Abstract
A total of 217 and 73 strains of Salmonella enterica serotype Typhi isolated from 1985 to 1997 in Hong Kong and in 2 months of 1989 and 1990 in Vietnam, respectively, were studied. These isolates were typed by plasmid profile analysis, plasmid fingerprinting, ribotyping with PstI, and total DNA fingerprinting with NarI. There appeared to be no major outbreak of typhoid fever in Hong Kong during the study period since there was considerable heterogeneity among the isolates. Isolates from Hong Kong were different from those from Vietnam. Thirty-seven percent of Vietnamese isolates belonged to two predominant clones, with the rest being heterogeneous in nature. Total DNA fingerprinting supplemented with ribotyping could be a reliable and rapid method for epidemiological typing of S. enterica serotype Typhi.
Typhoid fever remains an important cause of morbidity and mortality in many developing countries. It is also a problem in countries where the infection is rare but where local people travel to and acquire it in places where typhoid fever is endemic. Although typhoid fever is endemic in Hong Kong (average yearly incidence, 2.7 cases per 100,000 population), there has been no detailed epidemiological investigation of the source and spread of Salmonella enterica serotype Typhi in this locality. There were three studies on the incidence of Salmonella spp. isolated from hospitalized patients in Hong Kong, but these did not deal with the characterization of S. enterica serotype Typhi (7, 15, 18). Besides, there is a lack of rapid, reliable, and sufficiently discriminative methods for routine laboratory investigation of the epidemiology of typhoid fever. Several investigators have evaluated the potential of ribotyping (1, 24, 27), but this is not a rapid method, as it requires at least 4 days to complete. Other workers found pulsed-field gel electrophoresis (PFGE) to be useful in the typing of S. enterica serotype Typhi (9, 12, 14, 26, 30, 32–34). However, PFGE requires expensive equipment that may not be affordable by small laboratories. In the study described here we aimed to develop a simple and rapid molecular method for epidemiological typing of serotype Typhi in a busy laboratory and to use this method to investigate the epidemiology of this organism from Hong Kong and Vietnam.
MATERIALS AND METHODS
Bacterial strains.
Two hundred seventeen strains of S. enterica serotype Typhi isolated from blood or stools of patients in three hospitals in Hong Kong from 1985 to 1997 were studied. Seventy-three consecutive serotype Typhi strains isolated from blood or stools of patients in Cho Quan Hospital, Ho Chi Minh City, Vietnam, during January 1989 and October 1990 were also studied. All isolates were stored on nutrient agar slopes in the respective hospital laboratories before further work was performed in the microbiology laboratory of The Chinese University of Hong Kong. All were single patient isolates and were isolated by standard microbiological techniques (4). The isolates were identified with the API 20E system (bioMérieux SA, Montalieu Vercieu, France) and were serotyped by slide agglutination with specific antisera (Wellcome Research Laboratories, Beckenham, England).
Antimicrobial susceptibility testing.
The MICs of 18 antimicrobial agents (the β-lactams ampicillin, cephalothin, cefuroxime, cefamandole, ceftriaxone, cefoperazone, ceftazidime, aztreonam, and imipenem; the aminoglycosides gentamicin, tobramycin, and amikacin; the quinolones nalidixic acid, ciprofloxacin, and ofloxacin; and the other agents, trimethoprim-sulfamethoxazole, tetracycline, and chloramphenicol) were determined by the agar dilution method as described in our previous study (19).
Transferability of resistance plasmids.
Transfer of resistance by using a standard rifampin-resistant Escherichia coli recipient, JP995 (kindly supplied by M. H. Richmond, Bristol, United Kingdom), was performed by the method of Anderson and Threlfall (2).
Plasmid profile analysis and plasmid fingerprinting.
Plasmids were extracted from the S. enterica serotype Typhi isolates by the method of Kado and Liu (16) and were electrophoresed on 0.7% agarose gels as described previously (20). Plasmids of similar sizes were purified (16), digested with EcoRI or HindIII (Promega, Madison, Wis.) according to the manufacturer's instructions, and electrophoresed on 0.8% agarose gels. Individual plasmids in isolates harboring more than one plasmid were separated on agarose gels electrophorectically and were then purified with the Sephaglas BandPrep kit (Amersham Pharmacia Biotech AB, Uppsala, Sweden).
Extraction of total DNA.
Bacterial cells were pelleted from 10 ml of overnight broth and were then washed with 1 ml of 50 mM Tris-HCl–20 mM EDTA (pH 8). The cell pellet was then resuspended in 555 μl of 50 mM Tris-HCl–10 mM EDTA (pH 8), 12 μl of 10 mg of RNase per ml, 30 μl of 10% sodium dodecyl sulfate, and 3 μl of 20 mg of proteinase K per ml. The mixture was incubated for 1 h at 55°C and was extracted with equal volumes of phenol (once), phenol-chloroform-isoamyl alcohol (25:24:1) (twice), and chloroform-isoamyl alcohol (24:1) (once). The DNA was precipitated with 16 μl of 5 M NaCl and two parts ice-cold absolute alcohol at −70°C overnight, washed with 70% alcohol, and resuspended in 1 ml of distilled water.
Ribotyping.
About 0.3 μg of DNA was digested with PstI, ClaI, or KpnI (Promega) according to the manufacturer's instructions. The restricted fragments were separated by electrophoresis on 0.8% agarose (Sigma Chemical Co., St. Louis, Mo.) in a TBE (Tris-borate-EDTA) buffer system (10) for 42 h at 1 V/cm with a horizontal gel apparatus (GNA200; Amersham Pharmacia Biotech AB). The DNA fragments on the gel were transferred to a nylon membrane (Hybond N+; Amersham) by using an LKB VacuGene system (Pharmacia LKB). This was done by first depurinating the DNA with 0.25 M HCl for 30 min and then neutralizing with 0.4 M NaOH at a pressure of 50 to 55 mm Hg for 5 h. The membrane was washed for 10 min with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (29) and was hybridized as described by Grimont and Grimont (11) with a 16S-23S rRNA probe (Boehringer, Mannheim, Germany) labelled with [γ-32P]ATP (Amersham) by the exchange reaction of a 5′ end labelling kit (Boehringer).
