Significance
This paper reports an exciting breakthrough in dynamic biomaterials design mimicking the reversible interlocking and remoldable structure of extracellular matrix (ECM). Specifically, we realize a nature-derived molecular recognition event (i.e., the antibiotic glycopeptide vancomycin [Van] and the dipeptide d-Ala-d-Ala [AA] receptor–ligand interaction) as a reversible strategy for fabrication of dynamic biointerface and 3D ECM mimics. We believe that the specific but reversible Van–AA molecular recognition would be a strategy for dynamic biomaterial fabrication, and that the easy-handling merit, ECM-like remoldability, and inherent antibacterial activity will bring insights to biomaterial scaffold design in tissue engineering and regenerative medicine.
Keywords: biomimicry, dynamic biomaterial design, natural receptor–ligand interaction, cell regulation, tissue repair
Abstract
Dynamic biomaterials excel at recapitulating the reversible interlocking and remoldable structure of the extracellular matrix (ECM), particularly in manipulating cell behaviors and adapting to tissue morphogenesis. While strategies based on dynamic chemistries have been extensively studied for ECM-mimicking dynamic biomaterials, biocompatible molecular means with biogenicity are still rare. Here, we report a nature-derived strategy for fabrication of dynamic biointerface as well as a three-dimensional (3D) hydrogel structure based on reversible receptor–ligand interaction between the glycopeptide antibiotic vancomycin and dipeptide d-Ala-d-Ala. We demonstrate the reversible regulation of multiple cell types with the dynamic biointerface and successfully implement the dynamic hydrogel as a functional antibacterial 3D scaffold to treat tissue repair. In view of the biogenicity and high applicability, this nature-derived reversible molecular strategy will bring opportunities for malleable biomaterial design with great potential in biomedicine.
More than 2,500 y ago, the Chinese built their nail- and glue-less architectures (e.g., from Beijing's Forbidden City to Sichuan Province's Bao'en Temple) with a “dougong” structure, which is part of the network of wooden supports essential to the timber frame structure of the building. A typical dougong consists of a flat block of wood (dou), the top of which is fixed with an interlocking set of curved bows (gong), without the aid of nails or glue, to provide mechanical support to hold the three-dimensional (3D) network structure. Intriguingly, the microstructure of extracellular matrix (ECM) also demonstrates such reversible interlocking structure, which supports the integrity of tissues and organs. In natural ECM, the dougong structure occurs at the cell–ECM interfaces accompanied by a constant remodeling of the ECM network, giving rise to specific cell signaling, intracellular cascades, and subsequently, all relevant cell behaviors (1–3). Biomaterial designs based on the reversible interactions mimicking the cell–ECM interfaces are believed to boast distinct advantages, including the capability to modulate cell–biomaterial interactions, adapt to the development of cellular processes (1, 4–6), and facilitate the morphogenesis of tissues and organs (7, 8). Although the dynamic design of biomaterials is relatively complicated and challenging, this field attracts significant attention in building dynamic ECM mimics for regenerative medicine (1, 9–11).
To faithfully reproduce the dynamics of ECM in artificial matrices, various strategies, including congenitally reversible noncovalent interactions (e.g., hydrogen bonds, coordinate bonds, hydrophobic forces, π–π interactions, van der Waals forces, and electrostatic effects) and dynamic covalent bonds (e.g., reversible boronic esters and benzoic–imine bonds and photosensitive nitrophenyl and azobenzene groups), have been exploited. Currently, biomaterial interfaces with dynamically functionalized bioligands are mainly designed through reversible covalent phenylboronic esters or benzoic–imine bonds (12–16), deformable azobenzene bonds (17), DNA and peptide molecular assemblies (18, 19), cyclodextrins/cucurbiturils-based macrocycle host–guest supermolecules (20–22), metal–ligand coordination (23, 24), and other multiple noncovalent interactions (25–29). They can elicit controllable and reversible cell behaviors (e.g., adhesion, migration, differentiation, and apoptosis) on the biomaterial interfaces (1, 4, 8). Unfortunately, these dynamic ECM-mimicking strategies carry critical problems (13). First, most dynamic strategies are based on nonbiogenic chemical molecules, which are usually nonbiocompatible and probably harmful. Second, the dynamics of these strategies commonly rely on the nonbiological stimuli (e.g., ultraviolet [UV] light or toxic chemicals), which are potentially invasive to cells. Third, current studies on mimicking dynamic ECM are usually limited to either reversible bioligand presentation or remoldable network fabrication; few works focus on both. In this context, the exploration of biocompatible molecular means for recapitulation of both dynamic bioactivity and dynamic structure in ECM is highly anticipated.
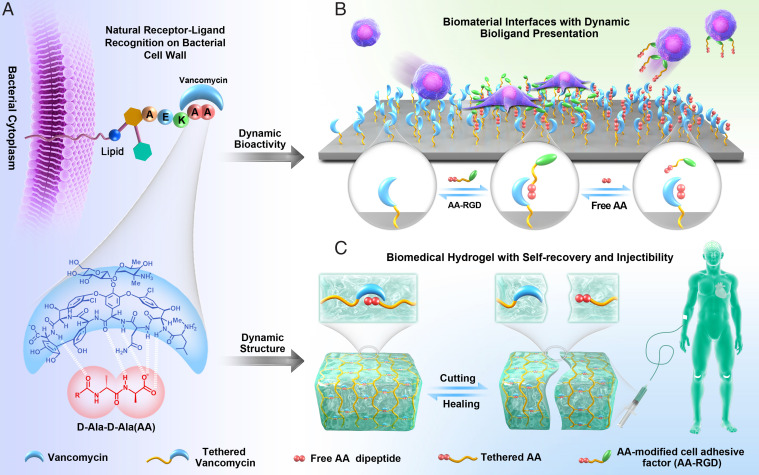
Here, we present a nature-derived reversible strategy inspired by the receptor–ligand molecular recognition for design of dynamic ECM-mimicking biomaterial. The receptor–ligand molecular recognition relies on multi-noncovalent interaction between two or more molecules with exquisite complementarity in their chemical groups and geometries (30). With this in mind, we focus our attention on a typical simple yet elegant receptor–ligand system (i.e., the glycopeptide antibiotic vancomycin [Van] and the dipeptide d-Ala-d-Ala [AA]). Produced by a bacterial species named Amycolatopsis orientalis, Van exhibits strong bactericidal effect by inhibiting cell wall biosynthesis via the specific binding (Kd = 1.6 μM) (31) toward the terminal AA dipeptide of the bacterial cell wall precursors (Fig. 1A). As a proof of concept, we employ the reversible Van–AA interaction for building both a reversible dynamic biointerface and a 3D hydrogel network (Fig. 1 B and C). Due to the specific but reversible AA–Van molecular recognition, the dynamic biointerface demonstrates excellent reversibility in binding to cell-adhesive tripeptide arginylglycylaspartic acid (RGD) and modulating adhesion of multiple cells, demonstrating our strategy’s general applicability. In addition, the 3D hydrogel network based on the reversible AA–Van molecular recognition demonstrates self-recovery and injectability. The inherently antibacterial activity of the Van–AA hydrogel well equips the 3D hydrogel network for treating infected open skin wounds; the hydrogel could adapt to the shape of wound sites, resist self-fragmentation, and inhibit proliferation of pathogenic bacteria while continuously supporting wound healing. We believe that the specific but reversible Van–AA molecular recognition would be a strategy for dynamic biomaterial fabrication, and the easy-handling merit, ECM-like remoldability, and inherently antibacterial activity involved in this dynamic system will bring insights to biomaterial scaffold design in tissue engineering and regenerative medicine.
