Abstract
The aims of this study were to investigate the prevalence of campylobacteria including Campylobacter jejuni subsp. jejuni (C. jejuni) and Campylobacter coli in human clinical samples and in samples from healthy individuals and to reevaluate the efficacies of conventional selective methods for isolation of Campylobacter spp. Two charcoal-based selective media, modified charcoal cefoperazone deoxycholate agar (mCCDA) and cefoperazone-amphotericin-teicoplanin (CAT) agar, were compared with Skirrow's blood-based medium and with a filter method (filter) applied to a yeast-enriched blood agar. A total of 1,376 specimens were tested on all four media, and the percentages of thermophilic Campylobacter-positive specimens isolated on Skirrow's medium, filters, CAT agar, and mCCDA were 82, 83, 85, and 95%, respectively. When additional samples were processed with the three selective media, mCCDA recovered significantly more thermophilic Campylobacter spp. than Skirrow's medium (P = 0.0034). No significant difference between Skirrow's medium and CAT agar was observed in this study. Another six taxa were identified, namely, Campylobacter concisus, Campylobacter curvus-like bacteria, Arcobacter butzleri, Arcobacter cryaerophilus, Helicobacter cinaedi, and Sutterella wadsworthensis. Most of these strains were isolated after 5 to 6 days of incubation by use of the filter technique. This paper provides evidence for the existence of S. wadsworthensis in human feces from clinical cases of gastrointestinal disorders and in feces from a healthy individual. Furthermore, C. concisus was isolated from a large number of diarrheal cases, particularly those at the extremes of age, but was additionally isolated from the feces of healthy people. Further investigations to establish the role of C. concisus and S. wadsworthensis in enteric disease is needed. We conclude that a range of campylobacteria may cause infections in Denmark.
The term “campylobacteria” may be used to refer to a range of fastidious, mainly spiral or curved-rod-shaped bacteria that includes members of the phylogenetically related genera Campylobacter, Arcobacter, and Helicobacter, among others (26, 37). In recent years, a number of species and/or subspecies have either been added to, or emended within, this group of organisms, and many taxa are associated with human disease, especially gastroenteritis (17, 26, 28). Within the genus Campylobacter, C. jejuni and C. coli are the species most frequently isolated from diarrheal illness in humans, and C. jejuni is considered the most common cause of sporadic bacterial enteritis worldwide (31, 34).
The true incidence of Campylobacter infections and the species distribution in human diseases are not known. When the diagnosis of infection is based exclusively upon culturing on selective media, it is found that approximately 85 to 95 and 5 to 10% of Campylobacter infections are caused by C. jejuni and C. coli, respectively, in developed countries (9, 24; F. J. Bolton, D. N. Hutchinson, and G. Parker, Letter, J. Clin. Pathol. 40:702–703, 1987). However, it has been suggested that other campylobacterial taxa, such as Campylobacter upsaliensis, C. jejuni subsp. doylei, Campylobacter fetus subsp. fetus, Campylobacter concisus, Arcobacter butzleri, Arcobacter cryaerophilus, Helicobacter fennelliae, and Helicobacter cinaedi, may be significantly underdiagnosed as causes of gastrointestinal disorders as a consequence of inappropriate isolation and identification methods (1–3, 7–9, 12, 18–20, 23, 26, 29, 38).
In general, campylobacteria other than C. jejuni, C. coli, and Campylobacter lari are too sensitive to the antibiotics in most conventional selective media to be isolated in routine laboratories. Some strains of C. jejuni and C. coli are also inhibited by antimicrobial agents, such as cephalothin, colistin, and polymyxin B, that may be present in selective media (9). In addition, species such as C. concisus, Campylobacter sputorum, Campylobacter curvus, Campylobacter rectus, and some strains of Campylobacter hyointestinalis also need incubation in a hydrogen-enriched microaerobic atmosphere to enable recovery (24). Moreover, accurate identification of these organisms is known to be problematic (26). Consequently, most clinical laboratories do not routinely identify campylobacteria to the species level, leaving the true prevalence of these taxa uncertain (26).
Numerous selective media for the isolation of campylobacters have been described, almost all containing several antibiotics as inhibitory agents (9, 11). A different approach, involving the passage of motile campylobacteria through a membrane filter onto a nonselective growth agar medium, is recognized as allowing the isolation of campylobacters sensitive to antibiotics incorporated into the selective media (1, 18, 32). The major drawback of this technique is the labor-intensive character of the method (2), the lower sensitivity of the medium compared to conventional selective media (12), and overgrowth of plates by competing fecal contaminants such as swarming Proteus spp. and Enterococcus spp. (2, 11, 20). As a consequence, the use of a selective agar and the filter method in combination is recommended to optimize recovery of campylobacteria from fecal samples (7).
The aim of the present study was to achieve an impression of the diversity of campylobacteria in Denmark and to reevaluate the comparative efficacies of conventional culturing methods for the isolation of Campylobacter spp. in order to optimize the recovery of Campylobacter spp.
