Abstract
Carya cathayensis, an important economic nut tree, is narrowly endemic to eastern China in the wild. The complete cp genome of C. cathayensis was sequenced with NGS using an Illumina HiSeq2500, analyzed, and compared to its closely related species. The cp genome is 160,825 bp in length with an overall GC content of 36.13%, presenting a quadripartite structure comprising a large single copy (LSC; 90,115 bp), a small single copy (SSC; 18,760 bp), and a pair of inverted repeats (IRs; 25,975 bp). The genome contains 129 genes, including 84 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. A total of 252 simple sequence repeats (SSRs) and 55 long repeats were identified. Gene selective pressure analysis showed that seven genes (rps15, rpoA, rpoB, petD, ccsA, atpI, and ycf1-2) were possibly under positive selection compared with the other Juglandaceae species. Phylogenetic relationships of 46 species inferred that Juglandaceae is monophyletic, and that C. cathayensis is sister to Carya kweichowensis and Carya illinoinensis. The genome comparison revealed that there is a wide variability of the junction sites, and there is higher divergence in the noncoding regions than in coding regions. These results suggest a great potential in phylogenetic research. The newly characterized cp genome of C. cathayensis provides valuable information for further studies of this economically important species.
Keywords: Carya cathayensis, chloroplast genome, genome skimming, phylogenetic relationship
1. Introduction
The genus Carya, belonging to the family Juglandaceae, comprises ~18 species and 4 varieties, which are distributed in the temperate and tropical regions of East Asia and eastern North America [1,2]. Carya species from East Asia and eastern North America are phylogenetically separated [2], while the relationships among some taxa within the genus have not been resolved yet.
Nuclear and plastid DNAs are the basics for phylogenetic reconstruction; the single- or low-copy nuclear genes are most suitable for systematic analyses [3]. Until now, several plastid (matK, rbcL-atpB, rpoC1, rps16, trnH-psbA, and trnL-F) and nuclear (ITS and phyA) DNA markers have been used for the phylogenetic study of the genus Carya. These nuclear genes were identified by ortholog screening, cloning, and sequencing; however, these methods can be costly and time-consuming. Compared with the nuclear genome, the chloroplast (cp) genome is an excellent alternative owing to its small size (75–250 Kb) [4], easily obtainable sequences by the low-cost next-generation sequencing (NGS) technique, and less interference from homologous regions. Besides the genic regions, the noncoding regions of cp genomes can also be harnessed for phylogenetic analysis due to a relatively high level of genetic variation resulting from the low selective pressure [5]. In addition, structural rearrangements, such as the loss of introns, genes, or even inverted repeats, extensively occur in the plastid genomes of many flowering plants [6,7,8,9,10,11]. Recently, the cp genomes of C kweichowensis [12], C. cathayensis [13], and C. illinoinensis (NBCI accession number: NC_041449.1) have been published, and the publication of more cp genomes of Carya species will facilitate the identification of genetic variations via sequence comparison, providing new insights into the evolutionary history and interspecific relationships among Carya species.
C. cathayensis (Chinese hickory) is naturally distributed in moist valleys at altitudes of 500–1200 m in Zhejiang, Jiangxi, and Anhui Provinces, China. Because of its high nutritional and economic values, C. cathayensis has been widely cultivated in Zhejiang Province, China [14]. C. cathayensis is an important economic nut tree and is vulnerable to abiotic factors [15,16], suggesting that suitable habitat is essential for its survival in the wild. In recent years, with the changes in climate and over-exploitation, the conservation of wild C. cathayensis populations has become an urgent task. The nuclear genome and cp genome of C. cathayensis have been released [13,17], although the cp genome has not been reported in detail. The cp genome of C. cathayensis is essential for the development of conservation and breeding strategies.
In this study, we present the whole plastome sequence of C. cathayensis and explore the utility of this new genomic resource and relationship with that of other Carya species. These results will lay the foundation for future phylogenetic and structural diversity studies of Carya.
2. Materials and Methods
2.1. DNA Extraction, Sequencing, and cp Genome Assembly
The young green leaves of C. cathayensis were collected from the nursery of Zhejiang A&F University (stored in the Institute of Botany, Chinese Academy of Sciences Mem, and the specimen accession number is PE00820836) and stored immediately at −80 ℃. Total genomic DNA was isolated from the leaves using a modified CTAB method [18]. After ensuring the quality of DNA, shotgun libraries (250 bp) were constructed in accordance with the standard protocol suggested by the manufacturer’s instructions (Illumina Inc., San Diego, CA, USA). Sequencing was performed with an Illumina Hiseq 2500 platform (Genepioneer Biotechnologies Co., Ltd.; Nanjing, China) with the PE150 strategy.
Quality control for the raw sequencing data was carried out using the package FastQC (version 0.11.8. Available online: http://www.bioinformatics.babraham.ac.uk/proje-cts/fastqc/, accessed on 8 September 2021). High-quality clean reads were obtained by removing the adapters and low-quality reads from the raw data using Trimmomatic (version 0.35) [19]. The C. cathayensis cp genome was assembled using the SPAdes pipeline [20] with the Cyclocarya paliurus cp genome as the reference (NCBI accession number: NC_034315).
2.2. Annotation of the C. cathayensis cp Genome
C. cathayensis cp genome annotation was performed via the CpGAVAS pipeline [21]. The annotated C. cathayensis genome was deposited to GenBank under accession number MN892516. The circular gene map was visualized in OGDRAWv1.2. Available online: http://ogdraw.mpimp-golm.mpg.de/, accessed on 12 September 2021). Relative synonymous codon usage (RSCU) was determined by CodonW version 1.4.4. Available online: http://codonw.sourceforge.net/, accessed on 15 September 2021).
2.3. Identification of Repeats
REPuter [22,23] was used to identify the repeat sequences [24,25] using the parameters reported by [7]. Then, the online microsatellite identification tool (MISA. Available online: https://webblast.ipk-gatersleben.de/misa/, accessed on 21 September 2021) [26] was applied to predict cpSSRs with default parameters.
2.4. Phylogenetic Analysis
To determine the phylogenetic relationships among Juglandaceae species, a Bayesian inference (BI) tree was inferred using protocols suggested by [27]. An alignment of 46 cp genomic sequences (See in ‘Data Availability Statement’ part) was created using the MAFFT online version [28,29] with default parameters.
2.5. Genomic Comparison with Related Species
The online tool Irscope [30] was employed to draw the genetic architecture of the IR/SSC and IR/LSC junctions. mVISTA [31] was used to compare the complete C. cathayensis cp genome to that of five related species including C. kweichowensis, C. illioninensis, C. paliurus, Juglans cathayensis, and Platycarya strobilacea. The shuffle-LAGAN mode was used in mVISTA [31], with the annotation of Quercus variabilis as the reference. The sequences were initially aligned using the MAFFT online version [28,29], the pi value of each gene was calculated through alignment of each gene CDS sequence of different species using vcftools, and the ratios of nonsynonymous (Ka) to synonymous (Ks) substitutions (Ka/Ks) in protein-coding genes were determined by KaKs_Calculator.