Total DNA fingerprinting.
Total DNA was digested with restriction enzyme NarI, EcoRV, or MluI (Promega) according to the manufacturer's instructions. Restriction fragments were separated on 0.4% agarose gels with GNA200 for 21 to 22 h at 2 V/cm and were then visualized under UV illumination.
Cluster analysis.
Cluster analysis of PstI ribotypes and NarI DNA types was performed by the unweighted pair group method with arithmetic averages (31) with the aid of SPSS, release 9.0, software (SPSS Inc., Chicago, Ill.). Types with 90% similarity were likely to be related and were grouped into a cluster (3).
RESULTS
A total of 290 strains of S. enterica serotype Typhi were studied: 217 isolated from patients in three hospitals in Hong Kong from 1985 to 1997 and 73 isolates from patients in Vietnam in 1989 and 1990.
The susceptibilities of Hong Kong strains of serotype Typhi to different antibiotics have been reported previously (21).
All serotype Typhi strains isolated from Vietnam were susceptible to all the β-lactams, aminoglycosides, and quinolones tested as well as to and trimethoprim-sulfamethoxazole, but for 45% (33 isolates) and 14% (10 isolates) of the isolates, chloramphenicol MICs were 512 mg/liter and tetracycline MICs were ≥128 mg/liter, respectively. All tetracycline-resistant isolates were also resistant to chloramphenicol.
The plasmid profiles of Hong Kong strains of serotype Typhi have also been reported (21).
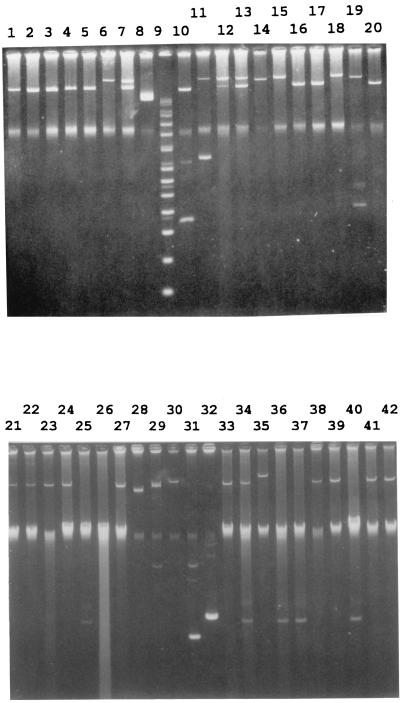
Fifty-two of the 73 (71%) serotype Typhi isolates from Vietnam harbored one to two plasmids ranging in size from 5 to 221 kb (Fig. 1). The 221-kb plasmid, either alone or in combination with a 98- or 5-kb plasmid, was found only in tetracycline- and chloramphenicol-resistant isolates. All chloramphenicol-resistant isolates harbored a 98-kb plasmid, while one had a 5-kb plasmid as well. Among the 40 fully sensitive serotype Typhi strains, 21 did not harbor any plasmid, while 12 harbored a 98-kb plasmid, 6 harbored a 5-kb plasmid, and 1 harbored a 151-kb plasmid.
FIG. 1.
Plasmid profiles of S. enterica serotype Typhi from Vietnam. Lanes 1 to 5, 16, 17, and 20, plasmids from chloramphenicol-resistant isolates; lanes 6, 7, 12 to 15, 18, and 19, plasmids from chloramphenicol- and tetracycline-resistant isolates. Size markers are given in lanes 8 (49 kb), 9 (supercoiled DNA ladder [GIBCO BRL, Gaithersburg, Md.]), 10 (91 and 4.4 kb), and 11 (221 and 9 kb). Lanes 21 to 26 and 33 to 42, plasmids from sensitive isolates. Size markers are given in lanes 27 (91 kb), 28 (58 kb), 29 (8.8 kb), 30 (119 kb), 31 (4.4 kb), and 32 (5.4 kb).
Thirty-three resistant isolates from Vietnam were tested for the transferability of their resistances. Nine of 10 tetracycline- and chloramphenicol-resistant isolates could transfer their resistances to the E. coli recipient at frequencies of 1.3 × 10−5 to 5.4 × 10−4 at 28°C and 8.0 × 10−10 to 9.2 × 10−8 at 37°C. Twelve of the 23 chloramphenicol-resistant isolates could transfer their resistances at frequencies of 4.1 × 10−10 to 5.8 × 10−7 at 28°C and 1.3 × 10−9 to 5.0 × 10−7 at 37°C. Chloramphenicol resistance was borne on plasmids of 98 kb, while tetracycline and chloramphenicol resistance was borne on plasmids of 221 kb. Plasmids that were not transferable were also not mobilizable.
Figure 2 shows the EcoRI restriction digest of 221- and 98-kb plasmids, and Fig. 3 shows the HindIII restriction digest of these plasmids. Eight of the nine transferable 221-kb plasmids had identical or similar EcoRI and HindIII restriction patterns (E2 or E2a and H2 or H2a, respectively), while one nontransferable 221-kb plasmid had different EcoRI and HindIII restriction patterns (E5 and H5, respectively) (Table 1). Thirty-four of the 37 (92%) 98-kb plasmids had the same EcoRI and HindIII restriction patterns (E1 and H1, respectively), irrespective of their other characteristics (Table 1), while 3 had two different restriction patterns.
FIG. 2.