(This paper was presented in part at the 9th International Workshop on Campylobacter, Helicobacter and Related Organisms, Cape Town, South Africa, 15 to 19 September 1997, abstr. A4, p. 51.)
MATERIALS AND METHODS
Selective agars.
The modified charcoal cefoperazone deoxycholate agar (mCCDA) (D. N. Hutchinson and P. J. Bolton, Letter, J. Clin. Pathol. 37:956–957, 1984) comprised a commercially supplied charcoal base (Oxoid Ltd., Basingstoke, United Kingdom) and cefoperazone (32 mg/liter) (Sigma, St. Louis, Mo.). The cefoperazone-amphotericin-teicoplanin (CAT) medium (2) comprised the same charcoal base and contained cefoperazone (8 mg/liter), amphotericin B (10 mg/liter; Fluka, Buchs, Switzerland), and teicoplanin (4 mg/liter; Astra Denmark, Albertslund, Denmark). The Skirrow's medium (30) comprised Campylobacter agar base (SSI Diagnostica, Hillerød, Denmark), 5% horse blood, vancomycin (10 mg/liter; Sigma), trimethoprim (5 mg/liter; GEA, Frederiksberg, Denmark), and polymyxin B (2,500 IU/liter; Kirsch Pharma, Roskilde, Denmark).
Filtration technique.
The technique used was similar to that described by Steele and McDermott (32). Sterile cellulose acetate membrane filters, each with a diameter of 47 mm and a 0.65-μm pore size (Sartorius AG, Goettingen, Germany) were placed on the surface of a (0.93% [wt/vol]) yeast-enriched 5% blood agar plate (SSI Diagnostica), and 8 drops of the fecal suspension were placed on the top of a membrane and allowed to filter passively for 45 min at 37°C under ambient atmosphere. After filtration, the filters were carefully removed with sterile forceps and discarded and the culture plates were incubated.
Incubation.
Inoculated plates were incubated at 37°C in a hydrogen-enriched microaerobic atmosphere (6% O2, 6% CO2, 3% H2, and 85% N2) and examined after 2 days. To assess the effect on the recovery of isolates after a prolonged incubation period, the first 2,201 samples were reincubated and the plates were reexamined after a total of 5 to 6 days.
Isolation studies.
Two studies were conducted consecutively. (i) Study A. A comparison of the efficacies of the three selective media and the filtration technique was performed with the first 1,376 of the 3,267 fecal samples. The study continued with the three selective media and the first 2,201 samples and concluded with only mCCDA and Skirrow's medium and another 1,066 samples.
(ii) Study B.
A total of 107 fecal samples from healthy individuals (no current or recent [less than 4 weeks] gastrointestinal symptoms) and 107 controls (clinical stool specimens submitted to the laboratory for culturing of enteric pathogens, as in study A) were processed by the filter method. The healthy population and controls were matched for sex and age (0 to 1 years old, +/−1 year; 2 to 4 years old, +/−2 years; >5 years old, +/−5 years).
Fecal specimens. (i) Study A.
A total of 3,267 stool specimens submitted to the laboratory for culturing of enteric pathogens were tested. The samples originated from both in- and outpatients and were sent by mail under ambient conditions without a transport medium. The majority of samples were tested within 24 h of collection. On arrival in the laboratory the samples were kept at ambient temperature until culture, which was usually performed within 4 to 5 h of receipt. To ensure that culture techniques were tested with a standardized inoculum, fecal samples were emulsified (approximately 1 g/ml) in sterile saline.
(ii) Study B.
One hundred and seven samples from healthy individuals and 107 controls (clinical samples) were tested for unusual Campylobacter spp. and related organisms. In addition, all samples were processed for C. jejuni and C. coli (with Skirrow's medium), Salmonella spp., Yersinia enterocolitica, Shigella spp., Plesiomonas shigelloides, Aeromonas spp., and Vibrio spp. to exclude causative bacterial agents among diarrheal cases and carrier states of enteric pathogens in the healthy population.
Strain identification.
The following methods were used, either separately or in combination, as required. (i) Conventional phenotypic tests. These tests were applied to all isolates as part of a standard identification scheme (24), principally for differentiation of thermophilic Campylobacter spp. The tests used were tests for motility, catalase, oxidase, nitrate reduction, hydrogen sulfide production in triple sugar iron medium, hydrolysis of hippurate and indoxyl acetate, sensitivity to nalidixic acid and cephalothin, and growth in anaerobic and aerobic conditions.
