Significance
Ecological disruption due to human impacts is evident worldwide, and a key to mitigation lies in characterizing the underlying mechanisms of species and ecosystem stability. Here we show that three extensive “supergenes” are maintained in Atlantic cod by stabilizing selection, tying these genes to the persistence of a keystone species distributed across the northern Atlantic Ocean. Removal of this species has caused severe ecosystem reshuffling in several areas of its range. Genomic inference of historic stock sizes further shows that cod has been under pressure in the North Sea system since the Viking period, in line with zooarchaeological records. Expansion of fisheries in Northern Europe through the past millennium is well documented and supports the inferred long-term declines.
Keywords: marine, exploitation, balancing selection, inversions, genomics
Abstract
Life on Earth has been characterized by recurring cycles of ecological stasis and disruption, relating biological eras to geological and climatic transitions through the history of our planet. Due to the increasing degree of ecological abruption caused by human influences many advocate that we now have entered the geological era of the Anthropocene, or “the age of man.” Considering the ongoing mass extinction and ecosystem reshuffling observed worldwide, a better understanding of the drivers of ecological stasis will be a requisite for identifying routes of intervention and mitigation. Ecosystem stability may rely on one or a few keystone species, and the loss of such species could potentially have detrimental effects. The Atlantic cod (Gadus morhua) has historically been highly abundant and is considered a keystone species in ecosystems of the northern Atlantic Ocean. Collapses of cod stocks have been observed on both sides of the Atlantic and reported to have detrimental effects that include vast ecosystem reshuffling. By whole-genome resequencing we demonstrate that stabilizing selection maintains three extensive “supergenes” in Atlantic cod, linking these genes to species persistence and ecological stasis. Genomic inference of historic effective population sizes shows continued declines for cod in the North Sea–Skagerrak–Kattegat system through the past millennia, consistent with an early onset of the marine Anthropocene through industrialization and commercialization of fisheries throughout the medieval period.
As ecosystems worldwide are facing disturbance, overexploitation, and environmental shifts, evidence that we have entered the geological epoch of the Anthropocene (i.e., “the age of man”) is compelling (1, 2). In terrestrial ecosystems early human impacts in the form of foraging and land use is well documented, and human expansions are closely tied to species declines and extinctions (3–5). In marine environments recordable impacts of human expansions are often delayed relative to terrestrial environments, and extinctions have been less frequently recorded. These observations are attributed both to the reduced capacity of humans to access the marine environment and to the generally higher mobility of harvested marine species compared with their terrestrial counterparts (6).
There are, however, pitfalls in assessing human impacts on biodiversity solely based on observed abundance or extinction rates. It has been postulated that the potential for stasis of species both spatially and temporally is determined by its standing portfolio of biological diversity (7–9), theoretically tying biodiversity, in terms of standing intraspecific variation, to stability (8, 9). In the face of current environmental challenges, species where intraspecific variation has been reduced due to human impacts may be particularly vulnerable. Additionally, the assessment of past ecological state, or baseline, is challenging in marine environments compared with terrestrial ones, as access to both faunal remains and ecological records is severely reduced. To assess the true extent of human impact on marine species and ecosystems, the use of proxies may thus be crucial. Genomic sciences now allow for the characterization of demographic history and molecular evolution at unprecedented time scales and resolutions, offering opportunities to improve our understanding of the trajectories of species and ecological assemblages.
Through the fossil record, ecological assemblages are often observed to develop in a stepwise manner (10) despite the broadly acknowledged potential for continuous evolution. An advocated explanation for this paradoxical observation (i.e., “the paradox of stasis”) is the presence of stabilizing selective forces (8, 9) that maintain species, and thus ecological stasis, over long timescales (11). One of the strongest traditional arguments against such an explanatory model is that stabilizing selection is rarely observed in nature relative to directional selection (12). The reasoning for this argument is that a major fraction of species in an assemblage should be stabilized by selection for such a mechanism to be considered as a driver of stasis. However, theoretical and empirical work proposing that ecological assemblages may be destabilized by the removal of a single keystone species call this assumption into question (13–18).
