Significance
The enormous complexity of metabolic pathways, in both their regulation and propensity for metabolite cross-talk, represents a major obstacle for metabolic engineering. Self-assembling, catalytically programmable and genetically transferable bacterial microcompartments (BMCs) offer solutions to decrease this complexity through compartmentalization of enzymes within a selectively permeable protein shell. Synthetic BMCs can operate as autonomous metabolic modules decoupled from the cell’s regulatory network, only interfacing with the cell’s metabolism via the highly engineerable proteinaceous shell. Here, we build a synthetic, modular, multienzyme BMC. It functions not only as a proof-of-concept for next-generation metabolic engineering, but also provides the foundation for subsequent tuning, with the goal to create a microanaerobic environment protecting an oxygen-sensitive reaction in aerobic growth conditions that could be deployed.
Keywords: bacterial microcompartment, formate assimilation, synthetic biology, metabolic engineering
Abstract
Formate has great potential to function as a feedstock for biorefineries because it can be sustainably produced by a variety of processes that don’t compete with agricultural production. However, naturally formatotrophic organisms are unsuitable for large-scale cultivation, difficult to engineer, or have inefficient native formate assimilation pathways. Thus, metabolic engineering needs to be developed for model industrial organisms to enable efficient formatotrophic growth. Here, we build a prototype synthetic formate utilizing bacterial microcompartment (sFUT) encapsulating the oxygen-sensitive glycyl radical enzyme pyruvate formate lyase and a phosphate acyltransferase to convert formate and acetyl-phosphate into the central biosynthetic intermediate pyruvate. This metabolic module offers a defined environment with a private cofactor coenzyme A that can cycle efficiently between the encapsulated enzymes. To facilitate initial design-build-test-refine cycles to construct an active metabolic core, we used a “wiffleball” architecture, defined as an icosahedral bacterial microcompartment (BMC) shell with unoccupied pentameric vertices to freely permit substrate and product exchange. The resulting sFUT prototype wiffleball is an active multi enzyme synthetic BMC functioning as platform technology.
The majority of sustainably produced bioproducts are derived from plant-based sugars (1), competing for resources and arable land that could otherwise be used for food crops (2). Therefore, alternative renewable feedstocks for biorefineries are highly desirable and should, ideally, be easily accessible or producible. One promising example is formate (3), which can be derived in many sustainable ways [e.g., photoreduction of CO2 (4)] and is frequently a bio-industrial waste product (3, 5). Although several naturally occurring metabolic pathways assimilate formate (5), organisms endowed with these pathways are largely unsuitable to be used in biorefineries because they are either difficult to cultivate or cannot be genetically modified (6, 7). To address this challenge, recent efforts have focused on engineering formate-assimilating metabolic pathways into model organisms or other microbes amenable to industrial processes (7). One promising approach has been to utilize the reverse direction of the glycyl radical enzyme (GRE) pyruvate formate lyase (PFL) to enable Escherichia coli to efficiently assimilate formate and acetate into the central metabolite pyruvate (8). PFL is a ubiquitous oxygen-sensitive enzyme that plays a central role in many organisms during glucose fermentation by supporting the production of three instead of two ATP molecules (9). Engineering formate assimilation through PFL to funnel the produced pyruvate toward a product will inevitably result in unwanted metabolic cross-talk due to the many distinct metabolic roles of pyruvate. Thus, spatial separation of the production pathway from PFL and downstream enzymes would increase efficiency by decreasing such unwanted deviation of intermediates.
In bacteria, many functionally diverse catabolic enzymes are naturally insulated from the surrounding metabolism by sequestration in bacterial microcompartments (BMCs), provisioning them with a private cofactor pool, minimizing metabolic cross-talk, and shielding the cytosol from toxic intermediates (10, 11). BMCs have evolved to offer metabolic flexibility to the hosting organism; up to six different BMC loci can be found in bacterial genomes (12), which are transcriptionally regulated presumably by their substrates (13). Many types of the BMCs carry out highly oxygen-sensitive reactions (11, 14). One particular group encapsulates GREs, a functionally diverse family of enzymes (15, 16). These GRE-associated microcompartments (GRMs) encapsulate propanediol dehydratase (17, 18) or choline trimethylamine-lyases (19); however, no GRM has been identified that naturally encapsulates the GRE PFL. By mimicking the naturally evolved BMCs and using them as blueprints, we aim to engineer a BMC encapsulating PFL that can function as a metabolic module for further engineering efforts. This platform can potentially be broadly used in production strains by addition of product biosynthesis pathway enzymes to be grown on the abundant feedstocks formate and acetate.
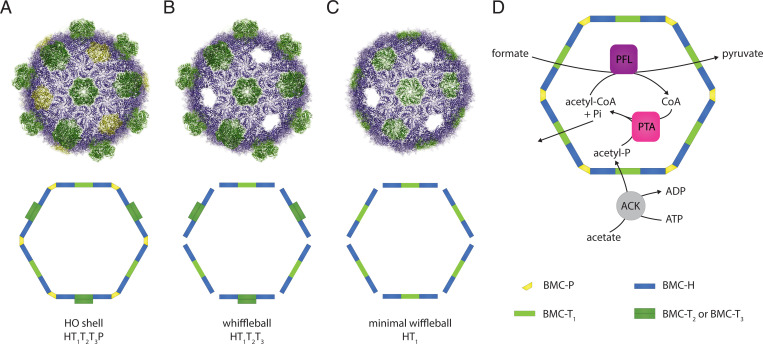
In contrast to eukaryotic organelles, BMCs are not enclosed by a phospholipid membrane but by a protein shell acting as a semipermeable barrier (10, 20). The BMC shell is formed by conserved families of proteins (10, 20) that assemble into three distinct oligomeric building blocks: BMC-H proteins, which assemble into hexagons (21); BMC-T proteins, which also form hexagons (22); and BMC-P proteins, forming pentagons (23). The BMC-Ts subdivide into two types: single-layer BMC-TS and double-layer BMC-TD (10, 20). These components tile together to form an icosahedral shell (24) that serves as a selectively permeable interface with the cytosol. Synthetic BMC shells can form without the presence of the BMC-P proteins (25–27) at the vertices, an architecture we define as “wiffleball.” The 12 6-nm-diameter gaps of wiffleballs permit free exchange of substrate and product across the shell, and therefore we chose this shell architecture for prototyping synthetic enzymatic cores.
Recently developed methods have made it possible to specifically load a BMC shell with a desired nonnative cargo (25, 28–30). One method adopted SpyTag/SpyCatcher bacterial split adhesin domains (31) to covalently bind cargo to the inside of the shell. SpyTag is a short peptide (13 amino acids) that forms an isopeptide bond upon encountering its protein partner, SpyCatcher (31). By incorporating SpyTag into a lumen-facing loop of the shell protein BMC-T and tagging the synthetic cargo with SpyCatcher, it is possible to precisely encapsulate proteins into a synthetic BMC (25). Here we expanded this approach to the SnoopTag/SnoopCatcher system (32), a molecular adhesin that has no cross-talk with SpyTag/SpyCatcher (32). With the ability to specifically target two distinct cargo proteins to the lumen, we built a synthetic GRE-containing wiffleball. It encapsulates active PFL and an active phosphotransacetylase (PTA) to utilize formate and acetyl phosphate as substrates to produce pyruvate, a versatile biosynthetic precursor. It has potential to enhance catalytic efficiency by scaffolding (33) and spatial organization into a BMC shell provides the enzymes with a private cofactor pool and minimizes metabolic cross-talk.
Results
Designing a Synthetic Formate and Acetate Utilizing BMC.
We used the model shell system derived from the BMC of the myxobacterium Haliangium ochraceum (HO-shell) for our synthetic formate utilizing BMC (sFUT) designs (Fig. 1A). This BMC shell can self-assemble without cargo (27) and has been characterized in detail structurally (24). Moreover, the pentamers at the vertices are not essential for shell assembly (25, 27) (Fig. 1B). We define this BMC architecture with unoccupied pentameric vertices as wiffleballs. The wiffleballs leave 6-nm-wide gaps in the shell to permit unrestricted substrate and product exchange, which facilitated testing of iterative sFUT designs. In the HO-shell facets there are three BMC-T proteins: single-layer T1, and double-layer T2 and T3 (Fig. 1). Loading the HO-shell with enzymes can be done using the previously developed method (25) that uses SpyCatcher/SpyTag (31) inserted into T1. When a BMC shell architecture is used that contains only T1 as the BMC-T component, there are 60 contact points to attach cargo into the lumen of the shell [referred to as a minimal shell (25)] (Fig. 1C). However, T1 can be diluted out of the shell by coexpression of T2 and T3 (Fig. 1B). This mitigates potential steric hindrance when assembling the shell with cargo (25).
