Significance
Acute stress involves the majority of brain areas, which can be sequentially organized in functional brain networks as shown by our study with foot shock in mice. We used whole-brain microscopy to investigate different spatial resolutions over time. From mesoscale region–based analyses, we identified the order of activation of brain areas. With single-cell analyses, we analyzed shifts in activation over time within small nuclei—a result impossible to achieve with functional MRI’s resolution. These findings required the development of a four-dimensional (4D) analytical pipeline, which is made available as an R package. This “atlas” of foot shock can be visualized in 4D in our interactive web portal.
Keywords: acute stress, whole brain, c-fos, foot shock, brain-wide analysis
Abstract
Acute stress leads to sequential activation of functional brain networks. A biologically relevant question is exactly which (single) cells belonging to brain networks are changed in activity over time after acute stress across the entire brain. We developed a preprocessing and analytical pipeline to chart whole-brain immediate early genes’ expression—as proxy for cellular activity—after a single stressful foot shock in four dimensions: that is, from functional networks up to three-dimensional (3D) single-cell resolution and over time. The pipeline is available as an R package. Most brain areas (96%) showed increased numbers of c-fos+ cells after foot shock, yet hypothalamic areas stood out as being most active and prompt in their activation, followed by amygdalar, prefrontal, hippocampal, and finally, thalamic areas. At the cellular level, c-fos+ density clearly shifted over time across subareas, as illustrated for the basolateral amygdala. Moreover, some brain areas showed increased numbers of c-fos+ cells, while others—like the dentate gyrus—dramatically increased c-fos intensity in just a subset of cells, reminiscent of engrams; importantly, this “strategy” changed after foot shock in half of the brain areas. One of the strengths of our approach is that single-cell data were simultaneously examined across all of the 90 brain areas and can be visualized in 3D in our interactive web portal.
Acute stress leads to the activation of multiple functional brain networks, as demonstrated in humans using functional MRI (fMRI) (reviewed in refs. 1 and 2). Yet, spatial resolution beyond the level of (networks of) nuclei is currently not possible with fMRI. This severely limits our ability to answer an important biological question. Which (single) cells belonging to brain networks are changed in activity over time after acute stress across the entire brain? Additionally, do all cells respond in the same way? This most likely is not the case. Previous rodent studies have established that even small areas, such as the basolateral amygdala (BLA), have a heterogeneous cellular composition (3) and contribute to a wide array of behaviors (4), presumably linked to long-range connectivity (3). Other studies in animals have highlighted that just a few cells within the dentate gyrus are greatly responsive to acute stress; this high degree of responsiveness was linked to demethylation of specific sites on the DNA (5, 6). Thus, studies confined to subparts of the brain point to heterogeneity in the cellular response to stress and emphasize the necessity of a whole-brain approach with cellular resolution.
In principle, whole-brain microscopy can be used to address these questions. This technique can provide a snapshot of transcriptional (7) cellular activity throughout the whole brain (8) by staining for immediate early genes (IEGs) (9). The analytical challenges are not trivial. Several tools have been developed to detect active cells and to register them to an atlas (an excellent review of open-source tools is in ref. 10). Most of these tools even offer built-in options for visualization, but to date, no study has thoroughly explored the subsequent steps of data analysis (i.e., dealing with missing values, batch effect corrections, normalization, and transformation). Yet, these steps are essential; they can influence results and the interpretation of findings (11), as previously shown in several other fields (for example, refs. 12–14). Therefore, before embarking on complex whole-brain analyses as well as introducing a time dynamic, we first tackled how to clean and preprocess the data.
Whole-brain microscopy has excellent spatial resolution [∼5 µm (8)] yet very poor time resolution, usually confined to a single time point. Solving this conundrum was the second step in our approach. For this, we used a method developed and commonly used by many laboratories before (for example, refs. 8 and 15–18) using the IEG c-fos (SI Appendix, Note 1) as a postmortem marker of cellular activity (19), to which we added a pseudotime. Previous studies report that c-fos messenger RNA (mRNA) can peak at different times across brain areas after swim or restraint stress (20). This suggests that there may be multiple waves of c-fos activation throughout the brain, which could be used to map the temporal dynamics across all brain areas up to the level of single cells and from minutes to hours after the initial stimulation. Determining shifts in activity during the different phases of the stress response can be a first step to clarify the temporal dimension of the stress response at a single-cell level across the whole brain, a topic that has received little attention so far (21). This approach moves the field beyond important earlier studies that looked at c-fos expression—some even brain wide (for example, refs. 16–18)—after a variety of stimuli and demonstrated both general patterns of activation that are typical for arousal as well as transcriptional changes that seem to be stressor specific, yet all confined to a single time point (22).
Overall, to understand how single cells across the three-dimensional (3D) brain adapt their activity at various time points after stress, we exposed adult male mice to a single stressful foot shock and charted cellular activity across 89 areas using c-fos staining as a proxy of cellular activity. A pipeline for data preprocessing and analysis at different spatial resolutions was developed, allowing investigation from the macro- (functional networks) to microscales (single-cell resolution) and with a pseudotime scale.
Results
Overview of the Pipeline and Quality Control.