Fig. 1.
Schematics showing the mechanism of dynamic biointerface and 3D ECM mimics based on a reversible dougong-structured natural receptor–ligand recognition. (A) The Van–AA molecular recognition on bacterial cell wall in nature. (B) Schematic illustration of the dynamic biointerface based on the reversible Van–AA interaction. Reversible bioligand presentation and controllable cell behaviors could be readily realized through the Van–AA interaction. (C) Schematic illustration of the dynamic hydrogels with remoldable network structure and its application in tissue repair.
Results
Dynamic Biointerface with Reversible Dougong Structure.
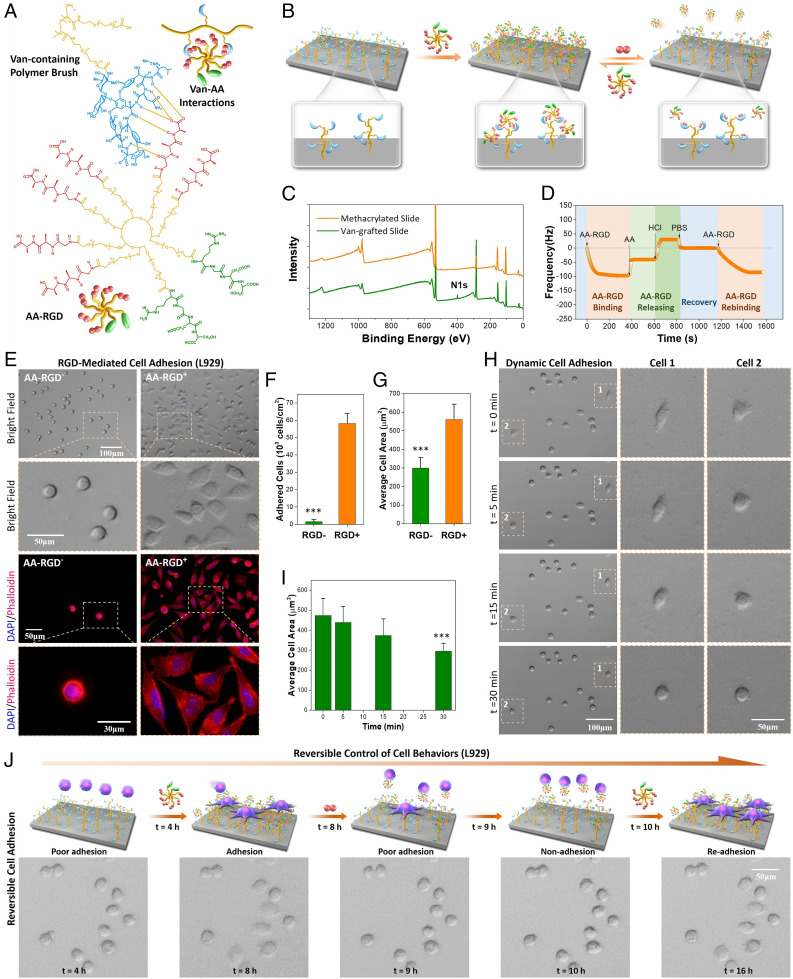
To reproduce the dynamic biointerface in natural ECM, we first fabricated a dynamic biomaterial surface by employing the reversible dougong-like Van–AA interaction for dynamic biomolecule functionalization. Since cell adhesion represents one of the most fundamental events and is the prerequisite for many other cell behaviors, a typical cell-adhesive factor RGD, which has been used to improve the cell adhesion property of a wide variety of biomaterials (32–35), was employed as the model bioactive ligand in this study. Specifically, for the fabrication of the dynamic biomaterial interface, a functionalized substrate with Van-containing polymer brushes and an AA-containing polymer with bioactive RGD (AA–RGD) were prepared (Fig. 2A). The Van-containing polymer brushes were grafted onto a quartz slide using a Van-modified polyethylene glycol (PEG) with polymerizable acrylamide terminal (Van–PEG) (SI Appendix, Fig. S1) through UV light–initiated surface polymerization (SI Appendix, Fig. S2). The hydrophilic PEG was chosen in our system to reduce the surface protein adsorption and nonspecific cell adhesion. The cell-adhesive molecule AA–RGD was synthesized by cografting RGD and AA (1:3 in feeding molar ratio) onto an N-hydroxysuccinimide (NHS)–terminated eight-arm PEG through NHS–amine coupling (SI Appendix, Fig. S3). In this case, the bioligand AA–RGD could be stably bound onto the Van-containing substrate to obtain an RGD-functionalized surface by virtue of multiple AA–Van molecular recognition (Fig. 2B). Dynamic binding of the bioligand AA–RGD could be handily achieved by adding medium with free AA dipeptides, which could replace and release the surface bound AA–RGD molecules (Fig. 2B). This peptide-triggered bioligand release reflects the biological responsiveness of our dynamic biointerface, which is believed to exhibit improved biocompatibility as compared with other dynamic systems relying on nonbiological chemicals or stimuli. More importantly, the reversible Van–AA molecular recognition allows for rebinding of bioligand to occur if the dynamic system is incubated again with AA–RGD, suggesting the potential for reversible bioligand presentation.
Fig. 2.
Design of dynamic biointerfaces with reversible RGD presentation and dynamic cell adhesion. (A) The multiple Van–AA interactions between the Van-containing polymer brush and cell-adhesive molecule AA–RGD. (B) Reversible presentation of bioactive AA–RGD on the dynamic biointerface. (C) XPS of the quartz slides before and after Van grafting Peak at 400.08 eV indicated the N1 signal. (D) Real-time QCM-D frequency (f) shifts of the Van-containing surface incubated with different solutions. (E) The bright-field and fluorescence images of L929 cells after culture for 4 h on the surfaces without (AA–RGD−) and with AA–RGD (AA–RGD+). (F) Quantitative result of the cell adhesion efficiency. (G) Quantitative result of the average cell-spreading area. (H) Time-dependent L929 cell detachment on the AA–RGD-bound surface after adding 20 mM AA dipeptide. The cell 1 and cell 2 images show a dramatic shrinking in 30 min. (I) The changes of average cell-spreading area during AA-triggered cell detachment. (J) Programmed manipulation of reversible cell adhesion on the dynamic biointerface. ***Statistically significant difference is indicated by P < 0.001 as compared with others.
