Abstract
Human babesiosis in the United States is caused predominantly by Babesia microti, a tick-transmitted blood parasite. Improved testing methods for the detection of infection with this parasite are needed, since asymptomatic B. microti infection represents a potential threat to the blood supply in areas where B. microti is endemic. We performed immunoscreening of an expression library of genomic DNA from a human isolate of B. microti (strain MN1). Among 17 unique immunoreactive clones, we identified 9 which represent a related family of genes with little sequence homology to other known sequences but with an architecture resembling that of several surface proteins of Plasmodium. Within this family, a tandem array of a degenerate six-amino-acid repeat (SEAGGP, SEAGWP, SGTGWP, SGTVGP) was found in various lengths between relatively well conserved segments at the N and C termini. In order to examine within-clone variation, we developed a PCR protocol for direct recovery of a specific bmn1-6 homologue directly from 30 human blood isolates, 4 corresponding hamster isolates, and 5 geographically corresponding Peromyscus leucopus (white-footed mouse) isolates. Isolates from the hamsters had the same sequences as those found in the corresponding human blood, suggesting that genetic variation of bmn1-6 does not occur during passage. However, clones from different patients were often substantially different from each other with regard to the number and location of the degenerate repeats within the bmn1-6 homologue. Moreover, we found that strains that were closely related geographically were also closely related at the sequence level; nine patients, all from Nantucket Island, Mass., harbored clones that were indistinguishable from each other but that were distinct from those found in other northeastern or upper midwestern strains. We conclude that considerable genetic and antigenic diversity exists among isolates of B. microti from the United States and that geographic clustering of subtypes may exist. The nature of the bmn1-6 gene family suggests a mechanism of antigenic variation in B. microti that may occur by recombination, differential expression, or a combination of both mechanisms.
Infection with Babesia represents one of the most common parasitic infections worldwide among wild and domestic animals and is second in prevalence only to trypanosomal infections. Members of the genus Babesia, along with their relatives the members of the family Theileridae, are called piroplasms because of their pear-shaped forms found in infected erythrocytes. The near ubiquitous distribution of Babesia spp. in nature has led to the identification of nearly 100 species, some of which are zoonotic. In general, Babesia spp. are minimally pathogenic in their reservoir hosts but may be highly pathogenic when transmitted to other species, including humans. Several species of Babesia are capable of causing infection in humans (10, 42). Of these, Babesia microti, a parasite of rodents, appears to be destined to have a significant public health impact in years ahead. In the United States, B. microti is transmitted by the deer tick Ixodes scapularis (also called Ixodes dammini), which acquires its infection from the white-footed mouse, Peromyscus leucopus (8, 35, 36). Its perpetuation in nature is thus similar to that of other tick-transmitted agents that are now known to exist within congruent zoonotic cycles, including the Lyme disease spirochete, Borrelia burgdorferi (25, 39), and the agent of human granulocytic ehrlichiosis (31). It is thus not surprising that coinfection with these agents exists in the mouse reservoir and occasionally in humans (14, 19, 29, 32, 41).
Little is known about the mechanisms of persistence of B. microti in vertebrate hosts. The white-footed mouse reservoir remains infected for the life of the animal, as do experimentally infected hamsters and mice (23, 40). Other species of Babesia are apparently capable of undergoing antigenic variation during persistent infection, presumably in association with immune responses mounted against parasite antigens (1). Our group has investigated the structure of immunodominant antigens of B. microti by expression cloning, followed by immunoscreening with a pool of high-titer mouse and human sera. In the course of these studies we identified a gene family that encodes related antigens comprising geographically variable immunodominant epitopes. Detection of immune responses to these proteins or amplification and characterization of the genes encoding them may be useful for the diagnosis and/or differentiation of babesial infection.
MATERIALS AND METHODS
Genomic DNA.
Infection with B. microti MN1 was established by intraperitoneal inoculation of 500 μl of cryopreserved, hamster blood into 3-week-old, 50- to 100-g, female Golden Syrian hamsters (SASCO; Charles River, Wilmington, Mass.). Infection was monitored by use of Giemsa-stained smears over a 2- to 3-week period. When the parasitemia levels reached 60 to 70%, the blood was harvested by cardiac puncture. Erythrocytes were then isolated with Histopaque 1077 (Sigma, St. Louis, Mo.) by diluting the whole blood 1:1 with 0.9% saline and layering it over a 1/3 volume of Histopaque. The samples were centrifuged at 500 × g for 40 min at room temperature. The upper layers were discarded, and DNA from the erythrocyte portion was extracted with the Isoquick Nucleic Acid Extraction Kit (Orca Research Inc., Bothell, Wash.).
Genomic expression library and screening.
The B. microti genomic expression library was constructed by sonication with a B. Braun (Allentown, Pa.) sonicator of 20 μg of total genomic DNA to generate fragments of approximately 0.5 to 5.0 kbp. DNA fragments were blunted with T4 DNA polymerase (Gibco BRL, Grand Island, N.Y.) and were ligated to EcoRI adaptors (Stratagene, La Jolla, Calif.). The adapted inserts were then phosphorylated with T4 polynucleotide kinase (Stratagene) and size selected with a Sephacryl S-400-HR column (Sigma). Insert DNA was ligated to Lambda ZAP II, EcoRI and calf intestine alkaline phosphatase-treated vector (Stratagene), and the ligation mixture was packaged with Gigapack II Gold packaging extract (Stratagene).
Expression screening.
Immunoreactive proteins were screened from approximately 3 × 105 PFU with nitrocellulose filters (Schleicher & Schuell, Keene, N.H.). Reactive plaques were assessed with Escherichia coli-adsorbed, B. microti-infected patient serum pools (a pool of high-titer sera from five patients from Minnesota, Nantucket, Mass., and Connecticut). Positive plaques were visualized with 125I-conjugated protein A (NEN Life Science Products, Boston, Mass.) or with an alkaline phosphatase-conjugated goat anti-human immunoglobulin G (IgG; heavy and light chains) secondary antibody (Zymed Laboratories Inc., South San Francisco, Calif.) and were developed with nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate (Gibco-BRL). Excision of the phagemid was done by the Lambda ZAP II protocol (Stratagene), and the resulting plasmid DNA was sequenced with an automated sequencer (Perkin-Elmer Applied Biosystems Division, Foster City, Calif.) with M13 forward, reverse, and internal DNA sequencing primers. Nucleic acid and protein homology searches were performed with DNA Star (Genetics Computer Group, Madison, Wis.) against the EMBL and GenBank (release 99) and the SwissProt, and Translated (release 97) databases. The predicted protein translocation sites were analyzed with the PSORT program (National Institute for Basic Biology, Okazaki, Japan).