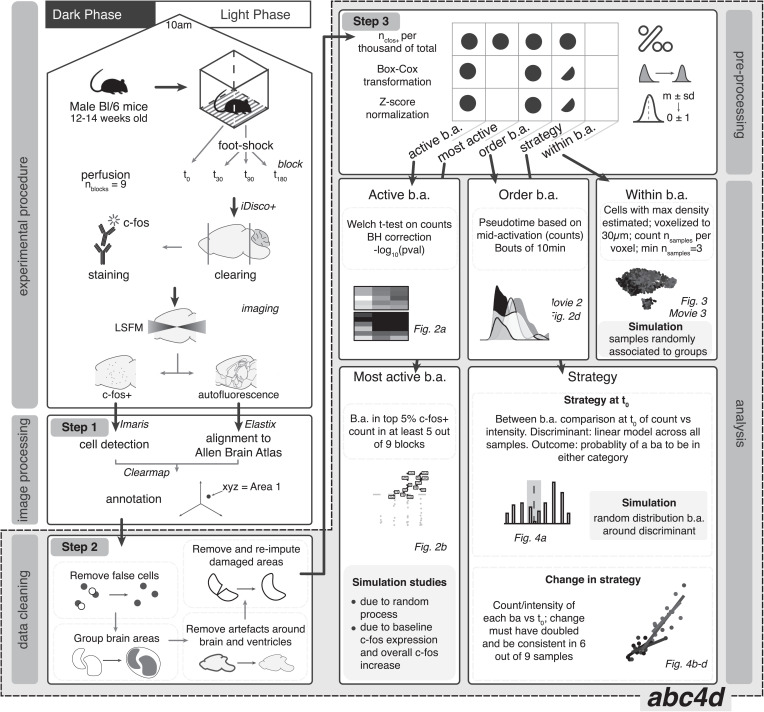
To approach our biological questions, we first optimized, combined, and expanded available methodological tools. Fig. 1 summarizes the main features of the pipeline, including experimental procedure, image processing (step 1), data cleaning (step 2), data preprocessing (step 3), and analysis.
Fig. 1.
Schematic overview of the pipeline. Animals were perfused at different time points (nanimals = ntime point × nblocks = 4 × 9 = 36) after foot shock. Whole-brain samples were processed with the iDisco+ protocol; c-fos+ cells imaged with light-sheet fluorescent microscopy (LSFM). Cells were detected with Imaris and annotated to the Allen Brain Atlas with Clearmap. Output yielded xyz coordinates per cell. Quality control (data cleaning) consisted of removal of various artifacts and of grouping brain areas (b.a.) to the spatial resolution of interest. Data preprocessing of b.a. was performed for each of the analyses. Circles indicate the step required; half-circles indicate the step recommended but not required. Strategy refers to t0 strategy categorization as well as change of strategy over time. Lower Right summarizes the analyses conducted and the main statistical decisions made. The image processing step uses software developed by others [i.e., Imaris (v9.2.0; Bitplane), Elastix (23), and Clearmap (8)]. The steps that we developed (data cleaning [step 2] and preprocessing [step 3] as well as analyses) are explained in detail in Materials and Methods and have been implemented in the abc4d package. BH, Benjamini–Hochberg.
In brief, cell detection (step 1) was performed with Imaris’s spot object (SI Appendix, Fig. S1A), which was then aligned to the Allen Brain Reference Atlas with Clearmap. Precision of alignment was assessed by comparing how sample images and template images would distort landmarks, which were previously manually placed (SI Appendix, Fig. S1B). The average absolute difference was 8.39 ± 5.88 µm (mean ± SD) in the horizontal plane and 10.92 ± 12.44 µm (mean ± SD; maximal displacement = 23.36 µm) in the sagittal plane, with more laterally placed landmarks being less precise (i.e., on average, an uncertainty of roughly one soma). Alignment did not differ per condition, suggesting that alignment error should not affect our results. Of note, we excluded from the analysis six brain areas because we deemed their size too small for a reliable quantification (SI Appendix, Table S1). Until this step, we adapted tools developed by others [Imaris, Elastix (23), and Clearmap (8)]; alternative tools [e.g., CellFinder (24) and WholeBrain (25)] could have also been used for the same purpose.
Subsequent steps in the pipeline, however, were not previously developed. Thus, during data cleaning (step 2), first unspecific binding was mitigated by removing background signals and applying a mask of three voxels (∼75 µm) around the borders of the brain and ventricles (Movie S1), and by removing cells with abnormally high intensity (ncells removed = 12). The background and mask step accounted for ∼97% of the removed cells. Second, across all samples, ∼5% of the brain areas showed some form of damage; these were removed from the analyses and reimputed. Ultimately, the number of cells removed during the quality control procedure did not differ between groups (SI Appendix, Fig. S2).
The next step in the pipeline is data preprocessing (step 3). As summarized in Fig. 1, Upper Right, data preprocessing is specific for each analysis type. Of note, we used a block design, meaning that a “block” (i.e., mice from the same cage, one animal for each time point, processed simultaneously to avoid isolation stress of the last mouse in the cage) was the experimental unit of randomization and processing of samples. This type of design is essential for effective batch effect correction. The data cleaning (step 2) and preprocessing pipelines (step 3) are available for similar future questions in the R package developed for the purpose, abc4d (“Analysis Brain Cellular activation in 4 Dimensions”), which is interoperable with several annotation/alignment tools [step 1; e.g., Clearmap (8) and CellFinder (24)].
Single-Cell Activity Is Increased after Foot Shock throughout the Brain but with Spatial and Temporal Specificity.
In answer to our biological question, we observed that—compared with control animals (see below)—the total number of c-fos+ cells (ncfos+) across the brain was increased 30 min (t30) after foot-shock induction and remained elevated at t90 and t180. It returned to t0 levels 300 min after foot shock (SI Appendix, Fig. S3). Across batches, ncfos+ was comparable with that of previous literature (8, 15). Of note, control animals (t0) were placed in a foot-shock chamber but did not receive a foot shock. As a consequence, they should be considered as a “mildly stressed” (novelty stressor) group rather than true baseline controls (more information on all control groups is in SI Appendix, Fig. S3).