The Atlantic cod (Gadus morhua) is a marine fish that has been extensively exploited for centuries. It is a keystone predator in the ecosystems it occupies, and its persistence is closely tied to stability of marine ecosystems across the northern Atlantic Ocean (13–15). The Atlantic cod genome harbors a set of highly divergent haplotypes, caused by chromosomal inversions, that extend several megabases (19, 20). These polymorphisms act as biallelic “supergenes” that segregate across the species’ distributional range, on both sides of the Atlantic (21), adding an additional level of complexity to the overall genomic stratification of the species.
Here we use whole-genome resequencing to show that stabilizing selection maintains the Atlantic cod supergenes, tying these extensive polymorphisms to species persistence, and potentially, to ecosystem stability. We further perform genomic inference to access historic effective population sizes, revealing continuing declines in the North Sea–Skagerrak–Kattegat system through the past millennium. These declines coincide with expansion and commercialization of North European fisheries through the past 1,000 y, starting at the intersect between the Viking period and the medieval period. Our results support an early onset of the marine Anthropocene in the North Sea system, with long-term human impacts shaping species and ecosystem trajectories.
Results
Genomic Complexity of Atlantic Cod in the North Sea System.
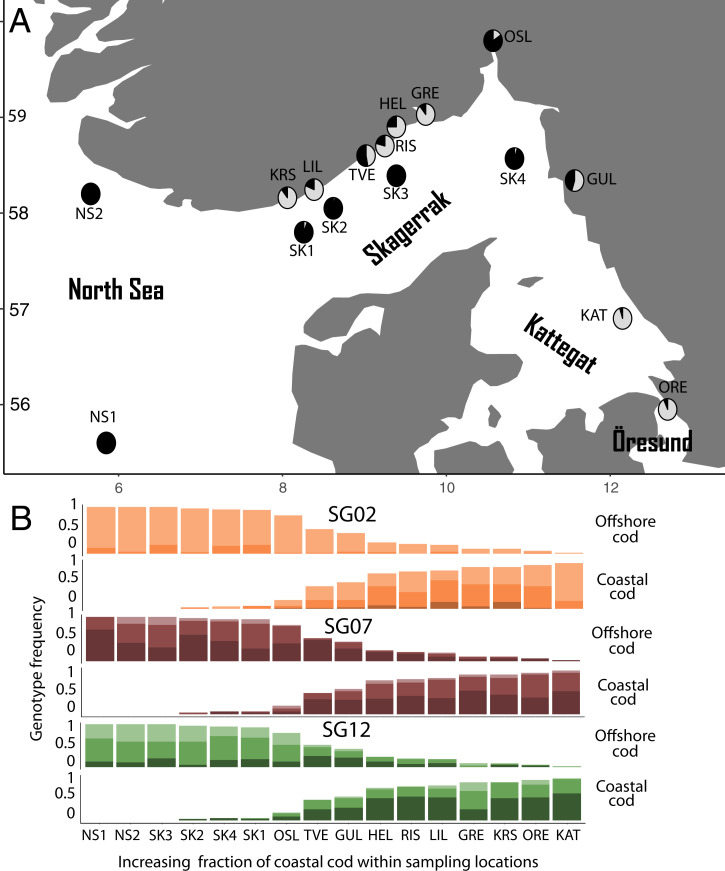
Our study system comprised 752 specimens of Atlantic cod from 16 coastal and offshore sampling locations in the North Sea–Skagerrak–Kattegat system (SI Appendix, Table S1). Discriminant analyses on principle components with 6,683 single-nucleotide polymorphisms (SNPs) identified two main genetic clusters of cod within the study area (Fig. 1A and SI Appendix, Fig. S1). These two clusters represent the ecotypes here denoted “coastal cod” and “offshore cod” (20, 22). Coastal cod tend to dominate inner-coast locations and were also prevalent in the Kattegat and Öresund region, while offshore cod dominated outer skerries and offshore locations in Skagerrak and the North Sea (Fig. 1).