Expression and purification of recombinant protein.
Expression of recombinant protein was achieved by amplifying the plasmid insert with Pfu polymerase (Stratagene) and clone-specific primers (25 to 30 nucleotides) which included a 5′ NdeI restriction site, an ATG initiation codon (underlined), and a nucleotide sequence coding for six histidines (in boldface type) (CAATTACATATGCATCACCATCACCATCAC) and a 3′ primer with a stop codon and an EcoRI restriction site. The amplification product was digested with the restriction enzymes NdeI and EcoRI (Gibco BRL), gel purified, and ligated into a pET17b plasmid vector (Novagen, Madison, Wis.) that had previously been cut with NdeI and EcoRI and dephosphorylated. The ligation mixture was transformed into competent XL1 Blue cells (Stratagene), and plasmid DNA was prepared for sequencing (Qiagen Inc., Valencia, Calif.). Recombinant protein was expressed by transforming plasmid DNA into competent BL21 pLysS cells (Novagen) and inducing a single colony cell culture with 2 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma). Recombinant protein was recovered from the cell lysate with Ni-NTA Agarose matrix (Qiagen) by following the manufacturer's instructions and was dialyzed in 10 mM Tris (pH 8.0). Recombinant protein was quality checked for purity by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by staining with Coomassie blue stain and by N-terminal protein sequencing (26) and was quantified by a Micro BCA assay (Pierce, Rockford, Ill.). Recombinants were also assayed for endotoxin contamination by the Limulus assay (BioWhittaker, Walkersville, Md.).
Immunoblots.
A crude B. microti lysate was prepared for immunoblotting by boiling a saponin-lysed erythrocyte pellet in 500 μl of SDS-PAGE buffer containing 6% β-mercaptoethanol, 0.02 mg of leupeptin per ml, and 2.0 mM phenylmethylsulfonyl fluoride. The lysate was then subjected to SDS-PAGE on a 10% polyacrylamide gel and was transferred to nitrocellulose (Schleicher & Schuell) with a mini-PROTEAN II (Bio-Rad Laboratories, Hercules, Calif.) transfer system. Serum samples for these immunoblots were diluted 1:200 in phosphate-buffered saline (PBS) containing 0.1% Tween 20. The blots were processed by the same protocol described below. Recombinant antigens (200 ng/lane) were subjected to SDS-PAGE analysis with 15% polyacrylamide minigels. The antigens were transferred to nitrocellulose BA-85 (Schleicher & Schuell) and were blocked for 1 h at room temperature with PBS containing 1% Tween 20. The blots were then washed three times (10 min each time) in PBS containing 0.1% Tween 20 and 0.5 M sodium chloride (wash buffer). Next, the blots were probed for 1 h at room temperature with serum diluted 1:500 in wash buffer followed by three 10-min washes. The blots were then incubated for 45 min at room temperature with a 1/20,000 dilution of protein A-horseradish peroxidase conjugate (Sigma) in wash buffer and were again washed three times for 10 min each time. Finally, the blots were incubated in ECL chemiluminescent substrate (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.) for 1 min and were exposed to X-ray film (Kodak XAR5; Eastman Kodak Co., Rochester, N.Y.) for 10 to 60 s, as required.
PCR amplification of B. microti-positive samples.
PCR primers were designed by using the regions flanking the six-amino-acid repeat region of the bmn1-3- and bmn1-6-related family of genes (Table 1). Genomic DNA that had been extracted by the Isoquick (Orca Research Inc.) protocol from 200 μl of patient whole blood that had previously tested positive with 18S rRNA-specific primers Bab1 and Bab4 (33) was used as the template for all PCRs. Five to 10 μl of DNA was added to a master mixture containing 1× buffer II (Perkin-Elmer Corp., Norwalk, Conn.), 2.0 mM MgCl2 (Perkin-Elmer Corp.), 200 μM (each) dATP, dGTP, and dCTP and 100 μM (each) dTTP and dUTP (Boehringer Mannheim Biochemicals, Indianapolis, Ind.), 1.5 μg of bovine serum albumin (fraction V; Sigma) per μl, 1.25 U of Taq Gold (Perkin-Elmer Corp.) per 50 μl, 50 pmol of each primer per 50 μl of the reaction mixture, and 2 drops of mineral oil. Amplification was performed in a Perkin-Elmer 480 thermal cycler by using the following profile: incubation at 94°C for 10 min and then 45 cycles at 94°C for 1 min and 55°C for 1 min, followed by 72°C for 5 min; the mixture was then held at 4°C. The PCR products were evaluated for size on 2% agarose gels (Seakem GTG; FMC Bioproducts, Rockland, Maine) and were then Southern blotted and probed with a digoxigenin-labeled oligonucleotide or digoxigenin-labeled PCR product (Boehringer Mannheim Biochemicals) from the bmn1-6 flanking or amino acid repeat region. Positive products were purified with the Qiaquick PCR purification kit (Qiagen Inc.) and were sequenced with forward and reverse primers on an automated sequencer (model 373A; Perkin-Elmer Applied Biosystems Division). Computer analyses of the sequence products were done with the Wisconsin package (Genetics Computer Group, Madison, Wis.). The Pileup program was applied to all sequences to create a dendrogram by using the unweighted pair-group method with arithmetic averages clustering strategy (38). See Fig. 4B for a dendrogram that represents the clustering of the sequences generated with Pileup.
TABLE 1.
