Abstract
We used PCR assays to determine the etiology of genital ulcers in patients presenting to a sexually transmitted disease clinic in Dakar, Senegal, and evaluated the ability of two PCR tests (groEL and recD) and two serological tests (adsorption enzyme immunoassay [EIA] and lipooligosaccharide [LOS] EIA) to detect current Haemophilus ducreyi infection. We found that in this population, H. ducreyi, T. pallidum, and herpes simplex virus HSV DNA were detected in 56, 15, and 13% of 39 genital ulcer specimens, respectively, and H. ducreyi DNA was detected in 60% (3 of 5) of samples from ulcerated bubos. Among 40 consecutive patients with genital ulcer disease and with sufficient sample for both PCR assays, the recD and groEL H. ducreyi PCR assays were 83% concordant, with the recD PCR assay detecting six (15%) additional positive specimens and the groEL assay detecting one (3%) additional positive specimen. Compared to PCR, the adsorption EIA and LOS EIA tests had sensitivities of 71 and 59% and specificities of 57 and 90%, respectively, for the diagnosis of current H. ducreyi infection. While these differences in specificity could be due either to previous infection with H. ducreyi or to the detection of cross-reacting antibodies, only 6% of patients from a nearby family planning clinic gave a positive reaction in both the adsorption EIA and LOS EIA assays, indicating that cross-reacting antibodies are not prevalent among clinic attendees in this city. Our studies indicate that the adsorption EIA detects both current and past infection, while the LOS EIA assay is more specific for current infection with H. ducreyi in this population.
Haemophilus ducreyi is a common cause of genital ulcer disease (GUD) in Morocco, Kenya, and Lesotho, where it has been shown to cause 49 to 62% of GUD in sexually transmitted disease (STD) clinic populations (29). In other developing countries, the prevalence of this pathogen in patients with GUD is much lower, ranging from 3% in Peru, to 11% in Tanzania, to 23% in the Dominican Republic, to 26% in the Central African Republic (29). Chancroidal disease is uncommon in the United States but is recognized as a significant cause of GUD in several outbreaks in inner cities of the United States, where it is typically associated with individuals who exchange sex for drugs or money and/or use crack cocaine (13, 18).
Besides H. ducreyi, other pathogens that cause GUD are Treponema pallidum, herpes simplex virus (HSV), and Chlamydia trachomatis, which cause syphilis, herpes, and lymphogranuloma venereum, respectively. Because clinical diagnosis is an unreliable means of distinguishing between these GUD etiologies (7, 11, 13, 14), laboratory tests are needed to ensure correct diagnosis and treatment. Early recognition and treatment is particularly important given the association of GUD, particularly chancroid, with the heterosexual transmission of human immunodeficiency virus (HIV) (23, 27, 30). The traditional strategy for managing GUD has relied on individual patient diagnosis and treatment. However, in resource-poor settings, syndromic management, in which treatment is selected based on the most common etiology of a clinical syndrome in a particular locale, can be utilized effectively (16, 26).
The use of PCR has revolutionized the laboratory diagnosis of chancroid because alternative methods of diagnosis such as Gram stain and culture are inaccurate and insensitive, respectively (4). Advantages of PCR include the ability to transport samples off-site for testing, as well as its increased sensitivity relative to culture, presumably due to the difficulty in growing this fastidious organism (18–20). The reliability of the PCR-based tests for H. ducreyi is demonstrated by the high concordance (>95%) of different PCR tests performed on the same samples (18, 20). Disadvantages include the high cost and the time between collection of the specimen and laboratory result, particularly if the PCR assay is performed off-site.
Several PCR-based tests for H. ducreyi have been developed, including those directed against genes for groEL (22) and an H. ducreyi-specific PCR assay that amplifies an undefined 1.1-kb target sequence (17). In addition to these assays, Orle et al. (20) have developed a multiplex PCR (M-PCR) assay which simultaneously detects H. ducreyi, T. pallidum, and HSV in a single specimen. Unfortunately, this M-PCR assay is not commercially available, although detailed procedures for the performance of this assay have now been described (21).
Several serological tests have been developed for the detection of antibodies to H. ducreyi, including the lipooligosaccharide (LOS) enzyme immunoassay (EIA), which uses purified H. ducreyi LOS, and the adsorption EIA, which uses whole-cell H. ducreyi antigens plus adsorption of the serum with different Haemophilus whole-cell extracts (1, 12). Because these EIA assays presumably use different target antigens, they should detect different H. ducreyi-specific antibodies in patient sera. In addition, these serologic assays might differ in their ability to detect previous infection with H. ducreyi or may detect antibodies that result from exposure to cross-reacting antigens.
The current study was performed in Senegal to determine the etiology of GUD in this country and to determine the sensitivity and specificity of serological tests relative to PCR for the detection of chancroidal disease. We tested genital ulcer specimens by PCR and found that chancroid was the most common cause of GUD in Dakar, Senegal. We first compared the sensitivity of two different H. ducreyi PCR assays to detect this organism in ulcers and bubos and then used these PCR results as a reference standard for comparing the ability of the adsorption EIA and the LOS EIA to identify those individuals with current H. ducreyi infection. In addition, serological test results from patients attending an STD clinic were compared to those of a low-risk family planning clinic population. The results of these studies will help us to evaluate the utility of using serological tests both for the diagnosis of current H. ducreyi infection and for screening populations for the prevalence of past or present infection with this organism.