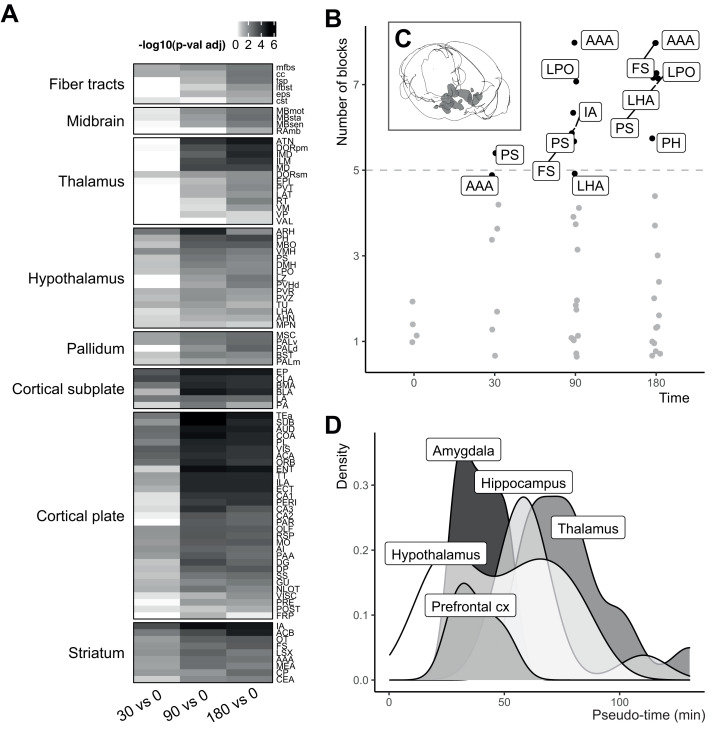
To test the extent of c-fos+ expression throughout the brain, we performed pairwise comparisons (Welch t test, one sided, Benjamini–Hochberg P value correction) between each foot-shock time point (t30, t90, t180) and t0 (Fig. 2A); 86 of the 89 brain areas had a significant increase in c-fos+ cells in at least one of the time points. Only three brain areas were not significantly changed (i.e., the medial preoptic nucleus, the ventral anterior–lateral complex of the thalamus, and the ventroposterior complex of the thalamus). The time point t180 had the highest number of significant brain areas (nsig brain areas = 85), followed by t90 (nsig brain areas = 79) and t30 (nsig brain areas = 40). The effect sizes (g ± SD) ranged between −0.32 ± 0.23 (midbrain raphe nuclei, t30 vs. t0) and 5.17 ± 0.96 (subiculum, t90 vs. t0), with a mean (±SD) of 1.62 ± 0.57.
Fig. 2.
Most brain areas are activated by foot shock, with spatial and temporal specificity. (A) Heat map of −log10 P values derived from pairwise comparisons of each foot shock time point (t30, t90, t180) against t0 for each brain area. White indicates P valueadj ≥ 0.05; gray shades indicate P valueadj < 0.05. The legend numbers correspond to the log values. (B) A set of hypothalamic areas was consistently found to have the highest (top 5% of the distribution) number of c-fos+ cells (per thousand total). The criterion for consistency was five of nine samples. Abbreviations are explained in SI Appendix, Table S1. (C) Cartoon of the brain areas identified in B [created with brainrender (50)]. (D) Functional order of brain areas’ c-fos activity following foot shock. Brain areas were ordered based on a pseudotime depending on c-fos+ activation across the time points and grouped based on functional categorization important for the stress response (26). Hypothalamic areas are the first to reach the midpoint of their activation followed by amygdalar, prefrontal, hippocampal, and lastly, thalamic areas. Of note, the functional order is based on the point of midactivation of brain areas rather than the first instance in which brain areas were activated. An interactive visualization of the single brain areas rather than the categorization is in Movie S2. AAA, anterior amygdala area; IA, intercalated amygdala nucleus; FS, fundus of striatum; LHA, lateral hypothalamic area; LPO, lateral preoptic area; PH, posterior hypothalamic nucleus; PS, parastriatal nucleus.
Since nearly all brain areas were active in at least one time point, we aimed to identify which brain areas were more active than others. To answer this question, we identified for each block (i.e., a unique set of each time point) the brain areas that had the highest (i.e., the top 5% of the distribution) c-fos+ cell count density (per thousand of total, ncfos+/tot). Under random circumstances, the same brain area would be in the top 5% in at least five of nine samples in about 1% of the cases, as illustrated by a simulation study (Materials and Methods). Being selected by at least five samples was, therefore, used as a criterion to define consistency of highly active brain areas. In our experimental data, the criterion was met by eight brain areas (Fig. 2B), which belonged mostly to the hypothalamus (Fig. 2C). This number (nhighly active = 8) was much higher than the 1% expected (nrandomly active = 1) by sheer randomness. With a simulation study, we confirmed that this activation could also not be attributed to the spatial localization of c-fos throughout the brain, as reported by the Allen Brain Atlas (SI Appendix, Fig. S4).
Next, we hypothesized that although in most brain areas, the number of c-fos+ cells is increased after foot shock, the peak of activation would not occur at the same time for every (network of) brain area(s). Brain areas are expected to be involved at different stages—and therefore, at different times—of the stress response as earlier proposed based on human fMRI studies (1). We organized brain areas on a pseudotime scale based on the time point in which a brain area (median across blocks) would reach the middle of its activation. This pseudotime should only be interpreted relatively. We visualized the order of activation (Fig. 2D) of (networks of) the brain areas using a functional categorization valuable to the stress response (26). Based on this classification, hypothalamic areas were found to be activated first followed by amygdalar and prefrontal, hippocampal, and finally, thalamic areas. Movie S2 shows a visualization of all brain areas over time.
Time-Dependent Wave of Activation within the BLA.
While the results so far confirm—in rodents—insights at the network level earlier obtained in humans with fMRI (1), our main goal was to investigate dynamic brain activity after foot shock with higher spatial resolution up to the single-cell level, which is a great advantage over, for example, fMRI studies. Rather than describing all areas, we here illustrate the findings for the BLA, an area key for the cognitive processing of a foot shock (27) and stressful conditions in general (28); for the remaining areas, we refer to an open-source dynamic database (https://osf.io/8muvw/) (29), with which one can browse through all other regions investigated.
