Significance
CD11c+ microglia enriched for osteopontin (OPN) expression appear at distinct stages of brain development, aging, and several neurodegenerative disorders. Whether coexpression of CD11c and OPN results from microglial activation or represents a part of a subset-specific genetic program is unknown. We find that this CD11c+ microglial subset is formed before birth upon uptake of apoptotic neurons. Our analysis also suggests that it represents a stable subset that requires OPN to mediate engulfment of synaptic proteins, proliferate, and develop a proinflammatory phenotype. Definition of OPN-producing CD11c+ microglia as a specialized microglial subset provides insight into the contribution of microglial differentiation to brain development and function in health and disease.
Keywords: osteopontin, CD11c microglia, inflammation, synaptic elimination
Abstract
Expression of Itgax (encoding the CD11c surface protein) and Spp1 (encoding osteopontin; OPN) has been associated with activated microglia that can develop in healthy brains and some neuroinflammatory disorders. However, whether CD11c and OPN expression is a consequence of microglial activation or represents a portion of the genetic program expressed by a stable microglial subset is unknown. Here, we show that OPN production in the brain is confined to a small CD11c+ microglial subset that differentiates from CD11c− precursors in perinatal life after uptake of apoptotic neurons. Our analysis suggests that coexpression of OPN and CD11c marks a microglial subset that is expressed at birth and persists into late adult life, independent of environmental activation stimuli. Analysis of the contribution of OPN to the intrinsic functions of this CD11c+ microglial subset indicates that OPN is required for subset stability and the execution of phagocytic and proinflammatory responses, in part through OPN-dependent engagement of the αVβ3-integrin receptor. Definition of OPN-producing CD11c+ microglia as a functional microglial subset provides insight into microglial differentiation in health and disease.
CD11c, also termed integrin alpha X (encoded by Itgax), is a defining marker for dendritic cells (DCs). When paired with β2-integrin, the heterodimeric receptor binds to complement iC3b and mediates phagocytosis (1). A subpopulation of central nervous system (CNS)–resident microglia that also expresses CD11c develops early in life and is a feature of microglial development in healthy brains and in murine models of neurodegenerative disease, including Alzheimer’s disease (AD) (2–4). Genes expressed by CD11c+ microglia include the Spp1 gene, which encodes osteopontin (OPN), a cytokine-like phosphoprotein that is a prominent feature of both protective and pathogenic immune responses in peripheral lymphoid tissues (4–8).
OPN is expressed as a secreted (OPN-s) or intracellular (OPN-i) isoform that originated from a single OPN messenger RNA (mRNA) precursor (9) after activation of immune cells. Regulation of immune responses by OPN includes promotion of proinflammatory responses following ligation of its canonical receptor, the αVβ3-integrin expressed on macrophages (10–12). Production of OPN by DCs also regulates the differentiation of T helper (TH) cell subsets (8), including TH17 cells, which contribute to the development of murine experimental autoimmune encephalomyelitis (13). More recently, microglial production of OPN has been implicated in diverse CNS pathologic disorders, including multiple sclerosis (3), spinal cord injury (14), and neurodegenerative disorders, including AD and amyotrophic lateral sclerosis (ALS) (4, 15). Although CD11c+ microglia are a major source of OPN production by activated microglia, whether coexpression of CD11c and OPN is a feature of microglial activation or marks a subset-specific genetic program is not known. This represents a central question in understanding microglial development, since it involves a choice between genetic mechanisms that regulate subset-specific differentiation rather than markers of a transient activation phenotype.
Here, we report that OPN-producing CD11c+ microglia represent a small (<5%) subset that differentiates from CD11c−OPN− precursors after engulfment of apoptotic neurons (ANs) in perinatal life and represent the sole microglial producers of OPN throughout life. The CD11c+OPN+ subset displays a stable phenotype in the steady state and expresses a core genetic program that is independent of microglial activation. Analysis of the contribution of OPN to CD11c+ microglial function indicates that OPN regulates microglial engulfment of synaptic proteins, proliferation and the development of a proinflammatory phenotype. Definition of OPN-producing CD11c+ microglia as a functional subset rather than a transient activation phenotype provides insight into the contribution of microglial differentiation to brain development and function in health and disease.
Results
CD11c+ Microglia Are Formed Early in Mouse Brain Development upon Engulfment of ANs Independent of Microglial Activation.
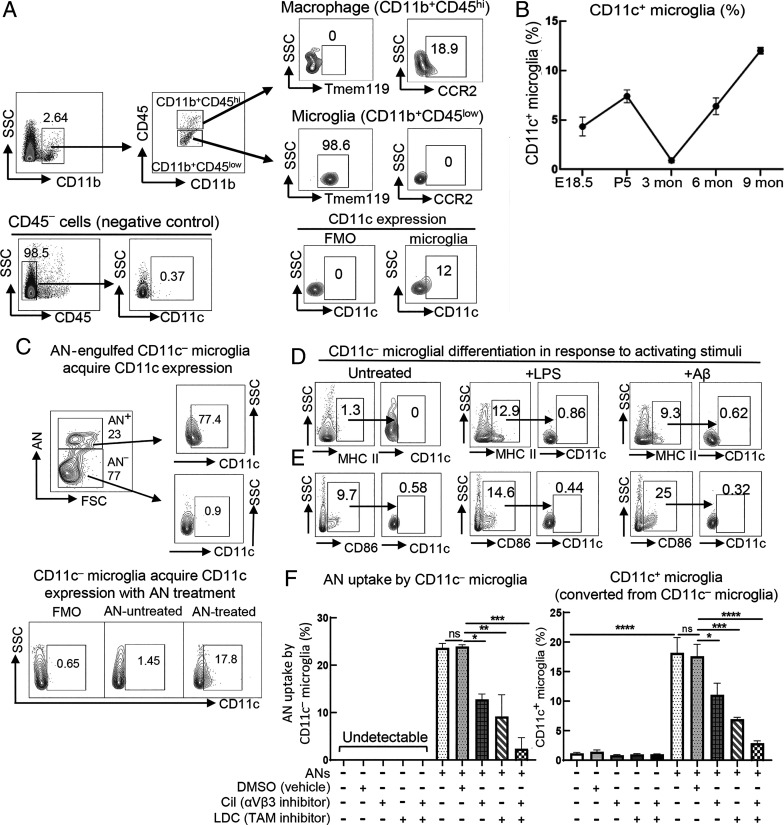
In DCs, the CD11c protein represents an essential part of the iC3b heterodimeric receptor that mediates phagocytosis (1). Microglia that express CD11c are detectable at birth but decrease dramatically by 3 mo of age (3). We therefore traced the development of CD11c+ microglia in healthy C57BL/6 mice from prenatal life through adulthood. We first validated microglial CD11c expression by flow cytometry. To distinguish microglia (CD11b+CD45low) and macrophages (CD11b+CD45high), we used the CCR2 marker expressed by blood-derived macrophages but not expressed by microglia (3, 16), and a microglia-specific marker, Tmem119 (17). The CD11b+CD45high subset that expresses CCR2 but not Tmem119 was confirmed as macrophages, while CD11b+CD45low cells that all express Tmem119 but do not express CCR2 were confirmed as microglia. Fluorescence minus one (FMO) negative controls and brain CD45− cells that do not express CD11c were included to confirm the specificity of CD11c staining in CD11b+CD45low microglial population (Fig. 1A). We note that CD11c+ microglia arise late in embryogenesis (embryonic day 18.5; E18.5), increasing to about 7 to 8% of total microglia by postnatal day 5 (P5) before receding to almost undetectable levels (<1%) in young adult life. However, CD11c+ microglia reemerge in older (6- to 9-mo-old) mice to represent about 10% of total microglia (Fig. 1B).
Fig. 1.