Nucleotide sequence and function of oligonucleotide primers used to evaluate genetic heterogeneity of the BMN1-6 family
| Primer designation | Nucleotide sequence | Function |
|---|---|---|
| BMN13-5′ | TTTGCAAGTGATACCGATCCC | PCR, RT-PCR (sense), sequencing |
| BMN13-3′ | TATAACTATTCTTCTAGAATACGG | PCR, RT-PCR (antisense), sequencing |
| BMN16-5′ | TTTGCAGGTGATACCGATCGCG | PCR, RT-PCR (sense), sequencing |
| BMN16-3′ | TATAACTATTCTTCTAGAATACCA | PCR, RT-PCR (antisense), sequencing |
| BMN16A-5′ | ATGAGCGGTGCTGTCTTTGCAGGTGAT | PCR, sequencing |
| BMN16B-3′ | AGATAAATAAATTTCATTAAATATAACTAT | PCR, sequencing |
| BMN112-3′ | TGTAACTATTCTTCTAGAATACGG | PCR, RT-PCR (antisense), sequencing |
| RCBB6B | GTTATGATATTGGAACGAGATAGGGTGAAC | Downstream BMN1 (sense) PCR, sequencing |
| RCBB6D | AATAGACTTCCATATATTACGGAAATGGTCAAA | Downstream BMN1 (antisense) PCR, sequencing |
FIG. 4.
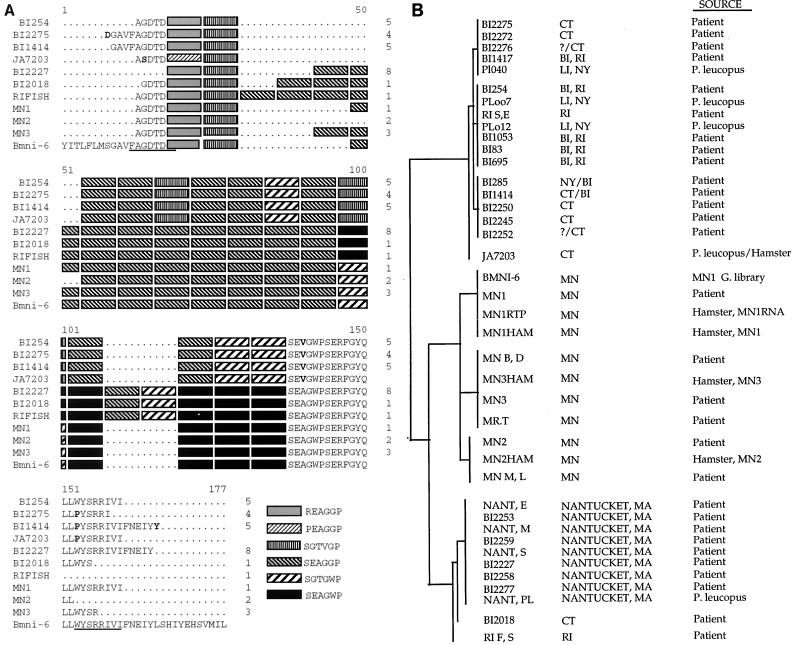
(A) Representative amino acid sequence alignment of B. microti from patients or animals amplified and sequenced with the BMN16-5′ and BMN16-3′ primers or, in some cases, the BMN16A-5′ and BMN16B-3′ primer set. Solid underlined regions indicate the former primer set. Boldface letters indicate amino acids that differ from the bmn1-6 consensus sequence. Numbers to the right designate the number of patients with the same motif. The key to the shaded sequences appears at the bottom. (B) Dendrogram generated from the Genetics Computer Group Pileup program demonstrating the clustering of all patient and animal isolate sequences generated from the bmn1-6 primer sets (BMN16-5′ and BMN16-3′ or BMN16A-5′ and BMN16B-3′). The left column is the patient or animal code. The center column is the known or probable location of infection (BI, Block Island; LI, Long Island; the other abbreviations represent states). The right column indicates the source of DNA used in the analysis.
RNA isolation and RT-PCR.
RNA was harvested from Histopaque (Sigma)-isolated infected hamster erythrocytes with Trizol LS (Gibco BRL) according to the manufacturer's instructions. RNA was then treated with DNase by the addition of an equal volume of DNase digestion buffer (20 mM Tris [pH 8.0], 10 mM MgCl2) and 1 U of RNase-free DNase I (Boehringer Mannheim Biochemicals) per μg of RNA, incubation at 37°C for 1 h, and then extraction with phenol-chloroform followed by ethanol precipitation. The reverse transcription (RT) reaction was performed in a 20-μl reaction volume containing 1.7 μg of RNA, 1× PCR buffer II (Perkin-Elmer Corp.), 3.0 mM MgCl2, 1 mM (each) dATP, dTTP, dGTP, and dCTP, 1 U of RNasin (Promega Corp., Madison, Wis.) per μl, 15 U of avian myelobastosis virus reverse transcriptase (Promega Corp.), and 50 pmol of antisense primer (Table 1). The reaction tubes were incubated at 42°C for 60 min, followed by incubation at 99°C for 5 min and 4°C for 5 min. PCR amplification was carried out in a total volume of 100 μl by the addition of a second mixture containing 0.8× PCR buffer II (Perkin-Elmer Corp.), 10% (vol/vol) glycerol, 25 μg of isopsoralen (IP-10; HRI Research, Concord, Calif.) per ml, 1.75 mM MgCl2, 50 pmol each of sense and antisense primers, and 2.5 U of Amplitaq (Perkin-Elmer Corp.). Amplification was performed in a Perkin-Elmer 480 thermal cycler by using the following profile: incubation at 94°C for 4 min and then 50 cycles at 94°C for 1 min and 55°C for 1 min, followed by a final extension step of 72°C for 5 min; the mixture was then held at 4°C. Before the tubes were opened they were exposed to UV light (320 to 400 nm) for 15 min to help eliminate future amplification of the same products. Controls without reverse transcriptase were run for each reaction to demonstrate that the amplification products were generated from an RNA template as opposed to a DNA template. RT-PCR products were visualized on 2% agarose gels, purified, and sequenced as described above.
RESULTS
Identification of the novel gene family (BMN1-6) and homology to known sequences.