3. Results
3.1. Genome Features of C. cathayensis
Filtering of the raw sequencing data yielded a total of 12,470,465 clean paired-end reads. There were 3.7 G bases, of which 89.47% of bases had a quality score higher than Q30. The whole cp genome of C. cathayensis is 160,825 bp in length, with a GC content of 36.13%. The genome assembly had an average read coverage of higher than 700×. The synteny was identified by comparing the C. cathayensis cp genome to the reference (Table S1), which showed that most of the sequences of the genomes were conserved.
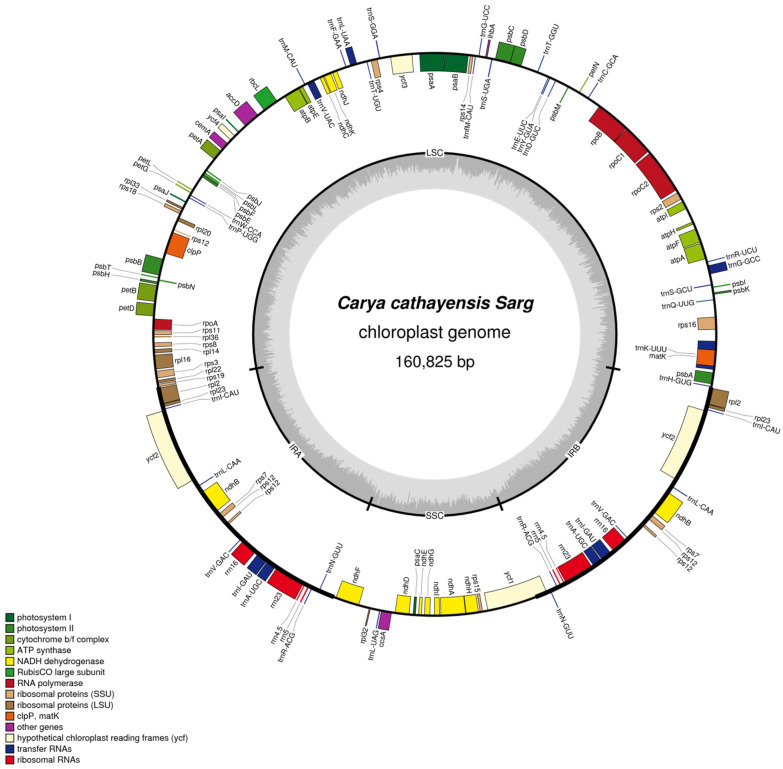
The genome of C. cathayensis displays a typical quadripartite structure, containing one large single copy (LSC; 90,115 bp) region, one small single copy (SSC; 18,760 bp) region, and two inverted repeat regions (IRs; 25,975 bp each) (Figure 1). The overall GC content is 36.13%. The IR regions have a relatively higher GC content compared with other regions (Figure 2). A total of 129 genes were identified, including 84 protein-coding genes, 37 transfer RNA (tRNA) genes, and 8 ribosomal RNA (rRNA) genes (Table 1). Seventeen genes are duplicated in IRs, including six protein-coding genes (rps7, rps12, rpl2, rpl23, ndhB, ycf2) (Table 1). In total, 18 intron-containing genes (12 protein-coding and 6 tRNA genes) were annotated (Table 2), among which there are only 3 protein-coding genes (rps12, ycf3, and clpP) with 2 introns and the others with 1 intron. Gene rps12 of C. cathayensis has its 5′-end exon situated in the LSC region and its 3′-end exons located in the IR region (Figure 1, Table 2).
Figure 1.
The complete C. cathayensis chloroplast (cp) genome. Genes shown outside the outer circle are transcribed clockwise, whereas those shown inside are transcribed counterclockwise. The gray plots in the inner circle represent GC contents. The circular gene map was drawn using OGDRAWv1.2.
Figure 2.
GC content of the C. cathayensis cp genome.
Table 1.
Annotated genes in the C. cathayensis cp genome.
| Category | Group of Genes | Name of Gene |
|---|---|---|
| Self-replication | Ribosomal RNA | rrn4.5 3, rrn5 3, rrn16 3, rrn23 3 |
| Transfer RNA | trnY-GUA, trnW-CCA, trnV-UAC 1, trnV-GAC 3, trnT-UGU, trnT-GGU, trnS-UGA, trnS-GGA, trnS-GCU, trnR-UCU, trnR-ACG 3, trnQ-UUG, trnP-UGG, trnN-GUU 3, trnM-CAU, trnL-UAG, trnL-UAA 1, trnL-CAA 3, trnK-UUU 1, trnI-GAU 1,3, trnI-CAU 3, trnH-GUG, trnG-UCC, trnG-GCC 1, trnfM-CAU 4, trnF-GAA, trnE-UUC, trnD-GUC, trnC-GCA, RNA-UGC 1,3 | |
| Small subunit of ribosome | rps2, rps3, rps4, rps7 3, rps8, rps11, rps12 2,3, rps14, rps15, rps16 1, rps18, rps19 | |
| Large subunit of ribosome | rpl2 1,3, rpl14, rpl16 1, rpl20, rpl22, rpl23 3, rpl32, rpl33, rpl36 | |
| RNA polymerase subunits | rpoA, rpoB, rpoC1 1, rpoC2 | |
| Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ | |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT | |
| Photosynthesis | Subunits of cytochrome | petA, petB 1, petD 1, petG, petL, petN |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF 1, atpH, atpI | |
| Large subunit of RuBisCO | rbcL | |
| Subunits of NADH | ndhA 1, ndhB 1,3, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Other gene | Maturase | matK |
| Envelope membrane protein | cemA | |
| Subunit of acetyl-CoA | accD | |
| C-type cytochrome synthesis gene | ccsA | |
| Protease | clpP 2 | |
| Unknown function | Conserved open reading frames | ycf1, ycf2 3, ycf3 2, ycf4, ihbA |
1 Gene containing a single intron; 2 gene containing two introns; 3 two gene copies in the IRs; 4 duplicated gene in the LSC region.
Table 2.