Fig. 1.
HO-shell architectures and design of the synthetic formate and acetate utilizing BMC. (A) The HO-shell is composed of three different types of building blocks: BMC-H (blue, hexamers), BMC-T (green, trimers/pseudohexamers), and BMC-P (yellow, pentamers) proteins. In the HO-shell are three BMC-T proteins, single-layer T1, and double-layer T2 and T3. (B) HO-shell without pentamers (wiffleball). Shells still form but leave 6-nm-wide vacancies for substrate and product exchange. (C) minimal HO-shell without pentamers (minimal wiffleball) containing only T1 and not T2 or T3. (D) Schematic of the enzymatic reactions of the sFUT for the conversion of formate and acetate into pyruvate. PFL and a PTA need to be coencapsulated to be able to cycle CoA between them; this creates a private cofactor pool for these enzymes.
The designed sFUT function is based on utilizing the reverse direction of the PFL for the conversion of formate and acetate to pyruvate (Fig. 1D). This requires three enzymatic reactions: an acetate kinase (ACK) converting acetate into acetyl phosphate, a PTA to produce acetyl-CoA from acetyl phosphate, and the PFL condensing formate and acetyl-CoA to pyruvate (Fig. 1D). Additionally, an activating enzyme for the PFL is required (PFL-AE) to generate the glycyl radical in the active site of the PFL. PFL-AE is theoretically only needed once to activate the enzyme, which can occur before/during sFUT BMC assembly. Thus, only two enzymes are strictly required to be encapsulated, PFL and the PTA, in order to cycle CoA between them (Fig. 1D). For the PTA, the endogenous E. coli PTA could be used; however, it has been reported to be a homohexamer (34). This oligomeric state could be problematic when encapsulation tags are added to the enzyme, because all six subunits would then function as contact points with the shell proteins and this might not necessarily match the geometry needed to form a complete shell. To decrease the chance of oligomerization interfering with the sFUT BMC assembly, we used the homodimeric E. coli PTA EutD (35). This enzyme has been reported to be a bidirectional PTA (35) and has in fact higher acetyl-CoA–forming activity than the PTA (35), making it the preferred candidate for building the sFUT BMC, structurally and enzymatically.
For our goal to build an active sFUT prototype platform, we used the wiffleball architecture. This simplifies activity testing of the iterate design-build-test-refine cycles to yield an active, context-independent prototype. Future work can build on this platform by engineering permeability, which will include closing the pentamer gaps, and metabolic contextualization into industrial relevant strains with potential to create a microanaerobic environment for the PFL inside the synthetic BMC. This concept of streamlining the building process of complex synthetic “organelles” using a wiffleball to shorten development periods can be applied broadly to BMC synthetic biology.
Specific Encapsulation of Two Cargo Proteins.
Initial versions of sFUT designs aimed to encapsulate a fusion protein of PFL-EutD, an approach that has been successful before encapsulating a synthetic three-domain fusion protein into a carboxysome (36). To identify functional PFL-EutD-adaptor domain fusion proteins for incorporation into a BMC shell, we tested variations of which enzyme carried the adaptor domain, the order in which the proteins were fused, the length of the glycine-serine linker (GS; 5x[GS]) between the fusion proteins, as well as which shell proteins were coexpressed with the cargo. However, disappointingly low expression of the PFL-EutD fusion protein and, presumably, steric hinderance obstructed SpyCatcher/SpyTag conjugation, resulted in no observed encapsulation.
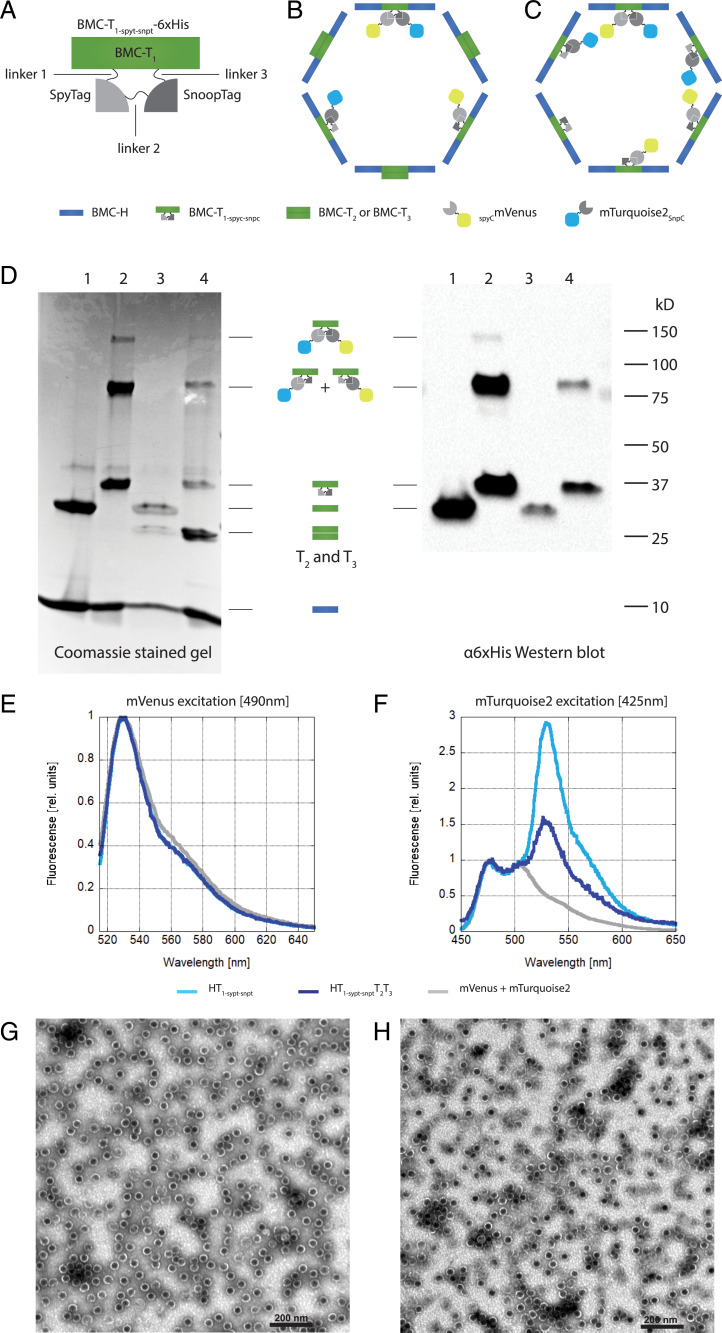
Because of the complications associated with the PFL-EutD fusion protein, we chose to modify the T1 shell protein to encapsulate two different enzymes simultaneously. To achieve this, we added a SnoopTag (32) to the lumen facing loop (between G84 and G86) in T1 that also displays SpyTag (KLGDIEFIKVNK), resulting in a double-tagged shell protein building block (T1-spyt-snpt-6xHis) (Fig. 2A). There are three linkers within the double-tagged T1-spyt-snpt-6xHis (Fig. 2A): the first is between the N-terminal region of T1 and SpyTag, the second between SpyTag and SnoopTag, and the third connecting SnoopTag to the C-terminal sequence of T1. We tested two different linkers in the second position, between SpyTag and SnoopTag: a flexible glycine-serine linker (2x[GGSGG]) and a stiff proline-lysine linker with a flexible GS part (GPKPKPKPKGGSGGGGSGG). The other two linker positions were kept invariable as short, stiff, proline-lysine peptides (5x and 4x [PK]). Encapsulation was tested with two distinct cargo proteins, mVenus (yellow) and mTurquoise2 (blue), fused to SpyCatcher and SnoopCatcher, respectively (Fig. 2 B and C). Both linker variants successfully encapsulated mTurquoise2-SnoopCatcher (mTurquoise2SnpC) and SpyCatcher-mVenus (SCmVenus).
Fig. 2.