Screening of the B. microti genomic library with the pool of high-titer sera from infected patients resulted in the recovery of 17 unique clones encoding immunodominant antigens. Analysis of the DNA sequences obtained from these clones showed that 9 of the 17 (bmn1-1, -2, -3, -5, -6, -7, -12, -13, and -16) were homologous to each other, and these were chosen for further analysis. The remaining eight clones were unique and will be described in detail elsewhere (M. J. Lodes, R. L. Houghton, E. S. Bruinsma, R. Mohamath, L. D. Reynolds, D. R. Benson, P. J. Krause, S. G. Reed, and D. H. Persing, submitted for publication). Clones bmn1-1 and bmn1-16 were determined to be partial clones of bmn1-3. Further analyses of the DNA sequences obtained from the seven unique clones identified them as members of a polymorphic multigene family. All contain a degenerate, arginine- and proline-rich repeat of six amino acids, with between 9 and 22 repeat units occurring in each antigen (Fig. 1). Seven unique hexapeptide variant repeats (REAGGP, PEAGGP, SGTVGP, SGTGWP, SEAGWP, SEAGGP, and SEAGWS) which occur in various combinations of repeat number and location of variant repeats were identified. These arrays of repeats show limited DNA homology and overall architecture similar to those of several gene families found in Plasmodium spp. Among those identified were the sequence encoding the mature erythrocyte surface antigen (MESA or PfEMP; e.g., bmn1-6 has 56.8% identity with P. falciparum [MESA] in a 623-nucleotide region) and the sequence encoding the merozoite surface antigen (MSA-2). When the deduced protein sequences were analyzed, however, the repeat region was found to be most closely related to various sequences of proteins involved in cytoskeletal structure (e.g., collagen and neurofilaments) even including conservation of the spacing of the proline residues within the sequence (e.g., BMN1-6 has 35.8% identity within a 109-amino-acid overlap with collagen derived from Bombyx mori [silkworm]). Similar to the P. falciparum MESA sequence, the repeat region is flanked on both ends by relatively well conserved sequences. There are two general motifs at the 5′ flanking region; six of the clones contain a serine (S) residue four nucleotides upstream from the tandem repeat region and 3 of the clones contain a glycine (G) (the positions of unique residues are marked by boldface lettering in Fig. 1). The 3′ end is well conserved, with two polymorphic sites: a tryptophan (W)-to-proline (P) change at residue 174 and an isoleucine (I)-to-threonine (T) change at residue 181, as shown in Fig. 1. The 5′ amino acid variations correlated with the 3′ variations in all cases, suggesting allelic forms of the gene family. Hydrophobicity analysis of the protein sequence predicted a hydrophobic N terminus which could represent a signal sequence, again similar to the MESA protein which displays cell-surface localization. Finally, a telomeric repeat sequence that is well conserved in a wide variety of organisms was found in five clones (bmn1-2, -5, -6, -7, and -16) (as reported elsewhere [Lodes et al., submitted]).
FIG. 1.
Amino acid sequence alignment of the BMN1-6-related family of genes recovered from the B. microti MN1 genomic expression library screened using B. microti-infected patient sera. The relatively well conserved amino and carboxy termini flank a variable length of pattern-encoded degenerate repeats composed of six amino acids. Oligonucleotide primers BMN16-5′, BMN16-3′, BMN13-5′, BMN13-3′, and BMN112-3′ (listed in Table 1) correspond to the solid underlined amino acids. Oligonucleotide primers BMN16A-5′ and BMN16B-3′ in Table 1 relate to the dashed overlined amino acids. Boxed amino acids indicate bmn1 downstream primers RCBB6B and RCBB6D in Table 1. Boldface letters indicate amino acids which differ from the consensus sequence. In the downstream sequence alignment, dots designate termination of the clone sequence and asterisks designate consensus with the bmn1-6 sequence. The key to the shaded sequences appears at the bottom.
Expression and immunoreactivity of BMN1 proteins.
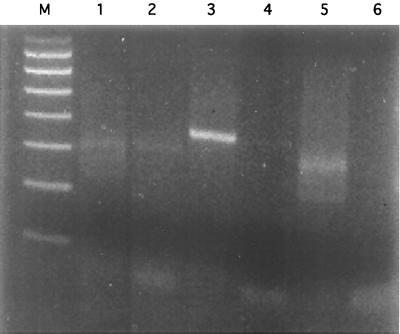
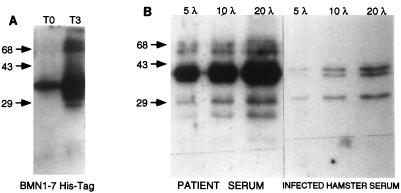
To examine the expression of the BMN1 proteins, RT-PCR was performed with RNA isolated from hamsters infected with B. microti MN1. Three primer sets were tested, and the results are shown in Fig. 2. Only the primer set comprising BMN16-3′ and BMN16-5′ (Table 1) amplified a product of the expected size (357 bp) (Fig. 2, lane 3). The expected products for the other two primers sets were 303 bp for BMN13-3′ and BMN13-5′ (Fig. 2, lane 1) and 267 bp for BMN112-3′ and BMN13-5′ (Fig. 2, lane 5). This finding would be consistent with predominant expression of the BMN1-6 homologue. However, attempts to generate adequate quantities of the BMN1-6 fusion protein to evaluate immune responses to this homologue were unsuccessful. Instead, a closely related clone (bmn1-7) was used to generate a fusion protein; this protein was expressed and purified and was then assessed for immunologic reactivity by Western blotting. Figure 3A shows the reactivity of the clone bmn1-7 culture lysate to the infected patient serum pool (used in the screening of the library) before and after induction with IPTG. The level of an immunoreactive protein of approximately 38 kDa showed a marked increase 3 h after induction compared to that at time zero. The reactivity of the recombinant protein to infected hamster serum was also tested, and significant serologic reactivity was observed (data not shown). The immunologic response to the recombinant protein was then compared with the reactivity to the B. microti crude lysate (Fig. 3B). For both antigen preparations, reactivity was observed in human and hamster sera (Fig. 3B and data not shown).