EcoRI digests of 221- and 98-kb plasmids of S. enterica serotype Typhi from Vietnam. Lanes 1 to 5, transferable 221-kb plasmid from chloramphenicol- and tetracycline-resistant isolates; lanes 7 to 12, nontransferable 98-kb plasmids from chloramphenicol-resistant isolates; lanes 14 to 20, transferable 98-kb plasmids from chloramphenicol-resistant isolates; lanes 22 to 28, 98-kb plasmids from sensitive isolates; lanes 6, 13, and 21, HindIII-digested bacteriophage λ DNA as a size marker.
FIG. 3.
HindIII digests of 221- and 98-kb plasmids of S. enterica serotype Typhi from Vietnam. The contents of the lanes are described in the legend to Fig. 2.
TABLE 1.
Characterization of S. enterica serotype Typhi strains from Vietnam
| Resistance pattern | Plasmid size (kb) | Plasmid fingerprint
|
Molecular type
|
Total
|
|||
|---|---|---|---|---|---|---|---|
| EcoRI | HindIII | Group | Ribotype and DNA type | No. of isolates | % of isolates | ||
| Chloramphenicol | 98 | E1 | H1 | 27 | P58N9022 | 11 | 15 |
| E1 | H1 | 1 | P904N9022 | 5 | 7 | ||
| E1 | H1 | 4, 37, 39, 176 | P904N9025, P58N9023, P58N9025, P85N9023 | 5 | 7 | ||
| E3 | H3 | 48 | P58N115 | 2 | 3 | ||
| Subtotal | 23 | 32 | |||||
| Tetracycline and chloramphenicol | 221 | E2 | H2 | 29 | P58N9026 | 2 | 7 |
| E2 | H2 | 66 | P33N108 | 3 | 4 | ||
| 221, 5 | E2 | H2 | 25, 46 | P904N101, P58N101 | 2 | 3 | |
| 221, 98 | E2, E1 | H2, H1 | 29 | P58N9026 | 1 | 1 | |
| E2a, E1 | H2a, H1 | 29 | P58N9026 | 1 | 1 | ||
| 221 | E5 | H5 | 68 | P33N114 | 1 | 1 | |
| Subtotal | 10 | 14 | |||||
| Sensitive | 5 | 6 groups | 6 | 8 | |||
| 98 | E1 | H1 | 27 | P58N9022 | 5 | 7 | |
| E1 | H1 | 1 | P904N9022 | 4 | 5 | ||
| E1 | H1 | 32, 77 | P58N907, P903N9022 | 2 | 3 | ||
| E4 | H4 | 4 | P904N9025 | 1 | 1 | ||
| 151 | —a | — | 67 | P33N110 | 1 | 1 | |
| Nil | 18 groups | 21 | 29 | ||||
| Subtotal | 40 | 55 | |||||
| Total (35 groups) | 73 | 100 | |||||
—, not done.
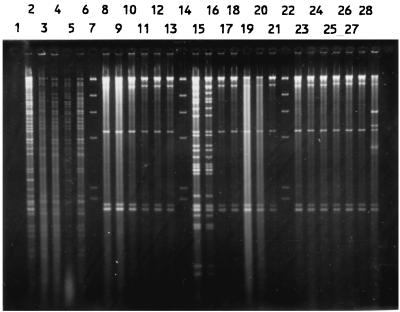
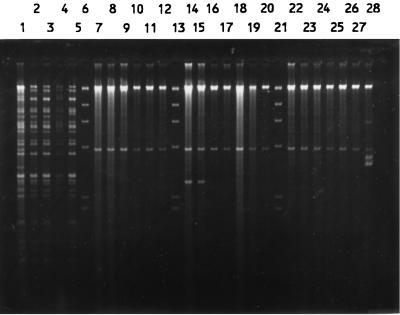
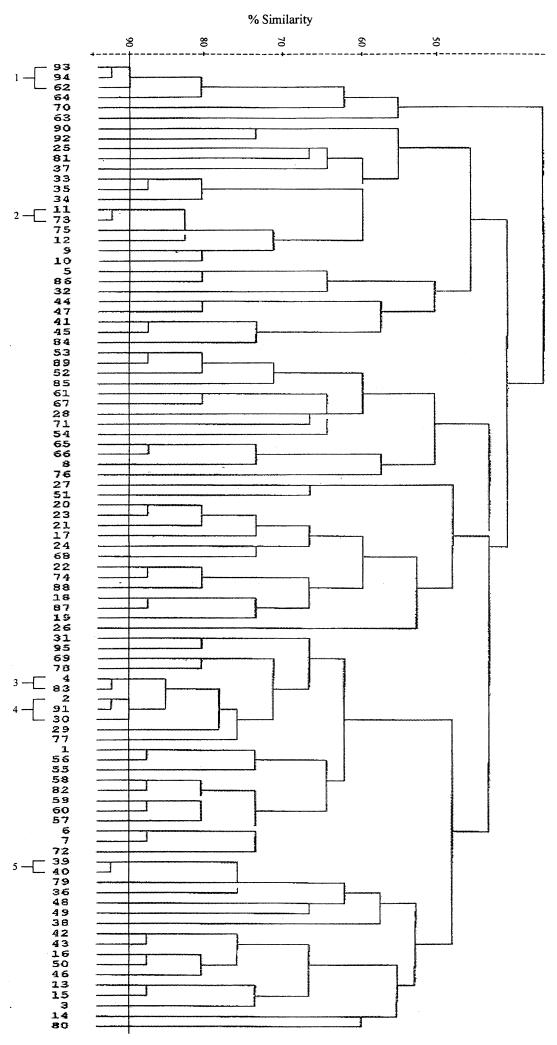
Total DNAs were digested with a restriction enzyme, PstI, ClaI, or KpnI, and the fragments were hybridized with a radiolabelled rRNA probe. Both PstI and ClaI digestions produced comparable patterns, while KpnI digestion resulted in very few fragments (data not shown). Thus, only the results of PstI digestion were used for analysis. The isolates were allocated to a total of 95 patterns or ribotypes, each with five to eight fragments ranging in size from 6.7 to >23.1 kb. Some of the ribotypes are shown in Fig. 4. The relatedness of the patterns is illustrated as a dendrogram in Fig. 5. Twelve ribotypes could be grouped into five clusters at 90% similarity, and the rest were of <90% similarity. Each ribotype was assigned a number following the letter P, while those with ≥90% similarity were numbered following the prefix P90. The ribotypes that contained the most number of isolates were P904 (21%) and P58 (19%). Three to 6% of isolates belonged to P902, P66, and P33, and the rest (46%) could be discriminated into 84 types.
