Abstract
Genital human papillomaviruses (HPVs) are commonly detected from clinical samples by consensus PCR methods. Two commonly used primer systems, the MY09-MY11 (MY09/11) primers and the GP5+-GP6+ (GP5+/6+) primers, amplify a broad spectrum of HPV genotypes, but with various levels of sensitivity among the HPV types. Analysis of the primer-target sequence homology for the MY09/11 primers showed an association between inefficient amplification of HPV types and the number and position of mismatches, despite accommodation of sequence variation by inclusion of degenerate base sites. The MY09/11 primers were redesigned to increase the sensitivity of amplification across the type spectrum by using the same primer binding regions in the L1 open reading frame. Sequence heterogeneity was accommodated by designing multiple primer sequences that were combined into an upstream pool of 5 oligonucleotides (PGMY11) and a downstream pool of 13 oligonucleotides (PGMY09), thereby avoiding use of degenerate bases that yield irreproducible primer syntheses. The performance of the PGMY09-PGMY11 (PGMY09/11) primer system relative to that of the standard MY09/11 system was evaluated with a set of 262 cervicovaginal lavage specimens. There was a 91.5% overall agreement between the two systems (kappa = 0.83; P < 0.001). The PGMY09/11 system appeared to be significantly more sensitive than the MY09/11 system, detecting an additional 20 HPV-positive specimens, for a prevalence of 62.8% versus a prevalence of 55.1% with the MY09/11 system (McNemar's χ2 = 17.2; P < 0.001). The proportion of multiple infections detected increased with the PGMY09/11 system (40.0 versus 33.8% of positive infections). HPV types 26, 35, 42, 45, 52, 54, 55, 59, 66, 73, and MM7 were detected at least 25% more often with the PGMY09/11 system. The PGMY09/11 primer system affords an increase in type-specific amplification sensitivity over that of the standard MY09/11 primer system. This new primer system will be useful in assessing the natural history of HPV infections, particularly when the analysis requires HPV typing.
L1 consensus primer PCR systems, particularly the MY09-MY11 (MY09/11) and GP5+-GP6+ (GP5+/6+) primer systems (1, 4, 9, 13), have been widely used to study the natural history of human papillomaviruses (HPVs) and their role in the development of genital cancer, particularly of the uterine cervix (8, 10, 18). The MY09/11 HPV DNA detection system was used to show convincingly for the first time that the determinants of infection with HPV were the same as those for cervical cancer, namely, the sexual behavior variables such as increased number of lifetime sexual partners (11). Furthermore, both consensus primer methods have been used in a number of important studies that show unequivocally the associated risk of infection with certain types of HPV with the development of cervical cancer (12, 15). The sensitivities of these methods and their ability to amplify and detect greater than 25 of the HPV genotypes known to infect the genital mucosa have provided researchers with an extremely valuable tool which has been considered a “gold standard” for HPV detection for the last several years. However, despite the progress toward the understanding of HPV-associated disease facilitated by the use of these consensus primer systems, limitations are still evident, particularly in regard to the variability of detection sensitivity among specific HPV types (17).
At the time that the MY09/11 primer system was designed, only 5 of the 20 or more known genital HPV genotype sequences had been reported; specifically, HPV types 6, 11, 16, 18, and 33 (13). The primers were thus designed in a conserved region of the L1 open reading frame with the intent of amplifying in a single reaction both the five genotypes whose sequences are known and, presumptively, other genital HPVs with shared sequence homology in this region. The chosen regions were not entirely homologous even among the five original HPV types, and positions with nucleotide base heterogeneity were accommodated by inclusion of degenerate base sites. The resultant degenerate primers comprised a mixture of 24 unique oligonucleotide sequences. Over the next decade studies with these primers for amplification and detection of HPV from genital samples demonstrated the ability of the primers to amplify a spectrum of more than 30 genital HPV types, albeit with various levels of sensitivity (2). Only a single modification to the original primer set was made, wherein an extra, sequence-specific oligonucleotide (HMB01) directed to the minus strand of HPV type 51 (HPV-51) was included to facilitate the amplification of this important, cancer-associated type of HPV (7). The MY09/11 system referred to in this paper is inclusive of the HMB01 primer.