MATERIALS AND METHODS
Strains and plasmids.
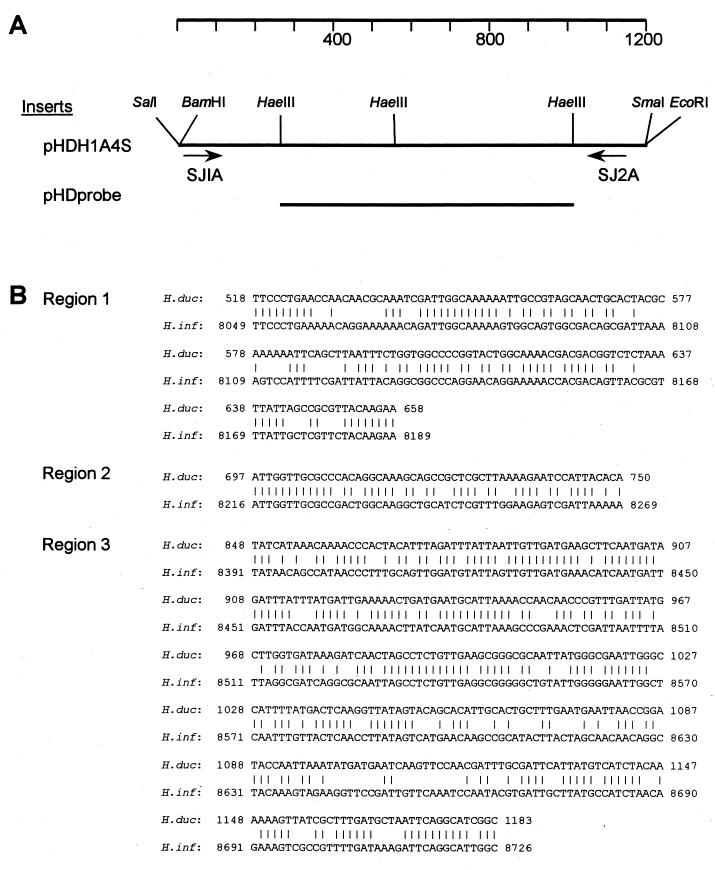
H. ducreyi CIP542 (type strain) was obtained from the stock culture collection at the Centers for Disease Control (CDC). Strains 35000 and R018 are H. ducreyi isolates from Winnipeg and Kenya, respectively, and are maintained at St. Boniface Hospital, Winnipeg, Manitoba, Canada. The recombinant plasmid pHD1A, which contains insert DNA derived from strain CIP542 by cloning, was originally selected by its homology to H. ducreyi but not to other organisms in the development of an H. ducreyi-specific PCR assay (17). This plasmid, along with subclones of this plasmid, pHDH1A4S and pHDprobe, were generously supplied by Steve Johnson (CDC, Atlanta, Ga.). Plasmid pHDH1A4S, a subclone of pHD1A, contains sequences homologous to primers SJ1A and SJ2A and the 1.1-kb sequence between these two primers, as shown in Fig. 1. Plasmid pHDprobe is an HaeIII subclone of pHDH1A4S containing a 707-kb insert internal to the SJ1A and SJ2A primer sequences (Fig. 1). All plasmids were obtained through subcloning whole-cell DNA from H. ducreyi CIP542 and not by PCR amplification to avoid errors which might be introduced into the sequence by the latter technique.
FIG. 1.
Characterization of insert in pHDH1A4S, the target sequence for the H. ducreyi recD PCR showing homology to H. influenzae. (A) Map of the insert of pHDH1A4S, showing the locations of HaeIII sites and the target sequence for primers SJ1A and SJ2A. The location in the insert of the pHD probe used to detect the H. ducreyi specific PCR product is also shown. (B) Nucleotide comparison of sequences in the H. ducreyi amplicon and H. influenzae genomic sequences (GenBank accession number HIU32811). Numbers to the left reflect nucleotides numbered from the start of SJ1A (H. ducreyi) and as published for the H. influenzae genome (15). Vertical lines indicate homology in nucleotides from H. ducreyi and H. influenzae. The sequence of the H. ducreyi amplicon has been deposited in GenBank under the accession number AF090193.
Plasmid isolation, DNA sequencing, and DNA analysis.
To determine the target DNA amplified by the H. ducreyi PCR described by Johnson et al. (17), termed recD PCR in the current paper, DNA was sequenced from the insert in plasmid pHDH1A4S and pHDprobe (Fig. 1). Plasmids used for sequencing were purified by using Wizard Minipreps DNA Purification System (Promega, Madison, Wis.). DNA sequencing was performed by using the DNA dye terminator cycle sequencing kit from Perkin-Elmer (Foster City, Calif.) according to the manufacturer's directions. The resulting template was purified from unincorporated primers by using CentriSep spin columns as described by the manufacturer (Princeton Separations, Adelphia, N.J.) and then subjected to automated sequencing at the Fred Hutchinson Biotechnology Center (Seattle, Wash.). Sequences from both DNA strands of pHDH1A4S were analyzed, and the products of different sequencing reactions were compared by using Genepro (Bainbridge Island, Wash.). DNA and amino acid homology searches were performed on the sequence of the full-length PCR product by using BLAST (3).