FIG. 4.
PstI ribotypes of S. enterica serotype Typhi (lanes 1 to 14 and 16 to 26) and HindIII-digested bacteriophage λ DNA as a negative control (lane 15).
FIG. 5.
Dendrogram showing relatedness of PstI ribotypes.
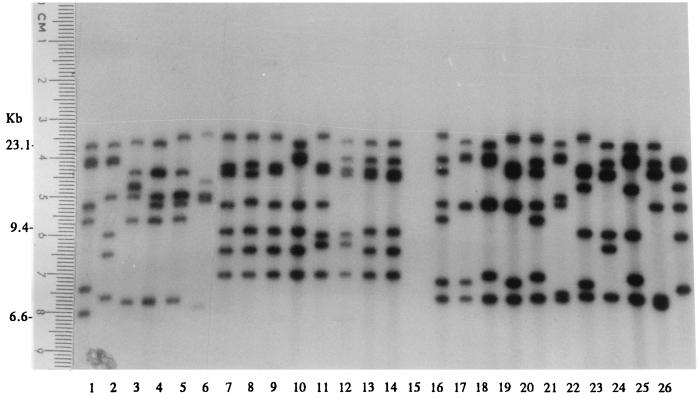
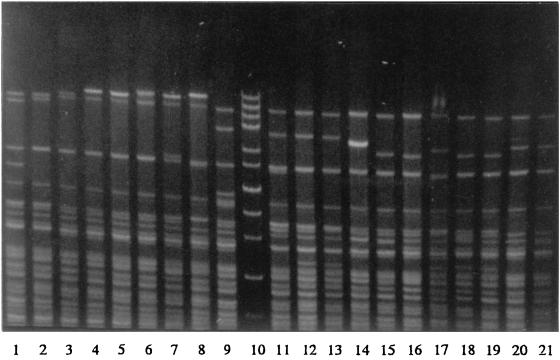
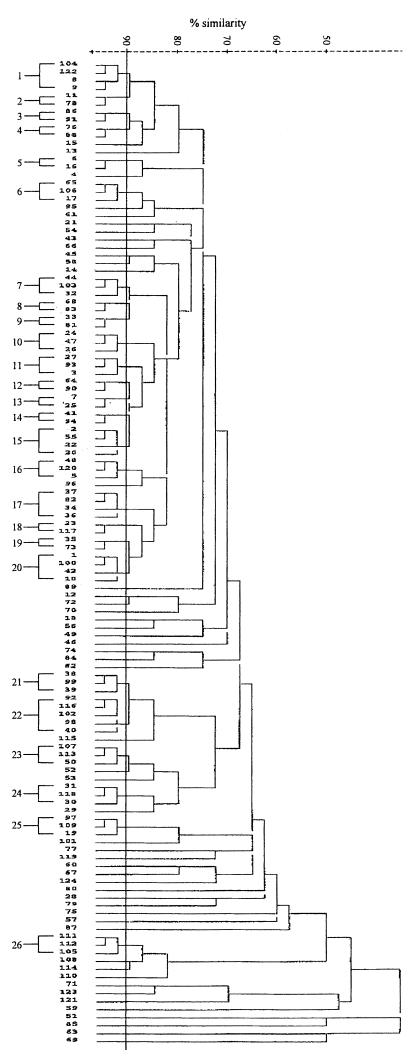
Three restriction enzymes were evaluated for epidemiological typing of serotype Typhi. Total DNAs were digested with a restriction enzyme, NarI, EcoRV, or MluI, and were electrophoresed under special conditions on agarose as described in Materials and Methods. Since EcoRV and MluI restriction resulted in very few fragments (data not shown), only NarI digestion patterns were analyzed. Restriction fragments greater than 12 kb in size were used for epidemiological analysis. NarI could differentiate the isolates into 124 patterns. The patterns consisted of 7 to 13 fragments ranging in size from 13.2 to 48.5 kb. Some of the DNA fingerprints are shown in Fig. 6. The relatedness of the patterns is illustrated in Fig. 7. Seventy-three DNA types could be grouped into 26 clusters at ≥90% similarity, while the rest were of <90% similarity. Each DNA fingerprint was assigned a number following the letter N, and fingerprints of ≥90% similarity were numbered with the prefix N90. The DNA fingerprint with the largest number of isolates was type N9022 (12%), while 3% or less of isolates belonged to the other types.
FIG. 6.
Total DNA fingerprints of S. enterica serotype Typhi obtained after NarI digestion (lanes 1 to 9 and 11 to 21) and high-molecular-size markers of 48.5, 38.4, 33.5, 29.9, 24.8, 22.6, 19.4, 17.1, 15, 12.2, and 10 kb from GIBCO BRL (Gaithersburg, Md.) (lane 10).
FIG. 7.
Dendrogram showing relatedness of NarI total DNA fingerprints.
When PstI ribotypes and NarI fingerprints were combined, the isolates could be differentiated into 183 groups (Table 2). Isolates within the two major ribotypes, P904 and P58, could be discriminated into 26 and 22 groups, respectively. Group 27 comprised the largest number of isolates (n = 19; 7%), followed by groups 1 (n = 10; 3%) and 2 (n = 9; 3%). Each of the other groups comprised one to six isolates.