FIG. 2.
Ethidium bromide-stained 2% agarose gel visualization of RT-PCR products. Lane M, 100- to 800-bp marker; lanes 1 and 2, primers BMN13-5′ and BMN13-3′ and MN1 RNA template with and without the addition of reverse transcriptase, respectively; lanes 3 and 4, primers BMN16-5′ and BMN16-3′ and the MN1 RNA template with and without the addition of reverse transcriptase, respectively; lanes 5 and 6, primers BMN13-5′ and BMN112-3′ and the MN1 RNA template with and without the addition of reverse transcriptase, respectively.
FIG. 3.
(A) Immunoblot showing mini-induction of clone bmn1-7 in the pET17b vector screened with the positive patient serum pool. T0 the culture lysate with an A560 of 0.5 at time zero; T3, the culture lysate 3 h after the addition of IPTG. (B) Immunoblot showing reactivity of infected patient serum pool or hamster serum with increasing amounts (5, 10, and 20 μl) of crude B. microti lysate. λ, microliters. Numbers on the left are in base pairs.
PCR analysis of patient samples.
Given the degenerate repeat structure of the gene family identified in these studies, we were interested in whether geographic variation in the gene family exists, as described for the MSA gene family of Plasmodium falciparum (6). To analyze variations within the clones, a PCR protocol was developed by using the conserved flanking regions (Fig. 1) as a basis for primer design. The two general motifs at the 5′ flanking region were represented along with three of the possible combinations from the 3′ end (underlined in Fig. 1). The spectrum of possible primer pairs from the clones was used in a trial-and-error process to find a set or sets of primers capable of generating a consistently amplifiable single PCR product that could be subjected to sequence analysis (data not shown). Ultimately, primers capable of amplifying the bmn1-6 allelic variant were chosen for further testing of specimens.
Recovery of bmn1-6 homologues directly from human, hamster, and mouse blood samples was then carried out by direct PCR amplification of DNA extracted from whole blood. Figure 4A shows an amino acid sequence alignment of the homologues recovered from enumerated patient samples, two P. leucopus samples, one hamster sample, and the bmn1-6 sequence as a reference. Ten variants were found, overall, together comprising 14 to 20 repeats in the tandem repeat region. The following six of the seven extant hexapeptides were represented in these variants: REAGGP, PEAGGP, SGTVGP, SGTGWP, SEAGWP, and SEAGGP. When duplicate PCRs were performed with one sample, these primers consistently generated the same PCR product, as determined by direct sequencing. Every isolate that generated a product with the bmn1-6 primers also generated appropriate PCR products, on the basis of their sizes and sequences, with the 18S rRNA primers Bab1 and Bab4 (33). However, only 24 of 48 samples that were positive for the 18S rRNA gene were positive with the bmn1-6 primer set. To test whether isolates potentially lacked the bmn1-6 sequence, we tested blood specimens that were PCR positive with the 18S rRNA primer set with PCR primers designed to detect the conserved C-terminal portion of the bmn1 sequence. Forty-four of 48 patient isolates tested were positive, including 20 from patients who were negative with the primer set designed to detect the variable region. This suggests that failure to detect the variable region of the bmn1-6 gene in some samples was not due to the absence of the gene but more likely was due to a lower efficiency of recovery of the variable region.
Differences observed between variants could be categorized into two general classes: heterogeneity in the tandem repeat region and heterogeneity in the flanking regions. The amplification product derived from strain MN1 was, as expected, closely related to bmn1-6 homologues recovered from strains MN2 and MN3 (also obtained from patients from Minnesota), which differed only by the number of SEAGGP repeats. The sequences of isolates RIFISH, BI2018, and BI2227 differed from that of bmn1-6 with additional repeats at positions 108 to 119 and a degenerate repeat at positions 96 to 101. Different numbers of SEAGGP repeats in these three samples also distinguished them as variants separate from one another. Some of the variants were also distinguished by changes in the more conserved regions. The sequences of BI254, BI2275, BI1414, and JA7203 differed from the bmn1-6 sequence with degenerate repeats at positions 66 to 71, 84 to 89, and 96 to the end of the repeat region, but they also harbored amino acid substitutions in the flanking regions. Furthermore, BI254, BI2275, and BI1414 harbored identical tandem repeat regions and could be differentiated only by substitutions in the flanking regions. Thus, the total array of sequence variation includes length polymorphism, amino acid substitutions in conserved flanking regions, as well as insertion of degenerate motifs into the repeat region. As a control for the reliability of our recovery technique, we amplified some samples several times, with consistent results. Furthermore, the isolates from hamsters had sequences indistinguishable from those found in the corresponding human blood samples. Taken together, these observations may have important implications for how B. microti genetic variation might (or might not) occur (see Discussion).
Geographic distribution of variants.
When these variants are analyzed with respect to the geographic location of the original sample, a clear distribution could be seen. Variants that were closely related geographically are also closely related at the sequence level. Figure 4B shows a dendrogram of several variants or homologues obtained from B. microti-positive samples. Each terminal cluster corresponds to a variant shown in Fig. 4A. The samples were from two general regions in the United States: the upper Midwest (Minnesota) and the northeastern United States. Both regions are known to be endemic for B. microti (42). Samples from the same geographic locations occurred in related clusters; e.g. the 11 samples from Minnesota fell into three separate variant groups or terminal clusters. Furthermore, these groups were found to be more related to one another than to any other variant. These variants were most similar to the bmn1-6 sequence and varied only by the number of SEAGGP repeats.
Samples from the northeastern states seemed to fall into two broad groups; those samples from Nantucket comprised one variant with closely related variants from Connecticut and Rhode Island. These variants had flanking regions with sequences identical to the bmn1-6 sequence but differed by the presence of additional and substituted repeats. Interestingly, the variant from Nantucket, Mass., was more closely related to the variants from Minnesota than to the geographically proximal variants in New York and Connecticut, which comprised a second northeastern group that contained four variants. The latter variants were distinguished primarily by amino acid substitutions in the flanking regions.