Patient population and sample collection.
Consecutive patients with GUD reporting to the STD clinic at the Public Health Institute in Dakar, Senegal, from May to September 1992 were included in this study. The patients were a subset of a larger study of 880 patients tested for genital human papillomavirus and STD infection among adults presenting to the STD clinic between February 1990 to June 1994. Informed consent was obtained, and the study was approved by the Human Subjects Committee at the University of Washington and the Minister of Public Health of Senegal. After patients were enrolled in the study, a genital ulcer specimen was obtained for analysis by PCR and 10 ml of blood was obtained by venipuncture for the serological assays. The patient's age, sex, and duration of symptoms were noted on a standardized questionnaire. Five patients with ulcerated inguinal bubos were also sampled as described below.
Genital ulcer and ulcerated bubos were cleaned with gauze that had been soaked with sterile saline and then a sterile swab was applied to the base of the ulcer or ulcerated bubo to obtain the sample. Swabs were then placed in 1 ml of Specimen Transport Medium (Digene Diagnostics, Inc., Silver Spring, Md.) and frozen at −20°C until analysis. At the same visit, venous blood was drawn. Serum was removed after centrifugation and frozen at −20°C until serological testing could be performed.
Sera from age-matched patients reporting to a nearby family planning clinic served as controls. These patients were expected to have a low risk of chancroid infection compared to the STD clinic population.
H. ducreyi PCR.
Two separate PCR assays for H. ducreyi were performed: the groEL PCR (22) and a PCR which amplifies an unidentified 1.1-kb fragment specific for H. ducreyi (17), both with modifications described in detail elsewhere (29) and summarized below. The designation of the latter PCR assay (17), previously designated S-PCR (18) due to its use of Southern blotting, is confusing since both the groEL PCR and S-PCR assays in the present study used Southern blots. Thus, in the present study we will designate the assay described by Johnson et al. (17) as the recD PCR, based on the homology of the amplified product to the recD gene (see below).
For the groEL PCR assay, samples were analyzed in two different laboratories. For the 36 samples tested in one laboratory (The Wadsworth Center, Albany, N.Y.), 300 μl of the patient sample was treated with 20 μg of proteinase K per ml for 2 h at 55°C and heated for 10 min at 95°C. DNA from this sample was then purified by adsorption to glass milk (Geneclean II Kit; Bio 101, Inc., La Jolla, Calif.) and suspended in 40 μl of sterile water. Next, 20 μl of this sample was added to each reaction mixture, and the PCR amplified products were analyzed by Southern blotting as described previously (22). Ten samples were amplified by groEL PCR in a second laboratory (University of Washington, Seattle). In these samples, prior to PCR, 200 μl of the sample was treated with proteinase K, ethanol precipitated, and suspended in 50 μl of Tris-EDTA buffer. Two PCR reactions were performed, one with 2 μl and the other with 5 μl of patient sample. The H. ducreyi amplicons from these reactions were detected by placing 10 μl of the PCR reaction product onto a nylon filter and then hybridizing to a biotin-labeled probe which was subsequently detected by using strepavidin and a chemiluminescent substrate (Amersham/Pharmacia, Piscataway, N.J.). In this sample set, all patients that were positive with a 5-μl sample were also positive with a 2-μl volume sample.
For the recD PCR, 200 μl of patient specimen was treated with proteinase K, ethanol precipitated, and then suspended in 100 μl of distilled H2O as described earlier (29). Two PCRs were performed on each treated patient sample, one with 2 μl and one with 10 μl of the treated patient specimen. These two concentrations of patient sample were selected to achieve the optimal amount of patient sample that was not inhibitory to the PCR reaction. The primers and thermocycler settings used in this PCR were performed as described by using primers SJ1A and SJ2A (17). Then, 20 μl of each PCR-amplified specimen was analyzed on agarose gels, followed by Southern blotting and hybridization to a DNA probe internal to the PCR product, plasmid pHDprobe. The 707-bp insert in this plasmid was amplified by PCR, purified, radiolabeled with 32P, and then used to probe Southern blots of PCR-amplified patient specimens as described previously (29). All samples that were positive with a 10-μl sample were also positive with a 2-μl volume sample. All samples that were positive were analyzed again with a second sample preparation, amplification, and detection. Discrepant samples were processed a third time and were considered positive if they were positive in two of the three PCR assays.
HSV and T. pallidum PCR.
Samples (2 and 5 μl) of each patient specimen treated with proteinase K and ethanol precipitation for the recD PCR were used for the HSV and T. pallidum PCR assays. For HSV DNA detection, primers HSVgB2a-1 and HSVgB2a-2 were used to amplify a 342-bp fragment of the glycoprotein B2a gene of both HSV-1 and HSV-2 as described elsewhere (9). Specific HSV PCR products were detected by dot blot hybridization as described above for the H. ducreyi samples but with the biotinylated oligonucleotide probe HSVgB2a-P (9).