(ii) C. concisus species-specific PCR. All isolates preliminarily identified as C. concisus by using conventional cultural and biochemical characteristics were confirmed by a species-specific PCR test described previously for presumptive detection of C. concisus (6). Template DNA was extracted from a 24-h subculture on a yeast-enriched 5% blood agar plate by picking colony material with a 1-μl inoculating loop and mixing it with 300 μl of a 20% slurry of Chelex-100 (Bio-Rad, Hercules, Calif.) in Tris-EDTA buffer (10 mM Tris [pH 8], 1 mM EDTA) and heating the mixture at 95°C for 10 min. The resin was pelleted by centrifugation at 10,000 rpm (Biofuge 13; Heraens Sepatech GmbH, Hanau, Germany) for 2 min. The following primers published by Bastyns et al. (6) were used for the PCR assay: MUC1 (forward), 5′-ATGAGTAGCGATAATTGGG-3′, and the combination of CON1 and CON2 (two reverse primers), 5′-CAGTATCGGCAATTCGCT-3′ (CON1) and 5′-GACAGTATCAAGGATTTACG-3′ (CON2). The forward primer was labelled with the FAM dye (DNA Technology, Aarhus, Denmark) during synthesis at the 5′ end to permit subsequent fragment analysis of the PCR products. One microliter of the supernatant, 9 μl of milliQ water, and a 15-μl dilution of a 25 M concentration of each primer in milliQ water from a master mixture were added to a Ready-To-Go PCR analysis bead (Pharmacia Biotech, Freiburg, Germany) containing premixed, predispensed AmpliTaq DNA polymerase, as well as all necessary buffer ingredients and nucleotides. The mixture was centrifuged for 10 s and subjected to PCR with a hot-start procedure in a PTC-200 Peltier thermal cycler (MJ Research Inc., Watertown, Mass.). The thermal cycling parameters consisted of an initial 4 min of denaturation at 94°C followed by 20 cycles each of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min. The reaction product was subjected for fragment analysis on an ABI PRISM 310 DNA genetic Analyzer (Perkin-Elmer Denmark, Allerød, Denmark) in accordance with the manufacturer's recommendations.
(iii) Extended phenotypic characterization.
Strains found not to resemble C. jejuni, C. coli, or C. concisus by the above methods were subjected to extensive phenotypic analysis by using 65 tests included in a previously described identification scheme (27).
(iv) Whole-cell protein profiling.
Whole-cell protein extracts of selected strains were prepared, electrophoresed in 12.5% polyacrylamide gels, and visualized by staining in Coomassie blue dye, as described previously (4). Field strains were identified by visual comparison of profiles with those derived from appropriate type strains of campylobacterial taxa.
(v) 16S rDNA sequence analysis.
Primers and methods used for DNA extraction, PCR amplification, and direct sequencing of 16S rRNA genes were as described previously (28), except that primer 1492r (5′-TAC GGY TAC CTT GTT ACG ACT T) was used in place of 1392r for the initial PCR amplification and subsequent sequencing of the PCR product. Sequences were obtained (both strands) over ∼1,450 bp (∼95%) of the 16S rRNA gene and were submitted for identification to the ribosomal database project (RDP) by using the “sequence match” tool on the World Wide Web server for the second release of the RDP (21). Sequence similarities were calculated after downloading the closest matching sequences from GenBank. Strains were considered to be identified when sequence similarities of field strains with Genbank entries exceeded 99%, provided no other GenBank sequences (representing different species) were greater than 98% related to the field strain.
Statistical analysis.
The comparative efficacies of plates for the recovery of Campylobacter spp. were tested by McNemar's test (P < 0.01).
RESULTS
Study A.
A total of 1,376 specimens were tested on all four media, and various campylobacterial species were recovered from 144 such samples. C. jejuni and C. coli accounted for 78 of the 144 strains isolated (Tables 1 and 2). The percentages of C. jejuni- or C. coli-positive specimens isolated by using Skirrow's medium, filter methods, CAT agar, and mCCDA medium were 82, 83, 85, and 95%, respectively. When a total of 2,201 fecal samples were processed, no significant difference in recovery of C. jejuni and C. coli between CAT and mCCDA was seen. However, since the results tended to favor mCCDA, the survey continued, for economical reasons, only with mCCDA against the hitherto-used medium in the routine laboratory, Skirrow's medium. When a total of 3,267 stool samples were cultured on Skirrow's medium and mCCDA, the differences between the two media were even more pronounced, with the latter recovering significantly more C. jejuni and C. coli after 2 days of incubation (12%; P = 0.0034). With a prolonged incubation period (5 to 6 days), another 7% of C. jejuni and C. coli could be recovered with mCCDA compared to 4% with Skirrow's medium.
TABLE 1.
Isolation of Campylobacter jejuni and C. coli from human stool specimens by using Skirrow's, CAT, or mCCDA media or the filter technique after 2 and 5 to 6 days of incubation
| Incubation time (days) | Na | No. (%) of specimens positive by using:
|
Sample size | |||
|---|---|---|---|---|---|---|
| Skirrow's medium | CAT agar | mCCDA medium | Filter technique | |||
| 2 | 76 | 62 (82) | 62 (82) | 68 (89) | 65 (86) | 1,376 |
| 5–6 | 78 | 64 (82) | 66 (85) | 74 (95) | 65 (83) | 1,376 |
| 2 | 138 | 109 (79) | 119 (86) | 124 (90) | 2,201 | |
| 5–6 | 143 | 114 (80) | 125 (87) | 134 (94) | 2,201 | |
| 2 | 173 | 138 (80) | 157 (91) | 3,267 | ||
N, number of thermophilic Campylobacter isolates isolated on any media. Numbers of thermophilic Campylobacter isolates (total: 177 isolates): C. jejuni, 167; C. coli, 6; not identified (isolates dead), 4.