The nature of the synthesis of a mixture of oligonucleotides with degenerate base sequences relies on the presumed random addition of one of two or more nucleotide bases at the position of degeneracy. The random insertion of bases at degenerate positions is not a controlled process, such that an equal proportion of each sequence combination cannot be guaranteed. Furthermore, no analytical method for the verification of sequence proportions was readily available for quality control purposes, so that functional testing with each HPV type as a template was required to ensure the comparability of different lots of primers. Results from our own laboratories (P. E. Gravitt and F. Coutlée, unpublished data) indicate differences in type-specific amplification efficiencies among separate syntheses of the MY09/11 degenerate primers (data not shown).
We sought to improve the reproducibility and sensitivity of the MY09/11 HPV amplification system by developing a set of oligonucleotide pools, PGMY09 and PGMY11, based on the same primer binding regions used for MY09/11. Rather than using the degenerate primer method, we grouped virus types together by sequence homology in each of the two primer binding regions. From these groupings, we designed a set of 5 upstream oligonucleotides comprising the PGMY11 primer pool and a set of 13 downstream oligonucleotides comprising the PGMY09 primer pool (PGMY09/11 primer system). These primers were used in amplification reactions similar to the standard MY09/11 PCR protocols, and we continued to coamplify HPV with the internal β-globin control using the primer pair PC04 and GH20. We compared the performance of this new set of L1 consensus primers to that of the standard MY09/11 system for the amplification and detection of HPV from cervical cell samples.
MATERIALS AND METHODS
HPV sequence alignment and primer design.
The L1 regions of all sequenced HPV genotypes were obtained through the Los Alamos National Laboratories HPV Database (http://hpv-web.lanl.gov/) and were aligned by using the Wisconsin Package (Genetics Computer Group, Madison, Wis.). The MY09/11 primer binding regions of each of these sequences were sorted into groups according to 3′ DNA sequence homology. The DNA sequence mismatches remaining within selected HPVs were chosen according to their stability (16) and were kept near the 5′ end of the oligonucleotide. Dissociation temperatures and duplex formation of each primer sequence were determined by using Oligo 5.0 (Molecular Biologic Insights, Inc., Cascade, Colo.). The criteria for redesigning the primers were as follows. The same primer binding regions in the target HPV types were used so that the same detection and genotyping methods could be retained. The broad-spectrum amplification that defines consensus PCR was accomplished with pools of oligonucleotides rather than the former addition of degenerate base sites in the MY09/11 primer sequences. The number of oligonucleotides for each primer pool (upstream and downstream primers) was kept to a minimum, such that the maximum numbers of HPV types were matched with a single primer.
Sample acquisition.
Cervicovaginal lavage specimens (10 ml) were collected as part of a large natural history study of HPV infection at Kaiser Permanente in Portland, Oreg. (18). A total of 1,421 cytologically normal women were seen twice during the enrollment period, and two specimens were collected at different visits for HPV testing. This convenience sample of multiply sampled women was included in a study of persistence of HPV infection. HPV testing was performed with all 1,421 specimens from the first visit by using MY09/11 consensus primers (18). Two hundred sixty-two women were positive for HPV DNA by L1 consensus PCR (MY09/11) at the time of study enrollment (i.e., at the first visit). The second specimens from these 262 women comprised the sample set for the present analysis. No clinical interventions were taken between the first and second samplings. This convenience sample set was selected on the basis of the assumption that at the second sampling point many of these women would still have detectable HPV DNA at the cervix, some would have cleared their infection and would be HPV negative, and others would have acquired a new HPV infection in the interim between the first and second samplings. This maximized the probability of a high HPV prevalence useful for meaningful HPV assay comparisons (i.e., expected 50% persistence or acquisition rate between the sampling time points, yielding approximately equal numbers of HPV-positive and -negative specimens). This second specimen was tested by PCR with both MY09/11 and PGMY09/11, and the results from each assay were blinded to the operators performing the analyses. All participating women gave informed consent.
Sample preparation.
The cervicovaginal lavage specimens were prepared for PCR by standard protocols (1, 18). In brief, each lavage specimen was digested for 1 h at 65°C in the presence of 200 μg of proteinase K per ml and 1% Laureth-12. The samples were spun briefly at maximum speed in an Eppendorf microcentrifuge to remove all condensation from the cap of the Eppendorf tube and were heated to 95°C for 10 min to heat denature the residual protease. The samples were centrifuged again briefly, and 5 μl was used for each PCR assay.