Characterization of the Van-grafted quartz substrate was further carried out by the static contact angle meter and X-ray photoelectron spectroscopy (XPS). Significant increases in surface hydrophilicity (SI Appendix, Fig. S4) and surface N and C element content on the Van-grafted substrates (Fig. 2C) confirmed the successful modification of poly(Van–PEG) brushes on the quartz surface. We then examined their potential for dynamic interlocking of bioligand (i.e., the cell-adhesive molecule AA–RGD). The reversible binding of AA–RGD on Van-grafted surface was investigated by quartz crystal microbalance with dissipation monitoring (QCM-D). As shown in Fig. 2D, the real-time frequency (f) of Van-grafted surface showed a sharp decrease after pumping AA–RGD solution into the QCM-D cell. We found that the Δf value of QCM-D was proportional to the mass of surface-bound molecules; the result reflected the rapid AA–RGD binding onto the Van-grafted surface through specific Van–AA molecular recognition. Upon the replacement of AA–RGD with free AA dipeptide, a significant increase in the f value appeared. This is mainly due to the molecular exchange process between AA and AA–RGD, in which the weight of released AA–RGD (Mw ∼ 23,000 Da) was significantly higher than that of the bound AA dipeptide (Mw = 217.3 Da). After further addition of HCl and phosphate buffered saline (PBS) in sequence, the bound AA dipeptide was released, and the surface recovered to its initial status owing to the acid-assisted destruction of hydrogen bonds in the Van–AA complex. When the recovered surface was reincubated with AA–RGD, a rebinding process was observed, indicating the recurrence of Van–AA molecular recognition at the dynamic biointerface. Altogether, the QCM-D monitoring results confirmed that the Van-grafted surface could reversibly bind to the AA-containing bioactive molecules, resulting in a dynamic biomaterial interface presenting reversible bioactivity.
Dynamic Biointerface for Reversible Control of Cell Behaviors.
Based on the reversible AA–RGD binding of Van-grafted surfaces, we assessed its potential to dynamically manipulate cell adhesion using L929 mouse fibroblasts (Fig. 2E). Without binding cell-adhesive AA–RGD, L929 cells on the Van-containing quartz substrate (bare surface) only exhibited a round shape (i.e., poor adhesion) after 4 h of culture in Roswell Park Memorial Institute (RP MI) 1640 medium. The cell-repellant property was probably due to the high hydrophilicity of surface-grafted poly(Van–PEG) brushes (SI Appendix, Fig. S4), which could efficiently reduce nonspecific protein adsorption and block cell adhesion. On the contrary, the adhesion of L929 cells on the AA–RGD-bound surface was significantly improved. We observed typically adhered L929 cells spreading on the AA–RGD-bound surface. On the other hand, cell adhesion on surfaces bound with a nonadhesive molecule AA–RGE (arginine–glycine–glutamic acid) was found to be as poor as that of bare surface (compare SI Appendix, Figs. S5 and S6), indicating the key role of bioactive RGD for cell adhesion. Quantitative analysis revealed that the number of adhered L929 cells (i.e., the spreading cells) on the bare surface was negligible, while almost all seeded cells on the AA–RGD-bound surface could achieve adhesion in 4 h (Fig. 2F). We also found that the average cell-spreading area on the AA–RGD-bound surface was 1.88-fold larger than that on the bare surface (Fig. 2G). This significantly improved cell adhesion and was further verified by cytoskeleton staining. We used DAPI and phalloidin to fluorescently stain the nuclei and filamentous actin in adhered L929 cells, respectively. Clearly, cells on the AA–RGD-bound surface exhibited typical focal adhesion patterns, whereas cells on the bare surface were scarce and only showed round shape with no presence of stress fiber (Fig. 2E). These results demonstrated that the specific molecular recognition between Van and AA could be used to interlock bioactive molecules and control specific cell responses.
Since the bound AA–RGD could be released by an AA-mediated molecular exchange (Fig. 2D), the detachment of adhered L929 cells was then examined to demonstrate the dynamic cell adhesion on the surface. L929 cells were first cultured on the AA–RGD-bound surface for 4 h; then, the adhered cells were incubated with competitive AA, and the cell morphology was recorded under a microscope. We observed a rapid transition of cell morphology from spreading to round shape upon the addition of free AA peptide (cell 1 and cell 2 in Fig. 2H, SI Appendix, Fig. S7, and Movie S1). After incubation for 30 min, almost all the L929 cells showed clear shrinking, and more than 85% of the cells could be easily removed with mild rinsing. The dramatic decrease (∼61%) of the cell-spreading area reflected a significant change of cell status from good adhesion to poor adhesion (Fig. 2I). The adhered L929 cells on tissue culture polystyrene plates (used as control) were also incubated with free AA peptide. In contrast, the cells maintained the spreading shape, and no significant change was observed in their morphology even after 1 h of incubation with the free AA peptide (SI Appendix, Fig. S8). These experiments jointly verified that the change of cell status on the dynamic biointerface was mainly due to the release of the cell-adhesive molecule AA–RGD triggered by AA-mediated molecular exchange, which resulted in a gradual reduction of cell adhesion force and subsequent cell detachment.
In addition to the unidirectional dynamics of cells from good adhesion to poor adhesion, the potential of this dynamic surface for reversible control of cell adhesion was further explored. L929 cells were found to be hard to adhere onto the bare surface after 4 h of culture in RPMI 1640 medium (t = 4 h) (Fig. 2J). Upon the addition of AA–RGD (20 mM) into the medium, the cells showed spreading and typical adhesion in another 4 h (t = 8 h). When the medium was replaced with an AA-containing RPMI 1640 medium (20 mM), the adhered cells shrank rapidly within 1 h (t = 9 h). To retrigger cell adhesion onto the surface, the medium was again changed with fresh RPMI 1640 medium and fed with AA–RGD (20 mM; t = 10 h). After incubation for another 6 h, the shrunken cells readhered onto the surface (t = 16 h). In a control experiment (cell adhesion to a surface with initial RGD ligands), we found that the adhesion status of cells on AA–RGD-bound surfaces did not undergo reversible changes, indicating the importance of reversible AA–RGD binding in the dynamic cell regulation (SI Appendix, Fig. S9). Taken together, these findings demonstrated that the dynamic biointerface, with reversible binding of bioactive molecules (i.e., RGD in this study), could be handily employed for reversible and programable manipulation of cell behaviors.
General Applicability and Biocompatibility of the Dynamic Biointerface.