DISCUSSION
We have identified a B. microti gene family that encodes highly variable immunodominant antigens. Two allelic forms of this gene family which are distinguishable by minor sequence variations in the amino- and carboxy-terminal fragments were identified. Located between the relatively well conserved amino- and carboxy-terminal fragments are a series of degenerate six-amino-acid repeats. Members of both supergroups vary with respect to the number of six-amino-acid repeats and the positions of degenerate motifs. The DNA sequence homology and the similarity in the overall structure of the bmn1-6 gene family to the MESA (also known as PfEMP) gene from Plasmodium could provide some insight into the potential role of the babesial homologues. MESA is synthesized in mature merozoites or parasites within erythrocytes and is then transported to the internal face of the erythrocyte membrane (4, 16), where it specifically but noncovalently binds to erythrocyte protein 4.1 (22). It is believed that MESA plays a role in altering the membrane of the infected erythrocyte; this ultimately aids in the persistence of the parasite but it is not required for in vitro growth or cytoadherence (34). The homology of the BMN1 protein sequence to various collagen and neurofilament sequences further supports this role in Babesia survival. All of these sequences have been shown or are predicted to fold into α-helical coiled coils which can form homopolymer filaments and which primarily have structural functions. It would seem reasonable to suppose that the primary function of BMN1 proteins might be structural, although they could play a more direct role in host cell invasion. Whether or not the specific role of BMN1 is similar to that of MESA remains to be seen. It is clear, however, that the overall similarity to MESA and several other multigene families from Plasmodium spp., Mycoplasma, and Neisseria identifies BMN1 as a family of proteins in which the ability to generate escape variants of immunodominant epitopes, presumably by intragenic or intergenic recombination, is important for survival.
The existence of multiple allelic variants within this gene family also suggests that differential expression of members of the gene family may occur. Several of the members of the bmn1-6 gene family were found to be located next to telomeric sequences, which is consistent with the findings that several other antigen-encoding genes are located in proximity to telomeric domains (15, 21). Telomeres can exert position effects on the transcription of nearby genes, resulting in either active expression or transcriptional repression, and these effects are under epigenetic control (11). Therefore, it is possible that changing the genomic location of the bmn1 clones can lead to differential expression. Indeed, clone bmn1-6, which was found to be expressed by RT-PCR, was located near telomeric repeats. The presence of apparently unexpressed clones bmn1-2, -5, -7, -16 near telomeres is not wholly inconsistent with the presence of antigenic gene families seen in other organisms. In trypanosomes, several copies of the gene encoding the variant surface glycoprotein (VSG) are found within telomeric expression sites, but only one site is active at a time (for reviews, see references 2 and 43). The mechanism of site switching is not known, but epigenetic regulation has been implicated. Further studies will be necessary to determine if telomeric location associates with expression of these genes, either directly or indirectly. It has been proposed that protozoan species such as Plasmodium spp. and Trypanosoma spp. might use the genetic flexibility of chromosome ends in immune evasion (21).
The finding that human isolates share the same antigen sequence variation as hamster isolates derived from corresponding blood specimens as well as isolates obtained from P. leucopus (the natural reservoir for B. microti) from the same geographic regions suggests that sequence variation in the bmn1-6 family, if it occurs, does not occur rapidly. Indeed, the finding that geographic variation exists among isolates derived from different geographic regions suggests that although variation does occur in this gene family, it probably occurs by recombination followed by clonal dissemination of a variant within a specific geographic region. In this regard, it is interesting that the isolates from eastern Long Island and Block Island share the same sequence motif and are distinct from isolates from Nantucket, mainland Connecticut, and the upper Midwest. These distinguishing features may become useful for epidemiologic studies in the future (19). Recent studies of B. microti infection in humans have indicated that in untreated patients, persistent infection is relatively common (18). Such persistence could be facilitated by a mechanism of antigenic variation involving differential gene expression or recombination.
Several other variant multigene families have been described in other species of Babesia such as variant erythrocyte surface antigen 1 (VESA1) from B. bovis (1, 30), variable surface merozoite antigen family (VSMA) from B. bovis (5, 13, 17), and rhoptry-associated protein 1 (RAP-1 [also known as Bv60, p58, Bo60, or Bc60]) from B. bovis, B. bigemina, B. divergens, B. ovis, and B. canis (7, 9, 28, 37). Some of these families also have tandem repeat regions which have been attributed as the source of antigenic variation in the expressed proteins, although the overall gene structure is not necessarily similar to that of the BMN1 family (7, 17, 20, 27). Like the BMN1 family, however, geographic variation is seen with the VSMA family (3, 17). In addition, several of the antigens generated from these multigene families have proved to be potentially valuable targets for immunization in cattle (24) and have even been shown to neutralize merozoites in vitro (12).
The identification of the bmn1-6 family offers opportunities for study of the location, function, antigenicity, and expression of the encoded proteins. In addition, it may be possible to study variation in these loci during long-term infection in mice. Such studies are now under way in the laboratory. Studies of this type will be useful in determining whether the bmn1-6 family will be useful as a diagnostic antigen and/or as a recombinant subunit vaccine.
ACKNOWLEDGMENTS
This work was supported by grants from the National Cancer Institute (Public Health Service grant AI 41103), the Centers for Disease Control and Prevention (CCU 513368-01), and the Corixa Corporation.