Genes with introns in the C. cathayensis cp genome.
| Gene | Region (bp) | Exon Ⅰ (bp) | Intron Ⅰ (bp) | Exon Ⅱ (bp) | Intron Ⅱ (bp) | Exon Ⅲ (bp) |
|---|---|---|---|---|---|---|
| atpF | LSC | 144 — | 762 | 411 — | ||
| clpP | LSC | 71 — | 847 | 292 — | 617 | 227 — |
| ndhA | SSC | 552 — | 1211 | 540 — | ||
| ndhB | IRB | 777 — | 686 | 762 — | ||
| ndhB | IRA | 775 + | 686 | 760 + | ||
| petB | LSC | 4 + | 822 | 640 + | ||
| petD | LSC | 6 + | 615 | 485 + | ||
| rpl16 | LSC | 9 — | 919 | 399 — | ||
| rpl2 | IRB | 390 — | 663 | 435 — | ||
| rpl2 | IRA | 388 + | 663 | 433 + | ||
| rpoC1 | LSC | 430 — | 843 | 1619 — | ||
| rps12 | IRB | 114 — | - | 229 + | 537 | 29 + |
| rps12 | IRA | 114 — | - | 231 — | 537 | 29 — |
| rps16 | LSC | 40 — | 894 | 230 — | ||
| trnA-UGC | IRB | 36 + | 801 | 40 + | ||
| trnA-UGC | IRA | 38 — | 801 | 42 — | ||
| trnG-GCC | LSC | 22 + | 715 | 45 + | ||
| trnI-GAU | IRB | 40 + | 950 | 33 + | ||
| trnI-GAU | IRA | 42 — | 950 | 35 — | ||
| trnK-UUU | LSC | 37 — | 2557 | 35 — | ||
| trnL-UAA | LSC | 35 + | 524 | 48 + | ||
| trnV-UAC | LSC | 38 — | 615 | 37 — | ||
| ycf3 | LSC | 126 — | 720 | 229 — | 793 | 151 — |
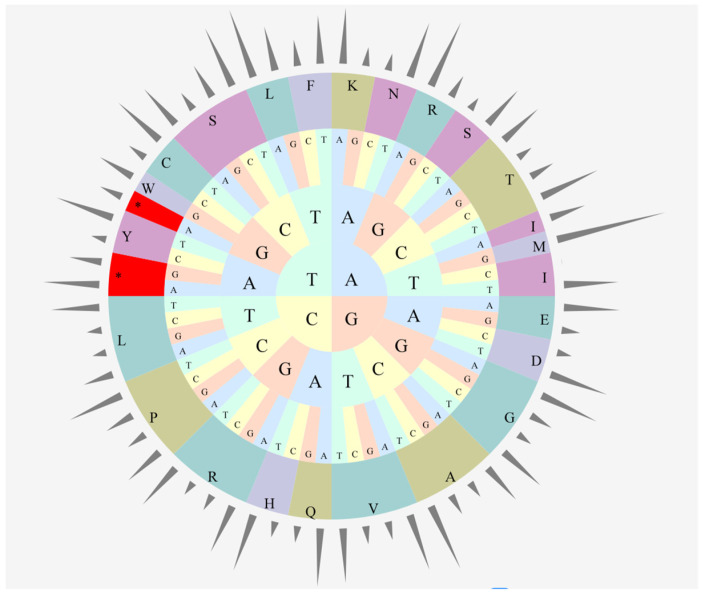
The relative frequency of synonymous codons of the C. cathayensis cp coding sequence was estimated. The results show that all genes are encoded by 26,476 codons, and the 4 most frequently used codons were AUU (isoleucine), AAA (lysine), GAA (glutamic acid), and AAU (asparagine), pertaining to 1145 (4.32%), 1066 (4.03%), 1040 (3.93%), and 1004 (3.79%) codons, respectively (Table S2 and Figure 3). The two most frequently used amino acids were leucine (2780) and isoleucine (2350); cysteine was the least abundant, with only 308 hits. A- and U-ending codons accounted for 70.62% among all codons.
Figure 3.
Codon usage frequency of the C. cathayensis cp genome.
3.2. Analysis of Long Repeats and Simple Sequence Repeats (SSRs)
We identified 24 forward, 9 reverse, 3 complement, and 13 palindrome repeats in the cp genome of C. cathayensis (Table S3). Most repeats ranged from 20 to 62 bp in length. The longest forward repeat with 62 bp resided in the LSC region. A total of 46, 5, and 4 long repeats were found in the LSC, SSC, and IR regions, respectively. Three forward repeats were found in the two IRs, including one repeat associated with the rpl14 and tRNA-UGC genes, one with the IGS genes, and one with the tRNA-CCA and tRNA-GUU genes.
A total of 252 SSRs were identified in the C. cathayensis cp genome (Table S4), among which 199, 12, 64, 2, and 1 were mono-, di-, tri-, tetra-, and pentanucleotide repeats, respectively. Mononucleotide SSRs were the richest (occupied 78.97%), and the mononucleotide A+T repeat units occupied the highest portion (75.00%).
3.3. Phylogenetic Analysis
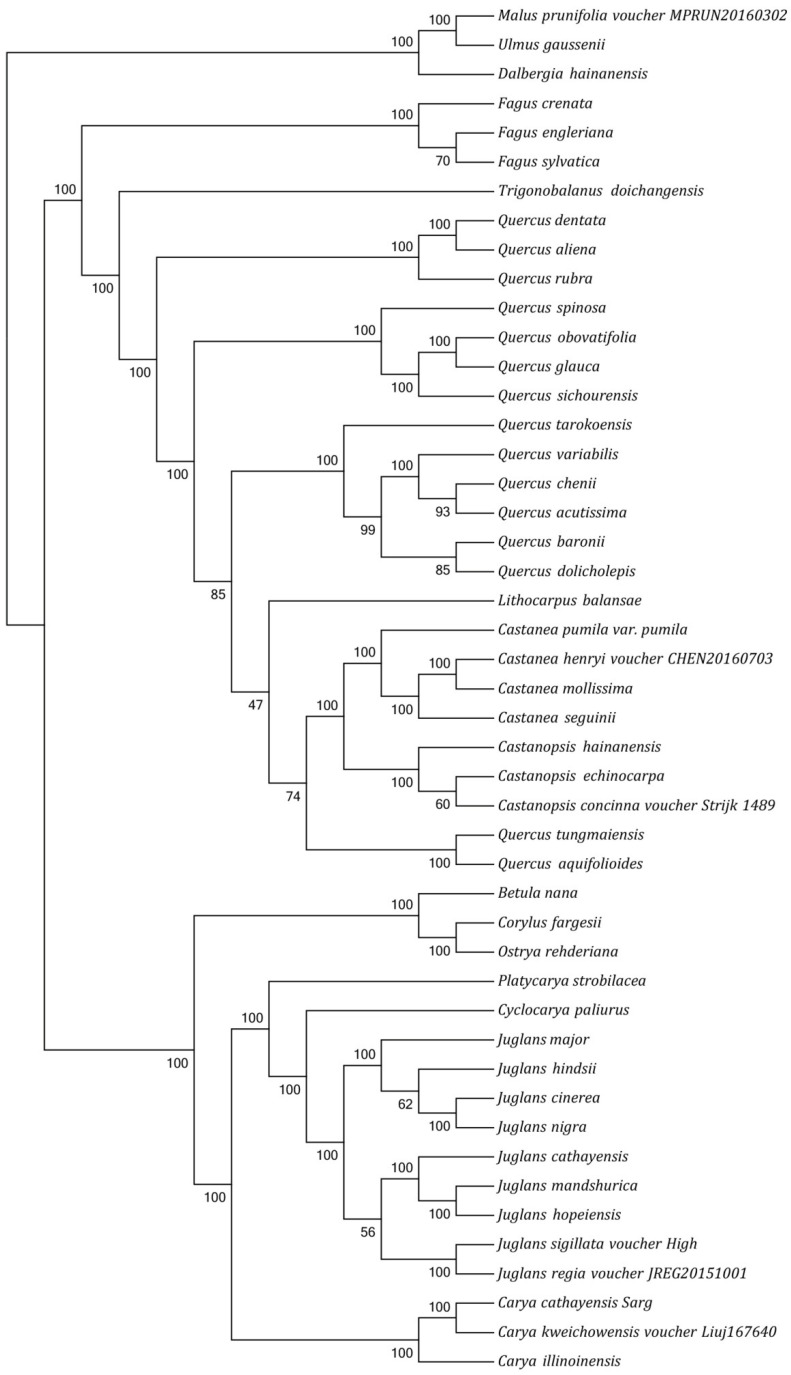
Phylogenetic analysis was carried out based on an alignment of the concatenated nucleotide sequences of all 46 angiosperm cp genomes (Figure 4). MAFFT was employed for multiple sequence alignment. The phylogenetic relationship was reconstructed using the GTR-γ model by RAxML, and Malus prunifolia, Ulmus gaussenii, and Dalbergia hainanensis were used as outgroups. Almost all relationships inferred from the cp genome data based on the maximum likelihood (ML) tree received strong support, with the support values ranging from 47 to 100. In addition, genera Betula, Corylus, and Ostrya were found to be sister to Juglans, whereas Platycarya and Cyclocarya were more closely related to Juglans (Figure 4). The well-supported phylogenetic tree (Figure 4) indicates that the genus Carya is monophyletic and is most closely related to the cluster formed by another genus of Juglandaceae. C. cathayensis is sister to C. kweichowensis, and they are sister to C. illinoinensis successively, with high support scores (bootstrap = 100; Figure 4).