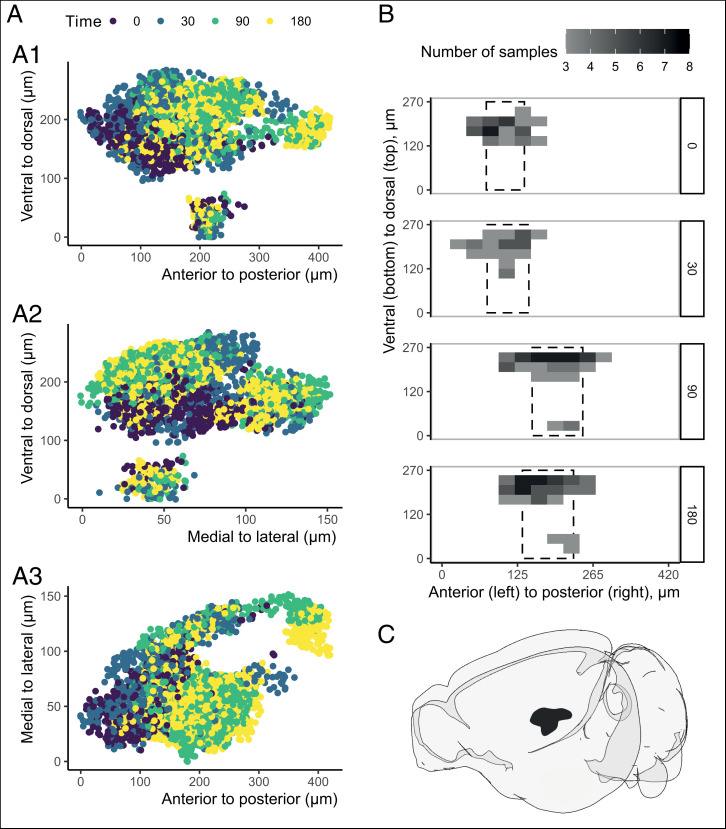
We hypothesized that the increase of c-fos+ cells was not uniform across the BLA; rather, it may be restricted to different subparts or cells. For each sample independently, we identified the most densely activated part of the BLA (i.e., the part with the highest number [density] of c-fos+ cells relative to the rest of the BLA). All samples considered, there are obvious regional distributions across time points (Fig. 3A and Movie S3), which are not evident when samples are randomly associated to the experimental groups (SI Appendix, Fig. S5A).
Fig. 3.
Changes in c-fos+ cell density within the BLA. (A) Cells in high-density regions of the right BLA. The 3D cell coordinates are represented as a set of three two-dimensional graphs, one for each couple of coordinates (xy, yz, xz in A1, A2, and A3, respectively). Each dot is a cell of a sample in a region with the highest density. The colors refer to the different time points. (B) The densest c-fos+ subpart of the BLA moved from more anterior (t0 and t30) to more posterior after foot shock (t90 and t180). The BLA has been voxelized (voxel size 30 × 30 × 30 µm), and the fill color refers to the number of samples with at least one cell in that voxel. The dashed boxes indicate the mean and SD per group along the posterior–anterior axis. (C) Cartoon visualization of the right BLA in the same orientation of A, 1 and B created with brainrender (50).
We voxelized the xyz coordinates (voxel size: ∼30 × 30 × 30 µm) and visualized per time point which voxels have at least one cell from three different samples. As shown in Fig. 3B, at later time points after foot shock (i.e., t90 and t180), the highest density of c-fos+ cells in the BLA was found to be more posterior (difference of ∼96 µm from anterior to posterior, 23% of BLA) than at t0 and t30.
Cells Use Different Strategies of Activation, Which Can Change after Foot Shock.
A third biological question is whether all cells use a comparable activation “strategy” at the various time points after foot shock. With whole-brain microscopy, one can count the ncfos+ but also, quantify the intensity of c-fos staining per cell. Among other parameters, a cell is considered c-fos+ if the intensity of c-fos is higher than the background (i.e., signal-to-noise ratio). As a consequence, one would expect the ncfos+ per brain area to strongly correlate with the average c-fos intensity. In other words, brain areas are expected to be normally distributed along the correlation line, as shown by our simulation (SI Appendix, Fig. S5B).
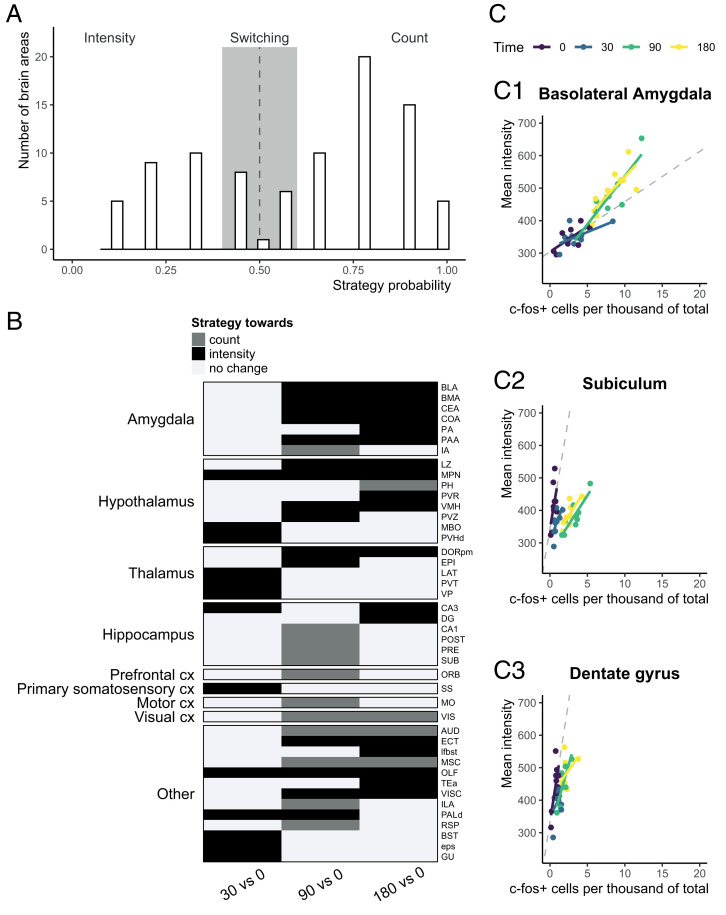
However, this was not the case (SI Appendix, Fig. S5C). Rather, different brain areas have a preferential strategy of activation: a few very active (i.e., low count, high intensity) cells vs. many lowly active cells (i.e., low intensity, high count). We categorized the brain areas of each t0 sample (corresponding to a very mildly stressful condition) based on their “strategy” (i.e., their preference for increasing in count or intensity). Across samples, we then calculated the probability of each brain area to belong to either categorization (Fig. 4A). The results showed a bimodal distribution (Fig. 4A), which is clearly different from the normal distribution expected under our hypothesis (SI Appendix, Fig. S5B). Therefore, whether a brain area is activated by increasing ncfos+ or by increasing the average intensity of c-fos per cell is unlikely to be the result of technical characteristics or of a random process.