Standard MY09/11 consensus PCR.
The protocol used as the gold standard to evaluate the new system was performed as described previously (6). Each sample was amplified with 5′ biotinylated MY09/11 (50 pmol of each primer) and HMB01, GH20, and PC04 (5 pmol of each primer) in the presence of 1× PCR Buffer II, 6 mM MgCl2, 200 μmol (each) dATP, dCTP, and dGTP, 600 μmol dUTP, and 7.5 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer, Foster City, Calif.). Amplifications were performed in a Perkin-Elmer TC9600 thermal cycler by using the ultrasensitive profile of AmpliTaq Gold activation at 95°C for 9 min and 40 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min. This was followed by a final extension at 72°C for 5 min, and the amplification reaction mixtures were stored at 4 to 15°C.
PGMY09/11 L1 consensus PCR.
The protocol for the L1 consensus PCR assay was optimized for the new primer pools PGMY09 and PGMY11. The MY09/11 primer set was replaced with 5′ biotinylated PGMY09 and PGMY11. An equimolar mixture of each primer was added to the PCR master mixture for a final concentration of 10 pmol of each oligonucleotide in the primer sets. The β-globin primers GH20 and PC04 were also biotinylated, and the concentration of each of these was reduced from 5 pmol per PCR, as in the standard L1 protocol, to 2.5 pmol per PCR mixture in the revised L1 protocol. Also, the final concentration of MgCl2 in the PCR mixture was reoptimized to a final concentration of 4 mM (reduced from 6 mM in the standard protocol). Otherwise, the PCR buffers, reagents, and amplification profiles were identical to those described above.
HPV genotyping.
The PCR products from both the standard and revised L1 consensus PCR assays are amenable to genotype discrimination by the recently described HPV immobilized probe assay (6). The protocol used for detection of products from both assays was performed as described previously (6), in which the PCR products were denatured in 0.4 N NaOH and were hybridized to an immobilized HPV probe array, with positive hybridization detected by streptavidin-horseradish peroxidase-mediated color precipitation at the probe site.
Statistical analysis.
Statistical analyses were performed with STATA 6.0 software (STATA, College Station, Tex.). Kappa statistics were calculated to measure the agreement between the primer systems beyond that expected by chance (5). Significance testing for the unequal distribution of discordant results was performed by McNemar's chi-square test for matched pair data when comparing dichotomous outcomes (3) and the Stuart-Maxwell test for marginal homogeneity when comparing multiple categorical outcomes (14).
RESULTS
In addition to the irreproducibility of the MY09/11 primer synthesis, amplification efficiency has been shown to vary systematically among the HPV genotypes when known target quantities of the genotype are analyzed and when amplification with the MY09/11 primer system is compared to that with another consensus PCR system (17). Analysis of the alignment of the MY09/11 primer binding regions for 19 of the 23 sequenced genital HPV genotypes (Table 1) revealed more destabilizing mismatches for the genotypes shown in our laboratories and others to amplify with poor efficiency (e.g., HPV types 26, 52, and 55) relative to the number of mismatches for the HPV types that amplified well (e.g., HPV types 16, 18, and 33). The efficiency of amplification appeared to be related to the number, position, and stability of the mismatch (data not shown). As expected, primers with greater than four mismatches to the target sequence tended to be less efficient (e.g., HPV types 42 [MY09], 26 [MY09], and 59 [MY11]). Primers with less than four mismatches overall but with one or more mismatches at the 3′ end of the oligonucleotide also tended to segregate with the less efficiently amplified HPVs (e.g., types HPV 39 [MY09], 45 [MY09], and 55 [MY09]).
TABLE 1.