TABLE 2.
Distribution of S. enterica serotype Typhi isolated from Hong Kong and Vietnam
| Molecular type
|
No. of strains isolated in Hong Kong in the following year:
|
No. of strains isolated in Vietnam | Total
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group | Ribotype | DNA type | 1985 | 1986 | 1987 | 1988 | 1989 | 1990 | 1991 | 1992 | 1993 | 1994 | 1995 | 1996 | 1997 | No. of isolates | % of isolates | |
| 1 | P904 | N9022 | 10 | 10 | 3 | |||||||||||||
| 2 | P904 | N9015 | 1 | 2 | 5 | 1 | 9 | 3 | ||||||||||
| 3 | P904 | N901 | 1 | 2 | 1 | 1 | 5 | 2 | ||||||||||
| 4 | P904 | N9025 | 1 | 3 | 4 | 1 | ||||||||||||
| 5 | P904 | N26 | 2 | 1 | 1 | 4 | 1 | |||||||||||
| 6–26 | P904 | 22 clusters | 4 | 6 | 3 | 2 | 4 | 2 | 2 | 1 | 1 | 4 | 1 | 30 | 10 | |||
| Subtotal | 62 | 21 | ||||||||||||||||
| 27 | P58 | N9022 | 2 | 17 | 19 | 7 | ||||||||||||
| 28 | P58 | N9021 | 4 | 1 | 5 | 2 | ||||||||||||
| 29 | P58 | N9026 | 4 | 4 | 1 | |||||||||||||
| 30–48 | P58 | 19 clusters | 1 | 3 | 1 | 2 | 7 | 1 | 2 | 2 | 8 | 27 | 9 | |||||
| Subtotal | 55 | 19 | ||||||||||||||||
| 49–57 | P902 | 9 clusters | 3 | 5 | 6 | 1 | 1 | 1 | 1 | 18 | 6 | |||||||
| 58 | P66 | N9014 | 2 | 3 | 1 | 6 | 2 | |||||||||||
| 59 | P66 | N9022 | 2 | 1 | 1 | 4 | 1 | |||||||||||
| 60–61 | P66 | 2 clusters | 2 | 1 | 1 | 4 | 1 | |||||||||||
| Subtotal | 14 | 5 | ||||||||||||||||
| 62–68 | P33 | 7 clusters | 1 | 1 | 1 | 7 | 9 | 3 | ||||||||||
| 69–183 | 115 types | 6 | 13 | 11 | 19 | 17 | 7 | 12 | 8 | 5 | 1 | 3 | 3 | 6 | 21 | 132 | 46 | |
| Total (183 groups) | 11 | 21 | 16 | 33 | 47 | 12 | 19 | 17 | 12 | 6 | 5 | 5 | 13 | 73 | 290 | 100 | ||
Hong Kong isolates were distributed among 150 groups. Six groups comprised four to nine isolates each (groups 2 [n = 9]; 58 [n = 6]; and 3, 5, 28, and 59 [n = 4 in each group]), while the rest comprised less than four isolates each. Except for group 28, which comprised all isolates from 1989, all other groups comprised isolates that were found from 1985 to 1997 (Table 2).
Eighty-nine Hong Kong isolates were phage typed in our previous study (21). Table 3 shows the correlation of phage types, DNA groups, and year of isolation for these isolates. Isolates of one phage type were not confined to a particular time period. Besides, they could be further differentiated into different types by ribotyping and DNA fingerprinting. Only three groups comprised isolates of different phage types; e.g., three isolates of group 2 were of phage types A and B1 and untypeable, two isolates of group 5 were of phage type N and untypeable, and two isolates of group 10 were of phage types A and 25.
TABLE 3.
Distribution of 89 S. enterica serotype Typhi strains that were phage typed
| Phage type | Molecular group | No. of strains isolated (yr) | Total no. of strains |
|---|---|---|---|
| 25 | 10 | 1 (1990) | 1 |
| 92 | 2 (1992) | 2 | |
| 44 | 135 | 1 (1990) | 1 |
| 51 | 86 | 1 (1989) | 1 |
| 56 | 27 | 1 (1986) | 1 |
| A | 2 | 1 (1989) | 1 |
| 10 | 1 (1992) | 1 | |
| 10 groups | 1 (1986), 1 (1987), 2 (1988), 2 (1989), 1 (1990), 2 (1992) | 9 | |
| B1 | 2 | 1 (1988) | 1 |
| 2 groups | 1 (1990), 1 (1991) | 2 | |
| C1 | 119 | 1 (1988) | 1 |
| C2 | 111 | 1 (1989) | 1 |
| C3 | 118 | 1 (1986) | 1 |
| D2 | 2 groups | 2 (1989) | 2 |
| E1 | 11 groups | 2 (1985), 1 (1986), 4 (1987), 2 (1988), 4 (1989), 1 (1990), 1 (1991), 1 (1992) | 16 |
| E9 | 4 groups | 2 (1985), 1 (1989), 1 (1990) | 4 |
| F6 | 70 | 1 (1987) | 1 |
| G1 | 120 | 1 (1988) | 1 |
| J1 | 141 | 1 (1988) | 1 |
| J4 | 85 | 1 (1987) | 1 |
| M1 | 3 groups | 1 (1986), 2 (1988) | 3 |
| M3 | 158 | 1 (1985) | 1 |
| N | 5 | 1 (1990) | 1 |
| UTa | 2 | 2 (1989) | 2 |
| 5 | 1 (1991) | 1 | |
| 27 groups | 3 (1985), 5 (1986), 1 (1987), 6 (1988), 4 (1989), 1 (1990), 6 (1991), 6 (1992) | 32 | |
| Total | 89 |
UT, untypeable.