For T. pallidum DNA detection, primers 47-1 and 47-2 were used to amplify a 658-bp fragment of the gene encoding the 47-kDa membrane immunogen (6). In these assays, the T. pallidum PCR products were detected by dot blot hybridization by using a biotinylated probe consisting of a 496-bp fragment internal to the amplified DNA (6).
β-Globin PCR.
Amplification of human β-globin gene, with detection of a 256-kb PCR product on a 7% acrylamide gel, was used to analyze the adequacy of the sample and to detect inhibitors in the PCR assay as described earlier (5) with slight modifications (29).
H. ducreyi serology.
The H. ducreyi adsorption EIA with whole-cell preparations from H. ducreyi 35000 and sorbents of H. influenzae, H. parainfluenzae, and H. parahemolyticus was performed as previously described (12) with minor modifications (8). The LOS EIA was performed as described earlier (1) using two separate assays, one using LOS purified from strain 35000 the other using LOS purified from strain R018.
RESULTS
Sequence of H. ducreyi PCR amplicon.
The sequence amplified by the H. ducreyi PCR described by Johnson et al. (17) was determined from plasmids pHDH1A4S and pHDprobe and was found to be homologous to the H. influenzae recD gene (Fig. 1B). This homology was most striking in three areas: region 1, positions 518 to 658 (61% identical); region 2, positions 697 to 750 (66% identical); and region 3, positions 848 to 1183 (66% identical). The amino acid homology of the putative RecD protein of H. ducreyi was 47% identical and 61% similar to H. influenzae RecD and 41% identical and 56% similar to Escherichia coli RecD. We thus renamed this H. ducreyi-specific PCR the recD PCR to distinguish it from the groEL PCR, which has a different target sequence.
While we easily identified the target sequence of SJ1A on plasmid pHDH1A4S, the target sequence of SJ2A was different from the published sequence (17) and was determined to be due to sequencing errors in the original study (Steve Johnson, personal communication). Thus, the first three bases of the target sequence are TTT rather than GCC, and there is an additional A in the middle of the published SJ2A primer sequence. The correct primer sequence should read TTT AGC CAG TG CGC CGA TGC C rather than GCC AGC CAG TGA CGC CGA TGC C. (The differences between the target sequence and the published sequence of primer SJ2A are underlined.) The sequence of SJ1A was correct as published.
Patient population.
There were 46 patients with GUD analyzed in this study: 41 men and 5 women. The average age of the patients was 28 years, with a range of 21 to 53 years. The mean duration of the ulcer was 34.5 days, with a range of 2 to 174 days.
H. ducreyi PCR.
Of the 46 genital ulcer samples analyzed by H. ducreyi PCR, 2 were negative for the β-globin PCR product and 4 had insufficient sample for analysis by both tests. Among the 40 specimens analyzed by both the recD and groEL PCR assays, 14 (35%) were positive in both PCR tests and 19 (48%) were negative in both PCR tests. Of the samples that gave divergent results, six (15%) were positive in the recD PCR assay only and one (3%) was positive by the groEL PCR assay only. Thus, the two PCR-based tests were 83% concordant. Although samples were tested using the groEL PCR assay in two laboratories, the agreement between samples tested with the recD PCR assay versus those tested with the groEL PCR assay was the same for both laboratories. In the recD PCR assay, testing samples in duplicate with two separate sample preparations revealed three more positive samples that were subsequently confirmed by a third sample prep and PCR assay. Of these three additional positive specimens which were discovered by duplicate analysis of specimens, two were negative by the groEL PCR assay and one was positive.
Bubo specimens were obtained from five patients with genital ulcers and were analyzed by both H. ducreyi PCR assays when the amount of patient specimen was sufficient for analysis (Table 1). In these tests, H. ducreyi DNA was detected in both the genital ulcer and bubo in two patients, in the bubo but not the genital ulcer in one patient, and in the ulcer but not the bubo in one patient. No H. ducreyi DNA could be detected in either the bubo or ulcer specimens from the one remaining patient. For further analysis, the etiology of the genital ulcer was considered to be H. ducreyi if either the bubo or genital ulcer was positive by either H. ducreyi PCR assay.
TABLE 1.
Detection of H. ducreyi DNA by PCR from bubos and ulcers compared to serologic tests from five patients with GUD
| Patient | PCR results
|
Serology results
|
||||
|---|---|---|---|---|---|---|
| Bubo
|
Ulcer
|
LOS | Adsorption EIA | |||
| groEL | recD | groEL | recD | |||
| 1a | − | − | + | − | − | + |
| 2 | + | + | − | + | + | + |
| 3 | + | + | − | NS | + | + |
| 4 | − | NSb | − | NS | − | + |
| 5 | + | NS | + | NS | + | + |
This was the only sample among the 42 tested by both PCR assays that was positive by the groEL PCR and negative by the recD PCR assay.
NS, specimen was not sufficient.
Prevalence of genital ulcer pathogens among consecutive patients in Senegal.
Sufficient sample was available to test for the presence of HSV and T. pallidum by PCR in 39 of the genital ulcer specimens (Table 2). T. pallidum was detected in six (15%) specimens, either as a sole pathogen and as a copathogen with H. ducreyi and HSV. In contrast, H. ducreyi was detected in 22 (56%) specimens, as the sole pathogen in 19 (49%) specimens and as a coinfection with T. pallidum or HSV in 3 (8%) specimens. HSV was detected in five (13%) specimens, as the sole agent in three (8%) specimens, and together with T. pallidum or H. ducreyi in two (5%) specimens. No etiologic agent could be detected in 10 (26%) specimens. H. ducreyi was the most common pathogen detected (76%; 22 of 29) in genital ulcer specimens with a defined etiology.