MY09/11 sequence alignmentsa
| Sequence name and HPV type |
Sequence (5′-3′) |
|---|---|
| MY09 | CGT CCM ARR GGA WAC TGA TC |
| HPV-6 | ... ... ... ... ... ... .. |
| HPV-11 | ... ... ... ... ... ... .. |
| HPV-16 | ... ..T ... ... ... ... .. |
| HPV-18 | ... ... ... ... ..T ... .. |
| HPV-26 | ..C ..T ..T ... ..T ... .. |
| HPV-31 | ..A ... ..T ... ... ... .. |
| HPV-33 | ... ... ... ... ... ... .. |
| HPV-35 | ..G ... ..C ... ... ... .. |
| HPV-39 | ... ... ... ..G ..T ... .. |
| HPV-40 | ... ..T ..T ... ..T ... .. |
| HPV-42 | .TA ..T ... ... ..T ... .. |
| HPV-45 | ..A ... ... ... ..T ... .. |
| HPV-52 | .TA ..T ... ... ... ... .. |
| HPV-53 | .TG ... ... ... ... ... .. |
| HPV-55 | .TA ... ... ... ..T ... .. |
| HPV-56 | .TA ... ..T ... ..T ... .. |
| HPV-58 | ... ... ... ... ... ... .. |
| HPV-59 | ... ... ... ... ... ... .. |
| MY11 | GCM CAG GGW CAT AAY AAT GG |
| HPV-6 | ... ... ... ... ... ... .. |
| HPV-11 | ..T ... ... ... ... ... .. |
| HPV-16 | ... ... ..C ..C ... ... .. |
| HPV-18 | ... ... ... ... ... ... .. |
| HPV-26 | ... ... ... ... ... ... .. |
| HPV-31 | ..T ... ... ..C ... ... .. |
| HPV-33 | ... ..A ... ... ... ... .. |
| HPV-35 | ... ..S ..C ... ... ... .. |
| HPV-39 | ... ... ..C ..C ... ... .. |
| HPV-40 | ... ... ..C ... ... ... .. |
| HPV-42 | ... ..A ... ..C ... ... .. |
| HPV-45 | ... ... ..C ... ... ... .. |
| HPV-52 | ..G ... ..C ..C ... ... .. |
| HPV-53 | ... ... ... ... ... ... .. |
| HPV-55 | ..G ... ..C ..C ... ... .. |
| HPV-56 | ... ..A ..C ... ... ... .. |
| HPV-58 | ... ..A ... ... ... ... .. |
| HPV-59 | ..T ... ... TTA ... ... .. |
The primer sequences are in boldface type with the corresponding HPV sequence alignments underneath. Nucleotide homology is indicated with a period, and mismatches are indicated with the nucleotide change in the corresponding sequence. The degenerate base code is as follows: M = A or C, W = A or T, Y = C or T, and R = A or G.
The MY09/11 consensus primers were redesigned in an attempt to correct both the irreproducibility of the degenerate primer synthesis and to increase the sensitivity of amplification to a 10-copy endpoint for each of the HPV genotypes commonly found in the genital tract. The primer sequences resulting from this analysis are shown in Table 2. The upstream primer pool, designated PGMY11, contains five oligonucleotide primers. The downstream primer pool, designated PGMY09, contains a total of 13 oligonucleotide primers. The amplification parameters were reoptimized in the presence of the new primer pools. The overall increase in stability of the new primers to their target sequences required a reduction in the total final MgCl2 concentration to 4 mM. This primer pool accommodates the efficient amplification of the following HPV genotypes to at least a sensitivity of 10 genomes per PCR: 6, 11, 16, 18, 26, 31, 33, 35, 40, 45, 51, 52, 56, and 59 (data not shown). Several other HPV genotypes, including HPV types 39, 42, 53, 54, 55, 58, 61, 62, 64, 66, 67, 68, 69, 70, 71, 72, 73, IS39, CP8304, CP6108, MM4, MM7, and MM8, were amplified as well as or better with PGM09/11 than with MY09/11, as determined from comparison of endpoint dilution amplifications (data not shown).
TABLE 2.
