). Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367 1814–1820. 10.1056/NEJMoa1211721 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., et al. (2020b). Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 80:eabd4570. 10.1126/science.abd4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jackson C. B., Mou H., Ojha A., Peng H., Quinlan B. D., et al. (2020a). SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 11:6013. 10.1038/s41467-020-19808-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Liu Z., Moncada-Velez M., Chen J., Ogishi M., Bigio B., et al. (2020c). Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 80:370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Liu F., Xie W., Zhou T. C., OuYang J., Jin L., et al. (2021). Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sens. Actuators B Chem. 327:128899. 10.1016/j.snb.2020.128899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. (2020). Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395 1054–1062. 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X. L., Wang X. G., Hu B., Zhang L., Zhang W., et al. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Hill C. S., Sarkar S., Tse L. V., Woodburn B., Schinazi R. F., et al. (2021). β-d-N4-hydroxycytidine inhibits SARS-CoV-2 through lethal mutagenesis but is also mutagenic to mammalian cells. J. Inf. Dis. 224 415–419. 10.1093/infdis/jiab247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Vedantham P., Lu K., Agudelo J., Carrion R., Nunneley J. W., et al. (2015). Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res. 116 76–84. 10.1016/j.antiviral.2015.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F. C., Guan X. H., Li Y. H., Huang J. Y., Jiang T., Hou L. H., et al. (2020). Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 396 479–488. 10.1016/S0140-6736(20)31605-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Snijder E. L., Gorbalenya A. E. (2000). Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81 853–879. [DOI] [PubMed] [Google Scholar]
- Zipeto D., Palmeira J. F., Argañaraz G. A., Argañaraz E. R. (2020). ACE2/ADAM17/TMPRSS2 interplay may be the main risk factor for COVID-19. Front. Immunol. 11:576745. 10.3389/fimmu.2020.576745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., et al. (2020). SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 382 1177–1179. 10.1056/NEJMc2001737 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.