TABLE 2.
Etiology of genital ulcers in 39 consecutive patients tested for H. ducreyi, T. pallidum, and HSV by PCRa
| Agent | No. (%) |
|---|---|
| H. ducreyi onlyb | 19 (49) |
| T. pallidum only | 3 (8) |
| HSV only | 3 (8) |
| H. ducreyi plus T. pallidum | 2 (5) |
| H. ducreyi plus HSV | 1 (3) |
| HSV plus T. pallidum | 1 (2) |
| PCR negative | 10 (26) |
| Total | 39 |
Of 46 patients tested by H. ducreyi PCR, 7 did not have samples available for PCR testing by T. pallidum and HSV.
Samples were considered positive for H. ducreyi if they were positive with either H. ducreyi PCR test in either the ulcer or the bubo sample.
Comparison of serological tests for H. ducreyi.
Serum specimens from 40 patients were analyzed by the two EIA tests to determine the antibody response to H. ducreyi. Initially, LOS from two strains, 35000 and R018, were tested for their reaction with patient sera. Seven serum specimens were positive in the R018 LOS EIA assays. These seven sera and an additional six serum specimens were positive with the 35000 LOS EIA. Thus, the results from the 35000 LOS assay were used in all subsequent analyses.
The adsorption EIA and the LOS EIA gave concordant results in 27 (68%) of 40 serum specimens studied; 11 sera (28%) were positive and 16 sera (40%) were negative in both EIA tests (Table 3). However, divergent results were obtained with 13 serum (33%) specimens; 2 serum specimens (5%) were positive in the LOS EIA and negative by the adsorption EIA assay, and 11 (28%) were positive in the adsorption-EIA and negative in the LOS EIA.
TABLE 3.
Comparison of the two H. ducreyi serology tests among the STD clinic patients and age-matched controls from the family planning clinic
| Adsorption EIA | LOS EIA, no. (%) of patients in each category
|
|||||
|---|---|---|---|---|---|---|
| Total casesa (n = 40)
|
Age-matched patients
|
|||||
| Casesb (n = 31)
|
Controlsc (n = 31)
|
|||||
| + | − | + | − | + | − | |
| + | 11 (28) | 11 (28) | 8 (25) | 9 (29) | 0 (0) | 2 (6) |
| − | 2 (6) | 16 (40) | 2 (6) | 12 (39) | 2 (6) | 27 (87) |
Consecutive cases with GUD reporting to the STD clinic.
Cases that could be age matched with controls.
Patients reporting to the family planning clinic.
To determine the prevalence of H. ducreyi antibodies among the STD clinic population compared to a population at low risk for H. ducreyi infection, serology tests were performed on serum obtained from patients attending a family planning clinic, and these were compared to a subset of the STD clinic patients that could be age matched (Table 3). In this analysis, the family planning clinic population had a much lower prevalence of antibodies reactive to H. ducreyi (6% in each assay) than the age-matched STD clinic population (32% [10 of 31] with the LOS assay; 55% [17 of 31] with the adsorption EIA assay).
Comparison of H. ducreyi serology and PCR.
To determine the sensitivity and specificity of the serologic tests for current infection with H. ducreyi, the results of the two serology tests were compared to those of the PCR from 38 patient samples analyzed with both serology and PCR tests (Table 4). In this analysis, patients were considered positive for current H. ducreyi infection if either of the PCR assays were positive by using either ulcer or bubo specimens. The sensitivity and specificity of the LOS EIA compared to PCR were 59% (10 of 17) and 90% (19 of 21); the sensitivity and specificity of the adsorption EIA were 71% (12 of 17) and 57% (12 of 21), respectively.
TABLE 4.
Comparison of PCR and serology in 38 consecutive patients with genital ulcers in Senegala
| Results of PCRb | Results of serologic tests, no. (%) of patients in each category
|
|||
|---|---|---|---|---|
| LOS EIA
|
Adsorption EIA
|
|||
| + | − | + | − | |
| + | 10c (26) | 7d (18) | 12e (32) | 5f (13) |
| − | 2 (5) | 19 (50) | 9 (24) | 12 (32) |
Of the 46 patients analyzed in this study, 2 had GUD specimens that were not amplifiable and 6 did not have serum specimens available for serology testing.
“PCR positive” was defined as positive in either of the PCR tests in either the bubo or the genital ulcer.
Of the 10 patients that were PCR and serology positive, 8 were positive in both PCR tests and 2 were positive in the recD PCR test only.
Of the seven patients that were PCR positive and serology negative, two were positive in both the recD and groEL PCR tests; one was positive in the groEL PCR test and negative in the recD PCR test; and four were positive in the recD PCR test and negative in the groEL PCR test.
Of the 12 patients that were PCR positive, 8 were positive in both PCR tests, 3 were positive in the recD PCR test and negative in groEL PCR test, and 1 was positive in the groEL PCR test and negative in the recD PCR test.
