Significance
Mycobacterium tuberculosis (Mtb) is the etiological agent of human tuberculosis (TB). Mtb can persist inside host macrophages by successfully adapting to intracellular conditions. Acquisition of balanced amounts of essential micronutrients is one such important process. Our studies have identified a metallophore produced on demand to restore Mtb zinc metabolic imbalance. These diacyl-diisonitrile lipopeptides, named kupyaphores, are specifically induced during infection and move in and out of cells to protect bacteria from host-mediated nutritional deprivation and intoxication. Furthermore, we identify an Mtb isonitrile hydratase homolog, expressed in low-zinc conditions, which probably facilitates zinc release from kupyaphores. Identification of this zinc acquisition strategy could provide opportunities in future to understand systemic zinc dysbiosis and associated manifestations in TB patients.
Keywords: zinc, metallophore, nutritional immunity, tuberculosis
Abstract
Mycobacterium tuberculosis (Mtb) endures a combination of metal scarcity and toxicity throughout the human infection cycle, contributing to complex clinical manifestations. Pathogens counteract this paradoxical dysmetallostasis by producing specialized metal trafficking systems. Capture of extracellular metal by siderophores is a widely accepted mode of iron acquisition, and Mtb iron-chelating siderophores, mycobactin, have been known since 1965. Currently, it is not known whether Mtb produces zinc scavenging molecules. Here, we characterize low-molecular-weight zinc-binding compounds secreted and imported by Mtb for zinc acquisition. These molecules, termed kupyaphores, are produced by a 10.8 kbp biosynthetic cluster and consists of a dipeptide core of ornithine and phenylalaninol, where amino groups are acylated with isonitrile-containing fatty acyl chains. Kupyaphores are stringently regulated and support Mtb survival under both nutritional deprivation and intoxication conditions. A kupyaphore-deficient Mtb strain is unable to mobilize sufficient zinc and shows reduced fitness upon infection. We observed early induction of kupyaphores in Mtb-infected mice lungs after infection, and these metabolites disappeared after 2 wk. Furthermore, we identify an Mtb-encoded isonitrile hydratase, which can possibly mediate intracellular zinc release through covalent modification of the isonitrile group of kupyaphores. Mtb clinical strains also produce kupyaphores during early passages. Our study thus uncovers a previously unknown zinc acquisition strategy of Mtb that could modulate host–pathogen interactions and disease outcome.
Infection of humans by Mycobacterium tuberculosis (Mtb) has been the leading cause of mortality and morbidity for more than a century (1). This debilitating chronic pathogenesis is attributed to the ability of Mtb to manipulate and survive within the phagocytic cells of the immune system (2). Mtb colonization of macrophages initiates a competitive conflict for utilization of host nutrients, including elements such as iron, zinc, copper, magnesium, and selenium (3, 4). By limiting transition metal availability to invading pathogens, the host is known to restrict pathogen proliferation, a physiological concept called “nutritional immunity” (5). The pathogen endures through these early phases of adaptation by employing mechanisms that facilitate continual access to host micronutrient supply. Paradoxically, excess free metal ions can exert toxic effects on microbial survival. In fact, studies suggest that human macrophages intoxicate intracellular Mtb with a burst of free zinc and copper (6–8). It is fascinating that these two strikingly opposing outcomes are resisted by intracellular pathogens like Mtb for the maintenance of zinc metallostasis.
The adaptive response strategy of bacterial pathogens to host-imposed zinc scarcity and poisoning is primarily governed by metal-sensing metalloregulatory proteins. As in most bacteria, Mtb contains two key zinc sensor proteins that coordinately function as uptake or efflux repressors. Mtb zinc uptake repressor (zur, Rv2359) regulates expression of several genes (9), including those involved in zinc uptake, while the efflux regulator (zntR, Rv3334) regulates transcription of genes encoding three types of exporters (10). Mtb has been shown to utilize P1-type ATPases to neutralize the toxic effects of zinc in macrophages (7). Mtb also employs zinc sparing to overcome zinc starvation, for example, remodeling of the ribosome 70S subunit (11). Similar mechanisms of metallostasis also govern the intracellular levels of iron during the host–pathogen interaction (12). In contrast, the general bacterial strategy of iron acquisition involving siderophore-mediated scavenging is yet to be recognized for other transition metals in Mtb. Mtb requirement of iron is accomplished by producing the siderophores—mycobactin and carboxymycobactin (13, 14). These siderophores move in and out of cells scavenging iron, releasing metal ions in the intracellular milieu, and then recycling the apo-siderophores. Ferric ion is released via reduction to ferrous ion or by modification of the siderophore scaffold using specific hydrolases (15). It is thus crucial to understand whether Mtb produces specialized iron siderophore-like small molecules on demand for zinc acquisition.
Staphylococcus aureus and Pseudomonas aeruginosa have been shown to produce staphylopine (16) and pseudopaline (17), respectively, for sequestering and importing metal ions, including zinc, from the host milieu to enhance the virulence and fitness of the intracellular bacteria (18). Although these molecules are not specific for zinc, opine-like metabolites have been referred to as zincophore and their biosynthesis is proposed to be widely distributed in bacteria (19). Recently, another class of low-molecular-weight metal chelating diisonitrile containing bacterial metabolites SF2768 and isonitrile lipopeptides have been identified from Streptomyces thioluteus and Mycobacterium marinum, respectively (20, 21). However, these metabolites could not be isolated from their natural hosts, but by heterologous overexpression of cognate biosynthetic gene clusters in host systems Streptomycetes lividans and Escherichia coli, respectively. The biosynthetic clusters of these diisonitrile-containing metabolites includes a nonribosomal peptide synthase (NRPS), fatty acyl-AMP ligase (FAAL), type II thioesterase, oxidoreductase and acyl carrier protein (ACP). Interestingly, a homologous five-gene biosynthetic cluster (Rv0097–Rv0101) can also be identified in the genome of Mtb, with the NRPS gene (Rv0101) being double the size (7,539 bp), as compared to other organisms. The mutant (Δnrps) Mtb strain has been shown to grow at a lower rate in immunocompetent and immunocompromised mice, causing less lung pathology, and was associated with significantly increased mice survival rates (22). Despite the role of this biosynthetic cluster in Mtb virulence, the metabolite produced is not known. Moreover, the metabolic function as well as significance of this large 10.8 kbp genomic locus in tuberculosis (TB) pathophysiology remains to be elucidated.
In this study, we have isolated and characterized diacyl-diisonitrile lipopeptides, named kupyaphores, from human pathogen Mtb. Kupyaphores are stringently regulated and can move in and out of cells scavenging zinc to maintain bacterial fitness. These metallophores are biosynthesized on demand during early phases of Mtb infection and here we delineate pathophysiological relevance of kupyaphores in zinc mobilization and redistribution. Further, we identify an isonitrile hydratase homolog in Mtb that is expressed in low-zinc conditions, and probably facilitates zinc release from these kupyaphores.
Results
Transcriptional Responses Maintain Mtb Zinc Homeostasis.
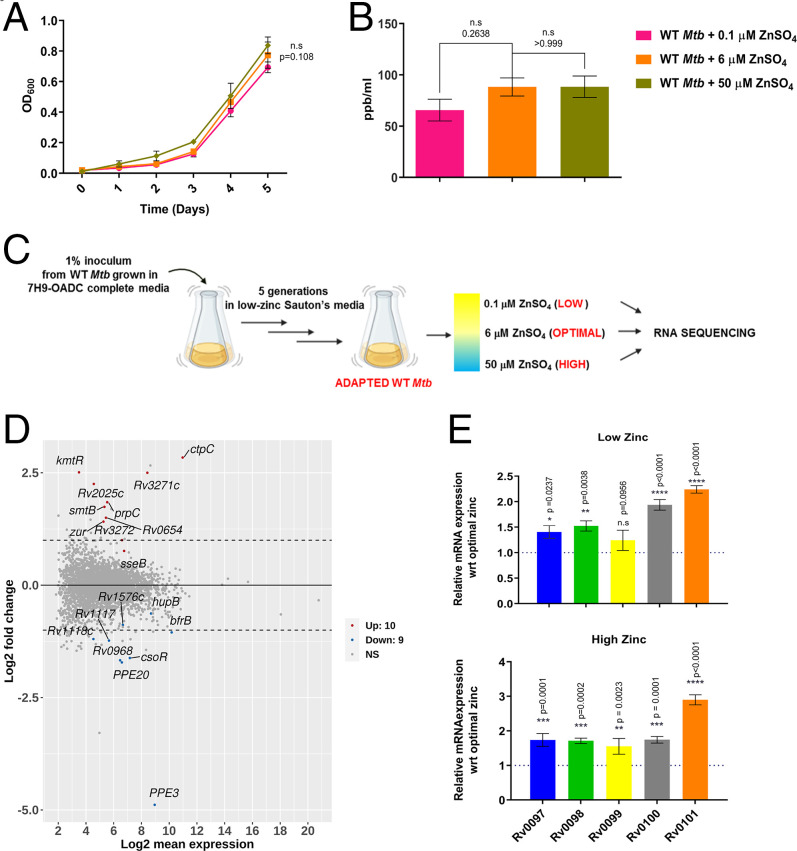
Analysis of genomic loci of the cryptic biosynthetic cluster Rv0097–Rv0101 suggested classic zur-binding sites in the putative promotor regions, 61 bp upstream of Rv0096 and 354 bp upstream of Rv0099. Previous studies with zur-deleted Mtb strain did not show significant differential regulation for any of the genes from the Rv0097–Rv0101 cluster (9). Since these studies were carried out in the metal-rich Middlebrook 7H9 medium, we decided to examine mycobacterial transcriptional adaptive response at varying zinc conditions in the defined minimal Sauton’s medium. The Middlebrook 7H9 medium used for culturing Mtb contains 6 μM concentration of ZnSO4 and we performed wild-type Mtb (WT Mtb) growth kinetics at log-order lower and higher zinc concentrations. WT Mtb growth curves at 0.1 μM (low), 6 μM (optimal), and 50 μM (high) of zinc showed no significant differences (Fig. 1A). WT Mtb intracellular total zinc levels, as measured by inductively coupled plasma mass spectrometry (ICP-MS), were also similar for all three conditions (Fig. 1B).