REFERENCES
- 1.Allred D R, Cinque R M, Lane T J, Ahrens K P. Antigenic variation of parasite-derived antigens on the surface of Babesia bovis-infected erythrocytes. Infect Immun. 1994;62:91–98. doi: 10.1128/iai.62.1.91-98.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borst P, Bitter W, Blundell P A, Chaves I, Cross M, Gerrits H, van Leeuwen F, McCulloch R, Taylor M, Rudenko G. Control of VSG gene expression sites in Trypanosoma brucei. Mol Biochem Parasitol. 1998;91:67–76. doi: 10.1016/s0166-6851(97)00184-9. [DOI] [PubMed] [Google Scholar]
- 3.Carson C A, Brandt H M, Jensen J B, Bailey C W, Allen G K. Use of random amplified polymorphic DNA analysis to compare Babesia bovis and B. bigemina isolates. Parasitol Res. 1994;80:312–315. doi: 10.1007/BF02351872. [DOI] [PubMed] [Google Scholar]
- 4.Coppel R L, Culvenor J G, Bianco A E, Crewther P E, Stahl H D, Brown G V, Anders R F, Kemp D J. Variable antigen associated with the surface of erythrocytes infected with mature stages of Plasmodium falciparum. Mol Biochem Parasitol. 1986;20:265–77. doi: 10.1016/0166-6851(86)90107-6. [DOI] [PubMed] [Google Scholar]
- 5.Cowman A F, Bernard O, Stewart N, Kemp D J. Genes of the protozoan parasite Babesia bovis that rearrange to produce RNA species with different sequences. Cell. 1984;37:653–660. doi: 10.1016/0092-8674(84)90397-0. [DOI] [PubMed] [Google Scholar]
- 6.Creasey A, Fenton B, Walker A, Thaithong S, Oliveira S, Mutambu S, Walliker D. Genetic diversity of Plasmodium falciparum shows geographical variation. Am J Trop Med Hyg. 1990;42:403–413. doi: 10.4269/ajtmh.1990.42.403. [DOI] [PubMed] [Google Scholar]
- 7.Dalrymple B P, Casu R E, Peters J M, Dimmock C M, Gale K R, Boese R, Wright I G. Characterisation of a family of multi-copy genes encoding rhoptry protein homologues in Babesia bovis, Babesia ovis and Babesia canis. Mol Biochem Parasitol. 1993;57:181–192. doi: 10.1016/0166-6851(93)90194-3. [DOI] [PubMed] [Google Scholar]
- 8.Etkind P, Piesman J, Ruebush II T K, Spielman A, Juranek D D. Methods for detecting Babesia microti infection in wild rodents. J Parasitol. 1980;66:107–110. [PubMed] [Google Scholar]
- 9.Goff W L, Davis W C, Palmer G H, McElwain T F, Johnson W C, Bailey J F, McGuire T C. Identification of Babesia bovis merozoite surface antigens by using immune bovine sera and monoclonal antibodies. Infect Immun. 1988;56:2363–2368. doi: 10.1128/iai.56.9.2363-2368.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorenflot A, Moubri K, Precigout E, Carcy B, Schetters T P. Human babesiosis. Ann Trop Med Parasitol. 1998;92:489–501. doi: 10.1080/00034989859465. [DOI] [PubMed] [Google Scholar]
- 11.Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 12.Hines S A, Palmer G H, Jasmer D P, Goff W L, McElwain T F. Immunization of cattle with recombinant Babesia bovis merozoite surface antigen-1. Infect Immun. 1995;63:349–352. doi: 10.1128/iai.63.1.349-352.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hines S A, Palmer G H, Jasmer D P, McGuire T C, McElwain T F. Neutralization-sensitive merozoite surface antigens of Babesia bovis encoded by members of a polymorphic gene family. Mol Biochem Parasitol. 1992;55:85–94. doi: 10.1016/0166-6851(92)90129-8. [DOI] [PubMed] [Google Scholar]
- 14.Hofmeister E K, Kolbert C P, Abdulkarim A S, Magera J M, Hopkins M K, Uhl J R, Ambyaye A, Telford III S R, Cockerill III F R, Persing D H. Cosegregation of a novel Bartonella species with Borrelia burgdorferi and Babesia microti in Peromyscus leucopus. J Infect Dis. 1998;177:409–416. doi: 10.1086/514201. [DOI] [PubMed] [Google Scholar]
- 15.Horn D, Cross G A. A developmentally regulated position effect at a telomeric locus in Trypanosoma brucei. Cell. 1995;83:555–561. doi: 10.1016/0092-8674(95)90095-0. [DOI] [PubMed] [Google Scholar]
- 16.Howard R J, Lyon J A, Uni S, Saul A J, Aley S B, Klotz F, Panton L J, Sherwood J A, Marsh K, Aikawa M, et al. Transport of an Mr approximately 300,000 Plasmodium falciparum protein (Pf EMP 2) from the intraerythrocytic asexual parasite to the cytoplasmic face of the host cell membrane. J Cell Biol. 1987;104:1269–1280. doi: 10.1083/jcb.104.5.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jasmer D P, Reduker D W, Hines S A, Perryman L E, McGuire T C. Surface epitope localization and gene structure of a Babesia bovis 44-kilodalton variable merozoite surface antigen. Mol Biochem Parasitol. 1992;55:75–83. doi: 10.1016/0166-6851(92)90128-7. [DOI] [PubMed] [Google Scholar]
- 18.Krause P J, Spielman A, Telford III S R, Sikand V K, McKay K, Christianson D, Pollack R J, Brassard P, Magera J, Ryan R, Persing D H. Persistent parasitemia after acute babesiosis. N Engl J Med. 1998;339:160–165. doi: 10.1056/NEJM199807163390304. [DOI] [PubMed] [Google Scholar]
- 19.Krause P J, Telford III S R, Spielman A, Sikand V, Ryan R, Christianson D, Burke G, Brassard P, Pollack R, Peck J, Persing D H. Concurrent Lyme disease and babesiosis. Evidence for increased severity and duration of illness. JAMA. 1996;275:1657–1660. [PubMed] [Google Scholar]
- 20.Kung'u M W, Dalrymple B P, Wright I G, Peters J M. Cloning and characterisation of members of a family of Babesia bigemina antigen genes containing repeated sequences. Mol Biochem Parasitol. 1992;55:29–38. doi: 10.1016/0166-6851(92)90124-3. [DOI] [PubMed] [Google Scholar]