TABLE 2.
Isolation of Campylobacter spp. other than C. jejuni and C. coli and related organisms from 1,376 human stool specimens by using Skirrow's, CAT, and mCCDA media and the filter technique
| Taxon | Na | No. of strains isolated by using:
|
Applied identification methods | |||
|---|---|---|---|---|---|---|
| Skirrow's medium | CAT agar | mCCDA medium | Filter technique | |||
| C. concisus | 45 | 8 | 0 | 1 | 43 | Phenotypic, 23S rDNA, SDS-PAGE, 16S rDNA |
| C. curvus-like | 3 | 0 | 0 | 0 | 3 | SDS-PAGE, 16S rDNA |
| A. butzleri | 1 | 0 | 1 | 1 | 1 | SDS-PAGE, 16S rDNA |
| A. cryaerophilus | 1 | 0 | 0 | 1 | 0 | Phenotypic, SDS-PAGE, 16S rDNA |
| H. cinaedi | 1 | 1 | 0 | 0 | 0 | SDS-PAGE, 16S rDNA |
| S. wadsworthensis | 6 | 0 | 0 | 0 | 6 | SDS-PAGE, 16S rDNA |
N, number of strains isolated on any media excluding duplicates.
For the isolation of campylobacteria other than C. jejuni and C. coli, the efficacies of the four media differed. Another six different campylobacterial taxa, including the newly described Sutterella wadsworthensis, were only isolated by the filter method (Table 2). The six S. wadsworthensis strains were isolated from six patients (median age, 20 years; range, 2 to 79 years) and exclusively after using the filter method of isolation with an incubation period of 5 to 6 days. One of the patients was coinfected with Y. enterocolitica. Three C. curvus-like strains were isolated by the filter method. They formed a discrete group by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) protein analysis, and the strains were identified as C. curvus by means of 16S rDNA gene sequencing. However, subsequent whole-cell protein analysis showed that these strains resembled, but were not identical to, the type strain of C. curvus. Furthermore, one strain of A. butzleri was recovered from the same fecal sample by the filter method and with CAT agar and mCCDA, while one A. cryaerophilus strain was obtained only on mCCDA. A single H. cinaedi strain was found only with Skirrow's medium.
A total of 52 isolates of C. concisus originating from 39 clinical cases of gastrointestinal disorders were identified. The majority of isolates (83%) were recovered by the filter method and mostly after 5 to 6 days of incubation (Table 3). All isolates were definitively identified by a combination of biochemical tests and 23S rDNA PCR. This process was necessary since, although a 308-bp fragment of the gene was amplified from all C. concisus isolates (data not shown), we consistently obtained a 308-bp fragment from type strains of Campylobacter showae and Wolinella succinogenes in preliminary setup experiments.
TABLE 3.
Isolation of C. concisus from human stool specimens by using Skirrow's, CAT, or mCCDA medium or the filter technique after 2 and 5 to 6 days of incubation
| Incubation time (days) | Na | No. (%) of isolates found by using:
|
Sample size | |||
|---|---|---|---|---|---|---|
| Skirrow's medium | CAT agar | mCCDA medium | Filter technique | |||
| 2 | 12 | 1 (8) | 0 (0) | 0 (0) | 11 (92) | 1,376 |
| 5–6 | 52 | 8 (15) | 0 (0) | 1 (2) | 43 (83) | 1,376 |
N, number of C. concisus isolates on any media.
In the original test description (6) no cross-reaction with any of the other taxa examined was noted. However, the report had no information on whether C. showae and W. succinogenes were included in the test panel for specificity. The identities of our C. concisus strains were confirmed by testing for oxidase production, nitrate reduction, and motility. In addition, a selection of the C. concisus strains was identified by whole-cell protein profiling (data not shown), and further, three of the strains were identified by means of 16S rRNA gene sequencing. Five of the C. concisus-positive clinical cases were coinfected with an established bacterial enteric pathogen (three with Salmonella enterica subsp. enterica serovar Enteritidis, one with Shigella sonnei, and one with Y. enterocolitica).
Study B.
C. concisus was recovered from 3 of 107 healthy individuals and from 5 of 107 clinical controls (statistically insignificant). In addition, S. wadsworthensis was isolated from one of the healthy individuals and from one of the controls. No coinfections with other bacterial enteric pathogens were registered.