CD11c+ microglia are differentiated from CD11c− precursors upon engulfment of ANs early in development independent of microglial activation. (A) Brain single-cell suspensions of 9-mo-old C57BL/6 (B6) WT mice were generated for validation of microglial CD11c expression by flow cytometry. We first gate on CD11b+ cells from single/live cells followed by gating of CD11b+CD45low populations as potential microglia. Almost all the cells in this population (∼99%) are Tmem119+ while CCR2 expression is not detectable. In contrast, CCR2 but not Tmem119 expression is detected in the CD11b+CD45high fraction, namely putative macrophage populations. Analysis of the CD11b+CD45low microglial population using FMO negative controls for these flow cytometry plots confirms specificity. Brain CD45− cells, mainly containing nonimmune cells (neurons, astrocytes, and oligodendrocytes but not microglia) that do not express CD11c were used as negative controls to further validate the specificity of this strategy. (B) The proportion of CD11c+ microglia in WT mice during early development and normal aging (n = 3) was determined by flow cytometry analysis. CD11c+ microglia were initially present at E18.5, peaked at P5, and gradually declined to marginal levels in young adulthood (3 mo) followed by reemergence and further expansion during aging. (C) CD11c− microglia were isolated from P5 WT mice by negative selection with anti-CD11c magnetic beads (purity >99%) followed by coincubation for 72 h in the presence or absence of pHrodo fluorescent dye–labeled ANs at a 1:1 ratio. (C, Upper) After a 72-h incubation, 23% of total CD11c− microglia had ingested ANs (AN+), of which 77.4% of these AN+ microglia were CD11c+. In contrast, the <1% of CD11c− microglia that had not ingested ANs (AN−, reflecting 77% of total microglia in culture at 72 h) expressed CD11c. (C, Lower) After incubation with ANs for 72 h, ∼18% of total CD11c− microglia express CD11c, while ∼98% of microglia incubated in the absence of ANs remain CD11c−. (D and E) CD11c− microglia isolated from P5 WT mice were incubated in the presence or absence of LPS (10 ng/mL) or Aβ1-42 (1 μM) for 24 h followed by analysis of microglial activation markers and CD11c expression. Expression of CD11c was assessed in CD86+ or MHC II+ activated microglia. Despite up-regulation of CD86 and MHC II in response to LPS and Aβ1-42, CD11c− microglial differentiation was not observed in response to these stimuli. Flow cytometry plots are representative data of three experiments. (F) CD11c− microglia were isolated from P5 WT mice by negative selection with anti-CD11c magnetic beads (purity >99%) followed by coincubation for 72 h in the presence or absence of pHrodo fluorescent dye–labeled ANs at a 1:1 ratio with or without the αVβ3-integrin inhibitor Cil (10 μM) and/or pan-TAM receptor inhibitor LDC1267 (1 μM). After a 72-h incubation, Cil or LDC1267 reduced AN uptake by CD11c− microglia by ∼50%, and the combination of these two inhibitors further down-regulated AN uptake to background levels. Stimulation by ANs induced CD11c expression by ∼20% of initial CD11c− microglia over 72 h, while CD11c expression was not detectable in the absence of AN stimulation during the same period. Inhibition of AN uptake by Cil or LDC1267 reduced acquisition of a CD11c+ phenotype by CD11c− progenitors by about 50 to 75% in each case and inclusion of both inhibitors fully prevented this phenotypic transition (n = 3). ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05 by one-way ANOVA with Bonferroni’s multiple-comparisons test; ns, not significant. Data are shown as mean ± SEM.
Since uptake of ANs may trigger changes in the microglial genetic program that include up-regulation of Itgax (encoding CD11c) gene expression (18), we asked whether up-regulation of CD11c expression by CD11c− precursors was a direct consequence of AN engulfment early in development. We enriched CD11c− microglia by negative isolation with anti-CD11c magnetic beads to yield CD11c− microglia (purity >99%) and incubated them with fluorescently labeled ANs for 72 h followed by a determination of the percentage of CD11c− microglia that expressed CD11c. We observed that 23% of total CD11c− microglia had ingested ANs (AN+), of which 77.4% of these AN+ microglia were CD11c+. In contrast, of the 77% of CD11c− microglia that had not ingested ANs, <1% expressed CD11c (Fig. 1 C, Upper). These findings also indicate that ∼18% of total CD11c− microglia go on to express CD11c after incubation with ANs, while <1.5% of the microglia incubated for 72 h in the absence of ANs become CD11c+ (Fig. 1 C, Lower).
Up-regulation of CD11c expression was not simply a consequence of microglial activation after, for example, phagocytosis or inflammation. Purified CD11c− microglia from P5 mice were treated with diverse activating stimuli that mimic CNS inflammatory disorders, including lipopolysaccharide (LPS) and the amyloid β1-42 (Aβ1-42) peptide. Although these stimuli provoked marked microglial activation, as judged by increased expression of both CD86 and major histocompatibility complex class II (MHC II) (19), activated CD11c− microglia did not up-regulate CD11c expression (Fig. 1 D and E). Collectively, these results suggest that the formation of CD11c+ microglia following ingestion of ANs at birth is independent of conventional microglial activation.
We then examined the interactions that might promote the formation of CD11c+ microglia in neonatal life. Phagocytic receptors expressed by developing microglia that may mediate uptake and clearance of apoptotic cells include TAM (Tyro3, Axl, MerTK) and the αVβ3-integrin receptors (20, 21). We analyzed the impact of specific inhibition of αVβ3 and TAM receptors on AN uptake and its potential impact on acquisition of a CD11c+ phenotype by CD11c− microglia. The αVβ3 inhibitor cilengitide (Cil) or the LDC1267 inhibitor of pan-TAM receptors reduced AN uptake of CD11c− microglia by ∼50%. A combination of these two inhibitors further reduced CD11c− microglial AN uptake to background levels (Fig. 1 F, Left). Consistent with the findings in Fig. 1C, we noted that stimulation by ANs induced CD11c expression by ∼20% of initial CD11c− microglia over 72 h, while CD11c expression was not detectable in the absence of AN stimulation during the same time period. Finally, inhibition of AN uptake by Cil or LDC1267 reduced acquisition of a CD11c+ phenotype by CD11c− progenitors by about 50 to 75% in each case and inclusion of both inhibitors fully prevented this phenotypic transition (Fig. 1 F, Right). These findings suggest that engulfment of apoptotic cells may trigger a CD11c−→CD11c+ transition following interaction of ANs with microglial receptors that include αVβ3-integrin and TAM.
The Stability of CD11c Expression by Microglia Is Regulated by OPN.
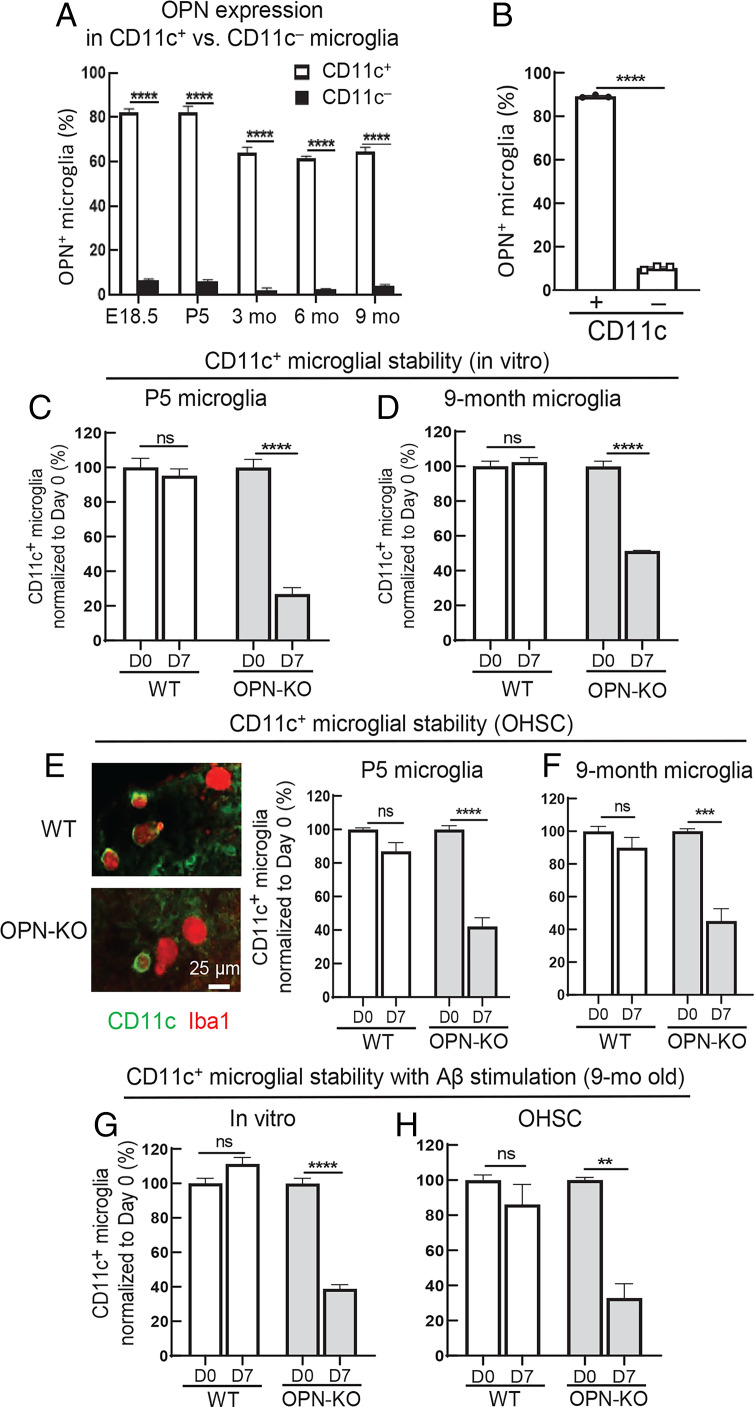
We validated microglial OPN expression by flow cytometry analysis. We used several controls, including an isotype control and, importantly, OPN knockout (OPN-KO) microglia as negative control cells and microglia that selectively express the intracellular isoform of OPN (OPN-i knockin [OPN-i-KI]) as a positive control. Wild-type (WT) microglia and OPN-i-KI microglia show similar levels of OPN staining, while staining of microglia from either OPN-KO donors or microglia stained with an isotype control does not yield a detectable signal (SI Appendix, Fig. S1). We noted that microglial OPN production is confined to CD11c+ microglia and is not significantly expressed by CD11c− microglia (Fig. 2A). Moreover, AN-induced differentiation of CD11c− to CD11c+ microglia was also accompanied by a sharp up-regulation of OPN production (Fig. 2B).