- 21.Lanzer M, Fischer K, Le Blancq S M. Parasitism and chromosome dynamics in protozoan parasites: is there a connection? Mol Biochem Parasitol. 1995;70:1–8. doi: 10.1016/0166-6851(95)00021-r. [DOI] [PubMed] [Google Scholar]
- 22.Lustigman S, Anders R F, Brown G V, Coppel R L. The mature-parasite-infected erythrocyte surface antigen (MESA) of Plasmodium falciparum associates with the erythrocyte membrane skeletal protein, band 4.1. Mol Biochem Parasitol. 1990;38:261–270. doi: 10.1016/0166-6851(90)90029-l. [DOI] [PubMed] [Google Scholar]
- 23.Lykins J D, Ristic M, Weisiger R M. Babesia microti: pathogenesis of parasite of human origin in the hamster. Exp Parasitol. 1975;37:388–397. doi: 10.1016/0014-4894(75)90008-9. [DOI] [PubMed] [Google Scholar]
- 24.Mahoney D F, Kerr J D, Goodger B V, Wright I G. The immune response of cattle to Babesia bovis (syn. B. argentina). Studies on the nature and specificity of protection. Int J Parasitol. 1979;9:297–306. doi: 10.1016/0020-7519(79)90078-x. [DOI] [PubMed] [Google Scholar]
- 25.Mather T N, Mather M E. Intrinsic competence of three ixodid ticks (Acari) as vectors of the Lyme disease spirochete. J Med Entomol. 1990;27:646–650. doi: 10.1093/jmedent/27.4.646. [DOI] [PubMed] [Google Scholar]
- 26.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 27.Mishra V S, McElwain T F, Dame J B, Stephens E B. Isolation, sequence and differential expression of the p58 gene family of Babesia bigemina. Mol Biochem Parasitol. 1992;53:149–158. doi: 10.1016/0166-6851(92)90017-e. [DOI] [PubMed] [Google Scholar]
- 28.Mishra V S, Stephens E B, Dame J B, Perryman L E, McGuire T C, McElwain T F. Immunogenicity and sequence analysis of recombinant p58: a neutralization-sensitive, antigenically conserved Babesia bigemina merozoite surface protein. Mol Biochem Parasitol. 1991;47:207–212. doi: 10.1016/0166-6851(91)90180-e. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell P D, Reed K D, Hofkes J M. Immunoserologic evidence of coinfection with Borrelia burgdorferi, Babesia microti, and human granulocytic Ehrlichia species in residents of Wisconsin and Minnesota. J Clin Microbiol. 1996;34:724–727. doi: 10.1128/jcm.34.3.724-727.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Connor R M, Lane T J, Stroup S E, Allred D R. Characterization of a variant erythrocyte surface antigen (VESA1) expressed by Babesia bovis during antigenic variation. Mol Biochem Parasitol. 1997;89:259–270. doi: 10.1016/s0166-6851(97)00125-4. [DOI] [PubMed] [Google Scholar]
- 31.Pancholi P, Kolbert C P, Mitchell P D, Reed K D, Jr, Dumler J S, Bakken J S, Telford III S R, Persing D H. Ixodes dammini as a potential vector of human granulocytic ehrlichiosis. J Infect Dis. 1995;172:1007–1012. doi: 10.1093/infdis/172.4.1007. [DOI] [PubMed] [Google Scholar]
- 32.Persing D H. The cold zone: a curious convergence of tick-transmitted diseases. Clin Infect Dis. 1997;25:S35–S42. doi: 10.1086/516170. [DOI] [PubMed] [Google Scholar]
- 33.Persing D H, Mathiesen D, Marshall W F, Telford III S R, Spielman A, Thomford J W, Conrad P A. Detection of Babesia microti by polymerase chain reaction. J Clin Microbiol. 1992;30:2097–2103. doi: 10.1128/jcm.30.8.2097-2103.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petersen C, Nelson R, Magowan C, Wollish W, Jensen J, Leech J. The mature erythrocyte surface antigen of Plasmodium falciparum is not required for knobs or cytoadherence. Mol Biochem Parasitol. 1989;36:61–65. doi: 10.1016/0166-6851(89)90200-4. [DOI] [PubMed] [Google Scholar]
- 35.Piesman J, Karakashian S J, Lewengrub S, Rudzinska M A, Spielman A. Development of Babesia microti sporozoites in adult Ixodes dammini. Int J Parasitol. 1986;16:381–385. doi: 10.1016/0020-7519(86)90118-9. [DOI] [PubMed] [Google Scholar]
- 36.Piesman J, Spielman A. Babesia microti: infectivity of parasites from ticks for hamsters and white-footed mice. Exp Parasitol. 1982;53:242–248. doi: 10.1016/0014-4894(82)90065-0. [DOI] [PubMed] [Google Scholar]
- 37.Skuce P J, Mallon T R, Taylor S M. Molecular cloning of a putative rhoptry associated protein homologue from Babesia divergens. Mol Biochem Parasitol. 1996;77:99–102. doi: 10.1016/0166-6851(96)02570-4. [DOI] [PubMed] [Google Scholar]
- 38.Sneath P H A, Sokal R R. Numerical taxonomy. The principles and practice of numerical classification. San Francisco, Calif.: W. H. Freeman & Company; 1973. [Google Scholar]
- 39.Spielman A. Human babesiosis on Nantucket Island: transmission by nymphal Ixodes ticks. Am J Trop Med Hyg. 1976;25:784–787. doi: 10.4269/ajtmh.1976.25.784. [DOI] [PubMed] [Google Scholar]
- 40.Spielman A, Etkind P, Piesman J, Ruebush II T K, Juranek D D, Jacobs M S. Reservoir hosts of human babesiosis on Nantucket Island. Am J Trop Med Hyg. 1981;30:560–565. doi: 10.4269/ajtmh.1981.30.560. [DOI] [PubMed] [Google Scholar]
- 41.Sweeney C J, Ghassemi M, Agger W A, Persing D H. Coinfection with Babesia microti and Borrelia burgdorferi in a western Wisconsin resident. Mayo Clin Proc. 1998;73:338–341. doi: 10.1016/S0025-6196(11)63699-9. [DOI] [PubMed] [Google Scholar]
- 42.Telford S R, III, Spielman A. Babesiosis of humans. In: Collier L, Balows A, Sussman M, editors. Topley and Wilson's microbiology and microbial infections. 9th ed. Vol. 5. London, England: Arnold; 1998. pp. 349–359. [Google Scholar]
- 43.Vanhamme L, Pays E. Control of gene expression in trypanosomes. Microbiol Rev. 1995;59:223–240. doi: 10.1128/mr.59.2.223-240.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]