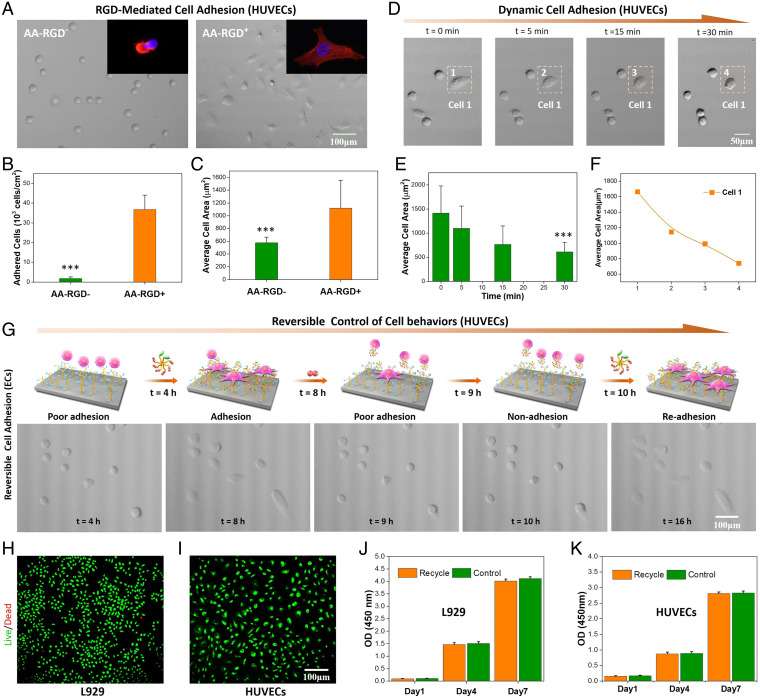
Human umbilical vein endothelial cells (HUVECs) were further employed to demonstrate the general applicability of our dynamic biointerface for reversible control of different cells. Likewise, the HUVECs could not adhere onto a bare surface without AA–RGD binding, while the AA–RGD-bound surface elicited significantly enhanced adhesion of HUVECs (Fig. 3A). Quantitative analysis revealed that the number of adhered HUVECs on the bare surface was almost negligible as compared with that of the AA–RGD-bound surface (Fig. 3B). Moreover, the average spreading area of HUVECs on the AA–RGD-bound surface was 1.94-fold larger than that of the bare surface (Fig. 3C). We further investigated the AA peptide–triggered HUVEC detachment after its adhesion onto the AA–RGD-bound surface (4 h). Similar to the dynamic adhesion of L929 cells, HUVECs also exhibited a gradual morphology transition from spreading to round shape in 30 min upon the addition of free AA (Fig. 3D and Movie S2). As shown in Fig. 3 E and F, the cell-spreading area at 30 min was less than 50% of the initially adhered HUVECs at 0 min, indicating the rapid and significant change from adhesion to nonadhesion. The programmed control of reversible cell adhesion was also shown by an alternative feeding cell culture medium with the bioactive cell-adhesive molecule AA–RGD and the inactive AA dipeptide. Similarly, we observed a reversible and successive transition of HUVEC adhesion onto the dynamic biointerface, which corresponded to the presence of surface RGD. These findings, together with the reversible control of L929 cell adhesion, indicated that the specific but reversible Van–AA interaction could be a general approach for dynamic bioligand presentation and reversible cell behavior modulation.
Fig. 3.
The general applicability and biocompatibility of the Van–AA-based dynamic biointerface for cell regulation. (A) Representative micrographs of HUVECs after culture for 4 h on the surfaces without (AA–RGD−) and with AA–RGD (AA–RGD+). Insets are representative fluorescence images of adhered HUVECs. (B) Quantitative result of the cell adhesion efficiency. (C) Quantitative result of the average cell-spreading area. (D) Time-dependent HUVEC detachment on the AA–RGD-bound surface after adding 20 mM AA dipeptide. (E) The change of the average HUVEC-spreading area during AA-triggered cell detachment. (F) The change of the cell 1–spreading area during the detachment. (G) Programmed manipulation of reversible HUVEC adhesion on the dynamic biointerface. (H and I) The live/dead cell-staining images of L929 and HUVECs. (J and K) The proliferation profile of the original (control) and recovered cells after one cycle of dynamic adhesion. OD, optical density. ***Statistically significant difference is indicated by P < 0.001 as compared with others.
More importantly, the reversible Van–AA interaction for cell regulation showed good biocompatibility. The recycled cells from Figs. 2J and 3G (including both HUVECs and L929 cells) were found to maintain good cell viability equivalent to that of the original cells (control) (Fig. 3 H–K). In other words, dynamic bioligand presentation and reversible cell behavior modulation mediated by the Van–AA interaction could be achieved with general applicability in a noninvasive manner.
Dynamic 3D ECM Mimics with Reversible Dougong Structure.
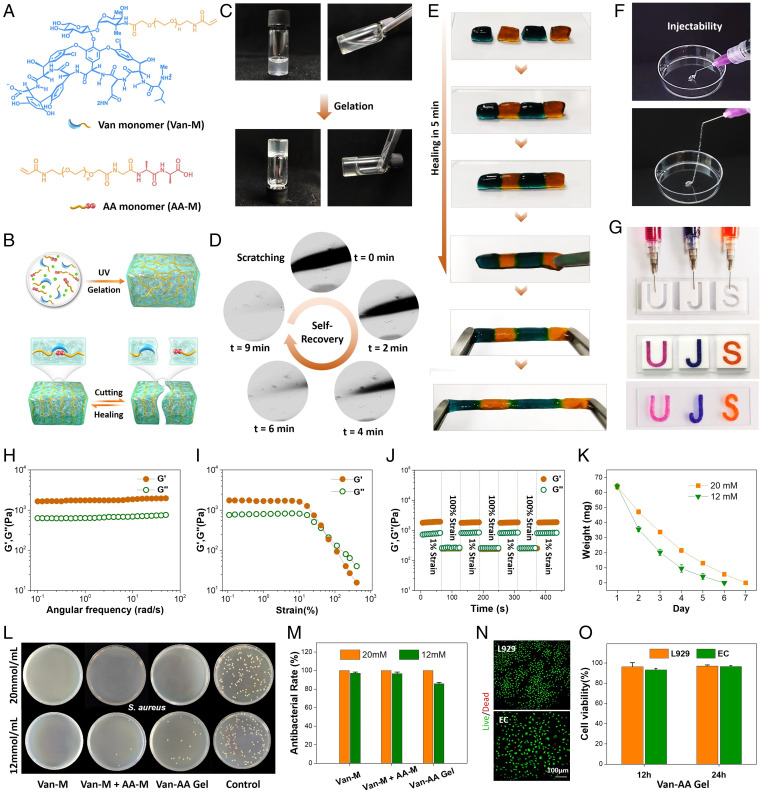
In addition to building a dynamic biointerface, the Van–AA molecular recognition has also been used to build dynamic 3D ECM mimics with reversible dougong structure. In this study, we synthesized two polymerizable acrylamide-based monomers (i.e., the vancomycin-containing monomer [Van-M] and the d-Ala-d-Ala–containing monomer [AA-M]) (Fig. 4A) by grafting Van or AA onto an acrylamide and NHS dual-terminated linear PEG through NHS–amine coupling (SI Appendix, Fig. S10). Typically, Van-M (20 mM) and AA-M (10 mM) monomers were mixed with 5 mg of photoinitiator 2-hydroxy-4′-(2-hydroxyethoxy)-2-methylpropiophenone in PBS solution and then, initiated by UV light (365 nm) for 30 min from a bulk hydrogel (Van–AA hydrogel) (Fig. 4 B and C). Generally, the polymerization of acrylamide-based monomers without cross-linkers (e.g., the bisacrylamide or diacrylate derivatives) cannot obtain gel-like structure (36, 37). However, in our system, the mixed two acrylamide–based monomers (Van-M and AA-M) could specifically recognize each other from a gel-like Van-M/AA-M complex. We also prepared Van–AA hydrogels with different Van-M concentrations (from 10 to 20 mM) and different molar ratios of Van-M/AA-M (1:1, 2:1, and 3:1). Gelation could be observed in all tests, indicating the efficient interlocking through the Van-M/AA-M complex. These hydrogels showed not only large porosity (61 to 71%) but also, controllable swelling ratio (SI Appendix, Fig. S11), indicating the flexibility as scaffold materials for cell growth in tissue engineering. Due to the dynamic nature of the Van-M/AA-M complex, we expect the Van–AA hydrogels to possess typical dynamic properties, such as self-healing, remoldability, and injectability.