Fig. 2.
Stability of CD11c expression by microglia is regulated by OPN. (A) Flow cytometry analysis of OPN expression in CD11c+ versus CD11c− microglia from WT mice at different stages of development and aging (n = 3). OPN production was confined to CD11c+ microglia during early development and normal aging. ****P < 0.0001 by two-way ANOVA with Bonferroni’s multiple-comparisons test. (B) CD11c− microglia were isolated from P5 WT mice followed by coincubation for 72 h with pHrodo fluorescent dye–labeled ANs at a 1:1 ratio. CD11c− microglia that had engulfed ANs differentiated into CD11c+ microglia accompanied by sharply increased expression of OPN (n = 3). ****P < 0.0001 by two-tailed Student’s t test. (C and D) CD11c+ microglia (>95% purity) were isolated from P5 and 9-mo-old WT and OPN-KO mice followed by incubation for 7 d in conventional microglial culture medium (Dulbecco’s modified Eagle’s medium–F12 with 10% FBS + 1% penicillin/streptomycin + 10 ng/mL rmM-CSF). In vitro analysis of CD11c+ microglial stability was evaluated by comparing the percentage of CD11c+ microglia on day 0 with day 7 (percentages were normalized to day 0). CD11c+ microglia of P5 and 9-mo-old WT mice were stable, while OPN-KO CD11c+ microglia displayed a significant loss of stability in P5 and 9-mo-old mice (n = 3). ****P < 0.0001 by two-way ANOVA with Bonferroni’s multiple-comparisons test; ns, not significant. (E and F) CD11c+ microglial stability was assessed using ex vivo OHSCs. Freshly prepared OHSCs were incubated for 24 h with 0.5 mg/mL clodronate liposomes at 35 °C to deplete endogenous microglia. Each microglia-free OHSC was replenished with 4 × 103 CD11c+ microglia isolated from P5 or 9-mo-old WT or OPN-KO mice (purity >95%). Representative images displaying CD11c+ microglia in hippocampal slices reconstituted with P5 WT or P5 OPN-KO CD11c+ microglia at day 7. CD11c+ microglia from P5 and 9-mo-old WT mice stably expressed CD11c, while OPN deficiency reduced CD11c expression by ∼60% (n = 3 or 4). (Scale bar, 25 μm) ****P < 0.0001, ***P < 0.001 by two-way ANOVA with Bonferroni’s multiple-comparisons test; ns, not significant. (G and H) CD11c+ microglia isolated from 9-mo-old WT and OPN-KO mice were incubated for 7 d in vitro or in OHSCs in the presence of 1 μM synthetic human Aβ1-42 peptide. CD11c+ microglia from 9-mo-old WT mice were stable, while OPN deficiency led to a substantial reduction of the CD11c phenotype in the presence of Aβ1-42 in vitro and in OHSCs, respectively (n = 3). ****P < 0.0001, **P < 0.01 by two-way ANOVA with Bonferroni’s multiple-comparisons test; ns, not significant. Data are shown as mean ± SEM.
To define the impact of OPN on the CD11c+ phenotype, we characterized CD11c+ microglia from OPN-KO and WT mice during early development and aging. OPN deficiency led to a ∼50% reduction in the proportions of CD11c+ microglia in WT mice at P5 and 6 and 9 mo of age, according to flow cytometry analysis (SI Appendix, Fig. S2A). In situ analysis of CD11c+ and CD11c− microglia in brain cryosections of P5 and 9-mo-old WT and OPN-KO mice confirmed reduced proportions of CD11c+ microglia from OPN-KO mice (50 to 75% reduction). CD11c-specific staining was validated using negative controls (SI Appendix, Fig. S2B).
We further defined the contribution of OPN to the stability of CD11c expression by microglia isolated from P5 and 9-mo-old WT and OPN-KO mice during a 7-d in vitro culture period. The numbers of CD11c+ microglia were comparable between P5 and 9-mo-old WT and OPN-KO mice at day 0 (SI Appendix, Fig. S3A). OPN-deficient neonatal (P5) CD11c+ microglia showed a 75% reduction in CD11c expression (Fig. 2C), while OPN-deficient CD11c+ microglia from 9-mo-old OPN-KO mice showed a ∼50% reduction in CD11c expression (Fig. 2D). We examined the stability of the microglial CD11c+ phenotype in a more physiological setting using organotypic hippocampal slice cultures (OHSCs). This system allows replacement of endogenous microglia after depletion of resident microglia from organotypic hippocampal tissue slices. We replenished these microglia-free hippocampal slices with CD11c+ microglia (>95% CD11c+) from WT or OPN-KO P5 and 9-mo-old donors. WT and OPN-KO mice showed identical numbers of CD11c+ microglia at day 0 in OHSCs (SI Appendix, Fig. S3B). The WT CD11c+ microglia retained their CD11c+ phenotype, while the majority of OPN-deficient CD11c+ microglia did not (Fig. 2 E and F).
We then assessed the response of CD11c+ microglia to Aβ, a pathogenic microglial stimulus that is a prominent feature of age-related neurodegenerative disease. We observed that CD11c+ microglia from 9-mo-old WT mice stably expressed CD11c after in vitro Aβ1-42 stimulation and in OHSCs in contrast to OPN-deficient CD11c+ microglia, which lost 60 to 75% of CD11c expression (Fig. 2 G and H). Taken together, these data suggest that stable expression of microglial CD11c may require coexpression of OPN.
Genetic Profiling of CD11c+ Microglia.
