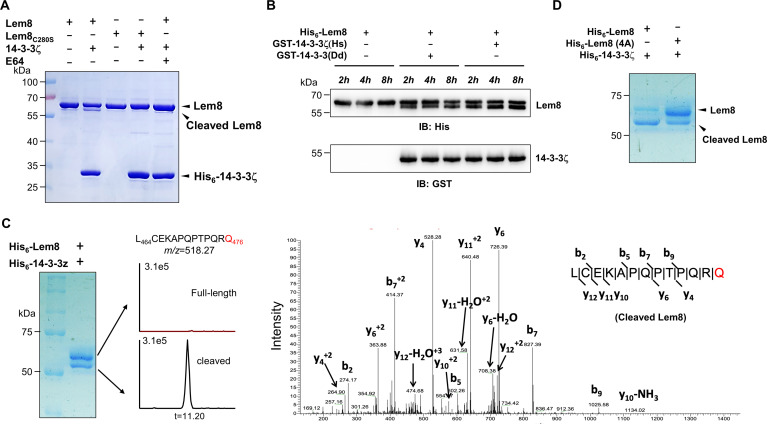
Figure 3. 14-3-3ζ induces Lem8 to undergo self-cleavage.
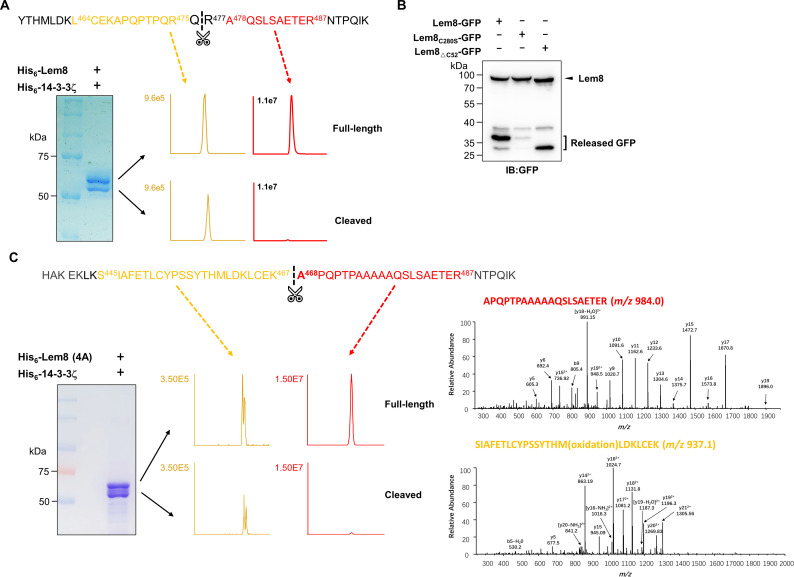
(A) Self-processing of Lem8 requires 14-3-3ζ. His6-Lem8 or His6-Lem8C280S was incubated with His6-14-3-3ζ for 2 hr, proteins resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) were detected by Coomassie brilliant blue staining. The cysteine protease inhibitor E64 was added to the indicated samples at a final concentration of 10 μM. Similar results were obtained from at least three independent experiments and the data shown here were from one representative experiment. (B) The 14-3-3 protein from D. discoideum induces the self-cleavage of Lem8. His6-Lem8 was incubated with GST-14-3-3ζ or GST-14-3-3Dd for the indicated time and the mixtures separated by SDS–PAGE were detected by immunoblotting with antibodies specific for Lem8 and GST, respectively. Similar results were obtained from at least three independent experiments and the data shown here were from one representative experiment. (C) Determination of the self-cleavage site of Lem8. His6-Lem8 was incubated with His6-14-3-3ζ for 16 hr, proteins were resolved by SDS–PAGE, stained with Coomassie brilliant blue. Protein bands corresponding to full-length and cleaved Lem8 band was excised, digested with trypsin and analyzed by mass spectrometry. The detection of the semitryptic peptide -L464CEKAPQPTPQRQ476- in cleaved samples suggested that the cleavage site lies between Gln476 and Arg477. (D) Mutations in cleavage site does not abolish Lem8 self-processing. Recombinant protein of Lem8 and the 4A mutant were each incubated with His6-14-3-3ζ for 4 hr. Proteins resolved by SDS–PAGE were detected by Coomassie brilliant blue. Similar results were obtained from at least three independent experiments and the data shown here were from one representative experiment.