Fig. 4.
Dynamic hydrogels based on reversible Van–AA interactions. (A) Structural formula of the Van-M and AA-M. (B) Schematic illustration of the Van–AA dynamic hydrogels. (C) The sol-to-gel transition after photo-initiated polymerization of Van-M (20 mM) and AA-M (10 mM). (D) The healing process of a scratched Van–AA hydrogel surface. (E) The self-healing ability of Van–AA hydrogel enables the integration of small hydrogel blocks. Block sizes: 0.5 × 1.0 cm. (F) The injectability of the Van–AA hydrogel. (G) Injection of Van–AA hydrogels for the formation of letter patterns. Teflon molds: 2.0 × 2.0 cm. (H) The dynamic oscillatory frequency sweeps (strain = 1%) of the Van–AA hydrogel. (I) The strain amplitude sweeps (frequency = 1 rad s−1) of the Van–AA hydrogel. (J) The step-strain sweeps (strain = 1 or 100%, frequency = 1 rad s−1) of the Van–AA hydrogel. Temperature: 25 °C. G′, storage modulus; G″, loss modulus. (K) The in vitro degradation profile. (L) Photos of agar plates after 24 h of incubation of S. aureus with different materials. (M) Quantitative analysis of the in vitro antibacterial efficiency of the Van–AA hydrogels. (N) The live/dead cell-staining images of L929 and HUVECs after incubation with Van–AA hydrogel for 12 h. (O) The 24-h proliferation efficiency of hydrogel-incubated cells. Cell viability of original cells was 100%. All of the above results were based on the hydrogel prepared with 20 mM Van-M and 10 mM AA-M (Van-M/AA-M = 2:1).
The dynamic properties of the Van–AA hydrogel composed of 20 mM Van-M and 10 mM AA-M were first examined by investigating their self-recovery ability against physical damage. A scratch was made on the hydrogel surface by a blade, and the healing process was recorded under a microscope (Fig. 4D). In the beginning (0 min), the scratch could be clearly observed on the surface. As expected, the scratch underwent a rapid healing process, and no trails were left on the hydrogel after 9 min (Movie S3). To confirm the self-healing ability, four blocks of the Van–AA hydrogels stained with methyl blue or methyl orange were staggered and placed in contact for self-healing. As shown in Fig. 4E, all the hydrogel blocks showed very fast healing at the contact interfaces within 5 min, and no obvious cracks could be observed. Moreover, the joint Van–AA hydrogel could be stretched over 200% of its original length, indicating its resistance against external tensile force. Apart from self-healing, our dynamic cross-linked hydrogels also feature a shear thinning behavior and exhibit injectability (Fig. 4G and SI Appendix, Fig. S12). This property was visually demonstrated by injecting the Van–AA hydrogels through a needle (22 gauge, 0.41-mm inner diameter). As shown in Fig. 4F and Movie S4, the hydrogel could be easily placed into a syringe and injected without fracture. By using letter-patterned Teflon molds, the injected Van–AA hydrogel could be padded to construct three intact hydrogel letters (Fig. 4G). Undoubtedly, the Van–AA hydrogel possesses typical characteristics of a dynamic hydrogel, specifically self-recovery, injectability, and moldability.
To assess the dynamism of the Van–AA hydrogel in depth, rheology examination was carried out. Dynamic oscillatory frequency sweep from 0.1 to 100 rad s−1 was first performed (Fig. 4H). The hydrogel showed a viscoelastic behavior with a higher storage modulus (G′) than the loss modulus (G″), suggesting its gel-like characteristic. It was worth noting that G′ and G″ were nearly unchanged over the frequency range, indicating that the dynamic but stable structure built by the Van–AA complex effectively restricted the polymer chain motion. Strain sweep was then performed to investigate the critical strain value to break the hydrogel network. As shown in Fig. 4I, the G′ and G″ values of the Van–AA hydrogels could stay constant under a lower strain, indicating the resistance to a moderate mechanical agitation. However, a reverse of the G′ and G′′ values was observed when the strain was further increased over 100%. This was mainly due to the breakage of the dynamic Van–AA interlocks and the collapse of the hydrogel network, leading to a transition from gel to sol state. The self-recovery of the Van–AA hydrogel was further examined using a step-strain sweep with alternative strain between 1 and 100% (Fig. 4J). The high oscillation excitation (100% strain)–caused network failure could recover immediately when the strain was reduced to 1%. Moreover, after a repeated dynamic strain sweep, both G′ and G′′ values recovered without significant loss. These rheology results well explained the dynamic phenomena (Fig. 4 D–G) of the Van–AA hydrogel under physical treatment, including self-healing property, injectability, and moldability.
In addition to the self-healing property, injectability, and moldability, the degradability, inherently antibacterial activity, and biocompatibility of the Van–AA hydrogel were further explored. We first evaluated the degradation of the Van–AA hydrogel (Fig. 4K and SI Appendix, Fig. S13). We found that the Van–AA hydrogel can be completed degraded in 1 wk in vitro and in 3 to 4 d in vivo. The accelerated degradation in vivo was probably due to the presence of enzymes as compared with that of the enzyme-free condition in vitro. In addition, increasing the monomer concentration would prolong the hydrogel degradation period. Furthermore, the inherently antibacterial activity of the Van–AA hydrogel was studied using gram-positive Staphylococcus aureus and gram-negative Escherichia coli (Fig. 4L and SI Appendix, Fig. S14). The Van–AA hydrogels with high (20 mM) and low (12 mM) concentration of Van-M both could exhibit significant bactericidal effects. Particularly, the Van–AA hydrogels with higher Van–AA concentration and higher Van-M ratio showed higher antibacterial activity (Fig. 4M). This finding implied the necessity of high Van content in the hydrogel to generate an efficient bacterial inhibition. Thus, in the subsequent experiments, we used the Van–AA hydrogel composed of 20 mM Van-M and 10 mM AA-M. In a further study, we also found that the bacterial inhibitory efficiency of our hydrogels was close to that of free Van and gentamycin (SI Appendix, Fig. S15). It is worth mentioning that, in addition to the antibacterial activity, the Van–AA hydrogel exhibited excellent biocompatibility. Cells cocultured with the Van–AA hydrogel could maintain high viability for 12 h (Fig. 4N). In addition, their proliferation rate was equivalent to that of the control cells without treatment. These results confirmed the great potential of our dynamic Van–AA hydrogel as a biocompatible biomaterial scaffold, especially for infection-prone tissue repair.