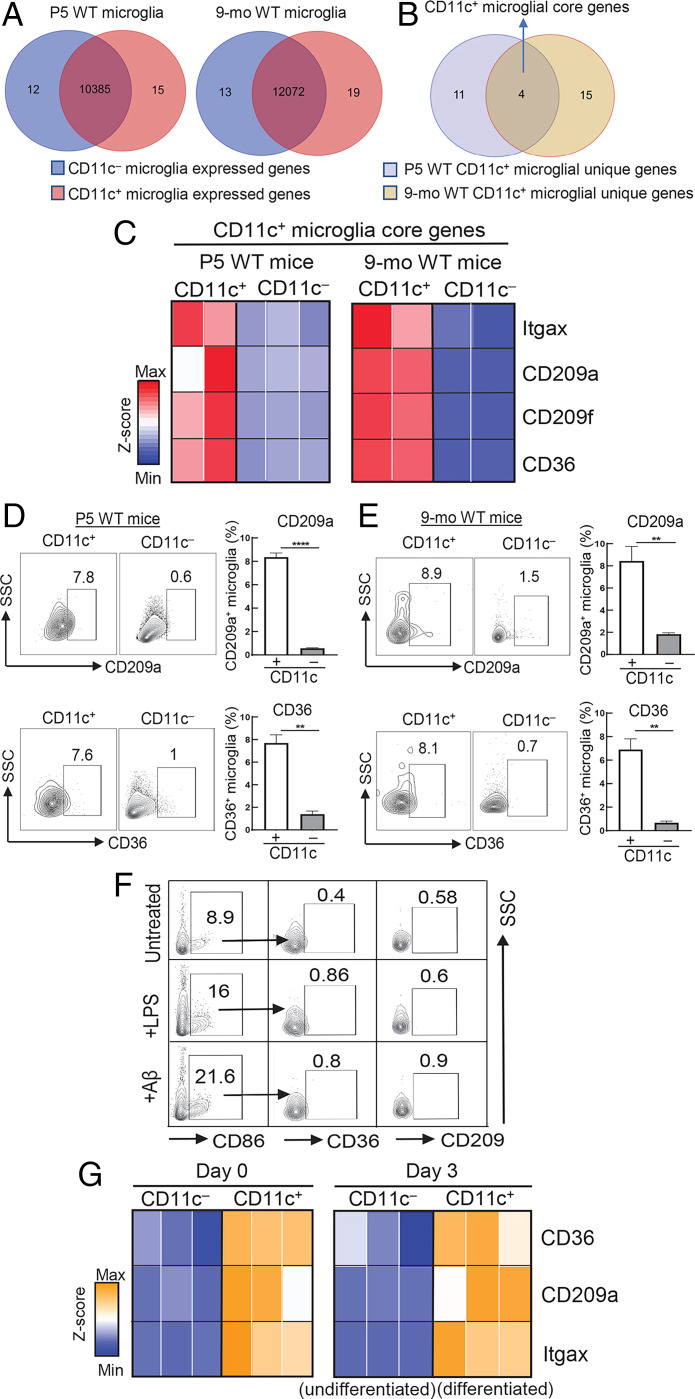
We then asked whether CD11c+ microglia represent a distinct subset that expressed a characteristic genetic profile. To that end, we performed RNA-sequencing (RNA-seq) analysis of fluorescence-activated cell sorting (FACS)–purified CD11c+ and CD11c− microglia using a cold isolation protocol followed by validation using flow cytometry (SI Appendix, Fig. S4). As expected, the transcriptomes of CD11c+ and CD11c− microglia from neonatal (P5) WT mice overlapped substantially, sharing 10,385 genes. However, 12 genes were uniquely expressed by CD11c− microglia and 15 genes were exclusively expressed by CD11c+ microglia. Likewise, CD11c+ and CD11c− microglia from 9-mo-old mice shared 12,072 genes, while 19 genes were solely expressed by CD11c+ microglia (Fig. 3A). We identified four genes that were shared by CD11c+ microglia by neonatal (P5) and aged (9-mo-old) mice (Fig. 3B) and were not expressed by CD11c− microglia from donors of either age (Fig. 3C). These represent genes selectively expressed by CD11c+ microglia that are retained from neonatal life to older adulthood in the absence of exogenous inflammatory or infectious stimuli. These genes include Cd36, an inflammatory response-modulating molecule (22), and Cd209a, a regulator of phagocytic activity (5, 23), and are expressed at both the RNA and protein levels (Fig. 3 D and E). Expression of these proteins by CD11c+ microglia was independent of activation stimuli, since deliberate activation of CD11c− microglia did not induce expression of these core proteins (CD209 and CD36) (Fig. 3F). Moreover, these signature genes were up-regulated by CD11c+ progeny of CD11c− precursors after engulfment of ANs, suggesting that up-regulation of these genes accompanies the formation of this subset in perinatal life (Fig. 3G).
Fig. 3.
Definition of an intrinsic genetic program of CD11c+ microglia. (A) Transcriptomic profiling of CD11c+ and CD11c− microglia was analyzed in P5 and 9-mo-old WT mice by RNA-seq. Venn diagrams show the number of genes expressed in P5 and 9-mo-old WT CD11c+ and CD11c− microglia and genes that are exclusively expressed by each microglial subset. CD11c+ microglial unique genes were identified within those genes that showed a fold change and raw counts in the top 0.15% and negatively expressed by their CD11c− counterparts. The threshold of negative expression was defined according to raw counts of genes with no expression, for example Itgax in CD11c− microglia. A similar strategy was used to identify CD11c− microglial unique genes. (B) The core genetic signature of CD11c+ microglia was identified as overlapping unique genes of P5 and 9-mo-old CD11c+ microglia. (C) Heatmap displaying four CD11c+ microglial core genes, including Itgax, Cd209a, Cd209f, and Cd36, in P5 and 9-mo-old CD11c+ microglia compared with CD11c− counterparts (FDR < 0.05). (D and E) Validation of core genes at the protein level. Core genes of CD11c+ microglia were validated at the protein level by flow cytometry analysis. Surface proteins CD36 and CD209a were solely expressed by CD11c+ microglia of P5 and 9-mo-old WT mice compared with CD11c− microglia (n = 3). ****P < 0.0001, **P < 0.01 by two-tailed Student’s t test. (F) CD11c− microglia isolated from P5 WT mice were incubated for 24 h with LPS (10 ng/mL) or Aβ1-42 (1 μM) followed by flow cytometry analysis of CD36 and CD209 expression. Protein expression of these core genes was barely detectable in CD86+CD11c− microglia in response to activating stimuli. Flow cytometry plots are representative of three experiments. (G) In vitro differentiation of CD11c+ microglia. CD11c+ and CD11c− microglia were freshly isolated from P5 WT mice. The CD11c+ microglial phenotype was validated by qPCR analysis of mRNA expression of representative core genes on day 0. Then, CD11c− microglia were incubated for 72 h with ANs to induce CD11c− microglial differentiation into CD11c+ microglia. The core genes were up-regulated in CD11c+ microglia (differentiated) compared with CD11c− microglia (undifferentiated) on day 3. Expression of core genes was normalized into a z score as shown in the heatmap (n = 3). P < 0.05. Data are shown as mean ± SEM.
OPN Regulates Intrinsic Functions of CD11c+ Microglia.
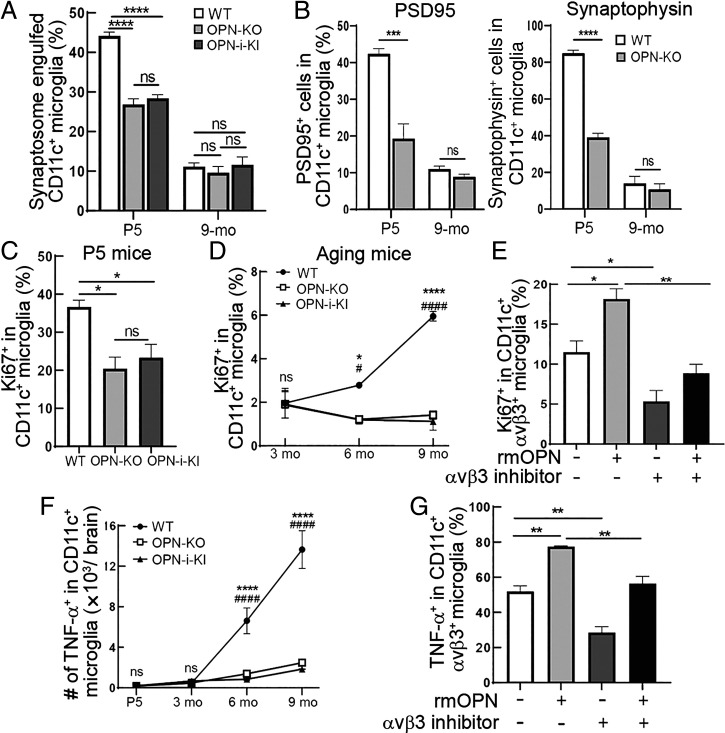
Microglia may contribute to the elimination of excessive neuronal synapses during neonatal brain development (24). We found that CD11c+OPN+ microglia from neonatal, but not from 9-mo-old, brains were highly phagocytic for synaptosomes (Fig. 4A). We asked whether OPN contributed to this microglial function. OPN is expressed as OPN-s or OPN-i isoforms that derive from different OPN translational initiation sites (25). We utilized OPN-mutant mice harboring different OPN isoforms to delineate the contribution of OPN isoforms to CD11c+ microglial functions. Full OPN deficiency resulted in a ∼45% decrease of synaptosome engulfment by CD11c+ microglia. OPN-i was not implicated in this process (Fig. 4A). Similarly, ex vivo analysis revealed that CD11c+OPN+ microglia from P5 but not 9-mo-old WT mice displayed robust engulfment of synaptic proteins, including synaptophysin and PSD95. Deficiency of OPN resulted in a ∼50% reduction of this CD11c+ microglial function (Fig. 4B).
Fig. 4.