Fig. 1.
Mtb transcriptional responses maintains zinc homeostasis. (A) Growth kinetics of WT Mtb grown in chelated Sauton’s medium supplemented with varying zinc concentrations: 0.1 μM, 6 μM, and 50 μM. (B) ICP-MS analysis of total intracellular zinc content of Mtb grown in these three conditions. (C) Experimental design for generation of low-metal adapted WT Mtb strain. (D) MA plot of differential gene expression analysis in a 50 μM zinc supplemented condition as compared to 0.1 μM zinc supplementation with log2 fold-change > 0.5 and false-discovery rate < 0.2 as cutoff. Genes marked in red are significantly up-regulated, while genes marked in blue are significantly down-regulated. (E) Gene-expression analysis of Rv0097–Rv0101 through qRT-PCR under low- and high-zinc conditions as compared to optimal zinc concentration. Data represent mean ± SEM (n = 3 biological replicates), P value indicated for each data point.
Rv0099 is a member of a previously identified family of FAALs, and in the genomic context are found adjacent to polyketide synthases (PKS) or NRPS. All of these genomic clusters in Mtb produce virulent lipidic metabolites and show modest regulation at the transcriptional level (23, 24). With an anticipation to obtain a robust transcriptional response, we carried out five passages of WT Mtb in the minimal Sauton’s medium treated with chelex X-100 resin (Fig. 1C). Adapted Mtb cells were then inoculated into low-, optimal-, and high-zinc conditions. RNA was isolated from log-phase cultures and transcriptome analysis was performed using an Illumina MiSeq sequencing platform for three biological replicates. After examining consistency of the raw reads (SI Appendix, Fig. S1A), we probed relatedness between these samples by using principal component analysis (SI Appendix, Fig. S1B). We observed tight clustering for all the three replicates of low- and high-zinc conditions, whereas one of the biological replicates showed greater variation from the optimal-zinc dataset.
To account for biological variation, we analyzed by including and excluding this dataset. This analysis yielded similar results for most of the significantly differentially expressed genes, many of which have been previously shown to be zinc (or metal) responsive genes. We compared differential expression analysis between 50 μM and 0.1 μM with cut off log2 fold-change > 0.5 and false-discovery rate < 0.2 and visualized the data using an MA plot (Fig. 1D). This analysis identified a small set of 19 genes to be differentially regulated. Up-regulated genes included zinc transcription repressors smtb and zur (9) and efflux zinc pump ctpC (7), Rv2025c (25), and cadI (26). Genes belonging to the esx-1 and esx-3 export system showed highest down-regulation at both high- and low-zinc, when compared to optimal level of zinc. Previous study had suggested a role of these clusters in metal-dependent regulation (27). Of the FAAL-PKS/NRPS gene clusters, some of the genes belonging to mycobactin, pthiocerol dimycocerosate and the biosynthetic cluster Rv0097–Rv0101, showed modest up-regulation at both low- and high-zinc conditions (SI Appendix, Fig. S1C). Expression changes of the genes from Rv0097 to Rv0101 were corroborated by carrying out qRT-PCR analysis. We observed statistically significant up-regulation for four of five genes at both low- and high-zinc conditions, when compared with optimal conditions (Fig. 1E). We therefore hypothesized that this cluster may have involvement in Mtb zinc metallostasis.
Significance of Mtb nrps in Mycobacterial Physiology.
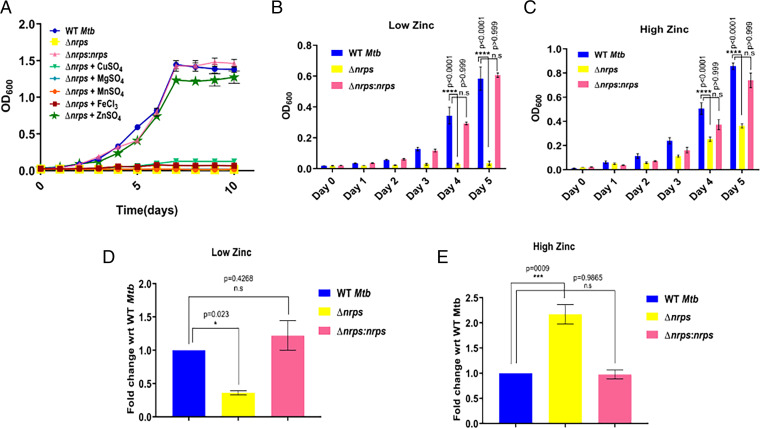
Mtb gene cluster Rv0097–Rv0101 spans 10.8 kbp, which over the years has been implicated in Mtb virulence, without exactly delineating the biochemical function (22, 28, 29). The largest open reading frame (Rv0101) of the Rv0097–Rv0101 gene cluster codes for a bimodular NRPS. We generated a NRPS knockout strain in Mtb (Δnrps) to assess the role of this gene cluster in metal homeostasis (SI Appendix, Fig. S2A). The mutant strain showed no growth profile differences under planktonic conditions in the 7H9 medium (SI Appendix, Fig. S2B). However, the Δnrps Mtb mutant exhibited differences under biofilm growth conditions as reported earlier, and formed a fragile film lacking the characteristic reticulation of WT Mtb biofilms (29). This mutant phenotype could be reversed on complementation with an integrative shuttle cosmid vector containing an Mtb H37Rv fragment spanning Rv0096 to Rv0109 (SI Appendix, Fig. S2 C and D). We then examined the growth profiles of WT Mtb and Δnrps strains in the metal-limited chelated Sauton’s medium. The Mtb Δnrps strain displayed defective growth in the Sauton’s medium, which contains only glycerol and asparagine as sole carbon and nitrogen sources. We then examined the rescue of this growth defect by adding various divalent metals one at a time at 6 μM concentration. Δnrps growth could be restored only upon zinc supplementation to Sauton’s medium and no reversal in growth phenotype could be obtained for other metals, such as copper, magnesium, manganese, and iron (Fig. 2A).
Fig. 2.
Role of Mtb nrps in mycobacterial physiology. (A) Growth kinetics of WT and Δnrps show mutant to be severely compromised in a nutrient-deficient condition of chelated Sauton’s medium. Addition of other key transition elements—copper, magnesium, manganese, or iron—to Sauton’s medium failed to revive mutant significantly. Only supplementation with 6 μM zinc could restore growth profile of Δnrps strain. (B and C) Titration of Sauton’s medium with different concentrations of zinc show compromised growth pattern of Δnrps under both low- (0.1 μM) and high- (50 μM) zinc concentrations as compared to WT Mtb. Complementation of Δnrps strain with a WT copy (Δnrps:nrps) reversed the phenotype. (D and E) ICP-MS analysis of WT, Δnrps, and Δnrps:nrps Mtb strains to measure intracellular levels of total zinc under low- and high-zinc conditions. Data represent mean ± SEM (n = 3 biological replicates) with P values indicated for each data point.