Dynamic Hydrogel for Infected Skin Wound Healing.
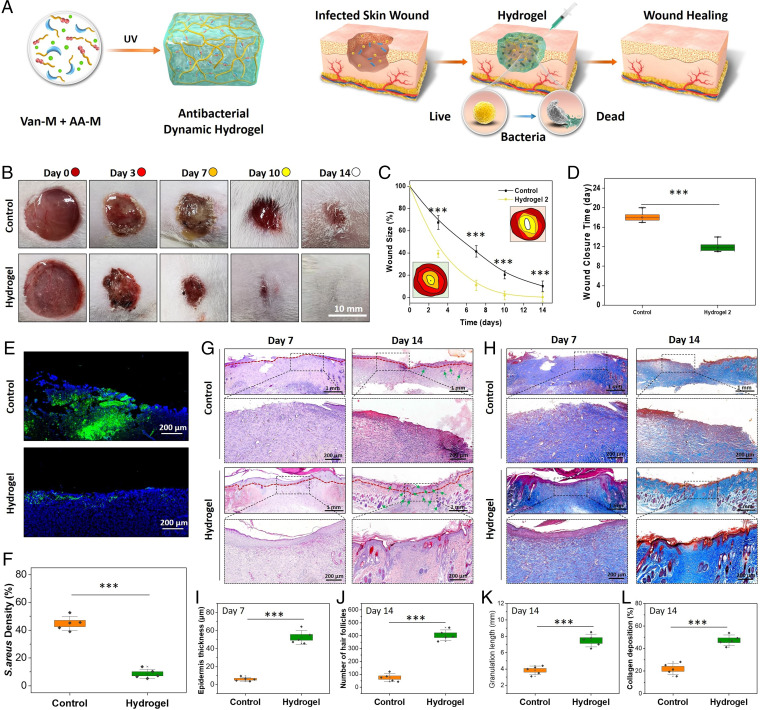
Clinically, skin wound infection may exhibit varied severity ranging from superficial paronychia to life-threatening bone infection (38–45). Moreover, skin wounds also face high risk of repeated damage and reinfection resulting from external mechanical irritation. Note that the Van–AA dynamic hydrogel features self-healing ability, injectability, remoldable property, and antibacterial activity, which are particularly appealing for infected open skin wound healing as the dynamic hydrogel can reduce self-fragmentation, adapt to the shape of the wound site, and wipe out the pathogenic bacteria to continuously support skin regeneration. In this context, full-thickness cutaneous wounds inoculated with S. aureus were established as the infected skin wound model on the dorsum panniculus carnosus of Sprague–Dawley rats. Two different treatments, injected with PBS (control) or with the typical Van–AA hydrogel, were employed to the infected wounds to demonstrate their wound-healing efficacy (Fig. 5A).
Fig. 5.
Dynamic antibacterial hydrogel for infected skin wound repair. The hydrogel was prepared with a molar ratio of Van-M and AA-M at 2:1. (A) Schematic illustration of the repair mechanism of the Van–AA hydrogel (composed of 20 mM Van-M and 10 mM AA-M) for infected skin wound healing. (B) Gross observation of S. aureus–infected wound healing in a Sprague–Dawley rat model. Wound size: 1.5 cm. (C) Quantitative analysis of the wound size on days 3, 7, 10, and 14. Insets show the traces of wound-bed closure. (D) The wound closure time. (E) Representative Immunofluorescence-staining images on day 3 using S. aureus antibodies (green) and DAPI counterstaining (nucleus; blue). (F) Quantitative analysis of the staining intensity indicating the significant bacterial inhibition of the Van–AA hydrogel in vitro. (G and J) Micrographs of H&E- and Masson’s trichome–stained tissue slices on days 7 and 14 (boundary of epithelium: red lines; hair follicle: green arrows). (H) Quantification of epidermis thickness in each group on day 7. (I, K, and L) Quantification of hair follicles, granulation length, and collagen deposition in each group on day 14 (n = 5). ***Statistically significant difference is indicated by P < 0.001 as compared with others.
The wound-healing and antiinfective efficacy of the Van–AA hydrogel was investigated over the course of 0, 3, 7, 10, and 14 d based on the photos captured with a digital camera (Fig. 5 B–D). At day 3, noticeable dark yellow pus was found dispersing on the wound bed in the control group, and the dark yellow pus could still be observed on day 7, indicating the severity of the infected wounds. However, this kind of pyogenic phenomena was hardly seen in the group of Van–AA hydrogels during the whole period, suggesting the high efficiency of antiinfection. The exudate from different wound sites was further collected and cultured on agar plates (SI Appendix, Fig. S16). As compared with the bacteria-covered agar plates in the control group, almost no bacteria strain was found in the hydrogel-treated group, confirming the high antibacterial activity of the Van–AA hydrogel. The hydrogel-treated wounds all appeared drier and smaller from day 3 (Fig. 5B), indicating that the external infection was effectively controlled and the tissue reconstruction process was initiated. In contrast, the wound-healing process in the control group was slower, presumably due to the uncontrolled infection that impeded the tissue healing. According to the quantitative analysis of the wound diameter, we found that almost all wounds in the Van–AA hydrogel-treated group could completely heal within 14 d, while within the same period, no wounds could heal in the control groups, and the average wound-healing ratio was only around 81%. Further investigation showed that the average wound closure time in the control group was 18 d (Fig. 5D), which was almost 1 wk longer than that of the Van–AA hydrogel-treated group. We further compared the wound-healing rate of our hydrogels with a commercially available product, the Tegaderm transparent film dressing. Infected skin wounds treated with our dynamic hydrogels showed faster healing rate compared with that of Tegaderm-treated ones (SI Appendix, Fig. S17), probably due to the high antibacterial activity and shape adaptability of our hydrogels to the wound bed. We also found that hydrogels with Van-M/AA-M molar ratio at 2:1 showed the optimal wound-healing efficacy compared with other hydrogels, probably due to the more balanced antibacterial activity and good mechanical property. These results together confirmed the promising efficacy of our dynamic Van–AA hydrogel on infected wound healing.