OPN regulates the inherent functions of CD11c+ microglia. (A) Microglia isolated from P5 and 9-mo-old WT, OPN-KO, and OPN-i-KI mice were incubated for 1 h with 136 μg pHrodo green–labeled synaptosomes per 1 × 105 cells followed by flow cytometry analysis of CD11c+ microglial engulfment of synaptosomes. CD11c+ microglia from P5, but not 9-mo-old, WT mice were highly phagocytic for synaptosomes in vitro. Deficiency of OPN-s significantly abrogated this function, while OPN-i was not implicated in this process (n = 3). ****P < 0.0001 by one-way ANOVA with Bonferroni’s multiple-comparisons test; ns, not significant. (B) Ex vivo analysis shows that CD11c+ microglia from P5, but not 9-mo-old, WT mice display robust activity in engulfment of synaptic proteins. P5 WT mice lacking OPN displayed substantially reduced engulfment of presynaptic protein synaptophysin or postsynaptic protein PSD95 by CD11c+ microglia (n = 3). ****P < 0.0001, ***P < 0.001 by two-tailed Student’s t test; ns, not significant. (C) CD11c+ microglia expressed high levels of the proliferation marker Ki-67 early in development (P5). OPN-s but not OPN-i deficiency decreased Ki-67 expression in CD11c+ microglia (n = 3). *P < 0.05 by one-way ANOVA with Bonferroni’s multiple-comparisons test; ns, not significant. (D) During aging, CD11c+ microglia showed relatively low levels of proliferative activity and CD11c+ microglial Ki-67 expression was substantially decreased in mice lacking OPN-s (n = 3). *WT vs. OPN-KO; #WT vs. OPN-i-KI. ****P < 0.0001, *P < 0.05, ####P < 0.0001, #P < 0.05 by two-way ANOVA with Bonferroni’s multiple-comparisons test; ns, not significant. (E) Microglia isolated from 9-mo-old WT mice were incubated for 24 h with or without 12.5 μg/mL recombinant mouse OPN (rmOPN) or 10 μM αVβ3 inhibitor (Cil) followed by flow cytometry analysis of the proliferation marker Ki-67. rmOPN enhanced Ki-67 expression in CD11c+αVβ3+ microglia of 9-mo-old WT mice, while this effect was fully abrogated by the αVβ3 inhibitor (n = 3). **P < 0.01, *P < 0.05 by one-way ANOVA with Bonferroni’s multiple-comparisons test. (F) The expression of tumor necrosis factor α (TNF-α) was barely detectable in CD11c+ microglia of P5 mice, while its expression gradually increased during aging. The age-dependent increase of TNF-α expression in CD11c+ microglia was markedly reduced in WT mice lacking OPN-s but not OPN-i (n = 3). *WT vs. OPN-KO; #WT vs. OPN-i-KI. ****P < 0.0001, ####P < 0.0001 by two-way ANOVA with Bonferroni’s multiple-comparisons test; ns, not significant. (G) Microglia isolated from 9-mo-old WT mice were incubated for 24 h with or without 12.5 μg/mL rmOPN or 10 μM αVβ3 inhibitor (Cil) followed by flow cytometry analysis of TNF-α expression. Up-regulation of TNF-α expression induced by rmOPN was fully abolished by the αVβ3 inhibitor in CD11c+αVβ3+ microglia of 9-mo-old WT mice (n = 3). **P < 0.01 by one-way ANOVA with Bonferroni’s multiple-comparisons test. Data are shown as mean ± SEM.
Examination of the contribution of OPN to proliferation of CD11c+ microglia revealed that selective deletion of OPN-s was sufficient to substantially reduce proliferation of CD11c+ microglia from both newborn and aging WT mice (Fig. 4 C and D). We then asked whether an interaction between OPN-s and its canonical receptors might contribute to this CD11c+ microglial function. Although the CD44 and αVβ5 OPN receptors (26, 27) are not expressed by microglia at any age, αVβ3 expression gradually increases during aging (SI Appendix, Fig. S5). Small molecule–mediated blockade of αVβ3 by the cyclic RGD peptide Cil markedly reduced OPN-dependent Ki-67 expression (Fig. 4E), suggesting that OPN-dependent proliferation of CD11c+ microglia reflects ligation of the αVβ3 receptor. Moreover, the robust proinflammatory response of CD11c+ microglia from 9-mo-old mice depended on OPN-s expression (Fig. 4F) and reflected engagement of the αVβ3-integrin receptor (Fig. 4G).
The age-dependent proinflammatory phenotype of this OPN-producing CD11c+ microglial subset suggests it might contribute to neuroinflammatory disorders. We therefore asked whether the expression of the signature genes of OPN-producing CD11c+ microglia might persist in the context of chronic inflammatory stimuli that develop in 5XFAD mice, which recapitulate amyloid pathology of AD. We observed that CD11c+ microglia from OPN-sufficient mice expressed the representative core genes Cd36 and Cd209a at the protein level compared with their CD11c− counterparts in the presence of 5XFAD pathology (SI Appendix, Fig. S6 A and B). Moreover, OPN production contributed to CD11c+ microglial stability, in contrast to strongly reduced stability of CD11c+ microglia from age-matched OPN-KO.5XFAD mice (∼40 to 80% reduction) in vitro and ex vivo (SI Appendix, Fig. S6C). The CD11c phenotype was also stably expressed by CD11c+ microglia from 9-mo-old 5XFAD mice in the presence of Aβ1-42, while OPN deficiency led to 40 to 80% reduction in vitro and in OHSCs, respectively (SI Appendix, Fig. S6D).
Discussion
Microglial expression of the CD11c receptor and production of OPN have been associated with microglial activation during the development of some neuroinflammatory diseases (3, 4, 7) and in response to exogenous stimuli (14). This may be a consequence of microglial activation or, alternatively, a part of the genetic program of a microglial subset that develops at or before birth and persists into late adulthood. Our studies support the latter view, namely that CD11c+ microglia represent a stable subset programmed to produce OPN rather than a transient activation phenotype.
We identify a small (<5%) OPN-producing CD11c+ microglial subset that differentiates from CD11c− at birth upon engulfment of ANs in the absence of external activation stimuli. After their initial formation in perinatal life, CD11c+ microglia recede to almost undetectable levels in young adult life before reemergence in late adult life to constitute 10 to 15% of total microglia. Both the perinatal CD11c+ microglia and late adult life CD11c+ microglia express a genetic signature that is independent of cellular activation in healthy mice.
Single-cell transcriptomics have described microglial subsets enriched for Itgax (encoding CD11c) at different ages and during development of neurodegenerative disease (4–7), following their description by the Owens group (3), who observed that CD11c+ microglial numbers peaked early after birth (P3 to P5) and were reduced to marginal levels by young adulthood (2 to 3 mo). Our analysis of CD11c+ microglia before birth and during normal aging revealed that CD11c+ microglia that appear during late embryogenesis (E18.5) transiently contract before reemergence and expansion into substantial numbers during normal aging.
Although alterations in the microglial gene transcription program after engulfment of apoptotic cells include up-regulation of Itgax gene expression (18, 28), studies of unselected microglial populations have not determined whether this reflects de novo expression of CD11c by CD11c− precursors or expansion of CD11c+ microglia from a smaller population. Analysis of the response of isolated CD11c− precursors to ANs revealed that induction of CD11c protein expression following AN engulfment by neonatal CD11c− precursors was accompanied by OPN expression, suggesting that this phenotype is a direct consequence of AN-induced differentiation. This view is supported by findings that formation of CD11c+ microglia is independent of nonspecific microglial activation, since deliberate activation of CD11c− precursors by various nonspecific stimuli failed to induce expression of the CD11c phenotype. It may be relevant that microglia within neonatal brain regions containing high amounts of apoptotic cells express high levels of Itgax/CD11c and phagocytosis-related genes (3, 6, 29, 30). Moreover, injection of ANs, but not live neurons, Escherichia coli, or zymosan particles, into C57BL/6 mouse brain induces a microglial phenotype that includes up-regulation of Itgax/CD11c (18), suggesting that microglial induction of CD11c is a tightly controlled response to cellular apoptosis. It may also be possible that conversion of adult microglia later in life can be efficiently promoted by other stimuli including Aβ either alone or as a synergistic mixture of ANs and Aβ.
Expression of the CD11c+ phenotype depended on OPN production, as judged from in vitro analyses and after transfer into microglia-free hippocampal tissues. Moreover, analysis of the CD11c+ microglial gene profile at birth and late adult life indicated a persistent genetic signature that was independent of conventional activation stimuli. Although selective expression of these signature genes by CD11c+ microglia was not mimicked by deliberate activation of CD11c− microglia, further transcriptomic profiling and single-cell transcriptomic analysis of CD11c+ and CD11c− microglia are required for a more detailed genetic description of this subset in healthy brain development and in the face of chronic inflammatory disorders. Expression of a stable phenotype and core genetic program that persists from birth to late adulthood independent of exogenous stimulation suggests that CD11c+ microglia may represent a microglial lineage. More direct probing using fate-mapping techniques to trace the genetic history of CD11c+ microglia in different reporter mouse models is needed to directly address this question (31, 32).