Isolates from Vietnam could be differentiated into 35 groups (Table 1). The groups with the largest number of isolates were 27 (22%) and 1 (12%). All isolates of these groups harbored a 98-kb plasmid of the same fingerprint (E1 and H1), but some were chloramphenicol resistant and some were chloramphenicol sensitive. Isolates that were tetracycline and chloramphenicol resistant (n = 10) were differentiated into six groups. Those without any plasmid were differentiated into 18 different groups.
The majority of isolates from Vietnam were of groups that were different from those of isolates from Hong Kong. Six groups (group 3, 4, 27, 28, 63, and 144) comprised isolates (a total of 86) from both Hong Kong and Vietnam. Of these isolates, only 13 (15%) were from Hong Kong.
DISCUSSION
Due to various constraints, strains from Vietnam were collected only in January 1989 and October 1990.
Our previous study has shown that chloramphenicol-resistant S. enterica serotype Typhi strains from Hong Kong were a rarity (21). However, almost 50% of serotype Typhi strains from Vietnam were chloramphenicol resistant, and a third of these were also resistant to tetracycline. Fortunately, all were susceptible to the β-lactams, trimethoprim-sulfamethoxazole, the aminoglycosides, and the quinolones.
Only 24% of Hong Kong serotype Typhi isolates harbored plasmids (21), while more than 70% of Vietnamese isolates harbored plasmids. The plasmid profiles of the isolates from the two different localities were also different.
Ninety percent of tetracycline- and chloramphenicol-resistant isolates and more than 50% of chloramphenicol-resistant isolates from Vietnam could transfer their resistances, which were borne on 221- and 98-kb plasmids, respectively. The tetracycline and chloramphenicol resistance plasmids could transfer more efficiently at 28°C than at 37°C, which suggested that they probably belonged to incompatibility group H. Previous studies have shown that chloramphenicol resistance plasmids in serotype Typhi usually belonged to incompatibility group H (13, 23, 25, 28), although other groups such as I1 (8, 17), B (17), N (17), FI (6), and FIme (28) have been reported. The location of chloramphenicol resistance on transferable plasmids would enable the resistance to spread rapidly among serotype Typhi strains. It is therefore important that there should be continuous surveillance for chloramphenicol-resistant S. enterica serotype Typhi.
Most of the 221-kb plasmids had identical or similar restriction patterns. However, these patterns were different from those of the 221-kb plasmid from Hong Kong (21). The patterns were also different from those of the 98-kb plasmid in that there were many more restriction fragments. Thirty-two of the 98-kb plasmids had identical restriction patterns, regardless of whether they were transferable. The patterns of these plasmids were also identical to those of the 12 plasmids that were present in those isolates that were sensitive to all the antibiotics tested.
Typhoid fever remains a major public health problem in many parts of the world. Large outbreaks frequently occur in developing countries but have also occurred in developed regions. Characterization of S. enterica serotype Typhi isolates traditionally depended on phage typing. However, phage typing would not provide useful epidemiological information when there were only a few predominating phage types (22). Besides, acquisition or loss of lysogenic phages might result in a change in the phage type (5). With the development of molecular biological techniques, it was shown that phage typing had to be supplemented by other methods to satisfactorily type serotype Typhi (12, 14). We therefore did not attempt to phage type all our isolates but used the phage typing results from our previous study (21) to evaluate the correlation of phage types with DNA types.
Various molecular biological techniques have recently been applied to the investigation of epidemic and endemic strains of S. enterica serotype Typhi. One of these was ribotyping, which was shown to be able to identify genomic variations among strains and which was thus regarded as a reliable and sensitive method that could complement phage typing for the further differentiation of strains (1). However, the variability of patterns and, thus, the usefulness of ribotyping depend largely on the restriction enzyme chosen for digestion of total DNA of strains of the bacterial species concerned (1).
Altwegg and colleagues (1) used EcoRI, SmaI, and PstI for ribotyping of S. enterica serotype Typhi and found PstI to provide the best discrimination among strains. Pang and colleagues (27) also found PstI to be superior to SmaI since PstI produced more different patterns than SmaI. Nastasi and colleagues (24) tried several restriction enzymes and found that ClaI provided the best discrimination among strains. Fica and colleagues (9) ribotyped their Chilean strains using PstI and ClaI digestion and found that both restriction enzymes were able to demonstrate that strains isolated during the epidemic in 1977 and 1981 were acquired from multiple sources.
Three enzymes, namely, PstI, ClaI, and KpnI, were used to produce ribosomal gene patterns in this study. Both PstI and ClaI were found to produce a large variety of patterns which correlated well with each other, while KpnI differentiated the isolates into a much smaller number of types (data not shown). Thus, the RNA gene restriction pattern produced by PstI was used as one of the means of evaluating the epidemiological association of typhoid bacillus in this study.
Three other restriction enzymes (EcoRV, MluI, and NarI) were used for DNA fingerprinting of serotype Typhi strains. NarI produced the largest number of restriction patterns that were easy to interpret because the patterns comprised well-separated bands. Thus, NarI was chosen as the restriction enzyme for the production of DNA restriction patterns for epidemiological evaluation.
Since ribosomal gene restriction patterns depend on the number and location of a specific restriction site and the copy number of the rRNA gene, only changes of restriction sites at different copies of the rRNA gene could be detected by ribotyping, while differences of restriction sites at regions other than the ribosomal operon would have to be detected by digestion of chromosomal DNA. Isolates which differed from each other at restriction sites other than that at the rRNA gene would have an identical ribosomal restriction pattern and appeared to belong to the same clone. It is therefore not reliable to study the epidemiology of a bacterial infection only by ribotyping. Total DNA restriction patterns should also be obtained and compared with ribotypes.