To further visualize and compare the antiinfection efficiency of different groups, immunofluorescence staining of the skin sections against S. aureus after 3 d postinjection was performed. The bright green fluorescence (S. aureus) with DAPI counterstains (nuclei; blue) in the control skin section indicated serious bacterial infection underneath the epidermal layer and the underlying dermis (Fig. 5E). In contrast, the S. aureus intensity was much lower and the bacteria were distributed only on the superficial skin layer in the hydrogel-injected group. Quantitative analysis revealed that 90% of the inoculated bacteria could be inhibited by the Van–AA hydrogel in 3 d, while the control group was still covered and even ingrown with plenty of bacteria (∼50% residue of the inoculated bacteria) (Fig. 5F). This finding confirmed that the Van–AA hydrogel possessed excellent antibacterial effect in vivo. This is advantageous as early bacterial elimination by the hydrogel will contribute greatly to the rapid healing of infected wounds. Further in our study, the skin samples with wounds at each time point were harvested and embedded into paraffin for hematoxylin and eosin (H&E) staining. As shown in Fig. 5G, relatively fewer inflammatory cells were observed in the Van–AA hydrogel group than in the control group on day 7. In addition, the skin defect in the hydrogel group was almost closed with highly regular epithelium, numerous hair follicles, and almost complete regeneration of dermal tissue after 14 d. In contrast, the control group showed a large area of immature granulation tissue deposited around the wound. As shown in Fig. 5 I–K, the epidermis thickness, the number of hair follicles, and the granulation length significantly increased in the hydrogel-treated group. We further applied Masson’s trichrome staining to assess collagen deposition in the defect sites (Fig. 5H). Compared with the control group, the collagen deposition was improved in the hydrogel group on day 14 (Fig. 5L). Moreover, the collagen fibers were denser, thicker, and better arranged in the hydrogel group.
To investigate the local inflammatory effect of the hydrogels on skin tissues, expression of proinflammatory cytokines, such as tumor necrosis factor–α (TNF-α) and interleukin-6 (IL-6), was examined. Subcutaneous injection of the hydrogels together with bacteria (S. aureus) was performed, and the tissue sections of injected regions were used for immunohistochemistry evaluation. As shown in SI Appendix, Fig. S18A, on day 7, the control group (with only bacteria injection) showed the highest expression of TNF-α and IL-6 and maintained the high level after 14 d. In contrast, the expressions of these proinflammatory factors were significantly decreased in the hydrogel-treated group. The results were consistent with the quantitative results of enzyme-linked immunosorbent assay (SI Appendix, Fig. S18B). We thus deduced that our dynamic antibacterial hydrogels could effectively reduce the inflammatory response under infected conditions, which contributed greatly to the improved wound repair effect.
Histological assessment of representative organs, including heart, liver, spleen, lung, and kidney, was also performed to evaluate the long-term biosafety of the Van–AA hydrogel (SI Appendix, Fig. S19A). In the Van–AA hydrogel group, no obvious histopathological abnormality or lesion was observed in the H&E-stained images. In addition, the serum biochemistry tests on the rats on day 14 revealed that the major indexes of kidney and liver functions remained in the normal range, indicating no detrimental effects caused by the hydrogel (SI Appendix, Fig. S19B). These positive in vivo results confirmed that the dynamic Van–AA hydrogels could be used as an easy-handling, remoldable, antibacterial, and biocompatible functional scaffold biomaterial for infective tissue repair.
Conclusion
In this study, we realized a nature-derived molecular recognition event (i.e., the Van–AA receptor–ligand interaction) as a reversible strategy for fabrication of a dynamic biointerface and 3D ECM mimics. The specific molecular recognition between Van and AA is based on multi-noncovalent interaction with exquisite complementarity in their chemical groups and geometry, exhibiting inherent dynamics and reversibility. Due to the specific but reversible Van–AA molecular recognition, the dynamic biointerface showed excellent reversibility in binding of cell-adhesive RGD and modulating cell adhesion. We also demonstrated the dynamic ECM mimicking molecular strategy’s general applicability for different cell types and the noninvasive stimulus responsiveness for cell regulation. Moreover, we constructed a 3D hydrogel network based on the reversible AA–Van molecular recognition with proven self-recovery and injectability. We surmised that these dynamic properties, as well as the inherently antibacterial activity of the Van–AA hydrogel, are particularly appealing for treating infected open skin wounds; the hydrogel could adapt to the shape of wound site, resist self-fragmentation, and kill the pathogenic bacteria to continuously support the wound healing. Note that the two-dimensional dynamic bioactivity and the 3D dynamic structure were demonstrated separately in this work. Further studies on the possibility of combining dynamic ligand presentation and dynamic structure in a promising 3D format are highly desirable. We believe that the specific but reversible Van–AA molecular recognition would be a promising strategy for dynamic biomaterial fabrication, and the easy-handling merit, ECM-like remoldability, and inherent antibacterial activity will bring insights to biomaterial scaffold design in tissue engineering and regenerative medicine.
Materials and Methods
The syntheses of Van-M and AA-M were performed through NHS–amine coupling methods using NHS-terminal PEG monomers. The dynamic Van–AA hydrogels were formed using photoinitiated polymerization of Van-M and AA-M. All the chemicals and materials as well as the monomer synthesis and the preparation and characterization of Van–AA hydrogels are available in SI Appendix, Materials and Methods. The in vitro and in vivo experimental details, including cell culture, antibacterial assay, animal experiment (approved by the Animal Investigation Ethics Committee of Soochow University , No. 202103A431), histological analysis, immunohistochemistry, and biosafety evaluation, are also available in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This work was financially supported by the National Key Research and Development Program of China Grants 2019YFA0112000 and 2016YFC1100203; National Natural Science Foundation of China Grants 21875092, 81925027, and 32130059; the Innovation and Entrepreneurship Program of Jiangsu Province; “Six Talent Peaks” Program of Jiangsu Province Grant 2018-XCL-013; and Research Grants Council of Hong Kong (General Research Scheme 15202119 and Early Career Scheme 25208218).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2117221119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Ma Y., Tian X., Liu L., Pan J., Pan G., Dynamic synthetic biointerfaces: From reversible chemical interactions to tunable biological effects. Acc. Chem. Res. 52, 1611–1622 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Hammer J. A., West J. L., Dynamic ligand presentation in biomaterials. Bioconjug. Chem. 29, 2140–2149 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Duan W., et al. , Combination of chondrocytes and chondrons improves extracellular matrix production to promote the repairs of defective knee cartilage in rabbits. J. Orthop. Translat. 28, 47–54 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uto K., Tsui J. H., DeForest C. A., Kim D.-H., Dynamically tunable cell culture platforms for tissue engineering and mechanobiology. Prog. Polym. Sci. 65, 53–82 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillette B. M., Jensen J. A., Wang M., Tchao J., Sia S. K., Dynamic hydrogels: Switching of 3D microenvironments using two-component naturally derived extracellular matrices. Adv. Mater. 