The genetic program of CD11c+ microglia includes genes associated with phagocytosis and inflammation (22, 23), suggesting that CD11c+ microglia are specialized to execute these microglial functions. Indeed, we note that CD11c+ microglia may contribute to neuronal synapse elimination by engulfing synaptic proteins in early development and mediate proinflammatory responses during aging, perhaps enabling elimination of defective or inactive synapses. These functions of the CD11c+ microglial subset are regulated by OPN, since engulfment of synaptic proteins by neonatal CD11c+ microglia is depressed in the absence of OPN, while the proliferative and proinflammatory responses of CD11c+ microglia in adult life reflect OPN engagement of αVβ3-integrin receptors. Analysis of OPN-mutant mice that specifically express OPN isoforms demonstrated that OPN-s, but not OPN-i, is responsible for these OPN-dependent functions. Promotion of inflammatory responses by CD11c+ microglia is reminiscent of the subset of DCs which express high levels of CD11c and carry out OPN-dependent inflammatory responses (33). Our observations that CD11c+ microglia from 5XFAD mice express the signature genes at the protein level and the OPN-dependent phenotype noted in healthy mice may allow definition of their potential contribution to disease development.
Taken together, we show that CD11c+ microglia, the sole producer of OPN in the brain, differentiate from CD11c− microglia after uptake of ANs at birth. We suggest that this OPN-producing CD11c+ microglial population represents a unique subset according to its stable phenotype and expression of a signature gene set at birth and late adult life that is independent of deliberate activation. The proinflammatory properties of CD11c+ microglia suggest that these cells may contribute to the development of neuroinflammatory diseases including AD and, potentially, ALS and Parkinson’s disease.
Materials and Methods
Mice.
WT C57BL/6 (B6) and B6.Cg-Tg (APPSwFlLon,PSEN1*M146L*L286V)6799Vas/Mmjax (5XFAD) mice were obtained from the Jackson Laboratory (MMRRC). Spp1flstop (OPN-KO) and Spp1flstopEllacre (OPN-i-KI) mice were generated by our laboratory as previously described (34). OPN-KO.5XFAD were bred by crossing Spp1flstop mice with 5XFAD mice. All the mice were housed in pathogen-free conditions. All experiments were performed in compliance with federal laws and institutional guidelines as approved by the Animal Care and Use Committee of the Dana-Farber Cancer Institute.
Microglial Isolation.
Isolation of microglia by magnetic-activated cell sorting.
Adult mice were anesthetized with isoflurane and transcardially perfused with ice-cold phosphate-buffered saline (PBS) before brains were removed and minced by scalpels. Tissues were subjected to enzymatic dissociation using collagenase (300 U/mL; Worthington) or papain (20 U/mL; Worthington) and DNase I (60 U/mL; Worthington) before a 30% Percoll gradient was performed to remove myelin, and pelleted cells were resuspended in magnetic-activated cell sorting (MACS) buffer (PBS, pH 7.2, 2 mM ethylenediaminetetraacetic acid [EDTA], and 0.5% bovine serum albumin [BSA]). Total microglia were obtained by magnetic isolation using CD11b microbeads (Miltenyi). For isolation of CD11c+ and CD11c− microglia, single-cell suspensions were incubated with CD11c microbeads (Miltenyi) and cells magnetically bound to columns using MACS were extensively washed before the CD11c+ fraction was eluted after lifting the magnetic field. The unbound fraction was then labeled with CD11b microbeads (Miltenyi) and separated using MACS isolation, and CD11c−CD11b+ cells that were bound to the column were eluted. MACS buffer was used according to the manufacturer’s suggested protocol. This standard method was used to isolate microglia for all of the experiments unless otherwise noted.
Microglial isolation and FACS for RNA-seq analysis.
Single-cell suspensions were prepared as described (6). Briefly, mice were anesthetized with isoflurane and transcardially perfused before brains were quickly dissected and minced using a scalpel on ice followed by Dounce homogenization in ice-cold Hank’s balanced salt solution (HBSS) ∼20 times. All tools used were prechilled and all isolation steps were carried out on ice to minimize microglial activation. Cell suspensions were transferred to prechilled 50-mL tubes and passed through a 70-μm cell strainer followed by transfer into a prechilled 15-mL tube and spun down at 500 × g for 5 min at 4 °C. Debris and myelin were then removed using a modified cold Percoll gradient and cell pellets were resuspended in 10 mL of ice-cold 40% Percoll (Sigma), diluted, and then spun for 30 min at 500 × g. This approach yielded a microglial pellet at the bottom of the 15-mL tube while Percoll and myelin were removed by vacuum suction. The cell pellet was washed with 10 mL of ice-cold HBSS and spun again for 5 min at 500 × g at 4 °C. All samples were then resuspended in ice-cold FACS buffer (0.5% BSA, 1 mM EDTA, in 1× PBS) for staining. Ghost Dye violet 510 (1:1,000; Tonbo Biosciences) was used to exclude dead cells. Fc receptors were blocked using anti-CD16/CD32 antibody (1:100; BD Biosciences) to avoid nonspecific staining. Single-cell suspensions were then stained with anti-CD11b (1:100), anti-CD45 (1:100), and anti-CD11c (1:50) antibodies (BioLegend) for 20 min on ice before samples were washed with ice-cold FACS buffer and spun down for 5 min at 500 × g. Cell pellets were resuspended in 5 mL of ice-cold FACS buffer before sorting on a BD FACSAria II using the 70-μm nozzle with purity mode at ∼10,000 events per second. After sorting, each sample was spun down and cell pellets were immediately stored at −80 °C until further processing.
For P5 mice, three biological replicates of CD11c+ microglia and CD11c− microglia were sequenced. Samples were pooled from 58 mice. For 9-mo-old WT mice, two replicates of CD11c+ microglia and CD11c− microglia were sequenced. Samples were pooled from 44 mice (22 male and 22 female).
AN Induction and Labeling for CD11c− Microglial Differentiation.
Primary mouse neurons were prepared from B6 embryos at E16.5 to E17.5. Cerebral hemispheres were isolated and freed from meninges before tissue digestion with 0.25% trypsin in HBSS for 15 min at 37 °C followed by titration to obtain single-cell preparations. Cell suspensions were filtered through a 70-μm cell strainer and cells were centrifuged at 600 × g for 5 min. Cell density was determined using a hemocytometer, and cells seeded in Neurobasal medium were supplemented with 1× B27 and 500 μM GlutaMAX (Invitrogen). Half of the medium was changed every 3 d. To induce apoptosis, primary cultured WT neurons were treated with 300 μM N-methyl-d-aspartate overnight before careful detachment from flasks by repeated washes with PBS followed by centrifugation, and the pellet was processed for labeling. Neurons (1 × 107) were carefully resuspended in 1 mL PBS and incubated in darkness for 2 h at room temperature with 100 μg of dissolved labeling dye (pHrodo iFL green; Life Technologies). To block and capture residual dye, cells were diluted with PBS, harvested by centrifugation, resuspended in 1 mL undiluted fetal bovine serum (FBS), and washed twice with PBS. Total apoptotic cell numbers were determined using trypan blue staining.
Flow Cytometry.
Microglial staining for flow cytometry analysis.
Microglia were stained with Ghost Dye violet 510 (1:1,000; Tonbo Biosciences) to exclude dead cells followed by Fc receptor blocking using an anti-CD16/CD32 antibody (1:100; BD Biosciences) to avoid nonspecific staining. Appropriate microglial surface markers were used for staining, including anti-CD11b (1:100; BioLegend), anti-CD45 (1:100; BioLegend), anti-CD11c (1:50; BioLegend), anti-CD36 (1:100; BioLegend), anti-CD209a (1:100; BioLegend), anti–αV-integrin (1:50; BioLegend), anti–β3-integrin (1:50; BioLegend), anti-CD86 (1:100; BioLegend), and anti–MHC II (1:100; BioLegend), followed by fixation and permeabilization for subsequent intracellular staining with anti-OPN (1:10; R&D Systems) and anti–TNF-α (1:50; BioLegend) and intranuclear staining with anti–Ki-67 (1:100; BioLegend).
Validation of microglial OPN expression by flow cytometry analysis.
Microglial OPN expression was validated in 9-mo-old WT mice using conventional intracellular staining protocols. Microglia were fixed and permeabilized with Intracellular Fixation & Permeabilization Buffer (eBioscience) followed by incubation with a phycoerythrin (PE)-conjugated anti-OPN antibody (1:10; IC808; R&D Systems) at 4 °C for 30 min. An isotype control (1:10; PE-conjugated goat immunoglobulin G [IgG]) and OPN-KO microglia were used as a negative control. Microglia that selectively express the intracellular isoform of OPN (OPN-i-KI) were used as a positive control.