We then evaluated growth of WT, the Δnrps mutant, and the complemented strain (Δnrps:nrps) at conditions of both low- (Fig. 2B) and high-zinc (Fig. 2C) levels. While the Δnrps Mtb mutant showed significantly reduced growth under both of these conditions, the complemented strain showed growth kinetics similar to WT Mtb. To understand whether the mutant strain had disrupted metal homeostasis, we measured metal ion concentrations within Mtb cells by using ICP-MS. At low-zinc conditions, the Δnrps mutant strain showed a significant decrease in zinc levels (Fig. 2D), whereas zinc accumulation could be observed at high-zinc conditions (Fig. 2E). The complemented Mtb strain (Δnrps:nrps) profiles were similar to WT cells. There was slight difference for copper levels between the mutant and WT (SI Appendix, Fig. S2E), and other metals—such as magnesium, cobalt, nickel, and manganese—showed no significant change across all three Mtb strains.
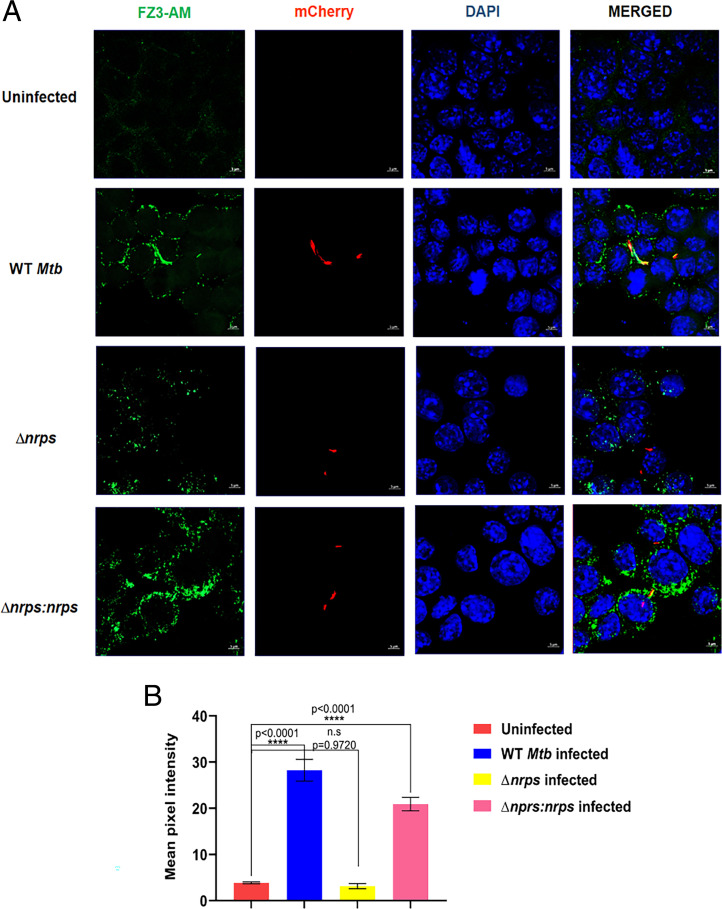
To understand the significance of the Rv0097–Rv0101 biosynthetic cluster in the context of Mtb pathogenesis, ex vivo infection experiments were performed with an immortalized murine bone marrow–derived RAW 264.7 macrophage cell line with WT and Δnrps Mtb strains. Within 2 h of infection, remarkable up-regulation of the Rv0097–Rv0101 gene cluster could be noted in WT Mtb cells (SI Appendix, Fig. S3A). This up-regulation response dampens in 24 h in the macrophage infection assays, suggesting an early role of the Rv0097–Rv0101 cluster during Mtb infection. Furthermore, the intracellular free-zinc levels for WT, Δnrps and complemented Mtb were measured using FluoZin 3-AM (FZ3-AM) dye, a zinc-selective indicator, 2 h postinfection. To observe Mtb cells in a fluorescence microscope, all the three strains were transformed with plasmid expressing mCherry. Macrophages infected with WT Mtb showed increased punctate signals for free-zinc, as compared with uninfected cells (Fig. 3A). Surprisingly, weak free-zinc signals could be observed in the mutant-infected macrophages, as measured by FZ3-AM. The complemented strain (Δnrps:nrps) showed similar free-zinc levels to the WT Mtb strain as quantitated by confocal microscopy (Fig. 3B). Interestingly, almost all WT Mtb cells colocalized with free-zinc, as reported earlier (7).
Fig. 3.
Mtb nrps modulates host free-zinc dynamics. (A) Free-zinc labeling using FZ3-AM dye for uninfected, mCherry-labeled WT, Δnrps, and Δnrps:nrps Mtb strains infected murine macrophages 2 h postinfection. (B) FZ3-AM signal quantitation from 75 cells for each condition. Unlike WT and Δnrps:nrps-infected macrophages, Δnrps-infected macrophages do not show elevated free-zinc levels as compared to uninfected macrophage. Data represent mean ± SEM (n = 3 biological replicates) with P values indicated for each data point.
To further understand these changes in zinc dynamics upon infection, we compared the zinc pools of macrophages and bacteria by performing differential lysis of WT and Δnrps infected cells. The total zinc levels were then measured by ICP-MS analysis. The total zinc levels of either of the infected macrophages (WT or Δnrps Mtb) were found to be comparable to uninfected macrophages (SI Appendix, Fig. S3B). Analysis of the total zinc pools of the internalized Mtb, however, interestingly revealed the zinc levels to be significantly low in Δnrps as compared to WT Mtb (SI Appendix, Fig. S3C). No significant difference could be observed between the two strains for any other metal ion (SI Appendix, Fig. S3 D and E). Together, the induction of Rv0097–Rv0101 biosynthetic pathway and altered intracellular zinc concentration in Δnrps Mtb strain provide evidence for the involvement of the unknown metabolite produced by this cryptic gene cluster in maintenance of bacterial zinc homeostasis.
Computational and Biochemical Analysis of Mtb nrps Biosynthetic Gene Cluster.
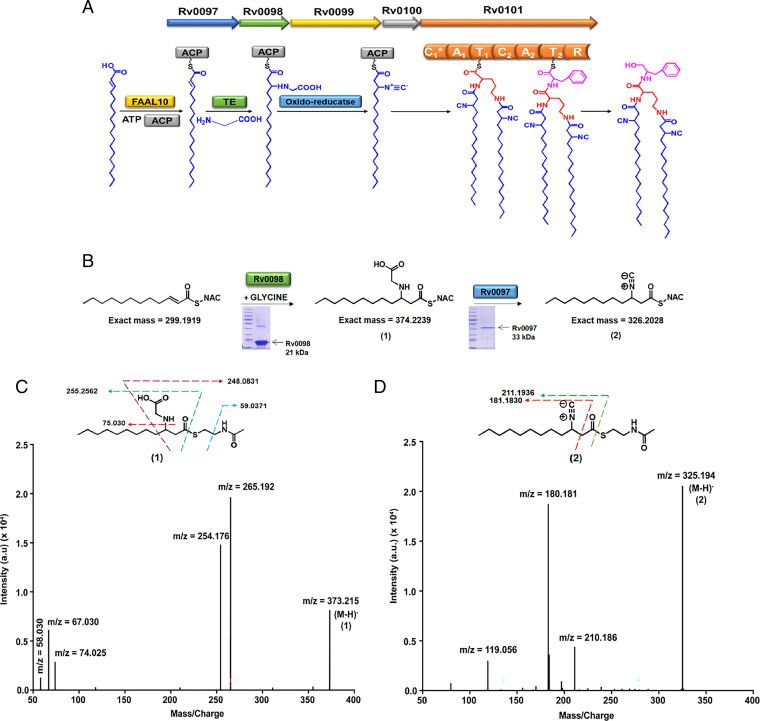
To predict the metabolite produced by the biosynthetic operon Rv0097–Rv0101, we carried out detailed in silico analysis and also confirmed biochemical functions for three enzymes by performing in vitro assays. Retro-biosynthetic analysis based on previous studies suggested a putative pathway wherein FAAL10 (Rv0099) activates α/β unsaturated long-chain fatty acids to corresponding acyl-adenylates, which are then transferred onto the thiol group of phosphopantetheine group of ACP (Rv0100) protein (30, 31). This ACP-bound lipid chain can be modified by Rv0097 and Rv0098, as shown previously (21, 32). Mtb NRPS (Rv0101) contains two modules (bimodular), each consisting of three enzymatic domains: condensation (C), adenylation (A), and thiolation (T) (33, 34). The second module of this NRPS protein contains a reductase (R) domain, which releases thioester-bound lipopeptides as corresponding alcohols (35) (Fig. 4A). Protein sequence analysis of the two modules of NRPS revealed the two C domains (C1 and C2) shows weak sequence homology (similarity/identity, 35/36%).
Fig. 4.