Fig. 4.
Brain areas have a preferential strategy of activation, which may change after foot shock. (A) Preferential strategy of brain areas based on t0 data (i.e., the relationship between intensity and count per brain area). Histogram of the number of brain areas across the strategy probability. If a brain area would be activated by indiscriminately increasing c-fos+ cells and expression, the distribution would be normal (SI Appendix, Fig. S5B), with µ around 0.5. The bimodal distribution suggests that certain brain areas preferentially increase the number of c-fos+ cells (probability > 0.5, count), whereas others increase the mean c-fos expression (probability < 0.5, intensity). (B) The strategy of brain areas can change after foot shock. Binary heat map of how strategy can change across brain areas for pairwise comparisons of time points. White corresponds to no change in strategy, black corresponds to a change toward intensity, and gray corresponds to a change toward count. Of note, a change toward intensity does not necessarily mean that the brain area does not increase count; rather, it means that the increase in intensity cannot be explained by the increase in count alone. (C) Representative examples of a brain area that after foot shock, changes strategy toward intensity (BLA; C, 1) or count (subiculum; C, 2). The dentate gyrus (C, 3) was added for literature validation (Discussion). Each dot represents one sample; the line represents that correlation between count and intensity per group. Dashed lines represent what one would expect based on t0 activation. cx, cortex.
Intensity and count are, therefore, expected to be related within brain areas rather than across the whole brain. This relationship should be constant across all groups; if not, foot shock must have induced transcriptional changes in specific subsets of cells. We, therefore, next examined whether the strategy of a brain area changes after foot shock, relative to t0. For each time point after foot shock (t30, t90, t180), we selected brain areas with a consistent (at least six of nine samples) change in either count and intensity and calculated to what extent count and intensity were increasing compared with each other. We categorized brain areas as “changing strategy” if they at least doubled the increase in one category. Forty-three brain areas met these criteria (Fig. 4B). Of these, 30 increased activation by means of intensity rather than c-fos+ cell count, especially in the amygdala, hypothalamus, and thalamus. Of note, the increase in intensity for the amygdalar nuclei was present only for the time points t90 and t180. Fig. 4C displays two brain areas (BLA and subiculum) as representative examples of the activation strategy toward intensity and count, respectively. We also added a visualization of the dentate gyrus (Fig. 4 C, 3) as a validation since this area has been described to increase intensity of IEG staining after stress in a very limited subset of cells (5, 6).
Discussion
Human fMRI studies over the past decades have shown that acute stress activates multiple functional brain networks, with hypothalamic and amygdalar areas being among the first to be activated, and areas linked to higher cognitive functions—such as prefrontal cortex and hippocampus—following in due course (1). Yet, beyond the scale of (networks of) nuclei (e.g., up to single cells), very little is known about stress-induced effects at a whole-brain level and over time. Are all cells among or within nuclei equally affected at various time points after stress? Animal studies focusing on specific areas [e.g., the dentate gyrus (5, 6)] suggest not.
To address this question, we first had to develop a thorough and robust analytical pipeline, investigating changes in cellular activity after a highly stressful foot shock in mice over time by staining for the IEG c-fos and introducing a pseudotime metric; this is summarized in Fig. 1. Although we adapted tools developed by others to transform images into numeric data (8, 23), we also outline how to then conduct the required subsequent steps of data analysis (i.e., data cleaning and preprocessing). Also, so far, analyses were limited to a region-based approach, where the number of active cells is calculated for each brain area separately. Voxel-based analyses similar to MRI have only recently been developed (30). Here, we took this one step further and suggest analyses for 1) the time dynamics and 2) the single-cell level. The resolution up to the level of single cells is certainly one of the major advantages of whole-brain microscopy. In the future, c-fos+ cells could be characterized in more detail, being able to distinguish excitatory from inhibitory neurons or neurons from glia cells, as the current findings confirm that c-fos staining is not confined to neurons only (for example, ref. 31). This could be achieved by multiple concurrent staining (for example, refs. 32 and 33) or by computationally categorizing cellular morphology (34). Restaining could also be an option [for example, by using SWITCH (35) rather than iDisco+]. Our pipeline can be applied independently of the type of clearing method or software used for alignment and annotation, and it can analyze 3D (i.e., whole-brain) or four-dimensional (4D; over time) experimental designs. Furthermore, it is interoperable with several other annotation/alignment tools. It is available in the newly developed R package abc4d, to which new improvements can be easily added in the future. abc4d also includes a framework of simulation studies, where the null hypotheses for different analyses can be investigated. An overview of the package is provided in the cheat sheet (SI Appendix, Fig. S6).
With this toolbox in place, we addressed our biological questions. Although foot shock increased the activation of 96% of the brain areas, distinct temporal dynamics in networks of brain areas stood out. Thus, foot shock first activated (cells in) hypothalamic areas followed by amygdalar and prefrontal, hippocampal, and lastly, thalamic areas. This is largely in line with the earlier human literature using fMRI (1). Importantly, while foot shock is not a common stressor in humans, it nevertheless captures crucial elements of stress exposure in humans, involving physical characteristics, such as discomfort, and psychological aspects, like uncontrollability. This lends credibility to the common patterns seen across species. The percentage of active brain areas is higher than previously reported after a single prolonged stress (16), which suggests that investigating multiple time points offers a more complete (dynamic) view of brain areas activated after stress. Furthermore, previous c-fos studies have identified common patterns of activation for rewarding and aversion stimuli, presumably linked to the aroused state (17, 18, 36). We here identified the same “aroused pattern” of nuclei previously observed (at t90), including—but not limited to—the cingulate cortex, nucleus accumbens, bed nucleus of the stria terminalis (BSNT), lateral hypothalamus, periventricular hypothalamic nucleus, and paraventricular thalamic. The absolute numbers of c-fos+ cells were comparable with the previous literature investigating whole-brain c-fos with other paradigms (8, 15). We also provide a temporal pattern of functional activation, as visible in Fig. 1.