Specific encapsulation of two cargo proteins. (A) Schematic of T1-spyt-snpt-6xHis depicting the position of the linkers 1 to 3, the SpyTag, and the SnoopTag. (B) Schematic of a HO-shell wiffleball with T1-spyt-snpt-6xHis T2 and T3 and conjugated cargo mTurquoise2SnpC (blue) and SCmVenus (yellow). (C) Schematic of a HO-shell minimal wiffleball with only T1-spyt-snpt-6xHis and conjugated cargo mTurquoise2SnpC (blue) and SCmVenus (yellow). (D) NiNTA purification of wiffleballs coexpressed with mTurquoise2SnpC and SCmVenus. Lane 1: BMC-H and T1; lane 2: BMC-H and T1-spyt-snpt-6xHis; lane 3: BMC-H, T1, T2 and T3; lane 4: BMC-H, BMC- T1-spyt-snpt-6xHis, T2 and T3. (Left) Coomassie stained SDS/PAGE gel. H T1-spyt-snpt-6xHis and H T1-spyt-snpt-6xHis T2T3 architectures show conjugations between T1-spyt-snpt-6xHis, mTurquoise2SnpC and SCmVenus, either individually (∼80 kD) or together (∼120 kDa). (Right) α6xHis Western blot. T1-spyt-snpt-6xHis contains a 6xHis tag used for detection. Conjugation bands between T1-spyt-snpt-6xHis and the fluorescent proteins can be observed. (E) Fluorescense emission spectrum after mVenus excitation at 490 nm. The emission spectra were normalized to mVenus emission intensity. (F) Fluorescense emission spectrum using mTurquoise2 excitation at 425 nm avoiding mVenus excitation. mVenus emission can be observed in the loaded wiffleballs due to FRET, while this isn’t present when only mVenus and mTurpuoise2 are expressed by themselves without being loaded into the wiffleballs (mVenus+mTurquoise2). Emission spectra were normalized to mTuriquise2 emission intensity. (G) TEM image of H T1-spyt-snpt-6xHis minimal wiffleballs. (Scale bar, 200 nm.) (H) TEM image H T1-spyt-snpt-6xHisT2T3 full wiffleballs. (Scale bar, 200 nm.)
Wiffleball NiNTA purifications are shown in Fig. 2D using a 6xHis tag on T1-spyt-snpt-6xHis. Wiffleballs were coexpressed with the cargo proteins mTurquoise2SnpC and SCmVenus, minimal wiffleballs (BMC-H and T1) with wild-type T1 (HT1), a minimal wiffleball containing the modified T1-spyt-snpt-6xHis (H T1-spyt-snpt-6xHis), and their respective full wiffleballs equivalents (HT1T2T3 and H T1-spyt-snpt-6xHisT2T3), in which not only T1 is present but also T2 and T3. The different plasmids expressing the shell proteins and their resulting shell architecture are summarized in Table 1 and operon schematics are shown in SI Appendix, Fig. S1. The protein bands of T1-spyt-snpt-6xHis conjugated to either one of the fluorescent proteins (∼80 kDa) can be identified on the Coomassie-stained SDS/PAGE gel for both the H T1-spyt-snpt-6xHis and H T1-spyt-snpt-6xHisT2T3 wiffleballs, and was verified by an α6xHis Western blot against the C-terminal 6xHis Tag of T1 and T1-spyt-snpt-6xHis. Additionally, tandem conjugations of SpyTag and SnoopTag with the respective fluorophores were also detected (Fig. 2D). The protein bands containing T1 (unconjugated T1-spyt-snpt-6xHis, T1-spyt-snpt-6xHis–SCmVenus or T1-spyt-snpt-6xHis–mTurquoise2SnpC) are much fainter for the H T1-spyt-snpt-6xHis T2T3 compared to the H T1-spyt-snpt-6xHis wiffleballs because the amount of T1-spyt-snpt-6xHis is lower in the full wiffleball architecture due to the presence of T2 and T3. Nevertheless, these results indicate that in minimal and full shell designs both fluorescent proteins are simultaneously encapsulated and, although less common, sometimes attached to the same T1-spyt-snpt-6xHis protein.
Table 1.
Acronyms of the HO-shell plasmids, the containing genes expressed in a synthetic operon and the resulting BMC architecture
| Plasmid name | Genes in the synthetic operon | BMC shell architecture |
| pHT1 | BMC-H; BMC-T1-6xHis | HT1 |
| pHT1T2T3 | BMC-H; BMC-T1-6xHis; BMC-T2; BMC-T3 | HT1T2T3 |
| pH T1-spyt-snpt-6xHis | BMC-H; BMC-T1-spyt-snpt-6xHis | H T1-spyt-snpt-6xHis |
| pH T1-spyt-snpt-6xHisT2T3 | BMC-H; BMC-T1-spyt-snpt-6xHis; BMC-T2; BMC-T3 | H T1-spyt-snpt-6xHisT2T3 |
Further evidence for the coencapsulation of both fluorescent proteins was provided by fluorescence emission spectra using excitation wavelengths specific for mVenus (490 nm) (Fig. 2E) and mTurquoise2 (425 nm) (Fig. 2F). For the H T1-spyt-snpt-6xHis as well as the H T1-spyt-snpt-6xHisT2T3 wiffleball sample, we noticed that the mTurquoise2 emission spectrum contains mVenus emission at around 540 nm, which is not present when both fluorescent proteins are mixed together without being loaded into wiffleballs (mVenus+mTurqouise2 sample) (Fig. 2F). This suggests FRET between mTurquoise2 and mVenus when loaded into the wiffleballs, consistent with both fluorophores being coencapsulated.
To investigate if complete wiffleballs formed incorporating T1-spyt-snpt-6xHis conjugated to both mTurquoise2SnpC and SCmVenus, we used transmission electron microscopy (TEM) (Fig. 2 G and H). The images revealed structures of about 40 nm in diameter, similar to the published HO-shell TEM images (24, 25) for the minimal (Fig. 2G) as well as the full wiffleballs (Fig. 2H). These results suggest that BMC wiffleballs can be specifically loaded with SpyCatcher- and SnoopCatcher-tagged cargo after addition of SpyTag and SnoopTag to T1 without impairing wiffleball formation.
Addition of Encapsulation Tags to sFUT Cargo.
PFL is a member of the GRE family, some of which are encapsulated in GRMs (15, 16). Most of the GREs in BMCs have an encapsulation peptide, which is not present in cytosolic GREs (15–17). The majority of the encapsulation peptides for these GREs are not found at the N or C terminus, but are located in an internal loop (15–17). Therefore, we tested if we could use the corresponding loop in the PFL to integrate the SpyCatcher domain into the PFL (between E551 and D552). Additionally, we created N- and C-terminal fusions of SpyCatcher to the PFL (SI Appendix, Fig. S2). We tested the expression level of the three different PFL variants: SpyCatcher loop insertion (PFLspyc), N-terminal SpyCatcher fusion (PFL-Nspyc), and C-terminal SpyCatcher fusion (PFL-Cspyc) (SI Appendix, Fig. S4A). Although the N-terminal fusion of the SpyCatcher to the PFL (PFL-Nspyc) did not express well, the two other variants and the unmodified PFL control expressed well and are visible on a Coomassie-stained gel (SI Appendix, Fig. S4A). However, most of the proteins are found in the insoluble fraction; only the wild-type PFL and PFL-Cspyc are found in significant amounts in the soluble fraction (SI Appendix, Fig. S4A), indicating that the PFL-Cspyc was the most promising variant to be used in the sFUT prototype. Additionally, we created and tested an N-terminal and C-terminal fusion of SnoopCatcher to EutD (EutD-Nsnpc and EutD-Csnpc, respectively) (SI Appendix, Fig. S3). Both variants expressed equally well (SI Appendix, Fig. S4B) and we proceeded to use the EutD-Csnpc for the sFUT construction.
Activity Assays of SpyCatcher Fusion Variants to the PFL.