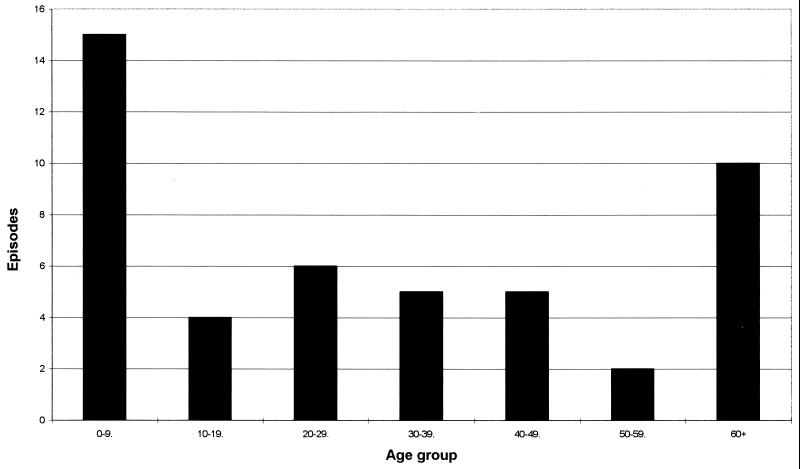
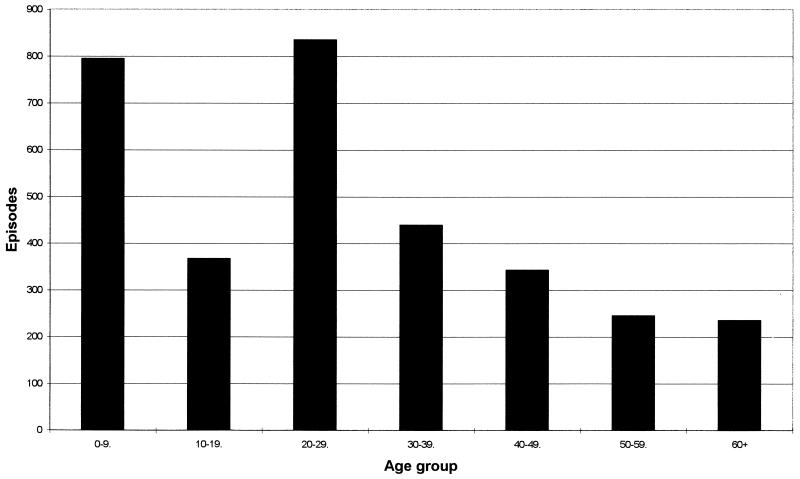
The distribution of C. concisus cases by age (studies A and B combined) is shown in Fig. 1. Ten of the 15 positive children in the age range of 0 to 9 years were less than 2 years old. For comparison, the general distribution of individuals positive for C. jejuni or C. coli by age as determined by our laboratory is shown in Fig. 2.
FIG. 1.
Episodes of C. concisus by age. The analysis was based on episodes from studies A and B.
FIG. 2.
Episodes of C. jejuni and C. coli by age. The analysis was based on 67,808 episodes (133,810 fecal samples) with 3,255 (4.8%) positive for C. jejuni or C. coli in our laboratory from January 1995 to September 1996.
DISCUSSION
In this study, mCCDA was significantly more effective than Skirrow's medium in recovering thermophilic Campylobacter spp. A prolonged incubation period for these two media resulted in only minor increases (7 and 4%, respectively) in recovery rates of thermophilic Campylobacter spp. This procedure, if applied routinely, would entail reading plates twice because clinical reports after a nominal 2-day incubation period are important for therapeutic reasons. Our results suggest that an extended incubation of media for confirmation of C. jejuni or C. coli infection is unnecessary.
C. lari and C. upsaliensis were not recovered in this study, even though a variety of media and methods principally designed for the isolation of these species were applied. In a recent study from Sweden (19), C. upsaliensis was the most common species next to C. jejuni among diarrheal children. Our finding is, however, supported by a recent report comparing CAT agar with mCCDA for the isolation of Campylobacter spp. from 7,000 human clinical samples in the United Kingdom, in which only five C. upsaliensis isolates were recovered (39). In the same study, the CAT agar successfully recovered a large number of C. upsaliensis cells from cat and dog feces, indicating that the infrequent isolation from humans was not a methodological artifact. The suspected principal sources for human C. upsaliensis infections are cats and dogs (8). Hald and Madsen (13) have recently shown that C. upsaliensis was recovered less frequently from healthy Danish puppies and kittens than from cats and dogs in the United Kingdom. We therefore conclude that gross, regional differences in the prevalence of C. upsaliensis may exist.
Several fastidious Campylobacter spp. are associated with the periodontal niche within the oral cavity, including C. concisus, C. gracilis, C. rectus (33), and C. showae (10). The role of these species in human diarrhea is uncertain. In addition, little is known of the role of Campylobacter spp. as components of the microflora of the lower gastrointestinal tracts of healthy humans (17).
We were able to isolate an unexpectedly large number of C. concisus and S. wadsworthensis cells by using the filter technique in combination with a yeast-enriched blood agar medium. An atmosphere enriched with hydrogen (6 to 7%) is generally advocated for the isolation of hydrogen-requiring Campylobacter spp. (11, 24). In our study, with a final hydrogen content of 3%, we were able to isolate a large number of hydrogen-requiring C. concisus strains, suggesting that this hydrogen content, if combined with a very potent nonselective medium, may suffice. Moreover, in contrast to the literature (11) in which C. concisus is reported to need 4 days of incubation for its isolation, 23% of our C. concisus strains were recovered after only 2 days of incubation. When subcultures of C. concisus, with the same medium and atmosphere, were performed, sufficient growth could be achieved within just 24 h of incubation. It is notable that eight and one C. concisus strains were isolated with Skirrow's and mCCDA selective media, respectively, suggesting that at least some strains of the species may be less fastidious than previously considered.