Fig. 1.
(A) For each of 16 sampling locations, pies show frequencies of coastal cod (gray) and offshore cod (black) as determined by discriminant analysis on principal components using genome-wide distributed SNPs (excluding all SNPs on chromosomes 2, 7, and 12). (B) Bars show genotype frequencies (I/I in light color, I/II in intermediate color, and II/II in dark color) for supergenes SG02 (orange, Top), SG07 (burgundy, Middle), and SG12 (green, Bottom) in coastal cod and offshore cod within 16 sampling locations. Bars are ordered from left to right by increasing fraction of coastal cod relative to offshore cod within each sampling location.
Oceanic modeling of Skagerrak and the North Sea showed that the water column was generally more stratified in terms of salinity at locations dominated by coastal cod than at locations dominated by offshore cod (SI Appendix, Fig. S2). Salinity stratification of the water column is also a characteristic of the Kattegat and Öresund region, which is influenced by both the low-salinity Baltic Sea and the high-salinity Skagerrak and North Sea (23).
In both coastal and offshore cod three extensive chromosomal inversions (19, 20), or supergenes, provide an added level of genomic complexity. The supergenes extend from 18.6 to 23.6 Mbp on chromosome 2, from 13.6 to 23 Mbp on chromosome 7, and from 0.4 to 13.4 Mbp on chromosome 12, and are here denoted SG02, SG07, and SG12, respectively. The two haplotypes for each supergene were denoted haplotypes I and II, following Sodeland et al. (20), and their genotypes I/I, I/II, and II/II.
For SG02, haplotype II was found in higher frequency in coastal cod relative to offshore cod across locations. For this supergene the II/II homozygote was absent in offshore cod and only observed at low frequencies in coastal cod (Fig. 1B). For SG07, intermediate frequencies of both haplotypes were present within both ecotypes at all sampling locations, and approximately equal genotype distributions were found within the two ecotypes (Fig. 1B). For this supergene, low frequencies of the I/I homozygotes were observed in both ecotypes. For SG12, higher frequencies of the I/I homozygote were observed in offshore cod relative to coastal cod, while higher frequencies of the II/II homozygote were observed in the coastal ecotype.
Each supergene spans over a large number of linked genes. From the gadMor2 genome assembly (24), a total of 10,866 unique annotated genes with available gene ontologies were retrieved. Supergenes SG02, SG07, and SG12 were found to span a total of 157, 182, and 303 of these genes, respectively. Comparison with the remainder of the genome showed that for supergenes SG07 and SG12, gene ontologies related to ion channel activity were overrepresented (SI Appendix, Table S2).
Selection on Atlantic Cod Supergenes.
Whole-genome resequencing of coastal cod from locations Lillesand (LIL) (n = 11) and Risør (RIS) (n = 8), and offshore cod from location North Sea 1 (NS1) (n = 22) (Fig. 1A and SI Appendix, Table S1) yielded a total of 1,204,973 SNPs, where 12,221 SNPs were positioned within supergene SG02, 30,937 in SG07, and 27,916 in SG12. The resequenced specimens were chosen from their respective sampling locations based on ecotype assignment from the SNP data. All resequenced LIL (n = 11) and RIS (n = 8) specimens were coastal cod, while all resequenced NS1 (n = 22) specimens were offshore cod. Their genotype frequencies for the three supergenes were similar to the frequencies shown in the larger genotyped panel from these three locations (SI Appendix, Table S3).