Figure 4.
ML phylogenetic tree of 46 complete cp genomes resolved by Raxml. Bootstrap values are shown near each node.
3.4. Comparative Analysis of Genome Structure
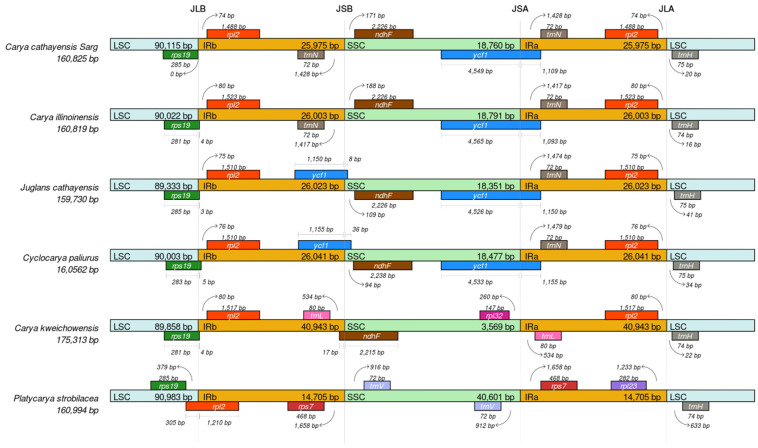
To further resolve the structural evolutionary history of the cp genomes of the genus Carya, we compared the IR/SSC and IR/LSC junctions across six selected Juglandaceae species, including C. cathayensis, C. illinoinensis, C. kweichowensis, Platycarya strobilacea, Cyclocarya paliurus, and Juglans cathayensis. The results of the IRscope analysis are presented in Figure 5. We observed a wide variability of the junction sites in these cp genomes. For example, in the genus Carya, C. cathayensis exhibited similar JLB, JSB, and JSA junction sites compared with its elder sister species C. illinoinensis (Figure 4 and Figure 5). All species used in this study had an IRa/b region of ~25,900 bp and an SSC region of ~18,700 bp. By contrast, C. kweichowensis, which is most closely related to C. cathayensis and C. illioninensis, displayed an extremely large IRa/b region of 40,943 bp. In addition, the C. kweichowensis cp genome showed some striking structural differences compared to its sister species. For example, the rps19 gene was shifted by 285 bp from the LSC to IRb at the LSC/IRb border, trnL was located in the IRa/b regions instead of the SSC region, and ycf1 was absent from the JSA site. Moreover, we observed variations in the IR/SSC and IR/LSC junction sites across other genera in the family Juglandaceae (Figure 5).
Figure 5.
Comparison of the LSC, SSC, and IR regions among six selected cp genomes in the family Juglandaceae. Genes are denoted by colored boxes. The gaps between the genes and boundaries are proportional to the distances in bps.
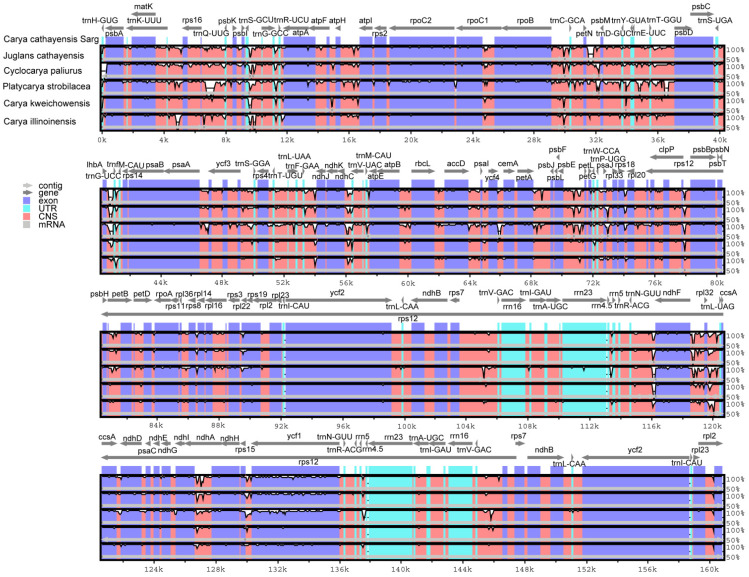
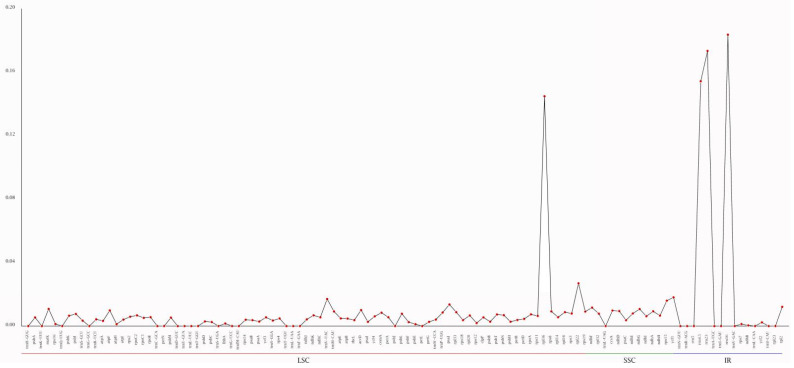
A cp genome identity analysis was performed on the six Juglandaceae species described above, with the C. cathayensis cp genome used as a reference (Figure 6). This analysis found a relatively higher level of divergence in the noncoding than in the coding regions. We also identified a considerable number of variations in the noncoding cp sequences, such as trnC-GCA, trnW-CCA, trnI-CAU, and trnI-UAG, of species in the genus Carya (Figure 6). Gene nucleotide variability (pi) values of six selected Juglandaceae species (including C. cathayensis, C. illinoinensis, C. kweichowensis, Platycarya strobilacea, Cyclocarya paliurus, and Juglans cathayensis) are shown in Figure 7, where the values of LSC.rpl36, IR. rrn4.5, rrn23, and rrn16 are higher than 1, while the values of other genes are lower than 0.03. The results show that there is lower nucleotide diversity among the six Juglandaceae species.