Of these five patients, three were positive for the recD PCR test and negative for the groEL PCR test and two were positive for the groEL PCR test and negative for the recD PCR test.
Patient analysis.
Various factors were analyzed to determine the association with a positive EIA test in the patient population analyzed. There was no association with the duration of ulcer and positivity, either in the adsorption EIA or the LOS EIA (with either the 35000 or the R018 antigens). The median for duration of ulcer for patients with ulcerated bubos was the same as that for patients without bubos (14 days). Of the five patients with ulcerated bubos, four were men and one was a woman.
DISCUSSION
In this study, we determined the etiology of genital ulcers in Dakar, Senegal, and defined the sensitivity and specificity of two serological tests for the detection of current infection with H. ducreyi by using detection of H. ducreyi in ulcer samples by either of two PCR assays as a “gold standard.” Among the GUD specimens from 39 patients analyzed by PCR assays, H. ducreyi, T. pallidum, and HSV were detected in 22 (56%), 6 (15%), and 5 (13%) patients, respectively. No pathogen was identified in 10 (26%) of these specimens, indicating a lack of sensitivity of the PCR assays, inadequacy of these samples for PCR, or alternative etiologies for GUD in these patients. Of the specimens from which a pathogen could be identified, H. ducreyi was the most common, being detected in 22 of 29 (76%) of the genital ulcer specimens.
We found that the two PCR assays, recD and groEL, were 83% concordant among the 40 patients analyzed by both methods. The agreement of these two PCR assays with each other is consistent with other studies in which the recD PCR assay compared well with the Roche Multiplex PCR test for genital ulcer pathogens, with greater than 95% concordance for H. ducreyi (18). Discordant specimens in our present study (six recD PCR-positive, groEL PCR-negative samples; one recD PCR-negative, groEL PCR-positive sample) could be due to differences in sample preparation, specificity of the primers used, sensitivity of detection by probes, possible variation within the sample, and duplicate sample processing for the recD PCR assay. Without the latter analysis, the recD and groEL PCR assays were more concordant (four recD PCR-positive, groEL PCR-negative samples; one recD PCR-negative, groEL PCR-positive sample). While we cannot rule out contamination as the cause of more positive samples in the recD PCR assay, this is unlikely since the samples were positive in two separate sample preparations and PCR amplifications.
The target sequence of one PCR assay for H. ducreyi (17) used here was sequenced and found to have homology with the recD gene of H. influenzae. Given its essential nature for housekeeping functions in bacteria, this gene might be expected to vary little between strains of the same organism, thus supporting its suitability for detection of all strains of H. ducreyi. We sequenced the original clone obtained from this organism and found that the target sequence for one of the primers, SJ2A, was different from the published sequence (17). However, in spite of these differences, this assay is very sensitive and specific for H. ducreyi DNA, both from isolated organisms and in patient specimens (17, 18, 28, 29).
Serological tests for H. ducreyi have been used to determine the prevalence of chancroid in several settings, including trucking company workers in Kenya (27%); female sex workers in Lagos, Nigeria (86%); and STD clinic patients in Jackson, Miss. (48 and 53% with two different EIA assays) (8, 10, 24). The availability of PCR testing for the detection of current infection with H. ducreyi provided an opportunity to reevaluate the sensitivity and specificity of these assays in Africa, in a way similar to an analysis in the United States (8).
In the current study, we chose the LOS EIA and the adsorption-EIA tests to detect antibodies to H. ducreyi in STD clinic patients because they presumably use two different target antigens (1, 12). We found that the sensitivity and specificity of these assays, compared to current infection with H. ducreyi detected by PCR, were 59 and 90% for the LOS assay and 71 and 57% for the adsorption EIA assay, respectively. These results are similar to those obtained in Mississippi (8), in which the LOS assay gave a slightly lower sensitivity (48 versus 53%) but a greater specificity (89 versus 71%) compared to the adsorption EIA assay based on current infection with H. ducreyi detected by PCR. As with a previous study (8), serologic assays for detection of current infection with H. ducreyi are insensitive compared to PCR. This low sensitivity could be due to a delayed immune response to H. ducreyi or to the use of antigens in the assay, which may differ between strains. Indeed, in our study, the assay with LOS from strain 35000 was more sensitive than a similar assay with LOS from strain R018. Similar results would be expected with adsorption EIA, since strain differences have also been observed in this assay (12, 25).
The low specificity of serologic tests relative to PCR could be due to several factors, including past infection with H. ducreyi, false-negative H. ducreyi PCR tests, or false-positive EIA tests. Because antibodies to H. ducreyi persist for many months postinfection (1, 2), antibodies to a previous infection with H. ducreyi may have been present in the PCR-negative, serology-positive patients in our high-risk STD population. Additionally, because 26% of patients in this study had no pathogen detected, some of these patients might have had a false-negative H. ducreyi PCR test. Finally, antibodies that cross-react with antigens from other organisms might account for some of the EIA-positive, PCR-negative results. However, the low background reactivity of the serological tests in the family planning clinic patients (6% by both assays) is consistent with the interpretation that antibodies to H. ducreyi are not prevalent in the general population. Furthermore, patients attending STD clinics are also more likely to have had a past infection with other STD pathogens and perhaps other commensal organisms that might cross react in these assays, making interpretation difficult.