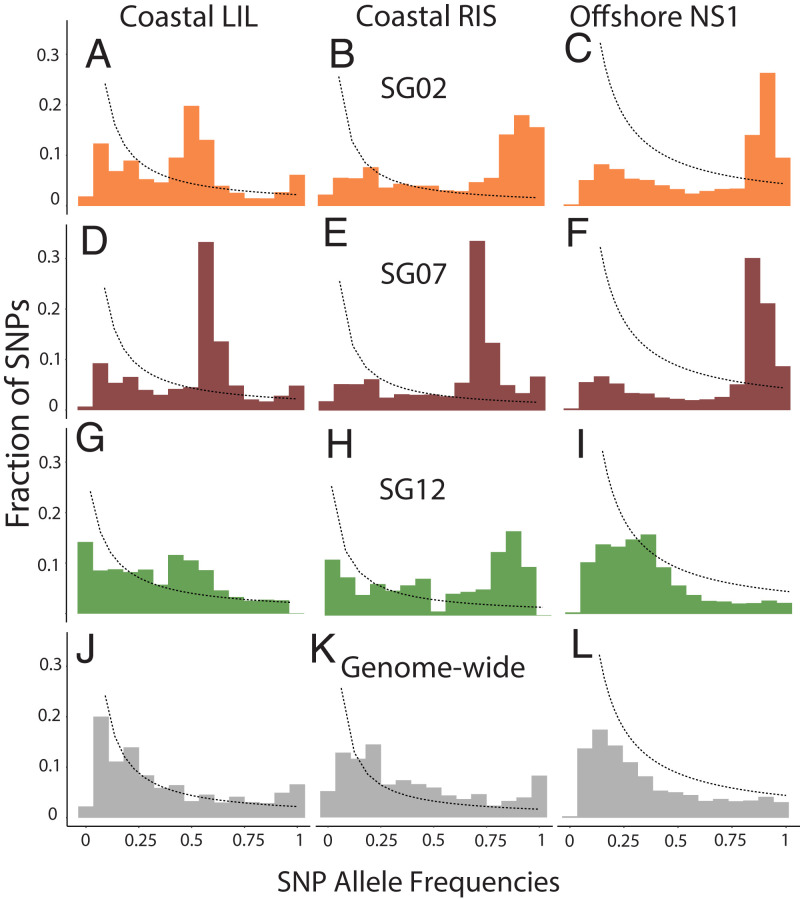
Allele-frequency spectra (AFS) were used to summarize distributions of derived alleles for SNPs within supergenes, as well as at the genome-wide level after excluding SNPs on chromosomes harboring supergenes (Fig. 2). For populations in equilibrium the AFS is theoretically expected to have an L shape under neutral conditions, with a high number of low-frequency–derived alleles (25). Both within supergenes and at the genome-wide level, a lack of low-frequency–derived SNP alleles in the AFS was observed (Fig. 2 A–L). The coastal LIL cod displayed peaks at intermediate frequencies of derived alleles for SNPs within supergenes SG02 (Fig. 2A) and SG07 (Fig. 2D). This pattern was also evident in the coastal RIS cod, but slightly shifted toward higher frequencies of derived alleles. Peaks at intermediate frequencies in the AFS for a genetic locus is a characteristic signature of balancing selection (26, 27), an evolutionary scenario where two or more alleles for a locus are maintained due to contributions to population fitness (28, 29). The offshore NS1 cod showed a marked excess of highly frequent derived SNP alleles in both SG02 (Fig. 2C) and SG07 (Fig. 2F), which was also apparent for coastal RIS cod for SG02 and SG12 (Fig. 2H). Excess of highly frequent derived alleles is a pattern that would result from directional selection (26, 27).
Fig. 2.
Allele-frequency spectrums for SNPs within supergenes (A–C) SG02 (orange), (D–F) SG07 (burgundy), and (G–I) SG12 (green) and (J–L) genome-wide (gray) distributed SNPs (excluding all SNPs on chromosomes 2, 7, and 12) for coastal cod from locations LIL (A, D, G, and J), RIS (B, E, H, and K), and offshore cod from location NS1 (C, F, I, and L). Derived SNP allele frequencies are shown on the x axis, with relative fraction of SNPs on the y axis. For comparison with observed allele frequencies a theoretical distribution of allele frequencies under neutral equilibrium conditions is included as stippled lines.