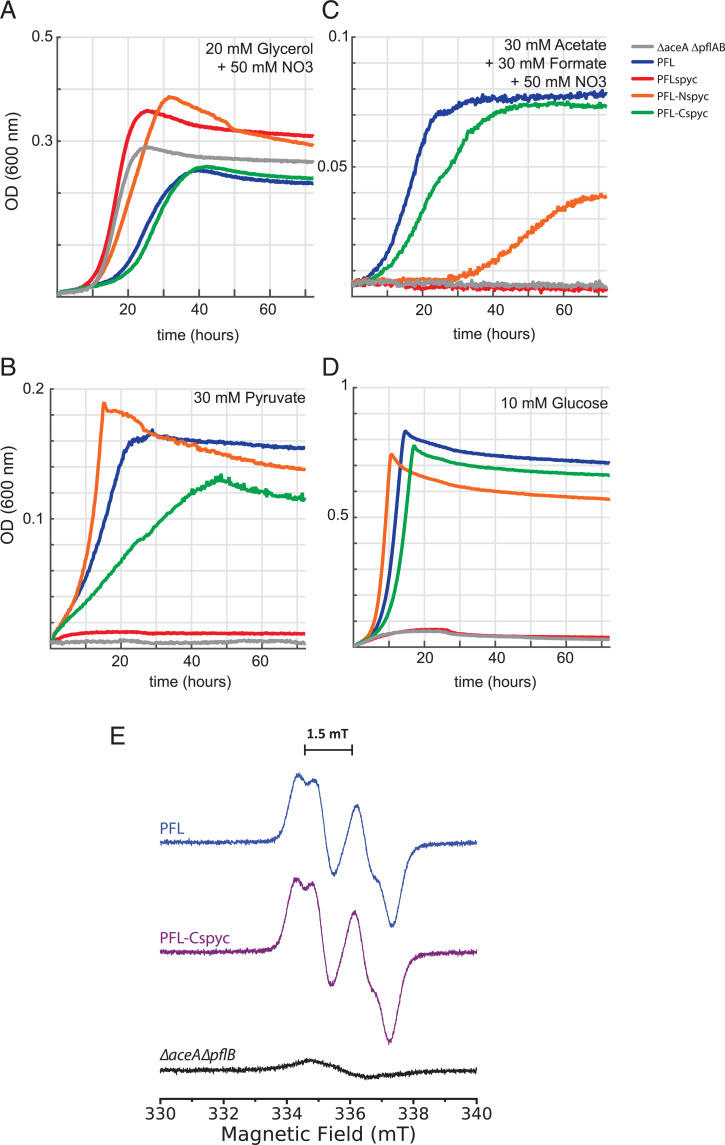
We tested enzymatic activity of our PFL fusion proteins in forward and reverse directions by expressing them from a plasmid in anaerobically grown E. coli lacking the endogenous PFL and its activating enzyme (ΔaceAΔpflAB) under different carbon sources (Fig. 3). The plasmids included genes for the PFL-AE as well as METK to synthesize the substrate S-Adenosyl methionine for the PFL-AE (SI Appendix, Fig. S2). Fig. 3A shows a positive control experiment where strains were grown anaerobically on glycerol using nitrate as an electron acceptor, a condition that doesn’t require PFL activity for growth, as the cells can use pyruvate dehydrogenase in the presence of an electron acceptor. Under these conditions, all strains were able to grow, demonstrating that none of the constructs caused a lethal phenotype when expressed. Anaerobic growth on pyruvate (Fig. 3B) and glucose (Fig. 3D) is only possible when the forward direction of the PFL is functioning. Under these conditions, strains with N- and C-terminal PFL fusions showed similar growth compared to the strain with unmodified PFL. However, the negative control (no PFL) and the PFL variant with the SpyCatcher insertion into the loop did not show any growth in these conditions, indicating the SpyCatcher insertion abolishes PFL activity in the forward direction. PFL activity in the reverse direction is tested by growth on acetate and formate (Fig. 3C). The N-terminal fusion shows lagging growth in this condition when compared to the strain expressing the wild-type PFL, which could be caused by lower expression, insolubility (SI Appendix, Fig. S4A), and partial inhibition of the enzyme by the presence of SpyCatcher. The internal loop insertion of SpyCatcher into the PFL again showed no growth, demonstrating complete loss of activity of this PFL variant in both directions. However, this experiment revealed that the strain expressing PFL-Cspyc has comparable growth kinetics to a strain expressing unmodified PFL, suggesting that this PFL variant is active to a similar degree as wild type PFL. Furthermore, we employed electron paramagnetic resonance (EPR) spectroscopy to confirm the PFL-Cspyc is successfully converted to its active glycyl radical-containing form by PFL-AE in vivo (Fig. 3E). Whole cells expressing either the PFL-Cspyc, or our unmodified PFL+ control, exhibited identical EPR spectra with a principal doublet splitting of ∼1.5 mT characteristic of the PFL glycyl radical (37). As expected, this radical signal was not detected in the PFL deletion strain (ΔaceAΔpflB). These experiments strongly suggest that PFL-AE is able to interact with PFL-Cspyc and generate the glycyl radical necessary for PFL activity, making this variant the best candidate to be used in the sFUT.
Fig. 3.
Selection for PFL activity in vivo. Growth curves of E. coli strains using the PFL deletion background (ΔaceAΔpflAB) and complementation with the PFL variants. (A) Positive control by growth on glycerol. (B) Growth on pyruvate, testing for the forward direction of the PFL. Negative control and PFLspyc (loop insertion) failed to grow. (C) Growth on acetate and formate, testing for the activity of the reverse direction of the PFL. Negative control and PFLspc failed to grow, PFL-Nspyc shows slow growth, while WT PFL and PFL-Cspyc show similar growth kinetics. As a negative control, all strains have also been grown in aerobic conditions using formate and acetate as carbon source; none of the strains were able to show any growth. (D) Growth on glucose, testing for activity of the forward direction of the PFL. Negative control and PFLspyc (loop insertion) failed to grow. (E) X-Band EPR spectra of E. coli ΔaceAΔpflB whole cells (black) expressing plasmid-encoded wild-type PFL (blue) or PFL-Cspyc (purple). Spectra were collected at 253 K and are the average of eight traces per sample.
Assembly of Active sFUT Synthetic BMCs.
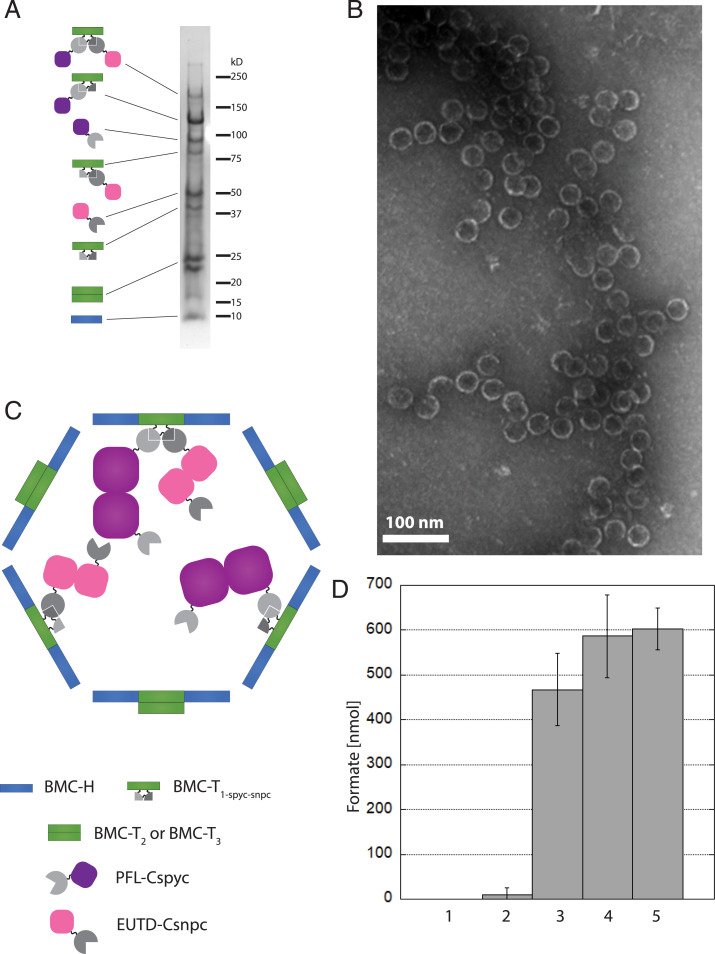
To build the complete sFUT wiffleballs, we coexpressed three synthetic operons: one containing the shell proteins (SI Appendix, Fig. S1), another containing the PFL-Cspyc as well as the PFL-AE and the METK (SI Appendix, Fig. S2), and the third expressing EutD-Csnpc and ACK (SI Appendix, Fig. S3). For the shell operon, we expressed two different versions: the minimal wiffleball (HT1-sypt-snpt) and a full wiffleball (H T1-spyt-snpt-6xHisT2T3); synthetic BMCs produced using both wiffleball versions were purified using the shell purification method described in the Materials and Methods. Although no sFUT BMCs based on the minimal HT1-sypt-snpt wiffleball architecture could be isolated, we were able to obtain a pure fraction of sFUT BMCs using full H T1-spyt-snpt-6xHisT2T3 wiffleballs (Fig. 4A). Estimated molecular weights of the bands on the Coomassie-stained SDS/PAGE match the predicted sizes for T1-spyt-snpt-6xHis in conjugation with PFL-Cspyc and EutD-Csnpc (∼170 kDa), T1-spyt-snpt-6xHis conjugated to PFL-Cspyc (∼120 kDa), PFL-Cspyc (∼95 kDa), EutD-Csnpc conjugated to T1-spyt-snpt-6xHis (∼80 kDa), EutD-CsnpC (∼50 kDa), T1-spyt-snpt-6xHis (∼40kD), T2 and T3 (22 and 23 kDa, respectively), as well as the BMC-H (10 kDa). We confirmed the presence of the proteins by mass spectrometry. To investigate if we indeed built complete sFUT BMCs, we used negative staining with TEM to image the purified assemblies (Fig. 4B).