PGMY primer sequences
| Primer designation | Primer sequence (5′-3′) |
|---|---|
| PGMY11-A | GCA CAG GGA CAT AAC AAT GG |
| PGMY11-B | GCG CAG GGC CAC AAT AAT GG |
| PGMY11-C | GCA CAG GGA CAT AAT AAT GG |
| PGMY11-D | GCC CAG GGC CAC AAC AAT GG |
| PGMY11-E | GCT CAG GGT TTA AAC AAT GG |
| PGMY09-F | CGT CCC AAA GGA AAC TGA TC |
| PGMY09-G | CGA CCT AAA GGA AAC TGA TC |
| PGMY09-H | CGT CCA AAA GGA AAC TGA TC |
| PGMY09-Ia | G CCA AGG GGA AAC TGA TC |
| PGMY09-J | CGT CCC AAA GGA TAC TGA TC |
| PGMY09-K | CGT CCA AGG GGA TAC TGA TC |
| PGMY09-L | CGA CCT AAA GGG AAT TGA TC |
| PGMY09-M | CGA CCT AGT GGA AAT TGA TC |
| PGMY09-N | CGA CCA AGG GGA TAT TGA TC |
| PGMY09-Pa | G CCC AAC GGA AAC TGA TC |
| PGMY09-Q | CGA CCC AAG GGA AAC TGG TC |
| PGMY09-R | CGT CCT AAA GGA AAC TGG TC |
| HMB01b | GCG ACC CAA TGC AAA TTG GT |
PGMY09-I and PGMY09-P are 18 bp in length. The first two 5′ bases were deleted to reduce the significant internal secondary structure of the oligonucleotide.
HMB01 is shifted 3′ from the downstream primer region of the other HPV genotypes to avoid secondary structure formation and internal priming.
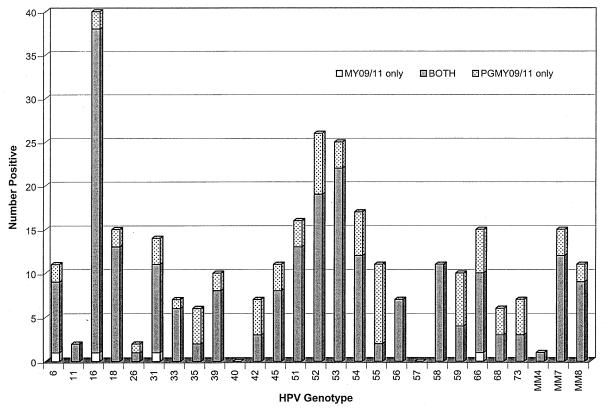
To verify the results of the analytic analyses, we conducted a parallel comparison of the MY09/11 and PGMY09/11 amplification systems with 262 cervical specimens. Of the 262 cervical specimens, a total of 15 were excluded from further analysis due to poor or no β-globin amplification, indicating either a lack of sufficient cellular material for PCR or the presence of polymerase inhibitors. Thirteen of these samples were negative for β-globin amplification by both the PGMY09/11 and the MY09/11 amplification systems, while two samples were excluded because of a lack of β-globin amplification by the PGMY09/11 system only. The general summary results for HPV prevalence for the remaining 247 samples are presented in Table 3. The overall percent agreement between the two methods was 91.5%, with a kappa value of 0.83 (P < 0.001). There was an increase in overall HPV prevalence with the PGMY09/11 system relative to that with the MY09/11 system (62.8 and 55.1%, respectively). Of the 21 samples with discordant HPV results, 20 were positive with the PGMY09/11 system only and 1 was positive with the MY09/11 system only (McNemar's χ2 = 17.19; P < 0.001). The additional positive specimens detected by the PGMY09/11 system comprised 17 samples with single infections with HPV types 16, 18 (2 samples), 35, 42, 51, 52, 54 (2 samples), 55, 59, 66 (four samples), MM7, and MM8 and 3 samples with multiple infections containing HPV type 51 and 42, HPV types 31, 54, and 66, and HPV types 33, 45, and 6. The one sample called positive only with the MY09/11 system contained HPV-31. The most notable differences between the two primer systems were seen when the abilities of the two systems to detect specific types as part of multiple infections were compared. The overall proportion of multiple infections detected with the MY09/11 primer system was 46 of 136 (33.8%), whereas that with the PGMY09/11 primer system was 62 of 155 (40%). A summary of the type-specific positive results is presented in Fig. 1. Figure 1 demonstrates graphically the absolute increase in the rate of detection of specific HPV types. The following cancer-associated HPV types were detected at least 25% more often with the PGMY09/11 primer system: HPV types 26, 35, 45, 52, 55, 59, 68, 73, and MM7. The following non-cancer-associated types were also detected at least 25% more often with the PGMY09/11 primer system: HPV types 42, 54, and 66. Table 4 shows a comparison of HPV detection results when categorized by cancer risk group as HPV negative, high-risk type positive (positive for at least one of the following HPV types: 16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 55, 56, 58, 59, 68, 73, MM4, and MM7), or low-risk type positive (positive for at least one of the following HPV types without concomitant coinfection with high-risk HPV types: HPV types 6, 11, 40, 42, 53, 54, 57, 66, and MM8). The agreement between the PGMY09/11 and MY09/11 amplification systems by risk group is 88.7% (kappa = 0.81; P < 0.001). The PGMY09/11 primers were more likely to reclassify samples into a higher risk group category compared with the risk assignment based on HPV typing with the MY09/11 primers (Stuart-Maxwell χ2 = 20.45; P < 0.001).