Biochemical analysis of Mtb Rv0097–Rv0101 gene cluster. (A) Schematic representation of putative biosynthetic steps for unknown metabolite production by Mtb Rv0097–Rv0101 gene cluster. Mtb harbors bimodular NRPS with seven catalytic domains. (B) Reaction scheme for enzymatic assays of purified Rv0098 and Rv0097 proteins with chemically synthesized C12-SNAC as substrate. (C) MS/MS spectra showing fragmentation products of glycine adduct of 2-dodecenoic-SNAC, 1, detected in the enzymatic assay with Rv0098 purified protein. No product could be detected in control reactions with no protein or no glycine. (D) MS/MS spectra of additional peak of m/z 326.2028 corresponding to isonitrile adduct of 2-dodecanoic-SNAC, 2, observed upon addition of Rv0097 purified protein to the Rv0098 assay conditions. The calculated masses for metabolites 1 and 2 are within 5-ppm mass error tolerance at MS1. All assays in C and D were performed in triplicates with reproducible results each time.
Recently, genome mining studies have revealed similar five-gene clusters from various Actinomycetes (20). We performed dendrogram-based analysis of C domains from NRPS proteins (unimodular or bimodular) of these five-gene clusters along with other atypical C domains. This analysis divided the C domains into two discrete clusters (SI Appendix, Fig. S4A), indicating that they may have distinctive catalytic functions. Interestingly, C2 domains from the bimodular NRPS clustered along with the typical C domains, whereas C1 domains formed a separate branch. Though both the domains are of similar length and share the conserved catalytic HHXXXDG motif, structural modeling of C1 domains show substantial homology to CoA-dependent acyltransferases. All C1 domains show unique conservation of amino acid residues around the active site pocket, suggestive of functional divergence from the cognate C domains (SI Appendix, Fig. S4 B and C) and we therefore refer to these as C1*. Based on reported studies of metabolites isolated from the heterologous overexpression systems for M. marinum and S. thioluteus (20, 21), we reasoned that this unusual C1* domain may be involved in the transfer of acyl chains on both the α-amino and ε-amino groups of a lysine.
To corroborate our computational prediction, we performed biochemical reconstitution of Rv0097 and Rv0098. Our previous studies showed FAAL10 (Rv0099) to activate long-chain fatty acids as acyl-adenylates and transfer them onto the thiol group of phosphopantetheine ACP (Rv0100) protein (35). We confirmed similar activity for α/β unsaturated long-chain fatty acids. We cloned and expressed Rv0097 and Rv0098 in E. coli and purified the proteins using affinity chromatography. Enzymatic assays were then performed by using phosphopantetheine mimics, N-acetyl cysteamine (NAC) thioesters of α/β unsaturated fatty acids of different carbon lengths: C4, C8, and C12 (Fig. 4B and SI Appendix, Fig. S5). Enzymatic activities were confirmed using liquid chromatography coupled to mass spectrometry (LC-MS) assay protocols (36, 37). Addition of glycine to NAC-thioester of dodecenoic acid in the presence of enzyme Rv0098 identified a molecular ion peak at m/z of 374.2239, corresponding to Michael addition of glycine across unsaturated fatty acyl thioester. MS/MS analysis of this ion resulted in fragment ions of m/z [M-H]− = 254.176, 74.025, and 58.030, providing confidence to the structure (Fig. 4C). Mass peak with m/z [M-H]− = 265.192 probably corresponds to the hydrated form of the fragment ion of m/z = 248.0831. No activity was observed for NAC-thioester of crotonoic and minor products were obtained with octenoic acids. When the purified protein Rv0097 was included in the above assays, a new peak corresponding to m/z of 326.2085 could be observed in high-resolution LC-MS (LC-HRMS). This molecular ion, along with fragment ions, corresponded to the isonitrile modification of the of C12 fatty acyl-SNAC moiety (Fig. 4D). Such an isonitrile modified C12 ACP-bound acyl chain could then condense with first amino acid, which is selected by the adenylation domain of the first module of the NRPS protein (C1*-A1-T1). Computational algorithms predict the adenylation A1 to activate lysine, while the second A2 domain shows specificity for phenylalanine (38, 39). Finally, the acyl chain would be reductively released to the corresponding alcohol, thus producing a novel isonitrile lipopeptide.
Identification and Characterization of Diacyl-Diisonitrile Lipopetides from Mtb.
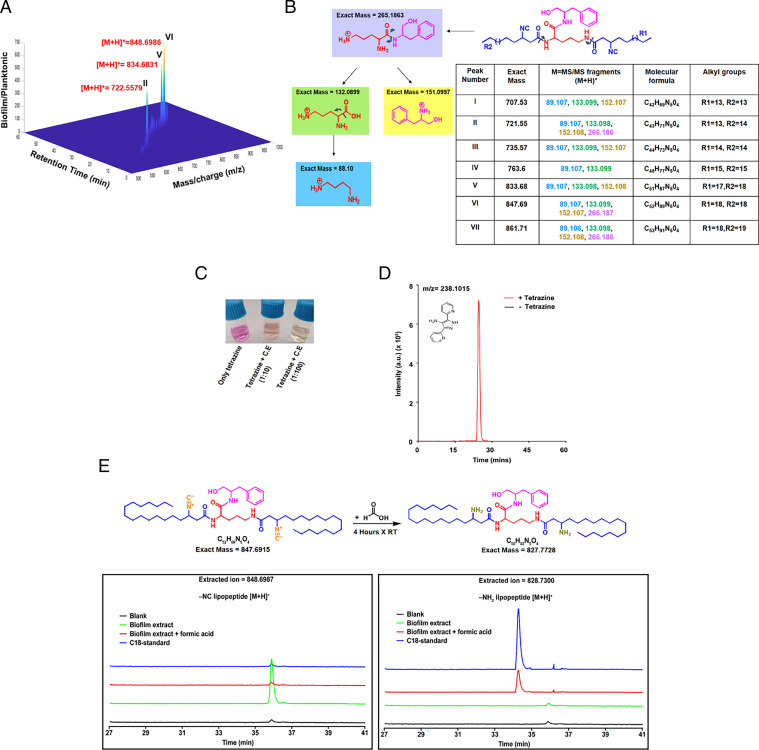
In order to isolate these molecules from Mtb, we extracted metabolites from Mtb cells grown in Middlebrook 7H9 medium and from biofilm using ethyl acetate. These samples were analyzed by LC-MS/MS using a recently developed method from our laboratory (36). Analysis of Mtb biofilm extract detected a cluster of unique peaks with m/z 707.5350, 721.5506, 735.5663, 763.5967, 833.6758, 847.6915, and 861.7071, which were absent in the Mtb planktonic cultures grown in Middlebrook 7H9 medium (Fig. 5A). The parent ion masses (I–VII) differed by a multiple of 14, typical of series with varying –CH2 group and thus can be attributed to different acyl chain lengths. The previously reported molecular formula C40H73N6O5 with m/z of 717.5612 was absent in our biofilm-analyzed samples (29). MS/MS fragmentation patterns of parent ions revealed a common set of fragments with m/z values of [M+H]+= 266.1863, 152.1080, 133.099, and 89.1073 (Fig. 5B). The m/z of 266.1863 along with the fragments of m/z 152.1080 and 133.099 can be assigned to phenylalaninol ion and ornithine ion, respectively. A peak of 89.1073 corresponds to decarboxylation of the ornithine ion, which suggests the backbone to be composed of a common core of ornithine and phenylalaninol dipeptide. Concomitantly, these peaks were absent in the biofilm metabolic extract of Δnrps but could be detected in the Δnrps:nrps strain (SI Appendix, Fig. S6A). Additionally, negative ion mode LC-MS analysis of the biofilm extract revealed two types of metabolites: symmetric (with identical acyl chains on the two –NH2 groups of ornithine) and asymmetric (with different acyl chains on the two –NH2 groups of ornithine) (SI Appendix, Fig. S6 B and C). Recently, click chemistry-based analytical detection of isonitrile using tetrazine has been reported (40). Tetrazine treatment of WT Mtb biofilm extract rapidly changed the color from pink to yellow (Fig. 5C) and the signals from MS analysis corresponding to I–VII diminished substantially (SI Appendix, Fig. S6D). Furthermore, the universal reaction product Py-aminepyrazoles with m/z [M+H]+= 238.1014 could be detected only in the tetrazine-treated samples (Fig. 5D).
Fig. 5.