Additionally, we demonstrated within the BLA—a key area in the processing of a stressful foot shock (27, 28)—a clear shift after foot shock from activation of cells in the lateral–anterior part toward a more posterior–medial subset of cells. Although we here present information about the BLA only, data about all other areas investigated are available for closer scrutiny. To dive deeper into any area of interest, we provide an interactive interface on the data at our web portal (https://osf.io/8muvw/) (29). For example, subparts of the bed nucleus of the stria terminalis (BSNT) were previously found to be similar in c-fos expression in two dimensions 60 min after a multimodal stress (18). Our data (visible on our web portal) indeed confirm that 90 and 180 min after foot shock, the c-fos activation is widespread across the BSNT. However, this was not the case for earlier time points (t0 and t30), where the highest distribution of c-fos+ cells was more ventral.
Lastly, our approach allowing single-cell investigation revealed that brain areas follow specific c-fos expression strategies that are skewed toward either an increase in the number of c-fos+ cells or c-fos intensity per cell. Importantly, in a subset of areas, the strategy changed after foot shock. The finding that brain areas use a distinct strategy in c-fos cell activation under mildly stressful conditions, such as a novel environment (in this case, the shock box at t0), compared with exposure to a very stressful situation, like an inescapable foot shock, is hypothesis-generating. One can currently only speculate about the functional relevance of the two main cellular strategies. Earlier studies showed that the expression of c-fos is proportional to the rate of firing of the cell (37). If so, one could hypothesize that in certain brain areas, many cells are slightly more activated after stress (i.e., express c-fos above the detection threshold), whereas other brain areas may have only a few cells that are very strongly excited, which would lead to an increase in their c-fos intensity. Several previous studies (for example, refs. 36, 38, and 39) suggested that c-fos expression may be different across cell populations. This could now be investigated brain wide with new experiments. Of note, in amygdalar areas, the increase in intensity was particularly observed at 90 and 180 min after foot shock, which is compatible with a gene-mediated, possibly glucocorticoid-dependent mechanism (21). This observation that a limited number of cells get highly activated fits extremely well with current views on engrams (40). For instance, engram cells in the dentate gyrus were shown to be powerful in reversing behavior caused by chronic stress (41). Moreover, using inducible IEG promotor approaches in areas of interest, others have shown that immediately after contextual fear conditioning—also involving foot-shock exposure—a consistent ∼10% of (baso-)lateral amygdala neurons become part of the engram, while participation of dentate granule cells is much lower (42). This resembles the ∼5% of BLA neurons with very high–intensity staining we observed 90 to 180 min postfoot shock and the far lower number of high-intensity c-fos+ cells in the dentate. The somewhat lower percentage of c-fos+ cells in the BLA we observed may be explained by the recency of the foot shock (42), the level of excitability prior to foot shock (43), and/or the fact that we used c-fos rather than Arc as IEG. Very high intensity of IEG staining in a small subgroup of dentate cells was also reported after swim stress, which was found to increase DNA demethylation in the dentate gyrus at specific CpG sites (i.e., sites where a cytosine nucleotide is followed by a guanine nucleotide) close to the c-fos transcriptional start site in the gene promoter region of early growth response protein 1 (5, 6). Overall, the fact that dentate cells indeed follow an “intensity strategy” lends credibility to our approach; the strength of our study is that we do not focus on a single area but can simultaneously examine and compare 89 regions.
There are some limitations to consider. The choice of c-fos as an activity marker is arguably appropriate in the case of acute stress exposure (19), but it is by no means the only IEG one could choose for the current approach. The cellular role of c-fos remains largely unknown (39); therefore, it is not possible to determine its exact function in our experimental setup. Other markers of cellular activity may afford additional insights into the circuits being activated after stress. More than 100 genes have been classified as IEG (39, 44), although only a subset is expressed in neurons (45). Arc and Egr1 were reported to be transcriptionally activated following acute stress in a multiomics approach (46). Egr1 has a high expression maintained already by normal ongoing cellular activity (47), so this gene could potentially be used to investigate deactivation of brain areas. Lastly, IEGs are not equally expressed by all cells in all brain areas (39). This means that the sensitivity of our method may not be equal across the whole brain; specifically, it may be lower for subcortical/thalamic structures. In our analysis, we moderated this limitation with normalization/standardization steps, which showcases the importance of proper data processing. We also performed simulations using the baseline mRNA c-fos distribution of the Allen Brain Atlas to validate our findings against the nonhomogeneity of c-fos across brain regions. Ideally, a combination of markers should be applied to get a more complete view. Another technical limitation is linked to the current size restraints of imaging with light-sheet microscopy. In our study, we trimmed the most frontal and most caudal parts of the brains, and we excluded brain areas of small volume (SI Appendix, Table S1) that could not be reliably measured. Researchers interested in small structures (especially laterally placed and without strong landmarks) may opt for other alignment methods that do not rely on autofluorescence, although to the best of our knowledge, no previous study has quantified the displacement due to alignment. Although this does not impact the current methodology and the main finding of between- and within-area cellular differentiation in response to stress, some brain areas involved in the acute stress response (e.g., locus coeruleus) are missing. A solution could be to divide the brain for scanning but to analyze the data together after the appropriate corrections. This would also be a solution for those interested in hemisphere-specific effects (i.e., lateralization). In our experimental design, we did not randomize the direction of the brain within the microscope chamber. The right/left hemispheres were always each scanned by the same laser. Although we took care in laser calibration, a “laser-specific” effect cannot be excluded. We, therefore, refrained from investigating lateralization, although it is plausible to occur after acute stress (48). A third consideration concerns the pseudotime approach. On the one hand, it overcomes the absence of high-frequency sampling (as possible with fMRI). On the other hand, it is a mere approximation of real-time processes. For example, it is likely that place cells (49) within the hippocampus are activated promptly when in a new environment. Our pseudotime metric is based on the midpoint of activation of a brain area rather than the instance when activation was first measured. As a consequence, it misses the temporal resolution to pick up the earliest changes. Thus, while the pseudotime metric might be an acceptable approximation of the activation phase, the method gives little insight in the gradual turning off of brain areas since this also depends on the half-life time of the c-fos protein. The half-life time may differ across brain areas (19), something that could be investigated with a meta-analytic approach.