TABLE 3.
Overall agreement in results for HPV with MY09/11 and PGMY09/11 for 247 cervicovaginal lavage specimensa
| PGMY09/11 result | No. of specimens with the following result with MY09/11:
|
||
|---|---|---|---|
| HPV positive | HPV negative | Total | |
| HPV positive | 135 | 20 | 155 |
| HPV negative | 1 | 91 | 92 |
| Total | 136 | 111 | 247 |
Percent agreement = 91.5%; kappa = 0.83 (P < 0.001); McNemar's χ2 = 17.19 (P < 0.001).
FIG. 1.
HPV type-specific positive results with PGMY09/11 and MY09/11. The total number of positive results by HPV type are plotted: open bar, detected with MY09/11 only; shaded bar, detected with both primer sets; stippled bar, detected with PGMY09/11 only. The type-specific results include positive results for HPV types from specimens infected with both single and multiple HPV types.
TABLE 4.
Agreement in HPV risk group assignment with MY09/11 and PGMY09/11 for 247 cervicovaginal lavage specimensa
| PGMY09/11 result | No. of specimens with the following result with MY09/11:
|
|||
|---|---|---|---|---|
| HPV negative | Low-risk type positive | High-risk type positive | Total | |
| HPV negative | 91 | 0 | 1 | 92 |
| Low-risk positive | 8 | 24 | 0 | 32 |
| High-risk positive | 12 | 7 | 104 | 123 |
| TOTAL | 111 | 31 | 105 | 247 |
Samples were hierarchically assigned to risk groups as follows: HPV negative if negative for any HPV genotype, HPV low-risk type positive if positive for one or more of the low-risk HPV genotypes (HPV type 6, 11, 40, 42, 53, 54, 57, 66, or MM8) without concomitant infection with a high-risk genotype (HPV type 16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 55, 56, 58, 59, 68, 73, MM4, or MM7), and HPV high-risk type positive if positive for at least one high-risk HPV genotype. Percent agreement = 88.7%; kappa = 0.81 (P < 0.001); Stuart-Maxwell χ2 = 20.45 (P < 0.001).
DISCUSSION
Overall, the MY09/11 L1 consensus primer system was improved, both practically, in terms of the elimination of the need for degenerate primer synthesis, and functionally, as demonstrated by the increased sensitivity of amplification across the genital HPV type spectrum. Each oligonucleotide comprising the PGMY09/11 pools is synthesized independently, allowing verification of the sequence of each primer. Also, the concentration of each primer can be ascertained and consistent proportions of primer in the PGMY09/11 pools can thus be maintained. This represents an important improvement in the quality assurance for the HPV consensus primers that was absent for the degenerate primer system. The sensitivity of type-specific amplification, particularly from samples infected with multiple HPV types, was substantially improved. The HPV types that were most affected in the clinical validation study were consistent with the prediction based on the mismatch analysis with target regions and MY09/11. There was a general increase in type-specific sensitivity with the PGMY09/11 system that was independent of the targeted type-specific improvements. It is not clear whether this was due to an overall increase in sensitivity due to the redesigned HPV primers or to less competition from β-globin product amplification. The β-globin primer concentration was reduced by one-half from that in the MY09/11 protocol in response to our observations that the coamplification of β-globin reduced the endpoint sensitivity of HPV detection for several HPV genotypes. It is clear from the plasmid amplifications that the major improvements to the types most affected by use of PGMY09/11 were not attributable to differences in β-globin primer concentrations.