Validation of microglial CD11c expression by flow cytometry analysis.
Brain single-cell suspensions from 9-mo-old WT mice were stained with Ghost Dye violet 510 (1:1,000; Tonbo Biosciences) and anti–CD16/CD32 antibody (1:100; BD Biosciences) followed by anti-CD11b (1:100; BioLegend), anti-CD45 (1:100; BioLegend), anti-CD11c (1:50; BioLegend), anti-TMEM119 (1:200; Abcam), and anti-CCR2 (1:100; BioLegend). CD11b+ cells were gated from single/live cells followed by subsequent gating of CD11b+CD45low as microglia and CD11b+CD45high as macrophages. To distinguish microglia and macrophages, the CCR2 marker expressed by blood-derived macrophages but not by microglia was used (3, 16). To further distinguish these two cell types, the microglia-specific marker Tmem119 (17) was also included. The CD11b+CD45high cells that express CCR2 but not Tmem119 were confirmed as macrophages, while the CD11b+CD45low cells that all express Tmem119 but do not express CCR2 were confirmed as microglia. FMO negative controls were included to confirm the specificity of CD11c staining in CD11b+CD45low microglial populations. Brain CD45− cells that mainly contain nonimmune cells (e.g., neurons, astrocytes, and oligodendrocytes) that do not express CD11c were also included as negative controls to further validate the specificity of this strategy.
Flow cytometry detection of microglial engulfment of synaptic protein.
After perfusion, mouse brains were harvested and dissected followed by myelin removal by centrifugation on a 30% Percoll gradient. Brain pellets were sequentially stained with Ghost Dye violet 510 (1:1,000; Tonbo Biosciences) followed by incubation with the microglial surface markers anti-CD11b, anti-CD45, and anti-CD11c for 30 min. Subsequently, stained samples were fixed and permeabilized using Intracellular Fixation & Permeabilization Buffer (eBioscience). Intracellular staining was performed for the presynaptic marker anti-synaptophysin (1:100; Invitrogen) or postsynaptic marker PSD95 (1:100; Invitrogen) followed by staining with Alexa Fluor 488–donkey anti-mouse IgG (H+L) secondary antibody (1:300; Invitrogen). Samples were acquired on a CytoFLEX (Beckman Coulter) flow cytometer followed by analysis with FlowJo v10 (Tree Star).
OHSCs.
OHSCs were prepared as described (35). Briefly, hippocampal slices were prepared from newborn (P3 to P5) C57BL/6 mice to a thickness of 350 μm before incubation at 35 °C in 5% CO2. Microglia were depleted from freshly prepared slice cultures using clodronate liposomes (FormuMax) and freshly prepared OHSCs were incubated with 0.5 mg/mL clodronate liposomes for 24 h at 35 °C. Subsequently, OHSCs were rinsed with warm PBS before replacement of medium (50% modified Eagle’s medium, 25% HBSS, 25% normal horse serum, 0.2 mM glutamine, 100 U/mL penicillin, 100 mg/mL streptomycin, and 4.5 mg/mL glucose). Microglia-depleted OHSCs were maintained for 7 d before experimentation. CD11c+ microglia were acutely isolated from P5 or 9-mo-old WT and OPN-KO mice or 9-mo-old 5XFAD and OPN-KO.5XFAD mice. After isolation, microglia were carefully resuspended in medium to a final concentration of 2,000 cells per microliter. Each microglia-free OHSC was replenished with 4,000 cells. OHSCs reconstituted with CD11c+ microglia were incubated in the presence or absence of 2 μL synthetic human Aβ1-42 peptide (AnaSpec; 15 μM stock) for 7 d. Aβ1-42 treatment was repeated four times every other day, and thus each slice was treated with a total of 8 μL Aβ1-42 peptide.
Immunofluorescent Staining.
Transcardial perfusion by cold PBS was performed on P5 and 9-mo-old WT and OPN-KO mice followed by brain removal and fixation in 4% paraformaldehyde (PFA) solution at 4 °C overnight. Then, the fixed brains were rinsed with PBS and dehydrated in 30% sucrose at 4 °C overnight. OCT compound (Sakura Finetek) was used to embed the brain tissues, and serial sagittal cryosections (10 μm) were cut using a cryostat (Leica). Brain cryosections were permeabilized with PBST buffer (PBS with 0.3% Triton X-100) for 1 h. OHSC slices were fixed in 4% PFA solution for 30 min and permeabilized with PBST buffer for 3 h. After a 1-h incubation in blocking solution containing 5% normal donkey serum (Jackson ImmunoResearch) in PBST to prevent nonspecific binding, cryosections or OHSC slices were incubated for 24 h with the appropriate primary antibodies: rabbit anti–Iba-1 (1:1,000; Wako) and biotin anti-mouse CD11c (N418; 1:50; BioLegend). To amplify CD11c signals, sections were washed with PBS and incubated with biotin anti-mouse CD11c overnight, followed by incubation with horseradish peroxidase–conjugated streptavidin for 1 h before incubation with Alexa Fluor 488–tyramides for 5 min to generate high-density Alexa Fluor 488 labeling of CD11c protein in situ using a Tyramide SuperBoost Kit (Invitrogen). Sections were then incubated for 1 h with Alexa 594–conjugated donkey anti-rabbit IgG (1:500; Invitrogen). Brain slides incubated without anti-CD11c primary antibody or tyramide signal amplification were included as negative controls to exclude nonspecific immunofluorescent signals. DAPI (Invitrogen) was used as a nuclear counterstain (10-min incubation) before samples were analyzed using an Olympus fluorescence microscope.
RNA Profiling.
RNA extraction.
Total RNA was extracted from FACS-sorted cell pellets using an RNeasy Plus Universal Mini Kit, according to the manufacturer’s instructions (QIAGEN). Library preparations, sequencing reactions, and bioinformatic analysis (gene hit counts) were conducted at GENEWIZ as follows.
Library preparation.
Extracted RNA samples were quantified using a Qubit 2.0 fluorometer (Life Technologies) and RNA integrity was checked using a TapeStation 4200 (Agilent Technologies).
PolyA selection strategy.
RNA-seq libraries of CD11c+ and CD11c− microglial samples of P5 WT mice were prepared using the NEBNext Ultra RNA Library Prep Kit for Illumina (NEB #E7530), per the manufacturer’s instructions. Briefly, mRNAs were first enriched with oligo(dT) beads before enriched mRNAs were fragmented for 15 min at 94 °C. First- and second-strand complementary DNAs (cDNAs) were subsequently synthesized and cDNA fragments were end-repaired and adenylated at the 3′ ends, and universal adapters were ligated to cDNA fragments followed by index addition and library enrichment by limited-cycle PCR. The sequencing libraries were validated on an Agilent TapeStation and quantified using a Qubit 2.0 fluorometer (Invitrogen) as well as by qPCR (KAPA Biosystems).
Ultra-low-input strategy.
Due to the very limited cell numbers obtained from adult mice, CD11c+ and CD11c− microglial samples of 9-mo-old WT mice were processed with a SMART-Seq v4 Ultra Low Input Kit for Sequencing for full-length cDNA synthesis and amplification (Clontech) and an Illumina Nextera XT library for sequencing library preparation. Briefly, cDNA was fragmented and adapter was added using transposase, followed by limited-cycle PCR to enrich and add index to the cDNA fragments. The final library was assessed with an Agilent TapeStation.
HiSeq sequencing.
The sequencing libraries were clustered on flowcell lanes before the flowcell was loaded onto an Illumina HiSeq instrument (4000 or equivalent) per the manufacturer’s instructions. Samples were sequenced using a 2 × 150-bp paired-end configuration, and image analysis and base calling were conducted by HiSeq Control Software. Raw sequence data (.bcl files) generated from Illumina HiSeq were converted into fastq files and demultiplexed using Illumina’s bcl2fastq 2.17 software. One mismatch was allowed for index sequence identification.
RNA-seq data analysis.
Mapping and gene counting were performed by GENEWIZ. After reviewing the quality of the raw data, sequence reads were trimmed to remove possible adapter sequences and nucleotides with poor quality using Trimmomatic v0.36. The trimmed reads were mapped to the Mus musculus reference genome (ENSEMBL) using STAR aligner v2.5.2b, a splice aligner that detects and incorporates splice junctions to align the entire read sequences. BAM files were generated, and unique gene hit counts were calculated using Counts (Subread package v1.5.2). Only unique reads that fell within exon regions were counted. Differential expression was considered significant with a false discovery rate (FDR)–adjusted P value < 0.05.
qPCR.