22, 686–691 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Robertus J., Browne W. R., Feringa B. L., Dynamic control over cell adhesive properties using molecular-based surface engineering strategies. Chem. Soc. Rev. 39, 354–378 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Rosso F., Giordano A., Barbarisi M., Barbarisi A., From cell-ECM interactions to tissue engineering. J. Cell. Physiol. 199, 174–180 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Li W., Yan Z., Ren J., Qu X., Manipulating cell fate: Dynamic control of cell behaviors on functional platforms. Chem. Soc. Rev. 47, 8639–8684 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Talebian S., et al. , Self‐healing hydrogels: The next paradigm shift in tissue engineering? Adv. Sci. (Weinh.) 6, 1801664 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosales A. M., Anseth K. S., The design of reversible hydrogels to capture extracellular matrix dynamics. Nat. Rev. Mater. 1, 1–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu X., et al. , Extracellular vesicles: Potential role in osteoarthritis regenerative medicine. J. Orthop. Translat. 21, 73–80 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian X., et al. , A magnetic dynamic microbiointerface with biofeedback mechanism for cancer cell capture and release. ACS Appl. Mater. Interfaces 11, 41019–41029 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Liu L., et al. , A versatile dynamic mussel-inspired biointerface: From specific cell behavior modulation to selective cell isolation. Angew. Chem. Int. Ed. Engl. 57, 7878–7882 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Pan G., et al. , Dynamic introduction of cell adhesive factor via reversible multicovalent phenylboronic acid/cis-diol polymeric complexes. J. Am. Chem. Soc. 136, 6203–6206 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Zeng X., et al. , A drug-self-gated mesoporous antitumor nanoplatform based on pH-sensitive dynamic covalent bond. Adv. Funct. Mater. 27, 1605985 (2017). [Google Scholar]
- 16.Zhao Q., Du X., Multi-scale adaptions of dynamic bio-interfaces. Smart Mater. Med. 3, 37–40 (2022). [Google Scholar]
- 17.Liu D., Xie Y., Shao H., Jiang X., Using azobenzene-embedded self-assembled monolayers to photochemically control cell adhesion reversibly. Angew. Chem. Int. Ed. Engl. 48, 4406–4408 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Li W., Wang J., Ren J., Qu X., Near‐infrared‐and pH‐responsive system for reversible cell adhesion using graphene/gold nanorods functionalized with i‐motif DNA. Angew. Chem. 125, 6858–6862 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Sur S., Matson J. B., Webber M. J., Newcomb C. J., Stupp S. I., Photodynamic control of bioactivity in a nanofiber matrix. ACS Nano 6, 10776–10785 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boekhoven J., Rubert Pérez C. M., Sur S., Worthy A., Stupp S. I., Dynamic display of bioactivity through host-guest chemistry. Angew. Chem. Int. Ed. Engl. 52, 12077–12080 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi P., et al. , Host–guest recognition on photo-responsive cell surfaces directs cell–cell contacts. Mater. Today 20, 16–21 (2017). [Google Scholar]
- 22.An Q., et al. , A supramolecular system for the electrochemically controlled release of cells. Angew. Chem. Int. Ed. Engl. 51, 12233–12237 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Kang H., et al. , An in situ reversible heterodimeric nanoswitch controlled by metal-ion-ligand coordination regulates the mechanosensing and differentiation of stem cells. Adv. Mater. 30, e1803591 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Pan G., et al. , Biomimetic design of mussel-derived bioactive peptides for dual-functionalization of titanium-based biomaterials. J. Am. Chem. Soc. 138, 15078–15086 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Liu H., et al. , Hydrophobic interaction-mediated capture and release of cancer cells on thermoresponsive nanostructured surfaces. Adv. Mater. 25, 922–927 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Pan G., et al. , An epitope-imprinted biointerface with dynamic bioactivity for modulating cell-biomaterial interactions. Angew. Chem. Int. Ed. Engl. 56, 15959–15963 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan G., Guo Q., Ma Y., Yang H., Li B., Thermo-responsive hydrogel layers imprinted with RGDS peptide: A system for harvesting cell sheets. Angew. Chem. Int. Ed. Engl. 52, 6907–6911 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Ma Y., et al. , Thermo-responsive imprinted hydrogel with switchable sialic acid recognition for selective cancer cell isolation from blood. Bioact. Mater. 6, 1308–1317 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guillaume‐Gentil O., et al. , Polyelectrolyte coatings with a potential for electronic control and cell sheet engineering. Adv. Mater. 20, 560–565 (2008). [Google Scholar]
- 30.Pan J., Chen W., Ma Y., Pan G., Molecularly imprinted polymers as receptor mimics for selective cell recognition. Chem. Soc. Rev. 47, 5574–5587 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Rao J., Lahiri J., Isaacs L., Weis R. M., Whitesides G. M., A trivalent system from vancomycin.D-ala-D-Ala with higher affinity than avidin.biotin. Science 280, 708–711 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Ren T., et al. , Dual responsive surfaces based on host-guest interaction for dynamic mediation of cell-substrate interaction and cell migration. Adv. Mater. Interfaces 4, 1500865 (2017). [Google Scholar]
- 33.Yan Z., Qin H., Ren J., Qu X., Photocontrolled multidirectional differentiation of mesenchymal stem cells on an upconversion substrate. Angew. Chem. Int. Ed. Engl. 57, 11182–11187 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Luo Y., Zheng X., Yuan P., Ye X., Ma L., Light-induced dynamic RGD pattern for sequential modulation of macrophage phenotypes. Bioact. Mater. 6, 4065–4072 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan J., et al. , Hyaluronic acid-based hydrogels with tobacco mosaic virus containing cell adhesive peptide induce bone repair in normal and osteoporotic rats. Biomater. Transl. 1, 89 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Billiet S., Hillewaere X. K. D., Teixeira R. F. A., Du Prez F. E., Chemistry of crosslinking processes for self-healing polymers. Macromol. Rapid Commun. 34, 290–309 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Ma Y., et al. , Dynamic colloidal photonic crystal hydrogels with self-recovery and injectability. Research (Wash D C) 2021, 9565402 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chastain C. A., Klopfenstein N., Serezani C. H., Aronoff D. M., A clinical review of diabetic foot infections. Clin. Podiatr. Med. Surg. 36, 381–395 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Xu Z., et al. , Bio-macromolecules/modified-halloysite composite hydrogel used as multi-functional wound dressing. Smart Mater. Med. 2, 134–144 (2021). [Google Scholar]
- 40.Lawson M. C., Shoemaker R., Hoth K. B., Bowman C. N., Anseth K. S., Polymerizable vancomycin derivatives for bactericidal biomaterial surface modification: Structure-function evaluation. Biomacromolecules 10, 2221–2234 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao Y., et al. , A versatile surface bioengineering strategy based on mussel-inspired and bioclickable peptide mimic. Research (Wash D C) 2020, 7236946 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen H., et al. , An injectable self-healing coordinative hydrogel with antibacterial and angiogenic properties for diabetic skin wound repair. NPG Asia Mater. 11, 1–12 (2019). [Google Scholar]
- 43.Zhao X., et al. , Cell infiltrative hydrogel fibrous scaffolds for accelerated wound healing. Acta Biomater. 49, 66–77 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun X., et al. , Electrospun photocrosslinkable hydrogel fibrous scaffolds for rapid in vivo vascularized skin flap regeneration. Adv. Funct. Mater. 27, 1604617 (2017). [Google Scholar]
- 45.Bai J., et al. , Biomimetic osteogenic peptide with mussel adhesion and osteoimmunomodulatory functions to ameliorate interfacial osseointegration under chronic inflammation. Biomaterials 255, 120197 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.