Figure 6.
Variable characters in homologous regions among C. cathayensis and five related species. The homologous regions are oriented according to their locations in the cp genome. The gray arrows above the alignment indicate the gene orientations. The Y-axis shows the identity from 50% to 100%.
Figure 7.
Gene nucleotide variability (pi) values of six Juglandaceae species. The Y-axis shows the pi values; the X-axis shows the genes.
To test whether the remaining cp genes in these six species of Juglandaceae have undergone selection, the synonymous (Ks) and nonsynonymous (Ka) substitution rates were calculated (Table S5). The Ka/Ks ratios were then categorized, with Ka/Ks < 1, Ka/Ks = 1, and Ka/Ks > 1 denoting purifying, neutral, and positive selections, respectively, in the context of a codon substitution model. The results show that only seven genes of C. cathayensis, namely, rps15, rpoA, rpoB, petD, ccsA, atpI, and ycf1-2, underwent positive selection compared with the other Juglandaceae species (Table S4). By contrast, most genes were shown to have undergone purifying selection, which was evidenced by a Ka/Ks ratio below 1 and the presence of negatively selected sites within some genes.
4. Discussion
Plant chloroplast genomes may have 63–209 genes, but most are concentrated between 110 and 130, with a highly conserved composition and arrangement, including photosynthetic genes, chloroplast transcriptional expression-related genes, and some other protein-coding genes [32]. As with other angiosperms, the cp genome of C. cathayensis displays a typical quadripartite structure [32,33], including a pair of inverted repeats (IRs; 25,975 bp each), separated by a large single copy (LSC; 90,115 bp) and a small single copy (SSC; 18,760 bp) region (Figure 1). In total, 129 genes, including 84 protein-coding genes, 37 tRNA genes, and 8 rRNA genes, were identified in our study. The overall GC content is 36.13%, which is similar to that observed for other Carya species (35.8–36.3%) [12,13,34]. It is obvious that the DNA G + C content of the IR region is higher than that of other regions (LSC, SSC) (Figure 2); this phenomenon is very common in other flowering plants [25,34,35]. GC skewness has been shown to be an indicator of DNA lead chains, lag chains, replication origin, and replication terminals, which is a very important indicator of species affinity [36]. The rps12 gene of C. cathayensis has its 5′-end exon situated in the LSC region and its 3′-end exons located in the IR regions (Figure 1); this result is similar to that for the congeneric species C. sinensis [34]. However, there is a certain difference with previous reports of the C. cathayensis cp genome, such as the length (160,666 bp), GC contents (36.2%), and annotated genes (86 protein-coding genes, 39 tRNA genes) of the whole cp genome [13]. The difference may be due to the geographical isolation or evolutionary differences of different plant populations from An’hui and Zhejiang Provinces, which facilitate the identification of genetic variations via sequence comparison, providing new insights into the evolutionary history of C. cathayensis.
The codon usage bias of cp genomes may be a result of selection and mutation [35]. The frequency of codon usage was estimated for the C. cathayensis cp genome in this study. We found that all genes are encoded by 26,476 codons, and the 4 most frequently used codons were AUU, AAA, GAA, and AAU; among these codons, A- and U-ending codons are common (Table S2 and Figure 3). This result is similar to the results reported in other angiosperms [6,7,24,37], and these features of codon usage preference can help to better decipher exogenous gene expression and the evolution mechanisms of the cp genome [24,25,38].
The cpSSR markers are excellent tools for phylogenetic research due to several characteristics, including non-recombination, haploidy, uniparental inheritance, and the low substitution rate [39]. They are especially valuable for intraspecific population genetic variation research [40,41] and interspecific evolutionary and identification studies [42,43,44,45,46]. A previous study reported that 213 SSRs and 44 long repeats were identified in the cp genome of C. illinoinensis [47], while 252 SSRs and 55 long repeats were identified in our study. This study found mononucleotide SSRs were the richest (occupied 78.97%), and the mononucleotide A+T repeat units occupied the highest portion (75.00%); these results are consistent with a previous study and verify the hypothesis that cpSSRs are generally composed of short polyadenine (polyA) or polythymine (polyT) repeats and rarely contain tandem guanine (G) or cytosine (C) repeats [38,48]. The cpSSRs are mainly distributed in the noncoding regions of the cp genome of C. cathayensis; a similar distribution preference of cpSSRs has been reported in other plants, such as Olea europaea, Salviamiltiorrhiza, and Avena sativa [47,49]. Dispersed repeats may facilitate intermolecular recombination and plastome diversity creation, because the genome regions with increased sequence diversity could be formed by repeat sequence abundance in prokarya and eukarya [50]. Hence, these cpSSR markers of C. cathayensis could be used to examine the genetic structure, diversity, differentiation, and maternity in Carya and provide a new avenue for the development of species protection and preservation strategies.
Phylogenetic analysis was completed on an alignment of all chloroplast genomes from 46 angiosperm species. The well-supported phylogenetic tree (Figure 4) indicates that the genus Carya is monophyletic and is most closely related to the cluster formed by another genus of Juglandaceae, which is consistent with previous studies [2,12]. The genus Quercus was polylogenetic in our analysis, resulting from the embedded branches of the genera Lithocarpus and Castanea; this result is consistent with previous results [6]. Phylogenetic relationships inferred that Juglandaceae is monophyletic, and that C. cathayensis is sister to C. kweichowensis and C. illinoinensis in our study. Previous studies reported that C. kweichowensis is one of the representative species of the Asian sect. Sinocarya, while C. illinoinensis is one of the representative species of the North American sect. Apocarya [47]. The C. cathayensis used in our study is native to China, in Asia. Thus, we speculated that the above factors led to C. cathayensis and C. kweichowensis falling into one clade, while C. cathayensis and C. illinoinensis fell into two clades.