RNA of CD11c+ and CD11c− microglia was extracted using an RNeasy Plus Universal Mini Kit per the manufacturer’s instructions (QIAGEN). cDNA was reverse-transcribed from 35 ng of RNA and prepared using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) per the manufacturer’s instructions. Real-time qPCR was performed using the QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems) for selected core genes using 5 μL cDNA, 4.92 μL PowerUp SYBR Green Master Mix (Applied Biosystems), and 0.08 μL primer (IDT; working concentration 200 nM) per reaction. The gene expression levels were compared using the ΔΔCt method normalized to β-actin.
In Vitro Synaptosome Engulfment Assay.
Synaptosome isolation and labeling.
Synaptosomes were isolated from WT mice using Syn-PER Synaptic Protein Extraction Reagent (Thermo Scientific), per the manufacturer’s instructions. For pHrodo labeling, dissolved pHrodo iFL green (Life Technologies) was incubated with synaptosomes on a shaker in PBS for 1 h at room temperature protected from light at the ratio of 20 μg pHrodo per 1 mg synaptosomes. Unconjugated pHrodo was removed by washing with PBS before pHrodo-conjugated synaptosomes were resuspended in PBS with 5% dimethyl sulfoxide, aliquoted, and stored at −80 °C until use.
Microglial engulfment of synaptosomes.
Microglia isolated from P5 and 9-mo-old WT, OPN-KO, and OPN-i-KI mice were incubated with 136 μg pHrodo green–labeled synaptosomes per 1 × 105 cells for 1 h followed by staining for 30 min with Ghost Dye violet 510 (1:1,000; Tonbo Biosciences) and anti-CD11c (1:50; BioLegend). CD11c+ microglial engulfment of synaptosomes was assessed by flow cytometry analysis.
Quantification and Statistical Analysis.
Data in figures are presented as mean ± SEM. Statistical analysis was performed using GraphPad Prism v9.0. Quantification of fluorescence microscopy images was performed using ImageJ. Statistical analysis to compare the mean values for multiple groups was performed using GraphPad Prism by one- or two-way ANOVA with Bonferroni’s multiple-comparisons test. Comparisons between two groups were analyzed with two-tailed Student’s t test. P value < 0.05 was considered statistically significant. All of the statistical details of experiments, including the statistical tests used and exact value of sample sizes, can be found in the figure legends.
Supplementary Material
Acknowledgments
We thank S. Carrillo and O. Oseghali for mouse genotyping and A. Angel for manuscript and figure preparation. This work was supported in part by research grants from the National Institute of Allergy and Infectious Diseases of the NIH under Awards R01AI037562 and R01AI048125 (to H.C.), a gift from the LeRoy Schecter Research Foundation (to H.C.), and an American Association of Immunologists Intersect Fellowship (to A.E.W.).
Footnotes
Reviewers: M.C., Washington University in St. Louis School of Medicine; M.S., Duke University School of Medicine; and L.S., Stanford University.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2116241119/-/DCSupplemental.
Data Availability
The RNA-seq data reported in this article have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) and are accessible through GEO Series accession no. GSE190713.
All study data are included in the article and/or SI Appendix.
References
- 1.Wu J., Wu H., An J., Ballantyne C. M., Cyster J. G., Critical role of integrin CD11c in splenic dendritic cell capture of missing-self CD47 cells to induce adaptive immunity. Proc. Natl. Acad. Sci. U.S.A. 115, 6786–6791 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haage V., et al. , Comprehensive gene expression meta-analysis identifies signature genes that distinguish microglia from peripheral monocytes/macrophages in health and glioma. Acta Neuropathol. Commun. 7, 20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wlodarczyk A., et al. , A novel microglial subset plays a key role in myelinogenesis in developing brain. EMBO J. 36, 3292–3308 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keren-Shaul H., et al. , A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290.e17 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Li Q., et al. , Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron 101, 207–223.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammond T. R., et al. , Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50, 253–271.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sala Frigerio C., et al. , The major risk factors for Alzheimer’s disease: Age, sex, and genes modulate the microglia response to Aβ plaques. Cell Rep. 27, 1293–1306.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shinohara M. L., Kim J. H., Garcia V. A., Cantor H., Engagement of the type I interferon receptor on dendritic cells inhibits T helper 17 cell development: Role of intracellular osteopontin. Immunity 29, 68–78 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shinohara M. L., Kim H. J., Kim J. H., Garcia V. A., Cantor H., Alternative translation of osteopontin generates intracellular and secreted isoforms that mediate distinct biological activities in dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 105, 7235–7239 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patarca R., et al. , Structural and functional studies of the early T lymphocyte activation 1 (Eta-1) gene. Definition of a novel T cell-dependent response associated with genetic resistance to bacterial infection. J. Exp. Med. 170, 145–161 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashkar S., et al. , Eta-1 (osteopontin): An early component of type-1 (cell-mediated) immunity. Science 287, 860–864 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Weber G. F., et al. , Phosphorylation-dependent interaction of osteopontin with its receptors regulates macrophage migration and activation. J. Leukoc. Biol. 72, 752–761 (2002). [PubMed] [Google Scholar]
- 13.Hur E. M., et al. , Osteopontin-induced relapse and progression of autoimmune brain disease through enhanced survival of activated T cells. Nat. Immunol. 8, 74–83 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Noristani H. N., et al. , RNA-seq analysis of microglia reveals time-dependent activation of specific genetic programs following spinal cord injury. Front. Mol. Neurosci. 10, 90 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiu I. M., et al. , A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep. 4, 385–401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizutani M., et al. , The fractalkine receptor but not CCR2 is present on microglia from embryonic development throughout adulthood. J. Immunol. 188, 29–36 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennett M. L., et al. , New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. U.S.A. 113, E1738–E1746 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krasemann S., et al. , The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 47, 566–581.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jurga A. M., Paleczna M., Kuter K. Z., Overview of general and discriminating markers of differential microglia phenotypes. Front. Cell. Neurosci. 14, 198 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fourgeaud L., et al. , TAM receptors regulate multiple features of microglial physiology. Nature 532, 240–244 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park S. Y., Kim I. S., Engulfment signals and the phagocytic machinery for apoptotic cell clearance. Exp. Mol. Med. 49, e331 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grajchen E., et al. , CD36-mediated uptake of myelin debris by macrophages and microglia reduces neuroinflammation. J. Neuroinflammation 17, 224 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulz D., Severin Y., Zanotelli V. R. T., Bodenmiller B., In-depth characterization of monocyte-derived macrophages using a mass cytometry-based phagocytosis assay. Sci. Rep. 9, 1925 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Favuzzi E., et al. , GABA-receptive microglia selectively sculpt developing inhibitory circuits. Cell 184, 4048–4063.e32 (2021). Correction in: Cell 184, 5686 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoue M., Shinohara M. L., Intracellular osteopontin (iOPN) and immunity. Immunol. Res. 49, 160–172 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weber G. F., Ashkar S., Glimcher M. J., Cantor H., Receptor-ligand interaction between CD44 and osteopontin (Eta-1). Science 271, 509–512 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Ruoslahti E., Pierschbacher M. D., New perspectives in cell adhesion: RGD and integrins. Science 238, 491–497 (1987). [DOI] [PubMed] [Google Scholar]
- 28.Anderson S. R., et al. , Developmental apoptosis promotes a disease-related gene signature and independence from CSF1R signaling in retinal microglia. Cell Rep. 27, 2002–2013.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White L. D., Barone S. Jr., Qualitative and quantitative estimates of apoptosis from birth to senescence in the rat brain. Cell Death Differ. 8, 345–356 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Hagemeyer N., et al. , Microglia contribute to normal myelinogenesis and to oligodendrocyte progenitor maintenance during adulthood. Acta Neuropathol. 134, 441–458 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stranges P. B., et al. , Elimination of antigen-presenting cells and autoreactive T cells by Fas contributes to prevention of autoimmunity. Immunity 26, 629–641 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madisen L., et al. , A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kourepini E., et al. , Osteopontin expression by CD103− dendritic cells drives intestinal inflammation. Proc. Natl. Acad. Sci. U.S.A. 111, E856–E865 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leavenworth J. W., Verbinnen B., Yin J., Huang H., Cantor H., A p85α-osteopontin axis couples the receptor ICOS to sustained Bcl-6 expression by follicular helper and regulatory T cells. Nat. Immunol. 16, 96–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hellwig S., et al. , Forebrain microglia from wild-type but not adult 5xFAD mice prevent amyloid-β plaque formation in organotypic hippocampal slice cultures. Sci. Rep. 5, 14624 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data reported in this article have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) and are accessible through GEO Series accession no. GSE190713.
All study data are included in the article and/or SI Appendix.