We conclude that, in our patient population, H. ducreyi caused the majority of GUD cases in Dakar, Senegal. In addition, our study indicates that serologic tests need further evaluation to determine their suitability to detect chancroid in a given community. The sensitivity and specificity of serologic assays may depend upon the strain of H. ducreyi used for detection and the patient exposure to cross-reacting antigens that might differ in different populations. Perhaps a more sensitive and specific serological test for H. ducreyi infection could be based on individual cellular components that are unique and conserved in this organism.
ACKNOWLEDGMENTS
We thank Alfred Waring and Dora Norn for technical assistance, Carol Langley for specimen collection, and Steve Hawes for data extraction.
This work was supported by National Institutes of Health grant CA50856 to N.B.K. and by a contract from the Division of AIDS, Sexually Transmitted Diseases, and Tuberculosis Laboratory Research, CDC, to P.A.T.
REFERENCES
- 1.Alfa M J, Olson N, Degagne P, Plummer F, Namaara W, MacLean I, Ronald A R. Humoral immune response of humans to lipooligosaccharide and outer membrane proteins of Haemophilus ducreyi. J Infect Dis. 1993;167:1206–1210. doi: 10.1093/infdis/167.5.1206. [DOI] [PubMed] [Google Scholar]
- 2.Alfa M J, Olson N, Degagne P, Slaney L, Plummer F, Namaara W, Ronald A R. Use of an adsorption enzyme immunoassay to evaluate the Haemophilus ducreyi specific and cross-reactive humoral immune response of humans. Sex Transm Dis. 1992;19:309–314. [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Ballard R, Morse S A. Chancroid. In: Morse S A, Moreland A A, Holmes K K, editors. Atlas of sexually transmitted diseases and AIDS. 2nd ed. Baltimore, Md: Mosby-Wolfe; 1996. pp. 47–63. [Google Scholar]
- 5.Bauer H M, Ting Y, Greer C E, Chambers J C, Tashiro C J, Chimera J, Reingold A, Manos M M. Genital human papillomavirus infection in female university students as determined by a PCR-based method. JAMA. 1991;265:472–477. [PubMed] [Google Scholar]
- 6.Burstain J M, Grimpel E, Lukehart S A, Norgard M V, Radolf J D. Sensitive detection of Treponema pallidum by using the polymerase chain reaction. J Clin Microbiol. 1991;29:62–69. doi: 10.1128/jcm.29.1.62-69.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapel T A, Brown W J, Jeffries C, Stewart J A. How reliable is the morphological diagnosis of penile ulcerations? Sex Transm Dis. 1977;4:150–152. doi: 10.1097/00007435-197710000-00008. [DOI] [PubMed] [Google Scholar]
- 8.Chen C, Mertz K J, Spinola S M, Morse S A. Comparison of enzyme immunoassays for antibodies to Haemophilus ducreyi in a community outbreak of chancroid in the United States. J Infect Dis. 1997;175:1390–1395. doi: 10.1086/516471. [DOI] [PubMed] [Google Scholar]
- 9.Cone R W, Hobson A C, Palmer J, Remington M, Corey L. Extended duration of herpes simplex virus DNA in genital lesion detected by the polymerase chain reaction. J Infect Dis. 1991;164:757–760. doi: 10.1093/infdis/164.4.757. [DOI] [PubMed] [Google Scholar]
- 10.Dada A J, Ajayi A O, Diamondstone L, Quinn T C, Blattner W A, Biggar R J. A serosurvey of Haemophilus ducreyi, syphilis, and herpes simplex virus type 2 and their association with human immunodeficiency virus among female sex workers in Lagos, Nigeria. Sex Transm Dis. 1998;25:237–242. doi: 10.1097/00007435-199805000-00004. [DOI] [PubMed] [Google Scholar]
- 11.Dangor Y, Ballard R C, da L. Exposto F, Fehler G, Miller S D, Koornhof H J. Accuracy of clinical diagnosis of genital ulcer disease. Sex Transm Dis. 1990;17:184–189. doi: 10.1097/00007435-199010000-00007. [DOI] [PubMed] [Google Scholar]
- 12.Desjardins M, Thompson C E, Filion L G, Ndinya-Achola J O, Plummer F A, Ronald A R, Piot P, Cameron D W. Standardization of an enzyme immunoassay for human antibody to Haemophilus ducreyi. J Clin Microbiol. 1992;30:2019–2024. doi: 10.1128/jcm.30.8.2019-2024.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiCarlo R P, Martin D H. The clinical diagnosis of genital ulcer disease in men. Clin Infect Dis. 1997;25:292–298. doi: 10.1086/514548. [DOI] [PubMed] [Google Scholar]
- 14.Fast M V, D'Costa L J, Nsanze H, Piot P, Curran J, Karasira P, Mirza M, Maclean I W, Ronald A R. The clinical diagnosis of genital ulcer disease in men in the tropics. Sex Transm Dis. 1984;11:72–76. doi: 10.1097/00007435-198404000-00004. [DOI] [PubMed] [Google Scholar]