The size variation in angiosperm plastid genomes is often accompanied by the expansion and contraction of the IR and SSC boundary regions [51,52]. It is well known that certain plastome regions show different mutation rates. To further resolve the structural evolutionary history of the cp genomes of the genus Carya, we compared the IR/SSC and IR/LSC junctions across six selected Juglandaceae species, including C. cathayensis, C. illinoinensis, C. kweichowensis, Platycarya strobilacea, Cyclocarya paliurus, and Juglans cathayensis. We observed a wide variability of the junction sites. The cp genomes of C. cathayensis exhibited similar JLB, JSB, and JSA junction sites. We observed variations in the IR/SSC and IR/LSC junction sites across other genera in the family Juglandaceae: for example, the rps19 gene was shifted by 285 bp from the LSC to IRb at the LSC/IRb border, trnL was located in the IRa/b regions instead of the SSC region, and ycf1 was absent from the JSA site (Figure 5). The LSC/IR and SSC/IR borders are relatively conserved among angiosperm plastomes, mostly positioned within rps19 or ycf1 [53]. Significant expansions have been reported in other plants, such as in Pelargonium × hortorum L.H. Bailey [54], Jasminum nudiflorum Lindl [55], and Avena sativa [49].
This study revealed a relatively higher level of divergence in the noncoding than in the coding regions, similar to what has been reported for the genus Quercus from the family Fagaceae [6], which is related to the family Juglandacea. We also identified a considerable number of variations in the noncoding cp sequences, such as trnC-GCA, trnW-CCA, trnI-CAU, and trnI-UAG, of species in the genus Carya (Figure 6). Hence, these noncoding sites may be useful for resolving the suspending phylogenetic relationships of Carya species [2]. Gene nucleotide variability (pi) values of LSC.rpl36, IR. rrn4.5, rrn23, and rrn16 were higher than 1, while the values of other genes were lower than 0.03. The results show that there is lower nucleotide diversity among the six Juglandaceae species. The results can provide reference for plastome marker selection, which should be carried out based on appropriate evolutionary rates (pi values) [49]. The plastid genome is typically conserved across most angiosperms [55]. Our results found that seven genes (rps15, rpoA, rpoB, petD, ccsA, atpI, and ycf1-2) of C. cathayensis underwent positive selection (Table S4); other genes were shown to have undergone purifying selection. These results indicate that there is selective pressure on plastid function, where genes encoding proteins for DNA maintenance underwent positive selection, and expression may be relaxed [49].
5. Conclusions
The diversification of C. cathayensis plastomes is explained by the presence of highly diverse genes, LSC intermolecular recombination, and the co-occurrence of tandem repeats. This study demonstrates that there is a wide variability of the junction sites between the cp genomes of six Juglandaceae species, and there is higher divergence in the noncoding regions than in coding regions in the cp genome of C. cathayensis. The genus Quercus was polylogenetic, resulting from the embedded branches of the genera Lithocarpus and Castanea. The characterization of the C. catayensis cp genome provides valuable genetic information for the phylogenetic study and the development of conservation strategies of the genus Carya.
Acknowledgments
The authors would like to thank Chuanbei Jiang (Genepioneer Biotechnologies Co., Ltd., Nanjing, China) for his technical assistance during the data analysis of this manuscript.
Abbreviations
LSC: large single copy; SSC: small single copy; IRs: inverted repeats; SSRs: simple sequence repeats; Ks: synonymous; Ka: nonsynonymous; pi: gene nucleotide variability; RSCU: relative synonymous codon usage.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13020369/s1, Table S1: Statistics of the synteny between the C. cathayensis and Cyclocarya paliurus cp genomes; Table S2: Codon usage of C. cathayensis cp genome from RSCU tools; Table S3: Long repeat sequences in the C. cathayensis cp genome; Table S4: Simple sequence repeats (SSR) in the C. cathayensis cp genome; Table S5: Ka/Ks ratios of the cp genes from C. cathayensis and five related species.
Author Contributions
Conceptualization, S.J. and J.S.; methodology, X.L. and J.S.; software, X.C.; validation, S.J. and J.S.; formal analysis, X.H. and X.L.; investigation, X.L. and S.J.; resources, S.J.; data curation, S.J. and J.S.; writing—original draft preparation, S.J. and J.S.; writing—review and editing, S.J. and J.S.; visualization, X.L.; supervision, S.J. and J.S.; project administration, S.J.; funding acquisition, S.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31971641), the National Key Research and Development Project (2019YFE0118900), the Zhejiang Provincial Natural Science Foundation of China (LY16C160011), and the Jiyang College of Zhejiang A&F University under grant (RQ1911B07).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data used in our study have been submitted to NCBI GenBank (accession number: MN892516). The related species and their GenBank accession numbers (website: https://www.ncbi.nlm.nih.gov/, accessed on 11 October 2021) in this study are listed as follows: Betula nana (KX703002), Castanopsis concinna (NC_033409), C. echinocarpa (NC_023801), C. hainanensis(NC_037389), Castanea henryi (NC_033881), C. mollissima (KY951992), C. pumila (KM360048), C. seguinii (NC_039749), Dalbergia hainanensis (NC_036961), Fagus crenata (NC_041252), F. engleriana (NC_036929), F. sylvatica (NC_041437), Juglans major (NC_035966), J. hindsii (NC_035965), J. cinereal (NC_035960), J. nigra (NC_035967), J. cathayensis (MF167457), J. mandshurica (MF167461), J. sigillata (MF167465), J. hopeiensis (NC_033894), J. regia (NC_028617), Lithocarpus balansae (NC_026577), Malus prunifolia (NC_031163), C. illinoinensis (NC_041449), C. kweichowensis (NC_040864), Cyclocarya paliurus (NC_034315), Platycarya strobilacea (NC_035413), Quercus acutissima (NC_039429), Q. aliena (NC_026790), Q. baronii (NC_029490), Q. chenii (NC_039428), Q. dentata (NC_039725), Q. dolicholepis (KU240010), Q. obovatifolia (NC_039972), Quercus rubra (JX970937), Q. sichourensis (NC_036941), Q. spinosa (NC_026790), Q. tarokoensis (NC_036370), Q. variabilis (KU240009), Trigonobalanus doichangensis (NC_023959), and Ulmus gaussenii (NC_037840).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lu A., Stone D., Grauke L. Juglandaceae. Flora China. 1999;4:277–285. [Google Scholar]