In order to evaluate methods for the epidemiological analysis of S. enterica serotype Typhi, isolates from Hong Kong and Vietnam were studied. These isolates had different resistance patterns, plasmid profiles, and fingerprints and would probably be different in their clonalities. In order to test that PstI ribotyping and NarI fingerprinting did not show any differences between epidemiologically related strains, we tested strains isolated at different times from the same patients and from carriers and found that they had the same patterns by PstI ribotyping or NarI fingerprinting (data not shown). Besides, the 16 isolates from Vietnam which harbored a 98-kb plasmid of the same fingerprint all had the same DNA type (DNA type N92, which was clustered with related types into N9022) and the same ribotype (ribotype P58) (Table 1).
Analysis of the combination of PstI and NarI types, phage types, plasmid profiles, and plasmid fingerprints showed that during the study period, typhoid fever in Hong Kong was caused by a large variety of strains. No single clone had become established. Although there were clusters of small numbers of cases, these might be regarded as infections acquired from the same sources but not as epidemics in the strictest sense. This is similar to the situation in other countries where typhoid fever is endemic (26, 27, 33, 35). Some strains had the same pattern combinations but were isolated in different years. It was difficult to assess in such a retrospective study how these patients acquired the infection from the same source over wide intervals of time. In contrast, almost 40% of isolates from Vietnam belonged to two groups (groups 1 and 27).
The majority of isolates from Hong Kong belonged to groups that were different from those of isolates from Vietnam. However, it is not surprising to find that some strains from the two localities shared the same ribotype and DNA pattern, as these might be a result of exchange of strains among areas in Southeast Asia where typhoid fever is endemic brought about by trade and travel.
Nine of the sensitive isolates from Vietnam that harboured a 98-kb plasmid identical to that in chloramphenicol-resistant isolates were of the same groups (groups 27 and 1) as chloramphenicol-resistant isolates. Thus, the chloramphenicol resistance gene probably was too small to be resolved by the present method and was picked up by or lost from the 98-kb plasmid that circulated among two different clones of serotype Typhi isolates. In contrast, Shanahan and colleagues (30) showed that resistant and sensitive isolates belonged to different clones.
To conclude, analysis of total DNA NarI restriction patterns might be a more rapid and discriminative method for use in a routine laboratory, complementing phage typing and ribotyping for the investigation of the epidemiology of typhoid fever. Serotype Typhi strains isolated in Hong Kong from 1985 to 1997 belonged to heterogeneous groups, and no obvious outbreaks had occurred, although there might be sporadic incidences of common-source infections. In contrast, there were two predominant clones in Vietnam.
ACKNOWLEDGMENTS
We thank M. L. Chin and T. H. S. Woo for technical assistance and Keith Arnold and Tran Tinh Hien for coordinating part of this study.
REFERENCES
- 1.Altwegg M, Hickman-Brenner F W, Farmer J J., III Ribosomal RNA gene restriction patterns provide increased sensitivity for typing Salmonella typhi strains. J Infect Dis. 1989;160:145–149. doi: 10.1093/infdis/160.1.145. [DOI] [PubMed] [Google Scholar]
- 2.Anderson E S, Threlfall E J. The characterization of plasmids in the enterobacteria. J Hyg. 1974;71:619–631. doi: 10.1017/s0022172400023718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arbeit R D. Laboratory procedures for the epidemiologic analysis of microorganisms. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: American Society for Microbiology; 1999. pp. 116–137. [Google Scholar]
- 4.Balows A, Hausler W J Jr, Herrmann K L, Isenberg H D, Shadomy H J, editors. Manual of clinical microbiology. 5th ed. Washington, D.C.: American Society for Microbiology; 1991. [Google Scholar]
- 5.Borecka J, Pucekova G, Bolchova S, Hlaucova Z, Suchanek M. To the question of stability of Salmonella typhi phage types. J Hyg Epidemiol Microbiol Immunol. 1973;17:202–206. [PubMed] [Google Scholar]
- 6.Chasseur-Libotte M L, Ghysels G, Pohl P. Characterization of resistance plasmids to antibiotics in two Salmonella typhi isolated in Belgium. Ann Microbiol (Institute Pasteur) 1983;134B:285–291. [PubMed] [Google Scholar]
- 7.Chau P Y, Huang C T. Salmonellosis in Hong Kong. Publ Health London. 1977;91:83–89. doi: 10.1016/s0033-3506(77)80005-x. [DOI] [PubMed] [Google Scholar]
- 8.Datta N, Richards H, Datta C. Salmonella typhi in vivo acquires resistance to both chloramphenicol and co-trimoxazole. Lancet. 1981;ii:1181–1183. doi: 10.1016/s0140-6736(81)92350-3. [DOI] [PubMed] [Google Scholar]
- 9.Fica A E, Prat-Miranda S, Fernandez-Ricci A, D'Ottone K, Cabello F C. Epidemic typhoid in Chile: analysis by molecular and conventional methods of Salmonella typhi strain diversity in epidemic (1977 and 1981) and nonepidemic (1990) years. J Clin Microbiol. 1996;34:1701–1707. doi: 10.1128/jcm.34.7.1701-1707.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greene P J, Betlach M C, Boyer H W, Goodman H H. The EcoRI restriction endonuclease. Methods Mol Biol. 1974;7:387–398. [Google Scholar]
- 11.Grimont N D F, Grimont P A D. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann Inst Pasteur Microbiol. 1986;137B:165–175. doi: 10.1016/s0769-2609(86)80105-3. [DOI] [PubMed] [Google Scholar]