- 15.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 16.Htun Y, Morse S A, Dangor Y, Fehler G, Radebe F, Trees D L, Beck-Sague C M, Ballard R C. Comparison of clinically directed, disease specific, and syndromic protocols for the management of genital ulcer disease in Lesotho. Sex Transm Infect. 1998;74(Suppl. 1):S23–S28. [PubMed] [Google Scholar]
- 17.Johnson S R, Martin D H, Cammarata V, Morse S A. Development of a polymerase chain reaction assay for the detection of Haemophilus ducreyi. Sex Transm Dis. 1994;21:13–23. doi: 10.1097/00007435-199401000-00004. [DOI] [PubMed] [Google Scholar]
- 18.Mertz K J, Weiss J B, Webb R M, Levine W C, Lewis J S, Orle K A, Totten P A, Overbaugh J, Morse S A, Currier M M, Fishbein M, St. Louis M E. An investigation of genital ulcers in Jackson, Mississippi: high prevalence of chancroid and human immunodeficiency virus infection. J Infect Dis. 1998;178:1060–1066. doi: 10.1086/515664. [DOI] [PubMed] [Google Scholar]
- 19.Morse S A, Trees D L, Htun Y, Radebe F, Orle K A, Danger Y, Beck-Sague C M, Schmid S, Fehler G, Weiss J B, Ballard R C. Comparison of clinical diagnosis, standard laboratory and molecular methods for the diagnosis of genital ulcer disease in Lesotho: association with HIV infection. J Infect Dis. 1997;175:583–589. doi: 10.1093/infdis/175.3.583. [DOI] [PubMed] [Google Scholar]
- 20.Orle K A, Gates C A, Martin D H, Body B A, Weiss J B. Simultaneous PCR detection of Haemophilus ducreyi, Treponema pallidum, and herpes simplex virus types 1 and 2 from genital ulcers. J Clin Microbiol. 1996;34:49–54. doi: 10.1128/jcm.34.1.49-54.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orle K A, Weiss J B. Detection of Treponema pallidum, Haemophilus ducreyi, and herpes simplex virus by multiplex PCR. In: Peeling R W, Sparling P F, editors. Methods in molecular medicine. 20. Sexually transmitted diseases: methods and protocols. Totowa, N.J: Humana Press; 1999. pp. 67–79. [DOI] [PubMed] [Google Scholar]
- 22.Parsons L M, Waring A L, Otido J, Shayegani M. Laboratory diagnosis of chancroid using species-specific primers from Haemophilus ducreyi groEL and the polymerase chain reaction. Diagn Microbiol Infect Dis. 1995;23:89–98. doi: 10.1016/0732-8893(95)00172-7. [DOI] [PubMed] [Google Scholar]
- 23.Plummer F A, Simonsen J N, Cameron D W, Ndinya-Achola J O, Kreiss J K, Gakinya M N, Waiyaki P, Cheang M, Piot P, Ronald A R, Ngugi E N. Cofactors in male-female sexual transmission of human immunodeficiency virus type 1. J Infect Dis. 1991;163:233–239. doi: 10.1093/infdis/163.2.233. [DOI] [PubMed] [Google Scholar]
- 24.Rakwar J, Jackson D, Maclean I, Obongo T, Bwayo J, Smith H, Mandaliya K, Moses S, Ndinya-Achola J, Kreiss J K. Antibody to Haemophilus ducreyi among trucking company workers in Kenya. Sex Transm Dis. 1997;24:267–271. doi: 10.1097/00007435-199705000-00006. [DOI] [PubMed] [Google Scholar]
- 25.Roggen E L, Hoofd G, Van Dyck E, Piot P. Enzyme immunoassays (EIAs) for the detection of anti-Haemophilus ducreyi serum IgA, IgG, and IgM antibodies. Sex Transm Dis. 1994;21:36–42. doi: 10.1097/00007435-199401000-00008. [DOI] [PubMed] [Google Scholar]
- 26.Ryan C A, Holmes K K. Editorial: how should clinical algorithms be used for syndromic management of cervical and vaginal infections? Clin Infect Dis. 1995;21:1456–1458. doi: 10.1093/clinids/21.6.1456. [DOI] [PubMed] [Google Scholar]
- 27.Telzak E E, Chaisson M A, Bevier P J, Stoneburner R L, Castro K G, Jaffe H W. HIV-1 seroconversion in patients with and without genital ulcer disease. Ann Intern Med. 1993;119:1181–1186. doi: 10.7326/0003-4819-119-12-199312150-00005. [DOI] [PubMed] [Google Scholar]
- 28.Totten P A, Stamm W E, Morse S A. How to detect Haemophilus ducreyi in genital ulcers and why you should care. Clinical Microbiol Newsl. 1996;18:145–152. [Google Scholar]
- 29.Totten P A, Kuypers J M, Morse S A. Haemophilus ducreyi: detection by PCR. In: Peeling R, Sparling P F, editors. Sexually transmitted diseases: methods and protocols. Vol. 20. Totowa, N.J: Humana Press; 1999. pp. 47–65. [DOI] [PubMed] [Google Scholar]
- 30.Wasserheit J N. Epidemiological synergy. Interrelationships between human immunodeficiency virus infection and other sexually transmitted diseases. Sex Transm Dis. 1992;19:61–77. [PubMed] [Google Scholar]