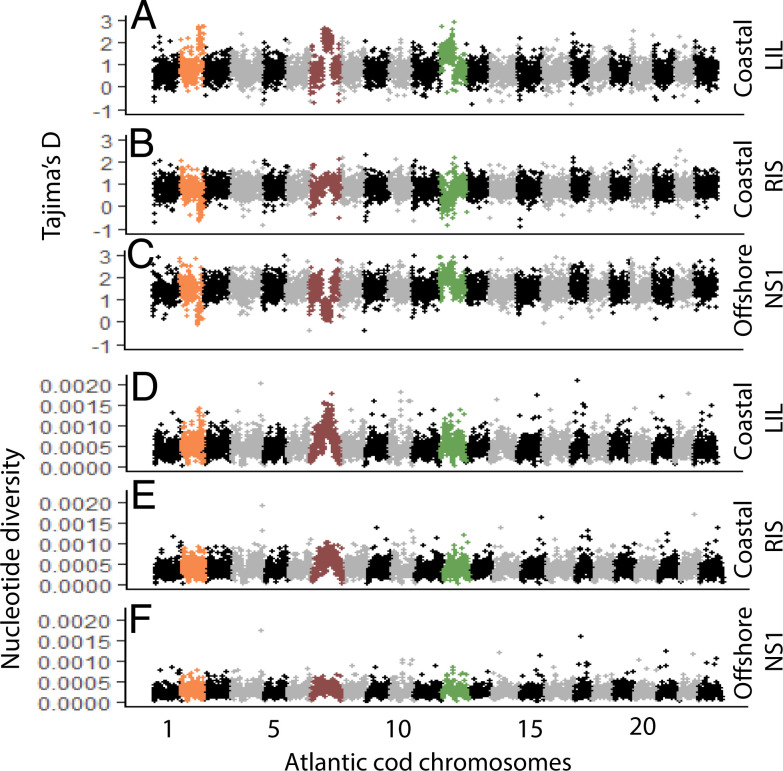
Deviations from expectations in the AFS were also evident from the neutrality statistic Tajima’s D (25), which distinguishes between neutrally developing genomic regions and genomic regions affected by selective or demographic processes. For the coastal LIL cod all three supergenes showed an elevation in Tajima’s D (Fig. 3A), which was also observed for the offshore NS1 cod for SG12 (Fig. 3C). Reduced Tajima’s D was observed for SG02 in both the coastal RIS cod (Fig. 3B) and the offshore NS1 cod (Fig. 3C) and for SG07 in the offshore NS1 cod (Fig. 3C). While elevated Tajima’s D is indicative of balancing selection, reduced Tajima’s D is indicative of directional selection (30, 31).
Fig. 3.
Sliding-window analysis of Tajima’s D (A–C) and nucleotide diversity (D–F) was conducted for bins of 100 kb for coastal cod from locations LIL and RIS and for offshore cod from location NS1 across the 23 Atlantic cod chromosomes. Chromosomes 2, 7, and 12, harboring supergenes SG02, SG07, and SG12, are highlighted in orange, burgundy, and green, respectively.
Besides intermediate allele frequencies and consequent elevation of Tajima’s D, increased genetic diversity at linked sites (26) is an expected signature of balancing selection. Here, nucleotide diversity was particularly elevated in coastal LIL and RIS cod in SG07 (Fig. 3 D and E), as well as for coastal LIL cod in SG02 and SG12 (Fig. 3D).
Genomic Inference of Historic Effective Population Sizes.