C. concisus is associated primarily with periodontal disease (33) but has also been isolated from patients with bacteremia, foot ulcers, and upper and lower gastrointestinal infections (14, 18, 19, 23, 35), as well as from healthy individuals, mostly children (17, 38).
This species is both genotypically and phenotypically diverse and can taxonomically be regarded as a complex (25). Our results showing C. concisus cells in nearly equivalent proportions in fecal samples from diarrhetic patients and healthy controls support the views expressed by other workers (17, 38) that this species, at present, should be considered a commensal of the human gut rather than a primary pathogen associated with gastrointestinal disease. We observed differences in the patient age distribution in which C. concisus was found, compared with that generally observed for C. jejuni and C. coli in the laboratory (Fig. 1 and 2). This was most noteworthy at both extremes of age. Although the difference in the total numbers of positive samples for the figures must be taken into account, these data could suggest that C. concisus is an important opportunistic pathogen in patients with compromised or immature immune systems.
We believe this to be the first report of the isolation of S. wadsworthensis from human stool samples. This species was first described in 1996 by Wexler et al. (40), who found asaccharolytic, bile-resistant microaerophilic Campylobacter gracilis-like organisms to represent a distinct taxon unrelated to the Campylobacter group. To date, S. wadsworthensis has mostly been isolated from patients with appendicitis, peritonitis, or rectal or perirectal abscesses (22). Molitoris et al. (22) suggested that S. wadsworthensis may be more likely to be involved in serious infections than C. gracilis. Using the filter isolation method, we found 7 of 1,483 (0.47%) clinical samples and 1 of 107 (0.93%) samples from healthy individuals to be positive. However, the small sample size in study B does not allow for any conclusion about the occurrence of S. wadsworthensis in healthy humans.
We recovered several other campylobacterial taxa from human diarrheal samples by using various isolation methods, including C. curvus-like strains (n = 3), A. butzleri (n = 1), A. cryaerophilus (n = 1), and H. cinaedi (n = 1) (Table 2). Each of these species has been described previously from cases of human disease, including gastroenteritis (15, 18, 36). The abilities of the methods used to isolate these taxa from fecal samples varied considerably. The filter method was the only technique that successfully recovered the C. curvus-like strains and also isolated an A. butzleri strain. However, A. butzleri was also isolated on both CAT and mCCDA media, while the single A. cryaerophilus strain detected was found only on mCCDA. Skirrow's medium did not detect any of these Campylobacter or Arcobacter spp. but was the only method which detected H. cinaedi.
These results emphasize the difficulty in screening human diarrhea samples for the presence of campylobacterial species other than C. jejuni, C. coli, and C. lari, since at present no single method will successfully isolate all taxa. It is nonetheless clear from this and other studies (18, 19, 23) that the application of comprehensive isolation and identification strategies offers a fundamentally important insight into the occurrence of different campylobacteria in diarreal samples and their possible role in human disease. Since many species are known to occur in food animals (including A. butzleri and A. cryaerophilus) (4, 5) and domestic pets (including H. cinaedi) (16), such results may ultimately prove of considerable value to public health protection. Although the pathogenic potential of C. concisus, S. wadsworthensis, and other campylobacteria requires clarification, it is clear that further studies regarding the prevalence, detection, and identification of such taxa are wholly justifiable.