Fig. 4.
Purification of complete assembled active sFUTs. (A) Coomassie-stained SDS/PAGE gel. T1-spyt-snpt-6xHis in conjugation with PFL-Cspyc and EutD-Csnpc (∼170 kDa), T1-spyt-snpt-6xHis conjugated to PFL-Cspyc (∼120 kDa), PFL-Cspyc (∼95 kDa), EutD-Csnpc conjugated to T1-spyt-snpt-6xHis (∼80 kDa), EutD-Csnpc (∼50 kDa), T1-spyt-snpt-6xHis (∼40 kDa), T2 and T3 (22 kDa and 23 kDa, respectively) as well as the BMC-H (10 kDa). Proteins have been identified by mass spectrometry. (B) TEM images of the sFUT compartments. Structures observed at ∼40 nm in diameter. (C) Model of a complete assembled sFUT wiffleball. PFL and EutD form dimers. Not all T1-spyt-snpt-6xHis conjugation points are occupied, probably due to steric hindrance. (D) Activity of sFUT wiffleballs. Lane 1: no sFUT control; lane 2: inactivated sFUT by exposure to oxygen after isolation; lane 3: sFUT; lane 4: sFUT + CoA; lane 5 sFUT + acetyl-CoA. Formation of formate is shown per sFUT wiffleball over a 15-min time frame when provided with 1 µmol pyruvate. Addition of 1 mM CoA boosts activity, as well as addition of 1 mM acetyl-CoA. Acetyl-CoA can only be used by the PFL if EutD converts it to acetyl phosphate and CoA, thus indicating EutD activity.
To investigate if the isolated sFUT wiffleballs are active, we measured the forward reaction of the enzymes producing formate in the presence of 1 μmol pyruvate. sFUT wiffleballs were able to produce about 0.5 to 0.6 μmol of formate, while an inactivated control showed only little formate present (Fig. 4D); 14 pmol sFUT wiffleballs were used in the reactions, which were loaded with an estimated 40 copies of the PFL and EutD enzymes based on the intensity of the protein bands in the SDS/PAGE gel (Fig. 4A). Considering that 500 to 600 nmol formate was produced, each enzyme carried out, on average at least 1,000 turnovers until activity was lost. This provides evidence that both enzymes are actively encapsulated and that CoA, which is presumably coencapsulated with the enzymes, can cycle between the enzymes. When the cofactor CoA was added to the sample, the activity improved slightly, by about 20%. Similarly, the activity could also be improved with the addition of acetyl-CoA, giving another indicating that EutD is active in the sFUT wiffleballs, because acetyl-CoA would need to be converted first to acetyl phosphate and CoA in order for the PFL to produce formate. The reported kcat for the PFL is between 105–770 s−1, assuming that the enzymes kinetics do not change significantly after addition of the encapsulation tag and incorporation into a wiffleball, it means that the reaction ended within seconds. This is consistent with our observation that we were not able to time-resolve this reaction with our experimental setup, which only allowed us to measure on a minute time scale.
Based on these results, we built a model of our assembled sFUT wiffleball (Fig. 4C) depicting a potential arrangement of the enzyme cargo inside a shell. It should be noted that the PFL as well as the EutD form homodimers, hence the presence of the unconjugated versions of these enzymes in the SDS/PAGE. Considering the SDS/PAGE and the TEM images, we therefore conclude that we successfully assembled active sFUT wiffleballs encapsulating the PFL-Cspyc as well as EutD-Csnpc.
Discussion
The overarching goal of the sFUT BMC design is to create a platform metabolic module that can integrate oxygen-sensitive metabolism into an aerobic host to produce, from a cheap feedstock, a central biosynthetic intermediate for the production of high-value compounds. We produced a prototype synthetic microcompartment core for the oxygen-sensitive enzyme PFL based on the HO-shell because of the availability of molecular tools to load the synthetic HO-shell with cargo and its potential to form wiffleball architectures. We modified this architecture with a purification tag that facilitates rapid isolation and the testing of enzymatic core designs. This decouples the design-test-refine cycle for the catalytic core, from optimization of shell permeability, by allowing unrestricted substrates and product exchange for activity measurements. However, for future designs, the BMC-P protein can be added to the shell system to form complete HO-shells, and therefore complete sFUT BMCs. This concept can be applied broadly, streamlining other engineering efforts aiming to design and optimize synthetic BMCs.
Core design and assembly constitute the first phase toward constructing a completely functional synthetic “organelle.” This required expanding the technique to specifically encapsulate cargo by covalent linkage developed by Hagen et al. (25) to include the addition of a new adaptor system (SnoopTag/SnoopCatcher). Heterologous enzymes have been targeted to the lumen of the BMC before (38, 39) by using encapsulation peptides (40, 41) on the cargo; however, this approach suffers from low efficiency, is hampered by aggregation, and it is unknown how the encapsulation peptides associate with the shell, making this method nonquantitative (27, 42–45). Our strategy enabled us to reliably, specifically, and independently target two different cargo proteins into the lumen of a BMC. Additionally, the expansion of the adaptor system effectively doubled the number proteins that can be encapsulated from 60 to 120 per BMC, compared to the previously described method (25). This can increase the overall efficiency of the synthetic BMC by increasing the metabolite flux and permitting faster substrate channeling due to a greater enzyme density.
This first-generation sFUT prototypes aimed to encapsulate the minimal number of proteins needed (PFL and EutD) to cycle CoA (Fig. 1C), which is a natural property of BMCs (46). Tagging the PFL with SpyCatcher for encapsulation was challenging because of low expression level, and if expressed it produced mostly inactive (Fig. 3) and insoluble protein (SI Appendix, Fig. S4A). However, we succeeded in creating an active version of the PFL with a C-terminal encapsulation adapter (PFL-Cspyc). EPR revealed the presence of a glycyl radical signal consistent with activated PFL, suggesting that PFL-AE was able to bind PFL-Cspyc and successfully activate the enzyme. Furthermore, our E. coli growth-based activity assays show that the PFL-Cspyc retains activity comparable to the unmodified PFL in both forward and reverse direction, making it suitable for constructing the sFUT wiffleball. It should be noted that because of the versatility of the SpyCatcher/SpyTag system, this C-terminal–tagged version of the PFL can potentially be used in other scaffolding or engineering efforts.