Identification and characterization of kupyaphores from Mtb (A) Three-dimensional heat map plot shows masses uniquely identified from biofilm extracts of Mtb cultures but not planktonic cultures at various retention times. Three peaks with the highest fold-change between biofilm and planktonic cultures corresponded to m/z [M+H]+= 722.55 (II), 834.68 (V), and 848.69 (VI), are marked here in red. (B) MS/MS fragmentation pattern revealed common dipeptide backbone of ornithine and phenylalaninol in all the seven kupyaphore species (I–VII) observed in the complete spectra. Common fragments identified in the positive ion mode for all these masses show identical pattern. The predicted fragments are color coded along with their masses in the table. (C) Color change of Py-tetrazine from pink to yellow observed upon addition of WT Mtb biofilm extract. (D) Extracted ion chromatogram (EIC) for [M+H]+ = 238.1014 corresponding to universal product, Py-aminepyrazole, in biofilm culture extract with and without tetrazine addition. (E) MS-based comparative analysis of EIC for biofilm organic extract and synthetic chemical standard at [M+H]+ = 828.7300 and 848.6987. Addition of formic acid to biofilm organic extract shifts the mass of parent ion (m/z-848.6987) to that of synthetic chemical standard of amino substituted lipopeptide (m/z-828.73) concomitant with conversion of isonitrile to amine upon acid hydrolysis. The observed masses for I–VII are within 10 ppm mass error tolerance at MS1 or MS/MS for each mass.
Furthermore, to confirm the isonitrile moiety and the overall chemical structure of the metabolite, we synthesized an analog of C18 lipopeptide with amine functionality at the β position. Previous studies had reported facile conversion of nitriles into amines on reaction with mild acid (41). Toward this end, protected β-amino fatty acid was coupled with L-ornithine and L-phenylalaninol sequentially, followed by deprotections resulting in the desired lipopeptide (SI Appendix, Fig. S7 A–K). Addition of formic acid to the biofilm metabolite extract resulted in disappearance of m/z for [M+H]+ ion 848.6987 (VI), having a retention time of 36 min, and a new peak could be observed at m/z for [M+H]+ ion 828.73 with a retention time at 34 min. This new peak corresponded to the chemical synthetic standard compound (Fig. 5E). Additionally, both the parent peak and synthetic standard had identical MS/MS fragments (SI Appendix, Fig. S7L). Our studies thus identify and characterize a series of diacyl-diisontirile lipopeptides from Mtb that consists of a dipeptide core of ornithine and phenylalaninol, where the amino-groups are acylated with isonitrile-containing fatty acyl chains ranging from C13 to C19. Based on their metal acquisition functionality, we propose to name these Mtb diacyl-diisonitrile lipopeptides as “kupyaphores” (Kupya in Sanskrit refers to rare metals and phores means carrier in Latin), and this gene cluster could be recognized as kupya A-E.
Characterization of Kupyaphores in Mtb-Infected Mice Lungs and Human Clinical Strains.
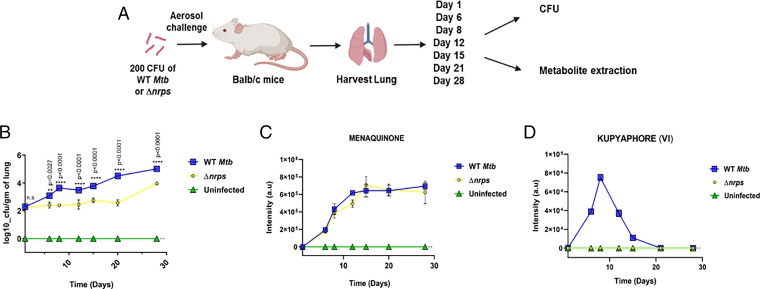
We next set out to understand the temporal regulation of this zinc acquisition machinery in the mouse infection model. Previous mice infection studies with this Δnrps mutant in immunocompetent and immunocompromised mice showed reduced lung pathology with significantly increased mice survival rates (22). This phenotype was attributed to impairment in the early infection events associated with the mutant Mtb strain. We therefore carefully dissected the early infection events from day 1 to day 28 (Fig. 6A). The mutant strain indeed showed more than one log-phase growth defect till week 3 postinfection, as compared to WT Mtb (Fig. 6B). This compromised growth of mutant Mtb highlights the significant role of this gene cluster in successful establishment of TB infection.
Fig. 6.
Kupyaphores are required for establishment of Mtb infection in vivo. (A) Study design for murine TB infection by WT and Δnrps Mtb strains. (B) Bacterial burden in the lungs of uninfected, WT and Δnrps Mtb strains infected mice as measured by CFU enumeration. (C) MRM-based LC-MS measurements of Mtb respiratory quinone, menaquinone, from the lungs of uninfected, WT, and Δnrps Mtb-infected mice. (D) Plot shows MRM-based semiquantitative LC-MS measurements of kupyaphore species: VI of m/z for [M+H]+ = 848.69 from lungs of uninfected, WT, and Δnrps Mtb-infected mice. Data represent mean ± SEM (n = 3 biological replicates per strain at each time point).
A portion of the lung tissue was also utilized for extraction of metabolites using ethyl acetate. LC-MS analysis of the tissue organic extracts of WT Mtb- and Δnrps-infected lung tissues showed clear signals at m/z of [M+H]+ = 787.6315, which corresponds to the Mtb-specific respiratory quinone, menaquinone (Fig. 6C). This confirms the presence of Mtb in the host lung tissue. Menaquinone peak was absent in the uninfected animal’s lung tissue extract. Furthermore, we performed semiquantitative measurements for kupyaphore masses using multiple reaction monitoring (MRM) LC-MS analysis. Signals for kupyaphores VI, II, and VII species could be detected only from the WT Mtb-infected lung tissues and these were absent in Δnrps mutant and uninfected lung tissue samples. Kupyaphore was absent on day 1 in WT Mtb and metabolite peaks built up from day 6 to day 15, then declined rapidly by day 21. Kupyaphores completely disappeared by day 28. Two series of kupyaphores could be identified based on their masses: one symmetrical corresponding to two C18 acyl chains of this lipopeptide and one asymmetrical, both modified with isonitrile groups (Fig. 6D and SI Appendix, Fig. S8 A and B). These studies suggest that kupyaphores are indeed an early response of Mtb to the host challenge and these metabolites disappear as the host–pathogen establish a more mutually coexistential harmony.
Furthermore, to recognize relevance of kupyaphores in pathophysiological context, we analyzed clinical strains obtained from TB patients for the production of these metallophores. MS analysis of ethyl acetate extract from four clinical Mtb strains showed robust expression of several of these kupyaphores (SI Appendix, Fig. S8C). These studies thus determine the significance of kupyaphores in Mtb pathogenesis.
Kupyaphores Are Secreted by Mtb for Zinc Acquisition.
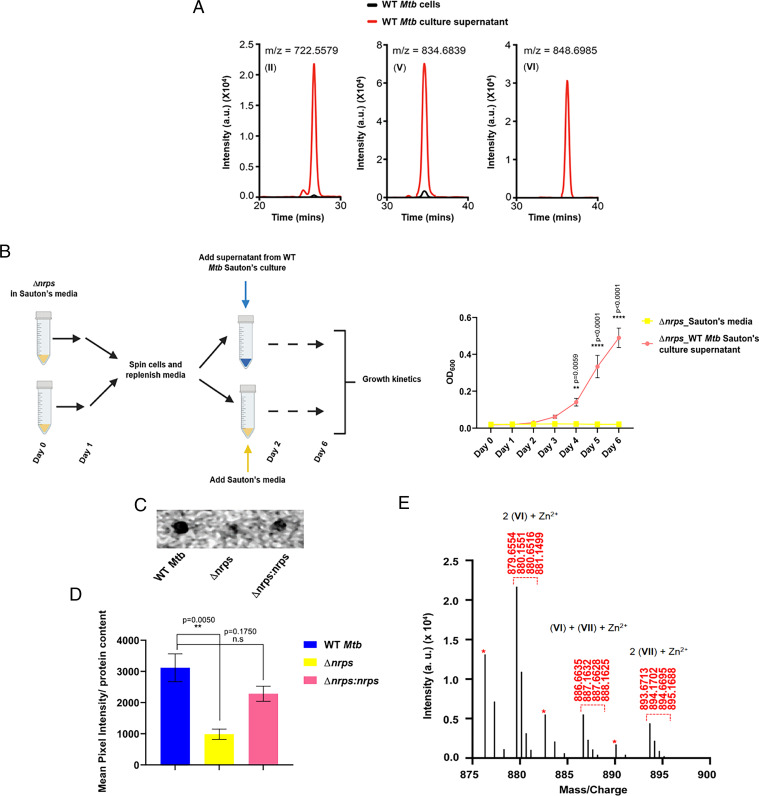
In order to capture zinc from the extracellular environment, kupyaphores would have to be secreted out of the Mtb cells. After chelating zinc, these zinc–kupyaphore complexes will have to be translocated across the Mtb cell envelope. To study kupyaphore trafficking, we compared the levels of kupyaphores in the Mtb cells grown in Sauton’s medium and in the supernatant of cultures. While we observed low levels of kupyaphore species I to VII in the cells, significant enrichment was observed in the media supernatant under both low- and high-zinc conditions (Fig. 7A). As expected, no kupyaphores could be identified from the Δnrps grown in Sauton’s media. Since Δnrps cells do not grow in the minimal medium, we then asked whether exogenous addition of kupyaphores, produced by WT Mtb, could rescue the Δnrps growth defect. Toward this end, we cultured the WT Mtb cells in Sauton’s medium for 7 d. The conditioned supernatant from this WT Mtb cells was collected and added to the Δnrps strain. We then followed the growth kinetics of the Δnrps strain for 6 d and indeed observed robust growth for the Δnrps culture supplemented with WT Mtb supernatant (Fig. 7B). As control, we had to use fresh medium, since the Δnrps strain does not grow any further than the initial OD in the Sauton’s medium. As expected, no growth was observed for Δnrps cells supplemented by fresh medium (Fig. 7B). Clearly, the presence of kupyaphores in WT Mtb supernatant is able to rescue the growth defect by transporting essential micronutrient zinc.