REFERENCES
- 1.Allos B M, Lastovica A J, Blaser M J. Atypical campylobacters and related organisms. In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press; 1995. pp. 849–865. [Google Scholar]
- 2.Aspinall S T, Wareing D R, Hayward P G, Hutchinson D N. Selective medium for thermophilic campylobacters including Campylobacter upsaliensis. J Clin Pathol. 1993;46:829–831. doi: 10.1136/jcp.46.9.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aspinall S T, Wareing D R A, Hayward P G, Hutchinson D N. A comparison of a new selective medium (CAT) with membrane filtration for the isolation of thermophilic campylobacters including Campylobacter upsaliensis. J Appl Bacteriol. 1996;80:645–650. doi: 10.1111/j.1365-2672.1996.tb03269.x. [DOI] [PubMed] [Google Scholar]
- 4.Atabay H I, Corry J E L, On S L W. Diversity and prevalence of Arcobacter spp. in broiler chickens. J Appl Microbiol. 1998;84:1007–1016. doi: 10.1046/j.1365-2672.1998.00437.x. [DOI] [PubMed] [Google Scholar]
- 5.Atabay H I, Corry J E L, On S L W. Identification of unusual Campylobacter-like organisms in poultry products as Helicobacter pullorum. J Appl Microbiol. 1998;84:1017–1024. doi: 10.1046/j.1365-2672.1998.00438.x. [DOI] [PubMed] [Google Scholar]
- 6.Bastyns K, Chapelle S, Vandamme P, Goossens H, De Wachter R. Specific detection of Campylobacter concisus by PCR amplification of 23S rDNA areas. Mol Cell Probes. 1995;9:247–250. doi: 10.1016/s0890-8508(95)90114-0. [DOI] [PubMed] [Google Scholar]
- 7.Bolton F J, Hutchinson D N, Parker G. Reassessment of selective agars and filtration techniques for isolation of Campylobacter species from faeces. Eur J Clin Microbiol Infect Dis. 1988;7:155–160. doi: 10.1007/BF01963069. [DOI] [PubMed] [Google Scholar]
- 8.Bourke B, Chan V L, Sherman P. Campylobacter upsaliensis: waiting in the wings. Clin Microbiol Rev. 1998;11:440–449. doi: 10.1128/cmr.11.3.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corry J E, Post D E, Colin P, Laisney M J. Culture media for the isolation of campylobacters. Int J Food Microbiol. 1995;26:43–76. doi: 10.1016/0168-1605(95)00044-k. [DOI] [PubMed] [Google Scholar]
- 10.Etoh Y, Dewhirst F E, Paster B J, Yamamoto A, Goto N. Campylobacter showae sp. nov., isolated from the human oral cavity. Int J Syst Bacteriol. 1993;43:631–639. doi: 10.1099/00207713-43-4-631. [DOI] [PubMed] [Google Scholar]
- 11.Goossens H, Butzler J P. Isolation and identification of Campylobacter spp. In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni: current status and future trends. Washington, D.C.: American Society for Microbiology; 1992. pp. 93–109. [Google Scholar]
- 12.Goossens H, De Boeck M, Colgnau H, Vlaes L, Van den Borre C, Butzler J P. Modified selective medium for isolation of Campylobacter spp. from feces: comparison with Preston medium, a blood-free medium, and a filtration system. J Clin Microbiol. 1986;24:840–843. doi: 10.1128/jcm.24.5.840-843.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hald B, Madsen M. Healthy puppies and kittens as carriers of Campylobacter spp., with special reference to Campylobacter upsaliensis. J Clin Microbiol. 1997;35:3351–3352. doi: 10.1128/jcm.35.12.3351-3352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson C C, Finegold S M. Uncommonly encountered, motile, anaerobic gram-negative bacilli associated with infection. Rev Infect Dis. 1987;9:1150–1162. doi: 10.1093/clinids/9.6.1150. [DOI] [PubMed] [Google Scholar]
- 15.Kiehlbauch J A, Brenner D J, Nicholson M A, Baker C N, Patton C M, Steigerwalt A G, Wachsmuth I K. Campylobacter butzleri sp. nov. isolated from humans and animals with diarrheal illness. J Clin Microbiol. 1991;29:376–385. doi: 10.1128/jcm.29.2.376-385.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiehlbauch J A, Brenner D J, Cameroun D N, Steigerwalt A G, Makowski J M, Baker C N, Patton C M, Wachsmuth I K. Genotypic and phenotypic characterization of Helicobacter cinaedi and Helicobacter fennelliae strains isolated from humans and animals. J Clin Microbiol. 1995;33:2940–2947. doi: 10.1128/jcm.33.11.2940-2947.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawson A J, Linton D, Stanley J. 16S rRNA gene sequences of ‘Candidatus Campylobacter hominis’, a novel uncultivated species, are found in the gastrointestinal tract of healthy humans. Microbiology. 1998;144:2063–2071. doi: 10.1099/00221287-144-8-2063. [DOI] [PubMed] [Google Scholar]
- 18.le Roux E, Lastovica A J. The Cape Town protocol: how to isolate the most campylobacters for your dollar, pound, franc, yen, etc. In: Lastovica A L, Newell D G, Lastovica E E, editors. Campylobacter, Helicobacter and related organisms. Cape Town, South Africa: Institute of Child Health; 1998. pp. 30–33. [Google Scholar]
- 19.Lindblom G B, Sjogren E, Westerberg J H, Kaijser B. Campylobacter upsaliensis, C. sputorum sputorum and C. concisus as common causes of diarrhoea in Swedish children. Scand J Infect Dis. 1995;27:187–188. doi: 10.3109/00365549509019006. [DOI] [PubMed] [Google Scholar]