Considering that both PFL and EutD form homodimers (35, 47–49), and the relatively large size of the PFL-Cspyc (calculated molecular mass of 95 kDa), we anticipated that sFUT wiffleballs cannot fully assemble when all 120 contact points are used for conjugating cargo, because of localized overcrowding of the sFUT wiffleball lumen. The consequent fragility may explain why we couldn’t isolate the minimal sFUT wiffleball architecture. To prevent steric hindrance while BMC self-assembly takes place, we diluted out the presence of T1-spyt-snpt-6xHis in the wiffleballs by coexpressing T2 and T3, which occupy the same geometric positions in the shell as T1-spyt-snpt-6xHis but don’t recruit cargo into the lumen. Using this approach, we were able to isolate completely assembled sFUT wiffleballs loaded with PFL and EutD. Our activity measurements of the forward reaction show that the enzymes are active and can undergo multiple turnovers cycling CoA between the enzymes. Although CoA is known to associate with cargo proteins before encapsulation in BMCs (45), we note that the sFUT wiffleball activity can be improved by adding CoA or acetyl-CoA to the reaction. Considering that isolation of sFUT wiffleballs is a lengthy process, including washes of the streptavidin column, we hypothesize that some CoA was released from the shell in this process; thus, its addition was able to slightly increase sFUT wiffleball activity. We have focused on the forward reaction for the activity assay because this is readily measurable, only requiring the addition of pyruvate to the sample, and we could follow the formation of formate. It should be noted that our in vivo experiments show that the modified enzymes have activity in both forward and reverse directions. However, we noted that the conversion of the provided pyruvate to formate does not reach the published equilibrium of the enzymatic reaction (50), suggesting that over time the enzymes get inactivated and there is not sufficient PFL-AE or Ado-Met available to reactivate the PFL. A kcat for the encapsulated PFL can be calculated roughly from the activity assay; using the estimated 40 PFL enzymes per sFUT wiffleball, it yields about 1.2 s−1. However, given that we could not time-resolve the enzymatic reaction because it presumably ends within seconds, and we could only measure on a minute timescale with our experimental setup, the kcat of the encapsulated PFL could be much closer to the published PFL kcat of 105–770 s−1 (51, 52).
In summary, we have built a synthetic BMC, directly targeting two enzymes to be encapsulated, one of which is extremely oxygen sensitive, and expressing three auxiliary enzymes (PFL-AE, METK, ACK), to enable its function in the cell. In contrast to the synthetic BMCs first pioneered (38, 39, 53), which were used for ethanol production, polyphosphate storage and hydrogen production, the sFUT prototype can be used as a platform in ambitious engineering projects to compartmentalize entire metabolic pathway for the production of a biomolecule of interest starting at the easily accessible feedstocks acetate and formate. Moreover, our prototype metabolic module is poised for shell permeability engineering to address the grand challenge of constructing devices for to compartmentalize oxygen-sensitive reactions for use in aerobic growth conditions.
Materials and Methods
Cloning Procedure.
The operon encoding for the focA (Uniprot accession no.: P0AC23), pflB (Uniprot accession no.: P09373), pflA (Uniprot accession no.: P0A9N4), and metK (Uniprot accession no.: P0A817) genes were PCR-amplified from E. coli K12 genomic DNA and cloning into pBbA2C (54) using a Gibson assembly (55). The pflB gene was then modified with additions of SpyCatcher, which was PCR-amplified using plasmids from Hagen et al. (25) and assembled into pBbA2C (54). The gene EutD (Uniprot accession no.: P77218) was ordered on a gBlock (IDT), PCR-amplified, and cloned along with a snoopCatcher PCR product (32), as well as the ackA gene (Uniprot accession no.: P0A6A3) PCR-amplified from E. coli K12 genomic DNA into pBbE2K (54) using a Gibson assembly (55). The intergenic region between cpcB and cpcA was PCR-amplified from the Synechocystis sp. PCC 6803 genome and used as an intergenic region between EutD and Ack in a synthetic operon via a Gibson assembly (55). Synthetic shell operons were modified by Gibson assembly (55) using plasmids from Hagen et al. (25) as templates to add a C-terminal 6x His-tag and an inserted snoopTag (32) to BMC-T1.
Protein Expression.
BL21(DE3) strains harboring the shell and or cargo plasmids were grown to OD600 0.6 to 0.8 in Luria-Bertani broth at 37 °C in the presence of 100 µg/mL ampicillin, 25 µg/mL chloramphenicol, and/or 50 µg/mL kanamycin, depending on the selectable marker in the plasmids and cold-shocked on ice for 5 min. The expression of the proteins was induced with 50 µM isfopropyl-β-d-thiogalactopyranoside (IPTG) and/or 5 ng/mL anhydrotetracycline, depending on the induction system of the plasmids, and then incubated at 18 °C for 16 to 20 h.
BMC Shell and Wiffleball Purifications.
Shells/wiffleballs were prepared according to the method described by Sutter et al. (24) with slight modifications. Briefly, the cell pellet from a 1-L culture expressing the HO-shell/wiffleballs and cargo plasmids was resuspended in phosphate-buffered saline (50 mM phosphate pH 7.5, 100 mM NaCl, referred to as 50/100 PBS), lysed by French press and centrifuged at 2,000 × g for 10 min to pellet unbroken cells. The supernatant was loaded on a sucrose cushion (20% sucrose [wt/vol]) and centrifuged at 26,000 rpm in a Beckman SW28 rotor. The pellet was resuspended in 50/100 PBS and then purified on an NiIDA affinity column, and in case of the purification of the complete sFUT applied onto a sucrose gradient 20 to 60% (wt/vol) after NiIDA purification. At around 55% sucrose, a clean sFUT fraction could be obtained. Wiffleball samples were buffer-exchanged to 50/100 PBS and concentrated with a 15 mL 100 kDa MWCO filter (Amicon); for storage 0.02% sodium azide as a preservative was added.
SDS/PAGE and Western Blot Analysis of Protein Preparations.
Protein preparations were normalized to A280 = 1, denatured in reducing sample buffer and loaded on 4 to 20% polyacrylamide gradient gel. Gels were washed and stained with Coomassie blue. Proteins were transferred to a nitrocellulose membrane via a tank transfer system and blocked in PBS + 5% (wt/vol) nonfat dry milk, 0.1% (vol/vol) Triton X-100. Cross-reactions with a monoclonal antibody (anti-6xHis, Invitrogen) was visualized by Supersignal West Pico Chemiluminiscent substrate detection system (Thermo Scientific) and imaged with ChemiDoc XRS+ System (Bio-Rad).
Fluorescence Measurements.
An M1000 or Spark plate reader (Tecan) was used to collect the fluorescence spectra. Samples were adjusted to total protein, 2.5 mg/mL and 100 µL of the samples were loaded into 96-well microplates to collect fluorescence intensity readings. Excitation and emission bandwidths were set to 5 nm; 425 nm was used for mTurquoise2 excitation and 490 nm for mVenus. The gain was kept at 100. The fluorescence intensity of sample containing the free fluorophores mTurquoise2 and mVenus without a shell was normalized to the HT1-sypt-snpt sample for comparison.
Transmission Electron Microscopy.
Purified shells were imaged by negative stained TEM on a JEOL JEM-1400Flash microscope. Accelerating voltage was 100 kV and images were taken with a “Matataki Flash” sCMOS camera. Purified shells were diluted 10-fold in HPLC-grade water and 5 μL of each sample was applied to 150-mesh carbon-coated copper grids (Electron Microscopy Sciences) for 30 s, wicked dry, stained for 15 s with 1% uranyl acetate, and again wicked dry before imaging.
PFL Activity-Dependent Growth Experiments.
ΔaceAΔpflAB E. coli cells (8) transformed with plasmids expressing PFL, PFL-spyc, PFL-Nspyc, and PFL-Cspyc. were grown in M9 minimal medium (47.8 mM Na2HPO4, 22 mM KH2PO4, 8.6 mM NaCl, 18.7 mM NH4Cl, 2 mM MgSO4 and 100 μM CaCl2), supplemented with trace elements (134 μM EDTA, 31 μM FeCl3·6H2O, 6.2 μM ZnCl2, 0.76 μM CuCl2·2H2O, 0.42 μM CoCl2·2H2O, 1.62 μM H3BO3, 0.081 μM MnCl2·4H2O). Precultures for growth experiments were incubated in aerobic conditions over night in 4 mL M9 medium containing 10 mM glucose and 2 nM anhydrotetracycline. Prior to inoculation cells were harvested by centrifugation (6,000 × g, 3 min, room temperature), washed three times in M9, and inoculated in M9 containing 10 mM glucose to a starting optical density (OD600 nm) of 0.01. Since the plasmids used the native PFL promoter, no inducer was needed, as the PFL variants were expressed when cells reached anaerobiosis. Nunc 96-well microtiter plates were used for cultivations (Thermo Scientific, 167008), each well contained 150-µL culture covered with 50 µL mineral oil (Sigma-Aldrich, M3516). To allow removal of dissolved oxygen, media were kept for at least 24 h in the anaerobic chamber. Microtiter plates were incubated at 37 °C in a Tecan Infinite 200 Pro plate reader (Tecan) in a vinyl anaerobic chamber (N2 with 10% CO2, 2.5% H2, model B, Coy Laboratory Products). OD600-nm measurements were followed by cycles consisting of three repeats of four shaking phases, 1 min of each: linear shaking, orbital shaking at amplitude of 3 mm, linear shaking, and orbital shaking at amplitude of 2 mm. Raw data from the plate reader were calibrated to cuvette values according to ODcuvette = ODplate/0.23.