Despite these limitations, the ready to use pipeline for 4D immunohistochemical whole-brain analysis presented in this report (and supported by an R package) revealed that stressors, like an acute foot shock, not only sequentially activate functional networks in the brain but also, specifically activate subsets of neurons using different strategies of activation.
Materials and Methods
An in-depth description of the methods is in SI Appendix, Methods. The protocol, data, scripts, acb4d R package, and additional experimental information are available at Open Science Framework, https://osf.io/8muvw/ (29). Data can also be interactively visualized at our web portal (https://osf.io/8muvw/). All animal procedures were approved by the Animal Ethical Committee at Utrecht University, The Netherlands (license: AVD1150020184806).
Experimental Design.
We used a block design (nblock = 9), where each block had an animal for each experimental group (ntime = 4) from the same cage. We used a total of nanimals = 36. Control animals were identical to experimental animals but did not receive the foot shock.
Brains were cleared with iDisco+ (8), stained for c-fos and imaged with light-sheet microscopy. c-fos+ cells were detected with Imaris and aligned to the Allen Mouse Brain Reference Atlas (25 mm) (50) with Elastix (18) via Clearmap (8).
Samples underwent a thorough quality control. Preprocessing was required for region-based analyses (Fig. 1 and SI Appendix, Methods).
Summary of Analyses.
To test activation from baseline, we used pairwise comparisons (Welch t test, one sided, alpha = 0.05, P value corrected with the Benjamini–Hochberg procedure) for t30, t90, and t180 against t0 on ncfos+/tot.
Brain areas were defined as “most active” if in at least five of nine blocks, they were in the top 5% of most activated areas.
To order brain areas, we considered the time points on a “continuum” of pseudotime, calculated per block the point of midactivation, and grouped each brain area to the closest 10-min bout (binning).
To identify the highest density within a brain area, we calculated how many samples (minimum of three) had at least one cell in each voxel (30 µm per side). In each xyz direction, we calculated per time point the median and interquartile of the voxels’ position.
To categorize the strategy, we calculated across samples the probability of a brain area to be toward count/intensity using a linear model on t0 (ncfos+ vs. mean intensity of each brain area) as a criterion. Brain areas “changed strategy” if their rate of change relative to t0 was at least doubled in either count or intensity.
We performed several simulation studies to exclude that our findings were due to chance.
Supplementary Material
Acknowledgments
We acknowledge the MIND facility (University Medical Center Utrecht Brain Center) for 3D imaging by light-sheet microscopy (https://mindresearchfacility.nl/) and Roger Koot for the IT infrastructure. We thank Kevin Kemna for suggesting the creation of the R package and the helpful feedback on the analyses. This work was supported by the Consortium of Individual Development and BRAINSCAPES, which are funded through the Gravitation Program of the Dutch Ministry of Education, Culture, and Science and Netherlands Organization for Scientific Research Grants 024.001.003 and 024.004.012.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114002119/-/DCSupplemental.
Data Availability
The protocol, data, scripts, acb4d R package, and additional experimental information have been deposited in the Open Science Framework (https://osf.io/8muvw/) (50).
References
- 1.Hermans E. J., Henckens M. J. A. G., Joëls M., Fernández G., Dynamic adaptation of large-scale brain networks in response to acute stressors. Trends Neurosci. 37, 304–314 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Berretz G., Packheiser J., Kumsta R., Wolf O. T., Ocklenburg S., The brain under stress—A systematic review and activation likelihood estimation meta-analysis of changes in BOLD signal associated with acute stress exposure. Neurosci. Biobehav. Rev. 124, 89–99 (2021). [DOI] [PubMed] [Google Scholar]
- 3.O’Leary T. P., et al. , Extensive and spatially variable within-cell-type heterogeneity across the basolateral amygdala. eLife 9, e59003 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janak P. H., Tye K. M., From circuits to behaviour in the amygdala. Nature 517, 284–292 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandramohan Y., Droste S. K., Arthur J. S. C., Reul J. M. H. M., The forced swimming-induced behavioural immobility response involves histone H3 phospho-acetylation and c-Fos induction in dentate gyrus granule neurons via activation of the N-methyl-D-aspartate/extracellular signal-regulated kinase/mitogen- and stress-activated kinase signalling pathway. Eur. J. Neurosci. 27, 2701–2713 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Saunderson E. A., et al. , Stress-induced gene expression and behavior are controlled by DNA methylation and methyl donor availability in the dentate gyrus. Proc. Natl. Acad. Sci. U.S.A. 113, 4830–4835 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clayton D. F., The genomic action potential. Neurobiol. Learn. Mem. 74, 185–216 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Renier N., et al. , Mapping of brain activity by automated volume analysis of immediate early genes. Cell 165, 1789–1802 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guzowski J. F., et al. , Mapping behaviorally relevant neural circuits with immediate-early gene expression. Curr. Opin. Neurobiol. 15, 599–606 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Tyson A. L., Margrie T. W., Mesoscale microscopy and image analysis tools for understanding the brain. Prog. Biophys. Mol. Biol. 168, 81–93 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steegen S., Tuerlinckx F., Gelman A., Vanpaemel W., Increasing transparency through a multiverse analysis. Perspect. Psychol. Sci. 11, 702–712 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Goh W. W. B., Wang W., Wong L., Why batch effects matter in omics data, and how to avoid them. Trends Biotechnol. 35, 498–507 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Haghverdi L., Lun A. T. L., Morgan M. D., Marioni J. C., Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 36, 421–427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tran H. T. N., et al. , A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 21, 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim Y., et al. , Whole-brain mapping of neuronal activity in the learned helplessness model of depression. Front. Neural Circuits 10, 3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azevedo H., Ferreira M., Mascarello A., Osten P., Guimarães C. R. W., Brain-wide mapping of c-fos expression in the single prolonged stress model and the effects of pretreatment with ACH-000029 or prazosin. Neurobiol. Stress 13, 100226 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho J.-H., Rendall S. D., Gray J. M., Brain-wide maps of Fos expression during fear learning and recall. Learn. Mem. 24, 169–181 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin X., et al. , c-Fos mapping of brain regions activated by multi-modal and electric foot shock stress. Neurobiol. Stress 8, 92–102 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovács K. J., c-Fos as a transcription factor: A stressful (re)view from a functional map. Neurochem. Int. 33, 287–297 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Cullinan W. E., Herman J. P., Battaglia D. F., Akil H., Watson S. J., Pattern and time course of immediate early gene expression in rat brain following acute stress. Neuroscience 64, 477–505 (1995). [DOI] [PubMed] [Google Scholar]