- 20.Lopez L, Castillo F J, Clavel A, Rubio M C. Use of a selective medium and a membrane filter method for isolation of Campylobacter species from Spanish paediatric patients. Eur J Clin Microbiol Infect Dis. 1998;17:489–492. doi: 10.1007/BF01691131. [DOI] [PubMed] [Google Scholar]
- 21.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molitoris E, Wexler H M, Finegold S M. Sources and antimicrobial susceptibilities of Campylobacter gracilis and Sutterella wadsworthensis. Clin Infect Dis. 1997;25(Suppl. 2):S264–S265. doi: 10.1086/516234. [DOI] [PubMed] [Google Scholar]
- 23.Musmanno R A, Russi M, Figura N, Guglielmetti P, Zanchi A, Signori R, Rossolini A. Unusual species of campylobacters isolated in the Siena Tuscany area, Italy. New Microbiol. 1998;21:15–22. [PubMed] [Google Scholar]
- 24.Nachamkin I. Campylobacter and Arcobacter. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of Clinical Microbiology. Washington, D.C.: ASM Press; 1995. pp. 483–491. [Google Scholar]
- 25.On S L W. Confirmation of human Campylobacter concisus isolates misidentified as Campylobacter mucosalis and suggestions for improved differentiation between the two species. J Clin Microbiol. 1994;32:2305–2306. doi: 10.1128/jcm.32.9.2305-2306.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.On S L W. Identification methods for campylobacters, helicobacters, and related organisms. Clin Microbiol Rev. 1996;9:405–422. doi: 10.1128/cmr.9.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.On S L W, Holmes B, Sackin M J. A probability matrix for the identification of campylobacters, helicobacters, and allied taxa. J Appl Bacteriol. 1996;81:425–432. doi: 10.1111/j.1365-2672.1996.tb03529.x. [DOI] [PubMed] [Google Scholar]
- 28.On S L W, Atabay H I, Corry J E L, Harrington C S, Vandamme P. Emended description of Campylobacter sputorum and revision of its infrasubspecific (biovar) divisions, including C. sputorum bv. paraureolyticus, a urease-producing variant from cattle and humans. Int J Syst Bacteriol. 1998;48:195–206. doi: 10.1099/00207713-48-1-195. [DOI] [PubMed] [Google Scholar]
- 29.Piersimoni C, Bornigia S, Curzi L, de Sio G. Comparison of two selective media and a membrane filter technique for isolation of Campylobacter species from diarrhoeal stools. Eur J Clin Microbiol Infect Dis. 1995;14:539–542. doi: 10.1007/BF02113436. [DOI] [PubMed] [Google Scholar]
- 30.Skirrow M B. Campylobacter enteritis: a “new” disease. Br Med J. 1977;2:9–11. doi: 10.1136/bmj.2.6078.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skirrow M B, Blaser M J. Campylobacter jejuni. In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press; 1995. pp. 825–848. [Google Scholar]
- 32.Steele T W, McDermott S N. The use of membrane filters applied directly to the surface of agar plates for the isolation of Campylobacter jejuni from feces. Pathology. 1984;16:263–265. doi: 10.3109/00313028409068535. [DOI] [PubMed] [Google Scholar]
- 33.Tanner A C R, Badger S, Lay C-H, Listgarten M A, Visconti R A, Socransky S S. Wolinella gen. nov., Wolinella succinogenes (Vibrio succinogenes Wolin et al.) comb. nov., and description of Bacteroides gracilis sp. nov., Wolinella recta sp. nov., Campylobacter concisus sp. nov., and Eikenella corrodens from humans with periodontal disease. Int J Syst Bacteriol. 1981;31:432–445. [Google Scholar]
- 34.Tauxe R V. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations. In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni: current status and future trends. Washington, D.C.: American Society for Microbiology; 1992. pp. 9–19. [Google Scholar]
- 35.Vandamme P, Falsen E, Pot B, Hoste B, Kersters K, De Ley J. Identification of EF group 22 campylobacters from gastroenteritis cases as Campylobacter concisus. J Clin Microbiol. 1989;27:1775–1781. doi: 10.1128/jcm.27.8.1775-1781.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vandamme P, Falsen E, Pot B, Kersters K, De Ley J. Identification of Campylobacter cinaedi isolated from blood and feces of children and adult females. J Clin Microbiol. 1990;28:1016–1020. doi: 10.1128/jcm.28.5.1016-1020.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vandamme P, De Ley J. Proposal for a new family, Campylobacteraceae. Int J Syst Bacteriol. 1991;41:451–455. [Google Scholar]
- 38.Van Etterijck R, Breynaert J, Revets H, Devreker T, Vandenplas Y, Vandamme P, Lauwers S. Isolation of Campylobacter concisus from feces of children with and without diarrhea. J Clin Microbiol. 1996;34:2304–2306. doi: 10.1128/jcm.34.9.2304-2306.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wareing D R A, Aspinall S T, Hayward P G, Hutchinson D N. Improved selective medium (CAT) for thermophilic campylobacters including Campylobacter upsaliensis. In: Lastovica A L, Newell D G, Lastovica E E, editors. Campylobacter, Helicobacter and related organisms. Cape Town, South Africa: Institute of Child Health; 1998. pp. 46–49. [Google Scholar]
- 40.Wexler H M, Reeves D, Summanen P H, Molitoris E, McTeague M, Duncan J, Wilson K H, Finegold S M. Sutterella wadsworthensis gen. nov., sp. nov., bile-resistant microaerophilic Campylobacter gracilis-like clinical isolates. Int J Syst Bacteriol. 1996;46:252–258. doi: 10.1099/00207713-46-1-252. [DOI] [PubMed] [Google Scholar]