The degree of the effect with the PGMY09/11 primer system is proportional to the amount of virus in the sample (as determined by plasmid titrations [data not shown]), reflective of the differences in sensitivities between the two primer systems. We have subsequently analyzed a set of HPV-containing swabs from women who were diagnosed by cytological analysis with low-grade squamous intraepithelial lesions and in whom the viral copy number is likely to be quite high, and we have seen a less dramatic increase in the additional number of specimens with type-specific positive results (data not shown). However, the types most affected were consistent with those presented here. Thus, the magnitude of the absolute differences in HPV type-specific results will vary depending on the viral burden of the samples being tested.
Because the increase in HPV type-specific detection with the PGMY09/11 primers is largely restricted to samples that are positive for multiple HPV types, we have considered the possibility that our results may be due to cross-reactivity among the HPV genotypes rather than true additional type-specific infections. Several characteristics of the system make this an unlikely explanation for the increase in type-specific prevalence observed in the present study. First, the only substantive difference between the PGMY09/11 and the MY09/11 assays is the primer sequence change. Both primer systems are designed to nondiscriminately amplify any genital HPV type present in the reaction mixture (essentially favoring primer-target cross-reactivity by design). Type-specific differences attributable to cross-reactivity would therefore be a consequence of probe cross-hybridization at the genotyping level. In this comparison, the detection system used for genotyping of the PGMY09/11 and MY09/11 amplification products was identical. There is a chance that a general increase in sensitivity with the PGMY09/11 primers could increase the total amount of product generated by PCR, which could in turn increase the rate of occurrence of false-positive signals due to cross-reactivity (as a function of total DNA concentration). However, such a result would show characteristic patterns of multiple infections (e.g., all strongly HPV-16-positive samples would be consistently coinfected with HPV-31), and we see no consistent patterns in our multiple infections. In addition, hybridization of amplification products from >106 input targets has shown no such cross-reactivity among the genotypes. Finally, the increase in type-specific detection is highly correlated with the types expected to be most affected by sequence analysis and analytic studies. We do not, therefore, attribute the improvement seen with the PGMY09/11 primer system to a nonspecific HPV cross-reactivity phenomenon.
Although the gross difference in HPV prevalence is only incremental, the overall increase in the type-specific positivity could be important in some natural history studies of HPV, particularly in terms of gaining a better understanding of viral persistence and host responses. Use of the PGMY09/11 system may offer a relatively simple change in current L1 consensus primer technology that will help to minimize the type-specific misclassification known to affect the standard HPV broad-spectrum assays.
REFERENCES
- 1.Bauer H M, Greer C E, Manos M M. Determination of genital human papillomavirus infection using consensus PCR. In: Herrington C S, McGee J O D, editors. Diagnostic molecular pathology: a practical approach. Oxford, United Kingdom: Oxford University Press; 1992. pp. 132–152. [Google Scholar]
- 2.Bernard H U, Chan S Y, Manos M M, Ong C K, Villa L L, Delius H, Peyton C L, Bauer H M, Wheeler C M. Identification and assessment of known and novel human papillomaviruses by polymerase chain reaction amplification, restriction fragment length polymorphisms, nucleotide sequence, and phylogenetic algorithms. J Infect Dis. 1994;170:1077–1085. doi: 10.1093/infdis/170.5.1077. [DOI] [PubMed] [Google Scholar]
- 3.Breslow N E, Day N E. Statistical methods in cancer research. Vol. 1. 1980. Classical methods of analysis of matched data; pp. 164–166. The analysis of case control studies. IARC Scientific Publications No. 32. International Agency for Research on Cancer, Lyon, France. [Google Scholar]