EPR Spectroscopy.
ΔaceAΔpflB E. coli cells (8) transformed with plasmids expressing wild-type PflB or C-terminally fused SpyCatcher (PFL-Cspyc) from a plasmid along with wild-type activating enzyme PflA were grown anaerobically and prepared for EPR analysis in an anaerobic chamber (Coy Laboratory Products) at room temperature under an atmosphere of 2.5% H2/97.5% N2. Cells were pelleted at 5,000 × g for 15 min and resuspended in 50/100 PBS containing 1 mM DTT to an OD600 = 100. Next, 300 μL of resuspended cells were transferred into 4-mm OD quartz X-band EPR tubes (Wilmad LabGlass) and removed from the anaerobic chamber and rapidly frozen in liquid nitrogen; these samples were stored in liquid nitrogen until data collection. EPR spectra were collected with a Bruker E680X spectrometer at X-band (9.4 GHz) using an ST4102 (TE102) cavity, 100-kHz magnetic field modulation frequency, 0.25 mT modulation amplitude, 0.2-mW microwave power, and a receiver gain of 70 dB. The time constant was 41 ms and conversion time was 164 ms. Next, 2,048 points were collected for each scan and eight scans were averaged for each sample using the built-in Xepr software (Bruker BioSpin). The sample temperature was maintained at 253 K using a Bruker liquid nitrogen temperature control system.
Formate Quantification.
Reactions were mixed together in an anaerobic atmosphere containing 1 µmol Pyruvate, 14 pmol isolated sFUT wiffleballs with or without the addition of 1 mM CoA or acetyl-CoA in 100 µL final volume in 50/100 PBS pH 7.5. The inactivated controls were mixed together in aerobic conditions. Samples were incubated for 15 min in a vinyl anaerobic chamber (N2 with 2.5% H2; model B, Coy Laboratory Products). Formate was then determined in aerobic conditions using the Formate assay kit (Sigma-Aldrich, MAK059) according to the manufacturer’s protocol.
Liquid Chromatography-Tandem Mass Spectrometry Analysis of SDS/PAGE Protein Bands.
Gel bend were digested in-gel as described in Shevchenko et al. (56) with modifications. Briefly, gel bands were dehydrated using 100% acetonitrile and then incubated in 10 mM dithiothreitol, 100 mM ammonium bicarbonate, pH ∼8, at 56 °C for 45 min. The gel bands were dehydrated again and incubated in the dark in 50 mM iodoacetamide, 100 mM ammonium bicarbonate for 20 min. Gel bands were washed with ammonium bicarbonate followed by dehydration. Gel bands were submerged in ∼100 μL of a 0.01 μg/μL sequencing grade modified trypsin solution in 50 mM ammonium bicarbonate and incubated at 37 °C overnight. Peptides were extracted from the gel pieces in a solution of 60% acetonitrile (ACN)/1% trifluoroacetic acid (TFA) using water bath sonication and then vacuum dried to ∼2 μL. Dried samples were resuspended to 20 μL in 2% ACN/0.1% TFA. 5 μL were automatically injected onto a Thermo Acclaim PepMap RSLC 0.1-mm × 20-mm C18 trapping column using a Thermo EASYnLC 1000 (https://www.thermofisher.com/cn/zh/home/brands/thermo-scientific.html). The column was washed for ∼5 min with buffer A (Buffer A = 99.9% water/0.1% formic acid). Bound peptides were then eluted over 35 min onto a Thermo Acclaim PepMap RSLC 0.075-mm × 250-mm resolving column with a gradient of 5% B to 40% B in 24 min, ramping to 90% B at 25 min and held at 90% B for the rest of the run duration (Buffer B = 80% acetonitrile/0.1% formic acid/19.9% water) at a constant flow rate of 300 nL/min. Column temperature was maintained at 50 °C using an integrated column oven (PRSO-V1, Sonation). Eluted peptides were sprayed into a ThermoScientific Q-Exactive mass spectrometer (https://www.thermofisher.com/us/en/home/brands/thermo-scientific.html) using a FlexSpray spray ion source. The resulting MS/MS spectra scan processed using Mascot Distiller, v2.7.1 (www.matrixscience.com), and searched against a database containing all E. coli K12 protein sequences with addition of common laboratory contaminants (downloaded from https://www.uniprot.org/ and https://www.thegpm.org/, respectively) using the Mascot searching algorithm, v2.7. The Mascot output was then analyzed using Scaffold, v5.0 (https://www.proteomesoftware.com/) to validate protein identifications.
Supplementary Material
Acknowledgments
We thank Dr. María Agustina Domínguez-Martín for her help with the immunoblotting; Dr. Lior Doron for his help with transmission electron microscopy imaging; Dr. Andrew Hagen, Dr. John F. C. Steele, and Dr. Markus Sutter for constructive discussions; Prof. John McCracken for use of his electron paramagnetic resonance spectrometer and helpful discussions; Dr. Douglas Whitten from the Proteomic Facility at Michigan State University for the liquid chromatography-tandem mass spectrometry analysis; and Dr. Gang Ren and Dr. Jianfanf Liu for initial transmission electron microscopy imaging. This work was supported by the National Science Foundation Award MCB 1733552 and the US Department of Energy, Basic Energy Sciences, Contract DE-FG02-91ER20021. The work at the Molecular Foundry was supported by the Office of Science, Office of Basic Energy Sciences, of the US Department of Energy under Contract DE-AC02-05CH11231.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2116871119/-/DCSupplemental.
Data Availability
All study data are included in the main text and SI Appendix.