- 2.Zhang J.-B., Li R.-Q., Xiang X.-G., Manchester S.R., Lin L., Wang W., Wen J., Chen Z.-D. Integrated fossil and molecular data reveal the biogeographic diversification of the Eastern Asian-Eastern North American Disjunct Hickory genus (Carya Nutt.) PLoS ONE. 2013;8:e70449. doi: 10.1371/journal.pone.0070449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naumann J., Symmank L., Samain M.S., Kai F.M., Wanke S. Chasing the hare—Evaluating the phylogenetic utility of a nuclear single copy gene region at and below species level within the species rich group Peperomia (Piperaceae) BMC Evol. Biol. 2011;11:357. doi: 10.1186/1471-2148-11-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raman G., Park V., Kwak M., Lee B., Park S.J. Characterization of the complete chloroplast genome of Arabis stellari and comparisons with related species. PLoS ONE. 2017;12:e0183197. doi: 10.1371/journal.pone.0183197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Böhle U.R., Hilger H., Cerff R., Martin W. Noncoding chloroplast DNA for plant molecular systematics at the infrageneric level. In: Schierwater B., Streit G.P., Desalle R., editors. Molecular Ecology and Evolution: Approaches and Applications. Birkhäuser; Basel, Switzerland: 1994. pp. 391–403. [Google Scholar]
- 6.Li X., Li Y., Zang M., Li M., Fang Y. Complete chloroplast genome sequence and phylogenetic analysis of Quercus acutissima. Int. J. Mol. Sci. 2018;19:2443. doi: 10.3390/ijms19082443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y., Sylvester S.P., Li M., Zhang C., Li X., Duan Y., Wang X. The complete plastid genome of Magnolia zenii and genetic comparison to Magnoliaceae species. Molecules. 2019;24:261. doi: 10.3390/molecules24020261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J., Xu Y., Xi L., Yang J., Chen H., Zhang J. Characterization of the chloroplast genome sequence of Acer miaotaiense: Comparative and phylogenetic analyses. Molecules. 2018;23:1740. doi: 10.3390/molecules23071740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng S., Zhou T., Han K., Yang Y., Zhao J., Liu Z.L. The complete chloroplast genome sequences of six Rehmannia species. Genes. 2017;8:103. doi: 10.3390/genes8030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu C., Dong W., Li W., Lu Y., Xie X., Jin X., Shi J., He K., Suo Z. Comparative analysis of six Lagerstroemia complete chloroplast genomes. Front. Plant Sci. 2017;8:15. doi: 10.3389/fpls.2017.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y., Zhou T., Duan D., Yang J., Feng L., Zhao G. Comparative analysis of the complete chloroplast genomes of five Quercus species. Front. Plant Sci. 2016;7:959. doi: 10.3389/fpls.2016.00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye L., Fu C., Wang Y., Liu J., Gao L. Characterization of the complete plastid genome of a Chinese endemic species Carya kweichowensis. Mitochondrial DNA Part B. 2018;3:492–493. doi: 10.1080/23802359.2018.1464414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhai D.-C., Yao Q., Cao X.-F., Hao Q.-Q., Ma M.-T., Pan J., Bai X.-H. Complete chloroplast genome of the wild-type Hickory (Carya cathayensis) Mitochondrial DNA Part B. 2019;4:1457–1458. doi: 10.1080/23802359.2019.1598815. [DOI] [Google Scholar]
- 14.Zhang R., Peng F., Li Y. Pecan production in China. Sci. Hortic. 2015;197:719–727. doi: 10.1016/j.scienta.2015.10.035. [DOI] [Google Scholar]
- 15.Grauke L.J., Wood B.W., Harris M.K. Crop vulnerability: Carya. Hortscience. 2016;51:653–663. doi: 10.21273/HORTSCI.51.6.653. [DOI] [Google Scholar]
- 16.Jin S.H., Huang J.Q., Li X.Q., Zheng B.S., Wu J.S., Wang Z.J., Liu G.H., Chen M. Effects of potassium supply on limitations of photosynthesis by mesophyll diffusion conductance in Carya cathayensis. Tree Physiol. 2011;31:1142–1151. doi: 10.1093/treephys/tpr095. [DOI] [PubMed] [Google Scholar]
- 17.Huang Y., Xiao L., Zhang Z., Zhang R., Wang Z., Huang C., Huang R., Luan Y., Fan T., Wang J., et al. The genomes of pecan and Chinese hickory provide insights into Carya evolution and nut nutrition. Gigascience. 2019;8:giz036. doi: 10.1093/gigascience/giz036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doyle J.J., Doyle J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- 19.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bankevich A., Nurk S., Antipov D., Gurevich A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu C., Shi L., Zhu Y., Chen H., Zhang J., Lin X., Guan X. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 2012;13:715. doi: 10.1186/1471-2164-13-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurt S. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2011;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurtz S., Schleiermacher C. REPuter: Fast computation of maximal repeats in complete genomes. Bioinformatics. 1999;15:426–427. doi: 10.1093/bioinformatics/15.5.426. [DOI] [PubMed] [Google Scholar]
- 24.Liu H.Y., Yu Y., Deng Y.Q., Li J., Huang Z.X., Zhou S.D. The chloroplast genome of Lilium henrici: Genome structure and comparative analysis. Molecules. 2018;23:1276. doi: 10.3390/molecules23061276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L., Wang Y., He P., Li P., Lee J., Soltis D.E., Fu C. Chloroplast genome analyses and genomic resource development for epilithic sister genera Oresitrophe and Mukdenia (Saxifragaceae), using genome skimming data. BMC Genomics. 2018;19:235. doi: 10.1186/s12864-018-4633-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beier S., Thiel T., Münch T., Scholz U., Mascher M. MISA-web: A web server for microsatellite prediction. Bioinformatics. 2017;33:2583–2585. doi: 10.1093/bioinformatics/btx198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou L.H., Huang J.X., Zhang G.Q., Liu Z.J., Zhuang X.Y. A molecular phylogeny of Aeridinae (Orchidaceae: Epidendroideae) inferred from multiple nuclear and chloroplast regions. Mol. Phylogenet. Evol. 2015;85:247–254. doi: 10.1016/j.ympev.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 28.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuraku S., Zmasek C.M., Nishimura O., Katoh K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013;41:W22–W28. doi: 10.1093/nar/gkt389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amiryousefi A., Hyvönen J., Poczai P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018;34:3030–3031. doi: 10.1093/bioinformatics/bty220. [DOI] [PubMed] [Google Scholar]
- 31.Mayor C., Brudno M., Schwartz J.R., Poliakov A., Rubin E.M., Frazer K., Pachter L.S., Dubchak I. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- 32.Jansen R.K., Raubeson L.A., Boore J.L., Depamphilis C.W., Chumley T.W., Haberle R.C., Wyman S.K., Alverson A.J., Peery R., Herman S.J., et al. Methods for obtaining and analyzing whole chloroplast genome sequences. Method Enzymol. 2005;395:348. doi: 10.1016/S0076-6879(05)95020-9. [DOI] [PubMed] [Google Scholar]
- 33.Daniell H., Lin C.S., Yu M., Chang W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu Y., Chen X., Feng X., Woeste K.E., Zhao P. Characterization of the complete chloroplast genome of the endangered species Carya sinensis (Juglandaceae). Conserv. Genet. Resour. 2016;8:467–470. doi: 10.1007/s12686-016-0601-4. [DOI] [Google Scholar]