Fig. 7.
Kupyaphores are secreted by Mtb for zinc acquisition (A) EIC of kupyaphore species II, V, and VI corresponding to m/z for [M+H]+ = 722.55, 834.68, and 848.69 detected from organic extracts of supernatant and cells of WT Mtb grown in Sauton’s medium. (B) Study design and growth kinetics of Δnrps supplemented with supernatant from WT Mtb cultures as compared to Δnrps grown in Sauton’s medium alone. Addition of WT Mtb supernatant rescued the growth defect observed in Δnrps. (C) Radioactive zinc-65 feeding of WT, Δnrps, and Δnrps:nrps Mtb strains. After 4 h, cells were lysed and spotted for estimating intracellular zinc-65 accumulation by autoradiography. (D) Quantitation of autoradiograph signals of three strains as mean pixel intensity in arbitrary units normalized to protein content estimated for each strain. (E) MS1 spectra showing doubly charged peaks corresponding to dimeric kupyaphore-zinc metabolites from WT Mtb extract supplemented with zinc. Putative zinc adducts for both homodimeric and heterodimeric kupyaphore species could be detected. Single charged ions are marked with red asterisks. The corresponding mass peaks were absent from WT Mtb supernatant extract, which was not supplemented with zinc. The calculated masses for metabolites are within 5 ppm mass error tolerance at MS1. Data represents mean ± SEM (n = 3 biological replicates).
Moreover, to assess whether secreted kupyaphores are involved in zinc uptake from the environment, we incubated WT, Δnrps, and Δnrps:nrps Mtb strains with 0.5 μM radioactive zinc-65 for 4 h. After 4 h, the internalized zinc-65 levels were measured in the cells. Significantly low radioactive counts were noted in Δnrps Mtb cells, when compared to WT and Δnrps:nrps Mtb strains, as measured by autoradiogram (Fig. 7 C and D). Together, these two studies directly demonstrate that absence of kupyaphores results in decreased zinc uptake efficiency, resulting in Δnrps mutant growth defect. We also attempted to characterize dimeric zinc-bound kupyaphores using MS. Supernatant of WT Mtb grown with zinc supplementation showed doubly charged peaks that correspond to the kupyaphore–zinc complex and were absent in WT Mtb cultures grown in the absence of zinc in chelated Sauton’s media (Fig. 7E). Our study thus demonstrates role of kupyaphore in restoration of bacterial zinc metabolic imbalance.
Putative Mechanism of Zinc Release from Kupyaphore by Mtb Isonitrile Hydratase.
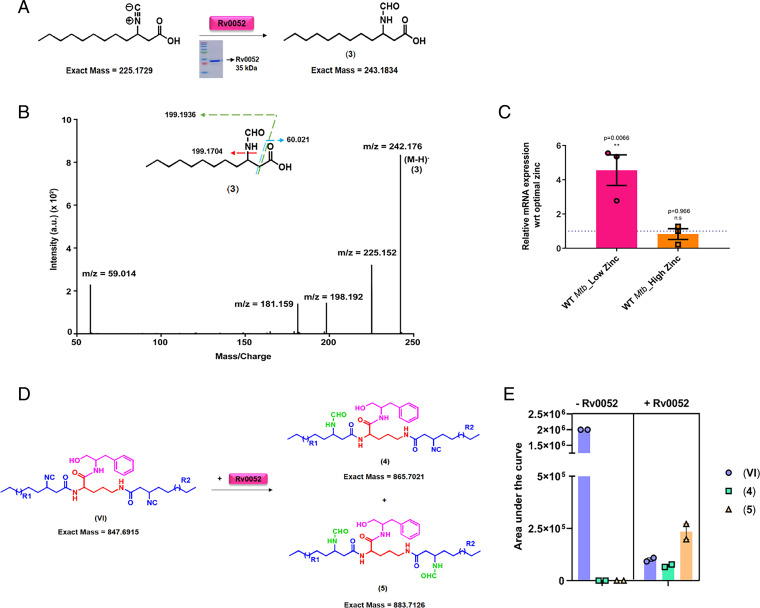
Since zinc is a redox-inert metal, the release of zinc from metallophores cannot follow the classic reductive release mechanism known for iron-siderophore. In the case of enterobactin, siderophore hydrolase is known to modify the scaffold mediating iron release (15). We therefore analyzed the Mtb genome to investigate the presence of putative isonitrile modifying enzyme. This enzyme activity (InhA) was first reported from Pseudomonas putida and was recently characterized (SfaF) from the SF2768 biosynthetic cluster of S. thioluteus (42, 43). Protein sequence analysis of the Mtb H37Rv genome coding sequence with InhA identified Rv0052 with an E-value score of 5e−31. Careful analysis of Rv0052 sequence with the three-dimensional structure (PDB ID code 3NON) indicated the absence of an important catalytic site (D17) from the annotated Rv0052. The gene Rv0052 is located between 57410 and 57973 bp in the genome of H37Rv. Analysis of upstream sequences revealed another start site 57 bp upstream in-frame with annotated Rv0052. A protein coding sequence of this open reading frame shows conservation of the catalytic residue D17. This protein was conserved across all MTBC strains and was absent from nonpathogenic mycobacterial species, where the kupya cluster is absent (SI Appendix, Fig. S9A).
To examine biochemical function, we cloned and expressed Rv0052 in BL21-DE3. The protein was purified and enzymatic assays were performed along with Rv0098 and Rv0097 (Fig. 8A). A clear peak corresponding to m/z of 243.176 for the N-formamide product could be observed only in the reactions supplemented with Rv0052 (SI Appendix, Fig. S9B). MS/MS fragmentation in the negative ion mode also provided further confidence to the hydration of isonitrile unit (Fig. 8B). Interestingly, Rv0052 expression was found to be significantly up-regulated by qRT-PCR only in low-zinc conditions and showed no induction under high-zinc conditions (Fig. 8C). Next, to investigate whether Rv0052 can directly modify kupyaphores, we incubated WT Mtb culture extract with purified Rv0052. Significant reduction in kupyaphore VI mass signals could be observed upon treatment with Rv0052, with concomitant appearance of new peaks corresponding to mono- and di-formamide kupyaphore analogs of m/z for [M+H]+ = 866.7021 and 884.7216, respectively (Fig. 8 D and E). Mass peaks corresponding to formamide and diformamide kupyaphore analogs could not be detected in the absence of Rv0052 (SI Appendix, Fig. S9C). These studies clearly demonstrate that Rv0052 can modify the isonitrile moiety of kupyaphore, which reduces the Lewis base character and destabilizes the coordinate complex bond with zinc. We thus propose that this could be a putative mechanism of zinc release from kupyaphores under low-zinc conditions.
Fig. 8.
Release of zinc from kupyaphores is mediated by Mtb isonitrile hydratase. (A) Reaction scheme for in vitro enzymatic assay of Rv0052. (B) MS/MS spectra with fragmentation pattern and products marked for expected product 3, confirming the presence of formamide product in the assay. The observed masses are within 10 ppm mass error tolerance at MS1 or MS/MS for each mass. (C) qRT based mRNA expression analysis of Rv0052 in WT Mtb under low- and high-zinc condition as compared to optimal-zinc growth condition. (D) Reaction scheme for Rv0052 enzymatic assay with kupyaphores. (E) Relative MS intensities of kupyaphore species VI of m/z for [M+H]+= 848.69, and corresponding monoformamide 4 and diformamide 5 analogs in WT Mtb extract treated with or without Rv0052. The calculated masses for metabolites are within 5 ppm mass error tolerance at MS1. All assays in B and E were performed in triplicates and duplicates, respectively, with reproducible results each time.