- 12.Hampton M D, Ward L R, Rowe B, Threlfall E J. Molecular fingerprinting of multidrug-resistant Salmonella serotype typhi. Emerging Infect Dis. 1998;4:317–320. doi: 10.3201/eid0402.980223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harnett N, McLeod S, AuYong Y, Brown S, Krishnan C. Emergence in Ontario, Canada, of multiresistant Salmonella typhi from South Asia. Lancet. 1992;340:177. doi: 10.1016/0140-6736(92)93253-j. [DOI] [PubMed] [Google Scholar]
- 14.Harnett N, McLeod S, AuYong Y, Wan J, Alexander S, Khakhria R, Krishnan C. Molecular characterization of multiresistant strains Salmonella typhi from South Asia isolated in Ontario, Canada. Can J Microbiol. 1998;44:356–363. [PubMed] [Google Scholar]
- 15.Huang C T, Chan-Teoh C H. Salmonella serotypes isolated in Hong Kong. J Trop Med Hyg. 1964;67:95–99. [PubMed] [Google Scholar]
- 16.Kado C I, Liu S-T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ling J, Chau P Y. Plasmids mediating resistance to chloramphenicol, trimethoprim and ampicillin in Salmonella typhi strains isolated in the Southeast Asian region. J Infect Dis. 1984;149:652. doi: 10.1093/infdis/149.4.652. [DOI] [PubMed] [Google Scholar]
- 18.Ling J, Chau P Y, Rowe B. Salmonella serotypes and incidence of multiply-resistant salmonellae isolated from diarrhoeal patients in Hong Kong from 1973–82. Epidemiol Infect. 1987;99:295–306. doi: 10.1017/s0950268800067777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ling J, Kam K M, Lam A W, French G L. Susceptibilities of Hong Kong isolates of multiply resistant Shigella spp. to 25 antimicrobial agents, including ampicillin and sulbactam and new 4-quinolones. Antimicrob Agents Chemother. 1988;32:20–23. doi: 10.1128/aac.32.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ling J M, Khin-Thi-Oo H, Hui Y-W, French G L. Antimicrobial susceptibilities of Haemophilus species in Hong Kong. J Infect. 1989;19:134–142. doi: 10.1016/s0163-4453(89)91889-6. [DOI] [PubMed] [Google Scholar]
- 21.Ling J M, Lo N W S, Ho Y M, Kam K M, Ma C H, Wong S C, Cheng A F. Emerging resistance in Salmonella enterica serotype Typhi in Hong Kong. Int J Antimicrob Agents. 1996;7:161–166. doi: 10.1016/s0924-8579(96)00316-0. [DOI] [PubMed] [Google Scholar]
- 22.Maher K O, Morris J G, Jr, Gotuzzo E, Ferreccio C, Ward L R, Benavente L, Black R E, Rowe B, Levine M M. Molecular techniques in the study of Salmonella typhi in epidemiologic studies in endemic areas: comparison with Vi phage typing. Am J Trop Med Hyg. 1986;35:831–835. doi: 10.4269/ajtmh.1986.35.831. [DOI] [PubMed] [Google Scholar]
- 23.Mourad A S, Metwally M, El Deen A N, Threlfall E J, Rowe B, Mapes T, Hedstrom R, Bourgeois A L, Murphy J R. Multiple-drug-resistant Salmonella typhi. Clin Infect Dis. 1993;17:135–136. doi: 10.1093/clinids/17.1.135. [DOI] [PubMed] [Google Scholar]
- 24.Nastasi A, Mammina C, Villafrate M R. rRNA fingerprinting as a tool in epidemiological analysis of Salmonella typhi infections. Epidemiol Infect. 1991;107:565–576. doi: 10.1017/s0950268800049268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nastasi A, Mammina C, Pontello M, Carrara C. Isolation of a multiresistant strain of Salmonella typhi in Italy. Infection. 1992;20:296–297. doi: 10.1007/BF01710803. [DOI] [PubMed] [Google Scholar]
- 26.Navarro F, Llovet T, Echeita M A, Coll P, Aladuena A, Usera M A, Prats G. Molecular typing of Salmonella enterica serovar typhi. J Clin Microbiol. 1996;34:2831–2834. doi: 10.1128/jcm.34.11.2831-2834.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pang T, Altwegg M, Martinetti G, Koh C L, Puthucheary S. Genetic variation among Malaysian isolates of Salmonella typhi as detected by ribosomal RNA gene restriction patterns. Microbiol Immunol. 1992;36:539–543. doi: 10.1111/j.1348-0421.1992.tb02053.x. [DOI] [PubMed] [Google Scholar]
- 28.Rowe B, Threlfall E J, Ward L R. Does chloramphenicol remain the drug of choice for typhoid? Epidemiol Infect. 1987;98:379–383. doi: 10.1017/s0950268800062142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Shanahan P M A, Jesudason M V, Thomson C J, Amyes S G B. Molecular analysis of and identification of antibiotic resistance genes in clinical isolates of Salmonella typhi from India. J Clin Microbiol. 1998;36:1595–1600. doi: 10.1128/jcm.36.6.1595-1600.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sneath P H A. Computer taxonomy. Methods Microbiol. 1972;7A:29–98. [Google Scholar]
- 32.Thong K-L, Cordano A, Yassin R M, Pang T. Molecular analysis of environmental and human isolates of Salmonella typhi. Appl Environ Microbiol. 1996;62:271–274. doi: 10.1128/aem.62.1.271-274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thong K-L, Cheong Y-M, Puthucheary S, Koh C-L, Pang T. Epidemiologic analysis of sporadic Salmonella typhi isolates and those from outbreaks by pulsed-field gel electrophoresis. J Clin Microbiol. 1994;32:1135–1141. doi: 10.1128/jcm.32.5.1135-1141.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thong K-L, Puthucheary S, Yassin R M, Sudarmono P, Padmidewi M, Soewandojo E, Handojo I, Sarasombath S, Pang T. Analysis of Salmonella typhi isolates from Southeast Asia by pulsed-field gel electrophoresis. J Clin Microbiol. 1995;33:1938–1941. doi: 10.1128/jcm.33.7.1938-1941.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yew F S, Goh K T, Lim Y S. Epidemiology of typhoid fever in Singapore. Epidemiol Infect. 1993;110:63–70. doi: 10.1017/s0950268800050688. [DOI] [PMC free article] [PubMed] [Google Scholar]