- 35.Morton B.R. The role of context-dependent mutations in generating compositional and codon usage bias in grass chloroplast DNA. J. Mol. Evol. 2003;56:616–629. doi: 10.1007/s00239-002-2430-1. [DOI] [PubMed] [Google Scholar]
- 36.Necsulea A., Lobry J. A new method for assessing the effect of replication on DNA base composition asymmetry. Mol. Biol. Evol. 2007;24:2169–2179. doi: 10.1093/molbev/msm148. [DOI] [PubMed] [Google Scholar]
- 37.Jian H.-Y., Zhang Y.-H., Yan H.-J., Qiu X.-Q., Wang Q.-G., Li S.-B., Zhang S.-D. The complete chloroplast genome of a key ancestor of modern Roses, Rosa chinensis var. spontanea, and a comparison with congeneric species. Molecules. 2018;23:389. doi: 10.3390/molecules23020389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen X., Wu M., Liao B., Liu Z., Bai R., Xiao S., Li X., Zhang B., Xu J., Chen S. Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules. 2017;22:1330. doi: 10.3390/molecules22081330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ebert D., Peakall R. Chloroplast simple sequence repeats (cpSSRs): Technical resources and recommendations for expanding cpSSR discovery and applications to a wide array of plant species. Mol. Ecol. Resour. 2009;9:673–690. doi: 10.1111/j.1755-0998.2008.02319.x. [DOI] [PubMed] [Google Scholar]
- 40.Provan J., Powell W., Hollingsworth P.M. Chloroplast microsatellites: New tools for studies in plant ecology and evolution. Trends Ecol. Evol. 2011;16:142–147. doi: 10.1016/S0169-5347(00)02097-8. [DOI] [PubMed] [Google Scholar]
- 41.Diekmann K., Hodkinson T.R., Barth S. New chloroplast microsatellite markers suitable for assessing genetic diversity of Lolium perenne and other related grass species. Ann. Bot. 2012;110:1327–1339. doi: 10.1093/aob/mcs044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh N., Pal A.K., Roy R.K., Tamta S., Rana T.S. Development of cpSSR markers for analysis of genetic diversity in Gladiolus cultivars. Plant Gene. 2017;10:31–36. doi: 10.1016/j.plgene.2017.05.003. [DOI] [Google Scholar]
- 43.Hu J.B., Li J.W., Zhou X.Y. Analysis of cytoplasmic variation in a cucumber germplasm collection using chloroplast microsatellite markers. Acta Physiol. Plant. 2009;31:1085–1089. doi: 10.1007/s11738-009-0325-0. [DOI] [Google Scholar]
- 44.Deng Q., Zhang H., He Y., Wang T., Sun Y. Chloroplast microsatellite markers for Pseudotaxus chienii developed from the whole chloroplast genome of Taxus chinensis var. Mairei (Taxaceae) Appl. Plant Sci. 2017;5:1600153. doi: 10.3732/apps.1600153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pan L., Li Y., Guo R., Wu H., Hu Z., Chen C. Development of 12 chloroplast microsatellite markers in Vigna unguiculata (Fabaceae) and amplification in Phaseolus vulgaris. Appl. Plant Sci. 2014;2:1300075. doi: 10.3732/apps.1300075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang J., Yang X., Zhang C., Yin X., Liu S., Li X. Development of chloroplast microsatellite markers and analysis of chloroplast diversity in Chinese Jujube (Ziziphus jujuba Mill.) and Wild Jujube (Ziziphus acidojujuba Mill.) PLoS ONE. 2015;10:e0134519. doi: 10.1371/journal.pone.0134519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mo Z., Lou W., Chen Y., Jia X., Zhai M., Guo Z., Xuan J. The chloroplast genome of Carya illinoinensis: Genome structure, adaptive evolution, and phylogenetic analysis. Forests. 2020;11:207. doi: 10.3390/f11020207. [DOI] [Google Scholar]
- 48.Wang L., Wuyun T.N., Du H., Wang D., Cao D. Complete chloroplast genome sequences of Eucommia ulmoides: Genome structure and evolution. Tree Genet. Genomes. 2016;12:12. doi: 10.1007/s11295-016-0970-6. [DOI] [Google Scholar]
- 49.Liu Q., Li X., Li M., Xu W., Heslop-Harrison J.S. Comparative chloroplast genome analyses of avena: Insights into evolutionary dynamics and phylogeny. BMC Plant Biol. 2020;20:406. doi: 10.1186/s12870-020-02621-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McDonald M.J., Wang W.C., Huang H.D., Leu J.Y. Clusters of nucleotide substitutions and insertion/deletion mutations are associated with repeat sequences. PLoS Biol. 2011;9:e1000622. doi: 10.1371/journal.pbio.1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dugas D., Hernandez D., Koenen E., Schwarz E., Straub S., Hughes C.E., Jansen R.K., Nageswara-Rao M., Staats M., Trujillo J.T., et al. Mimosoid legume plastome evolution: IR expansion, tandem repeat expansions, and accelerated rate of evolution in clpP. Sci. Rep. 2015;5:16958. doi: 10.1038/srep16958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Drescher A., Stephanie R., Calsa T., Carrer H., Bock R. The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J. 2000;22:97–104. doi: 10.1046/j.1365-313x.2000.00722.x. [DOI] [PubMed] [Google Scholar]
- 53.Downie S.R., Jansen R.K. A comparative analysis of whole plastid genomes from the Apiales: Expansion and contraction of the inverted repeat, mitochondrial to plastid transfer of DNA, and identification of highly divergent noncoding regions. Syst. Bot. 2015;40:336–351. doi: 10.1600/036364415X686620. [DOI] [Google Scholar]
- 54.Chumley T.W., Palmer J.D., Mower J.P., Fourcade H.M., Calie P.J., Boore J.L., Jansen R.K. The complete chloroplast genome sequence of Pelargonium × hortorum: Organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 2006;23:2175–2190. doi: 10.1093/molbev/msl089. [DOI] [PubMed] [Google Scholar]
- 55.Lee H.L., Jansen R.K., Chumley T.W., Kim K.J. Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol. Biol. Evol. 2007;24:1161–1180. doi: 10.1093/molbev/msm036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in our study have been submitted to NCBI GenBank (accession number: MN892516). The related species and their GenBank accession numbers (website: https://www.ncbi.nlm.nih.gov/, accessed on 11 October 2021) in this study are listed as follows: Betula nana (KX703002), Castanopsis concinna (NC_033409), C. echinocarpa (NC_023801), C. hainanensis(NC_037389), Castanea henryi (NC_033881), C. mollissima (KY951992), C. pumila (KM360048), C. seguinii (NC_039749), Dalbergia hainanensis (NC_036961), Fagus crenata (NC_041252), F. engleriana (NC_036929), F. sylvatica (NC_041437), Juglans major (NC_035966), J. hindsii (NC_035965), J. cinereal (NC_035960), J. nigra (NC_035967), J. cathayensis (MF167457), J. mandshurica (MF167461), J. sigillata (MF167465), J. hopeiensis (NC_033894), J. regia (NC_028617), Lithocarpus balansae (NC_026577), Malus prunifolia (NC_031163), C. illinoinensis (NC_041449), C. kweichowensis (NC_040864), Cyclocarya paliurus (NC_034315), Platycarya strobilacea (NC_035413), Quercus acutissima (NC_039429), Q. aliena (NC_026790), Q. baronii (NC_029490), Q. chenii (NC_039428), Q. dentata (NC_039725), Q. dolicholepis (KU240010), Q. obovatifolia (NC_039972), Quercus rubra (JX970937), Q. sichourensis (NC_036941), Q. spinosa (NC_026790), Q. tarokoensis (NC_036370), Q. variabilis (KU240009), Trigonobalanus doichangensis (NC_023959), and Ulmus gaussenii (NC_037840).