Discussion
Given that there is virtually no free-zinc in the cell and that this micronutrient cannot be produced de novo, it becomes mandatory for pathogens to acquire these from the host pools. Bacteria have therefore evolved rather intricate molecular mechanisms of metal ion sensing, uptake, efflux, and allocation to maintain homeostasis (44). We herein decipher a mechanism of zinc acquisition whereby Mtb secretes zinc-selective metallophores, named kupyaphores, to support its intracellular survival. Kupyaphores belong to the recently identified family of diacyl-diisonitrile lipopeptides and is probably unique as a metabolite characterized directly from the host. We show that kupyaphores are produced by Mtb during biofilm formation or when grown at low- or high-zinc conditions in minimal medium and also from the lungs of Mtb-infected mice, as well as from clinical strains during early passages. Interestingly, transcriptional induction of the operon is more dramatic in macrophages than low- or high-zinc. The biosynthetic five-gene cluster consisting of bimodular NRPS is conserved only in pathogenic MTBC species. It is kind of surprising that this pathway is absent in saprophytic mycobacteria, which otherwise contain two types of siderophores for iron acquisition.
The chemical features of kupyaphores are typical of Mtb lipidic metabolites. Long-fatty acyl chains facilitate these molecules to transverse the complex Mtb cell envelope. The electron-rich isonitrile functionality forms a coordinate bond and the affinity for chelation to zinc metal ions is probably governed by the structural architecture. In varying zinc conditions, kupyaphores shuttle in and out of the Mtb cells by using transporters that are yet to be characterized. Our studies show that the kupyaphore–zinc complex can be translocated inside. The intracellular release of zinc ions may be mediated by Mtb-encoded isonitrile hydratase (Rv0052) that modifies isonitriles to corresponding formamides. Surprisingly, we observed a stringent regulation of kupyaphores in both macrophages and in the lungs of infected mice, and these metabolites are present transiently during the early phase of infection. It will be interesting to identify mechanisms by which these metabolites are degraded. Recent study suggests that zinc-limited Mtb exhibit a global adaptive response that affects physiology, confers resiliency to oxidative stress, and possibly leads to increased virulence (45).
Kupyaphores show biological selectivity for the zinc and kupya biosynthetic cluster is transcriptionally activated under both low- and high-zinc levels. We therefore propose that kupyaphores may be potentially playing a role in both zinc acquisition, as well as quenching of toxic zinc levels (SI Appendix, Fig. S9D). Previous studies have indicated other isonitrile lipopeptides to chelate copper ions (20, 21, 46). Although at this stage we do not completely rule out the ability of kupyaphore to also bind to copper, the detailed binding affinities studies for various metal ions awaits total synthesis of kupyaphores. It is rather peculiar that the identification of these zinc chelating metallophores took this long, as the iron siderophores, mycobactins, of Mtb were discovered 70 y ago (47). The secretion of kupyaphores by WT Mtb actively augments free-zinc pools within phagosomes, as detected by zinc-selective dye. In contrast, the Δnrps mutant shows significantly reduced signals and the levels of free-zinc are similar to uninfected macrophages. However, the total zinc pools within the macrophages in the case of mutant infection are maintained, suggesting that the Δnrps mutant is unable to mobilize host zinc in the absence of kupyaphores. Our results thus suggest that kupyaphores can efficiently modulate host free-zinc redistribution and homeostasis; mechanisms underlying this triggered free-zinc redistribution require further investigation.
From a clinical perspective, low zinc levels have been reported in sera of TB patients as compared to healthy controls (48). Surprisingly, zinc supplementation does not restore zinc levels nor improve the clinical outcome in TB patients (49). Interestingly, studies with a mouse cachexia model of cancer have shown that proinflammatory responses modulate expression of zinc transporter, leading to a drop in serum zinc levels with concordant accumulation of zinc in tissues resulting in cachexia (50). With the discovery of kupyaphores as Mtb zinc acquisition machinery, it will be interesting to explore the dynamic processes of zinc mobilization and redistribution that could influence TB pathogenesis, and investigate relatively unexplored areas of TB-associated cachexia.
Methods
Bacterial Culture.
M. tuberculosis H37Rv were routinely grown in Middlebrook 7H9 culture medium supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC), 0.05% Tween-80 under shaking conditions or on Middlebrook 7H11 agar with 10% OADC at 37 °C. For metal studies, Mtb strains were grown in chelated Sauton’s medium, details of which are provided in the SI Appendix. Mtb biofilms were grown in Sauton’s medium (without Tween-80 or tyloxapol) by incubation without shaking at 37 °C for 5 wk under humidified conditions.
Determination of Total Metal Ions in In Vitro Bacterial Cultures.
Briefly, 0.8 to 1.0 OD WT Mtb cultures grown in chelated Sauton’s medium supplemented with ZnSO4 were lysed by boiling in 0.1% SDS and 0.2% HNO3 for 15 min and total metal concentrations were measured by ICP-MS (ThermoXcaliber II). Details provided in the SI Appendix.
Determination of Free Zinc in Macrophage Infection Studies.
Macrophage cultures uninfected or infected with mCherry labeled Mtb strains were stained with 0.5 μM FZ3-AM and fixed with 4% paraformaldehyde. Details of staining protocol are provided in the SI Appendix. Fixed cells were then mounted with DAPI. DAPI+, mCherry+, and FZ3 green+ cells were selected for each of the triplicate samples by confocal microscopy using Zeiss LSM980 and mean pixel intensity for FZ3 signals was analyzed using ImageJ.
Extraction and Analysis of Kupyaphores from Mtb Cultures.
Middlebrook 7H9 grown planktonic and biofilm cultures of WT Mtb, Δnrps, and Δnrps:nrps were harvested and weighed. Supernatant and cells of the WT Mtb strain from planktonic cultures of chelated Sauton’s medium supplemented with and without zinc were also collected separately. Low-molecular-weight molecules were then extracted by ethyl acetate and analyzed by an information dependent acquisition scanning on a Sciex X500R QTOF mass spectrometer fitted with an ExionLC UPHLC system using the SciexOS software using previously reported methods (36, 37). Details are provided in the SI Appendix.
Mtb Metabolite Isolation upon Murine TB Infection.
For Mtb metabolite extraction from mice, 0.1 g of lung tissue was taken from left apical lobe of uninfected and infected mice at indicated time points. Low-molecular-weight metabolites were extracted by homogenizing tissues in ethyl acetate (36, 37). All the species analyzed were quantified using the MRM-HR LC-MS method on a Sciex X500R QTOF mass spectrometer, details of which are provided in the SI Appendix.
Radioactive Zinc-65 Uptake Assay.
An equal number of WT, Δnrps, and Δnrps:nrps Mtb cells were incubated with 0.5 μM of radioactive zinc-65 for 4 h. After 4 h, cells were lysed. Intracellular radioactive count was then measured by autoradiography and quantitated using ImageJ analysis. Mean pixel intensity so obtained was then normalized to protein content estimated by BCA for each sample. Details are provided in the SI Appendix.
Statistical Analysis.
GraphPad Prism 8 software was used for statistical analysis. Statistical significance was analyzed by Student’s t test or one-way or two-way ANOVA, with P values mentioned for each data point when applicable. Data were plotted as the mean, with error bars representing SEM of three biological replicates for all experiments, unless stated otherwise.
Ethics Declaration.
All mouse studies described in this paper received formal approval from the National Institute of Immunology–Institutional Animal Ethics Committee (NII-IAEC 440/17) following the guidelines outlined by the Committee for the Purpose of Control and Supervision of Experiments on Animals, Government of India. Studies on clinical strains isolated from TB patients described in this paper have approval from Institutional Review Board (IRB) Christian Medical College, Vellore (IRB Min No.7239). The IRB reviewed and discussed the project after going through IRB application format, patient information sheet, consent form, and other documents. Informed consent was taken from all the participants involved in the study. All of these documents were submitted to Institutional Human Ethical Committee of the Council of Scientific and Industrial Research–Institute of Genomics and Integrative Biology.
Supplementary Material
Acknowledgments
We thank Dr. Apoorva Bhatt for providing us with Mycobacterium tuberculosis H37Rv cosmid spanning Rv0096 to Rv0109 for our complementation studies; the Tuberculosis Aerosol Challenge Facility staff at the International Centre for Genetic Engineering and Biotechnology (New Delhi, India) for their kind help; the Department of Biotechnology (DBT) for institutional support provided to National Institute of Immunology; and Saddam Shekh and Arnab Chakraborty (both from the Indian Institute of Science Education and Research Pune) for technical assistance. R.S.G. acknowledges support from a J. C. Bose fellowship and from DBT (Grant BT/PR20085/MED/291213/2017). This work was supported by DBT/Wellcome Trust India Alliance Grant IA/I/15/2/502058 (to S.S.K.); and a Department of Science & Technology–Funds for Improvement of S&T Infrastructure Development grant to the Department of Biology, Indian Institute of Science Education and Research, Pune. A.K.O. acknowledges support from NIH Grants AI132422, AI144474, and AI163599.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110293119/-/DCSupplemental.