References
- 1.Escobar J. C., et al. , Biofuels: Environment, technology and food security. Renew. Sustain. Energy Rev. 13, 1275–1287 (2009). [Google Scholar]
- 2.Naik S. N., Goud V. V., Rout P. K., Dalai A. K., Production of first and second generation biofuels: A comprehensive review. Renew. Sustain. Energy Rev. 14, 578–597 (2010). [Google Scholar]
- 3.Yishai O., Lindner S. N., Gonzalez de la Cruz J., Tenenboim H., Bar-Even A., The formate bio-economy. Curr. Opin. Chem. Biol. 35, 1–9 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Tamaki Y., Morimoto T., Koike K., Ishitani O., Photocatalytic CO2 reduction with high turnover frequency and selectivity of formic acid formation using Ru(II) multinuclear complexes. Proc. Natl. Acad. Sci. U.S.A. 109, 15673–15678 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bar-Even A., Formate assimilation: The metabolic architecture of natural and synthetic pathways. Biochemistry 55, 3851–3863 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Bar-Even A., Noor E., Flamholz A., Milo R., Design and analysis of metabolic pathways supporting formatotrophic growth for electricity-dependent cultivation of microbes. Biochim. Biophys. Acta. 1827, 1039–1047 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Yishai O., Goldbach L., Tenenboim H., Lindner S. N., Bar-Even A., Engineered assimilation of exogenous and endogenous formate in Escherichia coli. ACS Synth. Biol. 6, 1722–1731 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Zelcbuch L., et al. , Pyruvate formate-lyase enables efficient growth of Escherichia coli on acetate and formate. Biochemistry 55, 2423–2426 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Stairs C. W., Roger A. J., Hampl V., Eukaryotic pyruvate formate lyase and its activating enzyme were acquired laterally from a Firmicute. Mol. Biol. Evol. 28, 2087–2099 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Kirst H., Kerfeld C. A., Bacterial microcompartments: Catalysis-enhancing metabolic modules for next generation metabolic and biomedical engineering. BMC Biol. 17, 79 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerfeld C. A., Heinhorst S., Cannon G. C., Bacterial microcompartments. Annu. Rev. Microbiol. 64, 391–408 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Sutter M., Melnicki M. R., Schulz F., Woyke T., Kerfeld C. A., A catalog of the diversity and ubiquity of bacterial microcompartments. Nat. Commun. 12, 3809 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirst H., Kerfeld C. A., Clues to the function of bacterial microcompartments from ancillary genes. Biochem. Soc. Trans. 49, 1085–1098 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Axen S. D., Erbilgin O., Kerfeld C. A., A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLOS Comput. Biol. 10, e1003898 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zarzycki J., Erbilgin O., Kerfeld C. A., Bioinformatic characterization of glycyl radical enzyme-associated bacterial microcompartments. Appl. Environ. Microbiol. 81, 8315–8329 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferlez B., Sutter M., Kerfeld C. A., Glycyl radical enzyme-associated microcompartments: Redox-replete bacterial organelles. MBio 10, e02327-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zarzycki J., Sutter M., Cortina N. S., Erb T. J., Kerfeld C. A., In vitro characterization and concerted function of three core enzymes of a glycyl radical enzyme—Associated bacterial microcompartment. Sci. Rep. 7, 42757 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schindel H. S., Karty J. A., McKinlay J. B., Bauer C. E., Characterization of a glycyl radical enzyme bacterial microcompartment pathway in Rhodobacter capsulatus. J. Bacteriol. 201, e00343-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Craciun S., Balskus E. P., Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc. Natl. Acad. Sci. U.S.A. 109, 21307–21312 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerfeld C. A., Sutter M., Engineered bacterial microcompartments: Apps for programming metabolism. Curr. Opin. Biotechnol. 65, 225–232 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerfeld C. A., Protein structures forming the shell of primitive bacterial organelles. Science 309, 936–938 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Klein M. G., et al. , Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J. Mol. Biol. 392, 319–333 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Tanaka S., et al. , Atomic-level models of the bacterial carboxysome shell. Science 319, 1083–1086 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Sutter M., Greber B., Aussignargues C., Kerfeld C. A., Assembly principles and structure of a 6.5-MDa bacterial microcompartment shell. Science 356, 1293–1297 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagen A., Sutter M., Sloan N., Kerfeld C. A., Programmed loading and rapid purification of engineered bacterial microcompartment shells. Nat. Commun. 9, 2881 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sutter M., McGuire S., Ferlez B., Kerfeld C. A., Structural characterization of a synthetic tandem-domain bacterial microcompartment shell protein capable of forming icosahedral shell assemblies. ACS Synth. Biol. 8, 668–674 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lassila J. K., Bernstein S. L., Kinney J. N., Axen S. D., Kerfeld C. A., Assembly of robust bacterial microcompartment shells using building blocks from an organelle of unknown function. J. Mol. Biol. 426, 2217–2228 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Lee M. J., et al. , De novo targeting to the cytoplasmic and luminal side of bacterial microcompartments. Nat. Commun. 9, 3413 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferlez B., Sutter M., Kerfeld C. A., A designed bacterial microcompartment shell with tunable composition and precision cargo loading. Metab. Eng. 54, 286–291 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hagen A. R., et al. , In vitro assembly of diverse bacterial microcompartment shell architectures. Nano Lett. 18, 7030–7037 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zakeri B., et al. , Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. U.S.A. 109, E690–E697 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veggiani G., et al. , Programmable polyproteams built using twin peptide superglues. Proc. Natl. Acad. Sci. U.S.A. 113, 1202–1207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siu K. H., et al. , Synthetic scaffolds for pathway enhancement. Curr. Opin. Biotechnol. 36, 98–106 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Campos-Bermudez V. A., Bologna F. P., Andreo C. S., Drincovich M. F., Functional dissection of Escherichia coli phosphotransacetylase structural domains and analysis of key compounds involved in activity regulation. FEBS J. 277, 1957–1966 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Bologna F. P., Campos-Bermudez V. A., Saavedra D. D., Andreo C. S., Drincovich M. F., Characterization of Escherichia coli EutD: A phosphotransacetylase of the ethanolamine operon. J. Microbiol. 48, 629–636 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez-Esquer C. R., Shubitowski T. B., Kerfeld C. A., Streamlined construction of the cyanobacterial CO2-fixing organelle via protein domain fusions for use in plant synthetic biology. Plant Cell 27, 2637–2644 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Unkrig V., Neugebauer F. A., Knappe J., The free radical of pyruvate formate-lyase. Characterization by EPR spectroscopy and involvement in catalysis as studied with the substrate-analogue hypophosphite. Eur. J. Biochem. 184, 723–728 (1989). [DOI] [PubMed] [Google Scholar]
- 38.Lawrence A. D., et al. , Solution structure of a bacterial microcompartment targeting peptide and its application in the construction of an ethanol bioreactor. ACS Synth. Biol. 3, 454–465 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang M., Frank S., Lünsdorf H., Warren M. J., Prentice M. B., Bacterial microcompartment-directed polyphosphate kinase promotes stable polyphosphate accumulation in E. coli. Biotechnol. J. 12, 1600415 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Kinney J. N., Salmeen A., Cai F., Kerfeld C. A., Elucidating essential role of conserved carboxysomal protein CcmN reveals common feature of bacterial microcompartment assembly. J. Biol. Chem. 287, 17729–17736 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan C., et al. , Short N-terminal sequences package proteins into bacterial microcompartments. Proc. Natl. Acad. Sci. U.S.A. 107, 7509–7514 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai F., Bernstein S. L., Wilson S. C., Kerfeld C. A., Production and characterization of synthetic carboxysome shells with incorporated luminal proteins. Plant Physiol. 170, 1868–1877 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choudhary S., Quin M. B., Sanders M. A., Johnson E. T., Schmidt-Dannert C., Engineered protein nano-compartments for targeted enzyme localization. PLoS One 7, e33342 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jakobson C. M., Slininger Lee M. F., Tullman-Ercek D., De novo design of signal sequences to localize cargo to the 1,2-propanediol utilization microcompartment. Protein Sci. 26, 1086–1092 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Erbilgin O., Sutter M., Kerfeld C. A., The structural basis of coenzyme A recycling in a bacterial organelle. PLoS Biol. 14, e1002399 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kerfeld C. A., Aussignargues C., Zarzycki J., Cai F., Sutter M., Bacterial microcompartments. Nat. Rev. Microbiol. 16, 277–290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conradt H., Hohmann-Berger M., Hohmann H. P., Blaschkowski H. P., Knappe J., Pyruvate formate-lyase (inactive form) and pyruvate formate-lyase activating enzyme of Escherichia coli: Isolation and structural properties. Arch. Biochem. Biophys. 228, 133–142 (1984). [DOI] [PubMed] [Google Scholar]
- 48.Becker A., et al. , Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase. Nat. Struct. Biol. 6, 969–975 (1999). [DOI] [PubMed] [Google Scholar]
- 49.Becker A., Kabsch W., X-ray structure of pyruvate formate-lyase in complex with pyruvate and CoA. How the enzyme uses the Cys-418 thiyl radical for pyruvate cleavage. J. Biol. Chem. 277, 40036–40042 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Tanaka N., Johnson M. J., Equilibrium constant for conversion of pyruvate to acetyl phosphate and formate. J. Bacteriol. 108, 1107–1111 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.M. C. Andorfer, L. R. F. Backman, P. L. Li, E. C. Ulrich, C. L. Drennan, Rescuing activity of oxygen-damaged pyruvate formate-lyase by a spare part protein. J. Biol. Chem. 297, 101423 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.J. Knappe, H. P. Blaschkowski, P. Gröbner, T. Schmitt, Pyruvate formate-lyase of Escherichia coli: the acetyl-enzyme intermediate. Eur. J. Biochem. 50, 253–263 (1974). [DOI] [PubMed] [Google Scholar]
- 53.Li T., et al. , Reprogramming bacterial protein organelles as a nanoreactor for hydrogen production. Nat. Commun. 11, 5448 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anderson J. C., et al. , BglBricks: A flexible standard for biological part assembly. J. Biol. Eng. 4, 1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gibson D. G., et al. , Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Shevchenko A., Wilm M., Vorm O., Mann M., Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and SI Appendix.