- 4.de Roda Husman A M, Walboomers J M, van den Brule A J, Meijer C J, Snijders P J. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J Gen Virol. 1995;76:1057–1062. doi: 10.1099/0022-1317-76-4-1057. [DOI] [PubMed] [Google Scholar]
- 5.Fisher L D, van Belle G, editors. Biostatistics: a methodology for the health sciences. New York, N.Y: John Wiley & Sons, Inc.; 1993. Categorical data: contingency tables; pp. 256–259. [Google Scholar]
- 6.Gravitt P E, Peyton C L, Apple R J, Wheeler C M. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. J Clin Microbiol. 1998;36:3020–3027. doi: 10.1128/jcm.36.10.3020-3027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hildesheim A, Schiffman M H, Gravitt P E, Glass A G, Greer C E, Zhang T, Scott D R, Rush B B, Lawler P, Sherman M E, Kurman R J, Manos M M. Persistence of type-specific human papillomavirus infection among cytologically normal women. J Infect Dis. 1994;169:235–240. doi: 10.1093/infdis/169.2.235. [DOI] [PubMed] [Google Scholar]
- 8.Ho G Y, Bierman R, Beardsley L, Chang C J, Burk R D. Natural history of cervicovaginal papillomavirus infection in young women. N Engl J Med. 1998;338:423–428. doi: 10.1056/NEJM199802123380703. [DOI] [PubMed] [Google Scholar]
- 9.Jacobs M V, Snijders P J, van den Brule A J, Helmerhorst T J, Meijer C J, Walboomers J M. A general primer GP5+/GP6+-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in genital scrapings. J Clin Microbiol. 1997;35:791–795. doi: 10.1128/jcm.35.3.791-795.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kjaer S K, van den Brule A J, Bock J E, Poll P A, Engholm G, Sherman M E, Walboomers J M, Meijer C J. Human papillomavirus—the most significant risk determinant of cervical intraepithelial neoplasia. Int J Cancer. 1996;65:601–606. doi: 10.1002/(SICI)1097-0215(19960301)65:5<601::AID-IJC8>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 11.Ley C, Bauer H M, Reingold A, Schiffman M H, Chambers J C, Tashiro C J, Manos M M. Determinants of genital human papillomavirus infection in young women. J Natl Cancer Inst. 1991;83:997–1003. doi: 10.1093/jnci/83.14.997. [DOI] [PubMed] [Google Scholar]
- 12.Liaw K L, Glass A G, Manos M M, Greer C G, Scott D R, Sherman M, Burk R D, Kurman R J, Wacholder S, Rush B B, Cadell D M, Lawler P, Tabor D, Schiffman M. Detection of human papillomavirus DNA in cytologically normal women and subsequent cervical squamous intraepithelial lesions. J Natl Cancer Inst. 1999;91:954–960. doi: 10.1093/jnci/91.11.954. [DOI] [PubMed] [Google Scholar]
- 13.Manos M M, Ting Y, Wright D K, Lewis A J, Broker T R, Wolinsky S M. Use of polymerase chain reaction amplification for the detection of genital human papillomaviruses. Cancer Cells. 1989;7:209–214. [Google Scholar]
- 14.Maxwell A E. Comparing the classification of subjects by two independent judges. Br J Psychiatry. 1970;166:651–655. doi: 10.1192/bjp.116.535.651. [DOI] [PubMed] [Google Scholar]
- 15.Nobbenhuis M A, Walboomers J M M, Helmerhorst T J, Rozendaal L, Remmick A J, Risse E K, van der Linden H C, Voorhorst F J, Kenemans P, Meijer C J. Relation of human papillomavirus status to cervical lesions and consequences for cervical cancer screening: a prospective study. Lancet. 1999;354:20–25. doi: 10.1016/S0140-6736(98)12490-X. [DOI] [PubMed] [Google Scholar]
- 16.Petruska J, Goodman M F, Boosalis M S, Sowers L C, Cheong C, Tinoco I., Jr Comparison between DNA melting thermodynamics and DNA. Proc Natl Acad Sci USA. 1988;85:6252–6256. doi: 10.1073/pnas.85.17.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qu W, Jiang G, Cruz Y, Chang C J, Ho G Y, Klein R S, Burk R D. PCR detection of human papillomavirus: comparison between MY09/11 and GP5+/GP6+ primer systems. J Clin Microbiol. 1997;35:1304–1310. doi: 10.1128/jcm.35.6.1304-1310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schiffman M, Bauer H, Hoover R, Glass A G, Cadell D M, Rush B B, Scott D R, Sherman M E, Kurman R J, Wacholder S, Stanton C K, Manos M M. Epidemiologic evidence showing the HPV infection causes most cervical intraepithelial neoplasia. J Natl Cancer Inst. 1993;85:958–964. doi: 10.1093/jnci/85.12.958. [DOI] [PubMed] [Google Scholar]