Data Availability
Detailed codes used for the analysis have been submitted to the GitHub repository (https://github.com/viv3kanand/MTU-Manuscript/). The datasets generated during or analyzed during the current study can be accessed using the National Center for Biotechnology Information BioProject (ID PRJNA701877). All other study data are included in the main text and/or SI Appendix.
References
- 1.Rubin E. J., Troubles with tuberculosis prevention. N. Engl. J. Med. 370, 375–376 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Kaufmann S. H., Cole S. T., Mizrahi V., Rubin E., Nathan C., Mycobacterium tuberculosis and the host response. J. Exp. Med. 201, 1693–1697 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neyrolles O., Wolschendorf F., Mitra A., Niederweis M., Mycobacteria, metals, and the macrophage. Immunol. Rev. 264, 249–263 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zondervan N. A., van Dam J. C. J., Schaap P. J., Martins Dos Santos V. A. P., Suarez-Diez M., Regulation of three virulence strategies of Mycobacterium tuberculosis: A success story. Int. J. Mol. Sci. 19, 347 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hood M. I., Skaar E. P., Nutritional immunity: Transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 10, 525–537 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner D., et al. , Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell’s endosomal system. J. Immunol. 174, 1491–1500 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Botella H., et al. , Mycobacterial p(1)-type ATPases mediate resistance to zinc poisoning in human macrophages. Cell Host Microbe 10, 248–259 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolschendorf F., et al. , Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 108, 1621–1626 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maciag A., et al. , Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J. Bacteriol. 189, 730–740 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mikhaylina A., Ksibe A. Z., Scanlan D. J., Blindauer C. A., Bacterial zinc uptake regulator proteins and their regulons. Biochem. Soc. Trans. 46, 983–1001 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y., et al. , Zinc depletion induces ribosome hibernation in mycobacteria. Proc. Natl. Acad. Sci. U.S.A. 115, 8191–8196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez G. M., Voskuil M. I., Gold B., Schoolnik G. K., Smith I., ideR, An essential gene in mycobacterium tuberculosis: Role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70, 3371–3381 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chao A., Sieminski P. J., Owens C. P., Goulding C. W., Iron acquisition in Mycobacterium tuberculosis. Chem. Rev. 119, 1193–1220 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krithika R., et al. , A genetic locus required for iron acquisition in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 103, 2069–2074 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsen N. A., Lin H., Wei R., Fischbach M. A., Walsh C. T., Structural characterization of enterobactin hydrolase IroE. Biochemistry 45, 10184–10190 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Ghssein G., et al. , Biosynthesis of a broad-spectrum nicotianamine-like metallophore in Staphylococcus aureus. Science 352, 1105–1109 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Lhospice S., et al. , Pseudomonas aeruginosa zinc uptake in chelating environment is primarily mediated by the metallophore pseudopaline. Sci. Rep. 7, 17132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grim K. P., et al. , The metallophore staphylopine enables Staphylococcus aureus to compete with the host for zinc and overcome nutritional immunity. MBio 8, e01281-e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morey J. R., Kehl-Fie T. E., Bioinformatic mapping of opine-like zincophore biosynthesis in bacteria. mSystems 5, e00554-e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L., et al. , Diisonitrile natural product SF2768 functions as a chalkophore that mediates copper acquisition in Streptomyces thioluteus. ACS Chem. Biol. 12, 3067–3075 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Harris N. C., et al. , Biosynthesis of isonitrile lipopeptides by conserved nonribosomal peptide synthetase gene clusters in Actinobacteria. Proc. Natl. Acad. Sci. U.S.A. 114, 7025–7030 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhatt K., et al. , A nonribosomal peptide synthase gene driving virulence in Mycobacterium tuberculosis. MSphere 3, e00352-e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trivedi O. A., et al. , Dissecting the mechanism and assembly of a complex virulence mycobacterial lipid. Mol. Cell 17, 631–643 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Baral B., Akhgari A., Metsä-Ketelä M., Activation of microbial secondary metabolic pathways: Avenues and challenges. Synth. Syst. Biotechnol. 3, 163–178 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell D. R., et al. , Mycobacterial cells have dual nickel-cobalt sensors: Sequence relationships and metal sites of metal-responsive repressors are not congruent. J. Biol. Chem. 282, 32298–32310 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Festa R. A., et al. , A novel copper-responsive regulon in Mycobacterium tuberculosis. Mol. Microbiol. 79, 133–148 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tinaztepe E., et al. , Role of metal-dependent regulation of ESX-3 secretion in intracellular survival of Mycobacterium tuberculosis. Infect. Immun. 84, 2255–2263 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giovannini D., et al. , A new Mycobacterium tuberculosis smooth colony reduces growth inside human macrophages and represses PDIM Operon gene expression. Does an heterogeneous population exist in intracellular mycobacteria? Microb. Pathog. 53, 135–146 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Richards J. P., Cai W., Zill N. A., Zhang W., Ojha A. K., Adaptation of Mycobacterium tuberculosis to biofilm growth is genetically linked to drug tolerance. Antimicrob. Agents Chemother. 63, 1213–1219 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arora P., et al. , Mechanistic and functional insights into fatty acid activation in Mycobacterium tuberculosis. Nat. Chem. Biol. 5, 166–173 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong H. C., Liu G., Zhang Y. M., Rock C. O., Zheng J., The solution structure of acyl carrier protein from Mycobacterium tuberculosis. J. Biol. Chem. 277, 15874–15880 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Harris N. C., et al. , Isonitrile formation by a non-heme iron(II)-dependent oxidase/decarboxylase. Angew. Chem. Int. Ed. Engl. 57, 9707–9710 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walsh C. T., Insights into the chemical logic and enzymatic machinery of NRPS assembly lines. Nat. Prod. Rep. 33, 127–135 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Kapur S., Khosla C., Biochemistry: Fit for an enzyme. Nature 454, 832–833 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Chhabra A., et al. , Nonprocessive [2 + 2]e- off-loading reductase domains from mycobacterial nonribosomal peptide synthetases. Proc. Natl. Acad. Sci. U.S.A. 109, 5681–5686 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelkar D. S., et al. , A chemical-genetic screen identifies ABHD12 as an oxidized-phosphatidylserine lipase. Nat. Chem. Biol. 15, 169–178 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khandelwal N., et al. , Fatty acid chain length drives lysophosphatidylserine-dependent immunological outputs. Cell Chem. Biol. 28, 1169–1179.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ansari M. Z., Yadav G., Gokhale R. S., Mohanty D., NRPS-PKS: A knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res. 32, W405–W413 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bachmann B. O., Ravel J., Chapter 8. Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol. 458, 181–217 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Huang Y. B., Cai W., Del Rio Flores A., Twigg F. F., Zhang W., Facile discovery and quantification of isonitrile natural products via tetrazine-based click reactions. Anal. Chem. 92, 599–602 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ames B. D., et al. , Crystal structure and biochemical studies of the trans-acting polyketide enoyl reductase LovC from lovastatin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 109, 11144–11149 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goda M., et al. , Isonitrile hydratase from Pseudomonas putida N19-2. Cloning, sequencing, gene expression, and identification of its active acid residue. J. Biol. Chem. 277, 45860–45865 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Zhu M., et al. , Tandem hydration of diisonitriles triggered by isonitrile hydratase in Streptomyces thioluteus. Org. Lett. 20, 3562–3565 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Subramanian Vignesh K., Deepe G. S. Jr., Immunological orchestration of zinc homeostasis: The battle between host mechanisms and pathogen defenses. Arch. Biochem. Biophys. 611, 66–78 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dow A., et al. , Zinc limitation triggers anticipatory adaptations in Mycobacterium tuberculosis. PLoS Pathog. 17, e1009570 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Y., Tan D. S., Total synthesis of the bacterial diisonitrile chalkophore SF2768. Org. Lett. 21, 8731–8735 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snow G. A., Isolation and structure of mycobactin T, a growth factor from Mycobacterium tuberculosis. Biochem. J. 97, 166–175 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghulam H., et al. , Status of zinc in pulmonary tuberculosis. J. Infect. Dev. Ctries. 3, 365–368 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Visser M. E., et al. , The effect of vitamin A and zinc supplementation on treatment outcomes in pulmonary tuberculosis: A randomized controlled trial. Am. J. Clin. Nutr. 93, 93–100 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Wang G., et al. , Metastatic cancers promote cachexia through ZIP14 upregulation in skeletal muscle. Nat. Med. 24, 770–781 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Detailed codes used for the analysis have been submitted to the GitHub repository (https://github.com/viv3kanand/MTU-Manuscript/). The datasets generated during or analyzed during the current study can be accessed using the National Center for Biotechnology Information BioProject (ID PRJNA701877). All other study data are included in the main text and/or SI Appendix.