- 21.Joëls M., Sarabdjitsingh R. A., Karst H., Unraveling the time domains of corticosteroid hormone influences on brain activity: Rapid, slow, and chronic modes. Pharmacol. Rev. 64, 901–938 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Mukherjee D., et al. , Salient experiences are represented by unique transcriptional signatures in the mouse brain. eLife 7, e31220 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein S., Staring M., Murphy K., Viergever M. A., Pluim J. P. W., elastix: A toolbox for intensity-based medical image registration. IEEE Trans. Med. Imaging 29, 196–205 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Tyson A. L., et al. , A deep learning algorithm for 3D cell detection in whole mouse brain image datasets. PLoS Comput. Biol. 17, e1009074 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fürth D., et al. , An interactive framework for whole-brain maps at cellular resolution. Nat. Neurosci. 21, 139–149 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henckens M. J. A. G., et al. , Stress-induced alterations in large-scale functional networks of the rodent brain. Neuroimage 105, 312–322 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Silva B. A., Gross C. T., Gräff J., The neural circuits of innate fear: Detection, integration, action, and memorization. Learn. Mem. 23, 544–555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharp B. M., Basolateral amygdala and stress-induced hyperexcitability affect motivated behaviors and addiction. Transl. Psychiatry 7, e1194 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.V. Bonapersona, Project information: The mouse brain after foot-shock. Open Science Framework. https://osf.io/8muvw/. Deposited 8 December 2021. [Google Scholar]
- 30.Vandenberghe M. E., et al. , Voxel-based statistical analysis of 3D immunostained tissue imaging. Front. Neurosci. 12, 754 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Condorelli D. F., et al. , Induction of protooncogene fos by extracellular signals in primary glial cell cultures. J. Neurosci. Res. 23, 234–239 (1989). [DOI] [PubMed] [Google Scholar]
- 32.Brignani S., et al. , Remotely produced and axon-derived Netrin-1 instructs GABAergic neuron migration and dopaminergic substantia nigra development. Neuron 107, 684–702.e9 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Belle M., et al. , Tridimensional visualization and analysis of early human development. Cell 169, 161–173.e12 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Tyson A. L., Margrie T. W., Mesoscale microscopy for micromammals: Image analysis tools for understanding the rodent brain. arXiv [Preprint] (2021). https://arxiv.org/abs/2102.11812 (Accessed 24 February 2021).
- 35.Murray E., et al. , Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell 163, 1500–1514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson Z. V., Revis A. A., Burdick M. A., Rhodes J. S., A similar pattern of neuronal Fos activation in 10 brain regions following exposure to reward- or aversion-associated contextual cues in mice. Physiol. Behav. 99, 412–418 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fields R. D., Eshete F., Stevens B., Itoh K., Action potential-dependent regulation of gene expression: Temporal specificity in Ca2+, cAMP-responsive element binding proteins, and mitogen-activated protein kinase signaling. J. Neurosci. 17, 7252–7266 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moench K. M., Breach M. R., Wellman C. L., Chronic stress produces enduring sex- and region-specific alterations in novel stress-induced c-Fos expression. Neurobiol. Stress 10, 100147 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallo F. T., Katche C., Morici J. F., Medina J. H., Weisstaub N. V., Immediate early genes, memory and psychiatric disorders: Focus on c-Fos, Egr1 and Arc. Front. Behav. Neurosci. 12, 79 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rao-Ruiz P., Yu J., Kushner S. A., Josselyn S. A., Neuronal competition: Microcircuit mechanisms define the sparsity of the engram. Curr. Opin. Neurobiol. 54, 163–170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramirez S., et al. , Activating positive memory engrams suppresses depression-like behaviour. Nature 522, 335–339 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kitamura T., et al. , Engrams and circuits crucial for systems consolidation of a memory. Science 356, 73–78 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gouty-Colomer L. A., et al. , Arc expression identifies the lateral amygdala fear memory trace. Mol. Psychiatry 21, 364–375 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheng M., Greenberg M. E., The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron 4, 477–485 (1990). [DOI] [PubMed] [Google Scholar]
- 45.Sheng H. Z., Lin P. X., Nelson P. G., Combinatorial expression of immediate early genes in single neurons. Brain Res. Mol. Brain Res. 30, 196–202 (1995). [DOI] [PubMed] [Google Scholar]
- 46.von Ziegler L. M., et al. , Molecular roadmap of the healthy stress response in the mouse hippocampus. bioRxiv [Preprint] (2021). 10.1101/2021.03.26.436970 (Accessed 26 March 2021). [DOI]
- 47.Worley P. F., et al. , Constitutive expression of zif268 in neocortex is regulated by synaptic activity. Proc. Natl. Acad. Sci. U.S.A. 88, 5106–5110 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ocklenburg S., Korte S. M., Peterburs J., Wolf O. T., Güntürkün O., Stress and laterality: The comparative perspective. Physiol. Behav. 164, 321–329 (2016). [DOI] [PubMed] [Google Scholar]
- 49.O’Keefe J., Dostrovsky J., The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat. Brain Res. 34, 171–175 (1971). [DOI] [PubMed] [Google Scholar]
- 50.Claudi F., et al. , Visualizing anatomically registered data with Brainrender. eLife 10, e65751 (2021). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The protocol, data, scripts, acb4d R package, and additional experimental information have been deposited in the Open Science Framework (https://osf.io/8muvw/) (50).