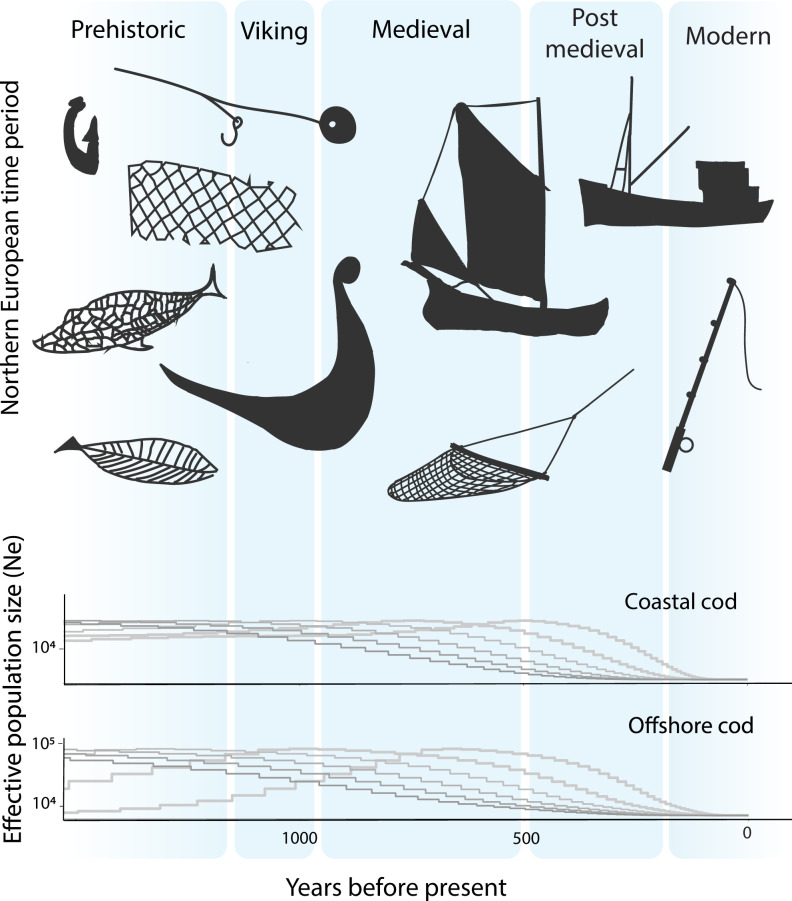
To infer the historic state of species and ecosystems predating modern ecological studies, proxies that are informative over larger temporal and spatial scales are needed (15). Here inferences of historical effective population sizes were conducted with whole-genome resequencing data by Markovian coalescent implemented in SMC++ software (32). This analysis showed declines starting ∼230 generation ago in coastal cod and 280 generations ago in offshore cod. Distributionwide, observed generation times for Atlantic cod range from 2 to 7 y, showing both geographic and temporal variation (33). Historic development of effective population sizes based on this range of generation times is shown in Fig. 4 for both coastal and offshore cod, together with development of fisheries through prehistoric and historic time periods in Northern Europe. Declines in effective population size for both coastal and offshore cod during the past millennia were further supported by model-based AFS diffusion approximation using Dadi software (34), which inferred onset of declines to ∼530 generations ago in coastal cod and 100 generations ago in offshore cod (SI Appendix, Fig. S3 and Table S4).
Fig. 4.
Historic effective population sizes (Ne) were inferred from whole-genome resequencing data by Markovian coalescent for coastal cod (Middle) and offshore cod (Bottom) in the North Sea–Skagerrak–Kattegat system. Inference of historic Ne for generation times in the range of 2 to 7 y is shown (higher generation times depicted in darker shades of gray). Years before present are given on the x axis. (Top) Prehistoric and historic time periods in Northern Europe are indicated, with illustrations portraying the development of fisheries in the region.
Discussion
Biological diversification, in terms of standing intraspecific variation, has been postulated to determine the spatial and temporal potential of stasis both at the species and ecosystem level (7–9). In the Atlantic cod, a keystone ecosystem species distributed across the northern Atlantic, great genomic complexity is observed. In this species, genomic stratification between inshore and offshore ecotypes is observed on both sides of the Atlantic (20, 22, 35), with extensive and highly divergent supergenes representing an additional level of complexity across the species distribution (26).
In the highly heterogeneous North Sea–Skagerrak–Kattegat system, the allelic frequencies at which these supergenes occur vary greatly between sampling locations and between ecotypes (Fig. 1), indicating variation in selective pressure. Heterogeneity in the selective pressure acting on these supergenes is confirmed by the allele frequency distributions of SNPs positioned within the supergenes (Fig. 2), as well as in deviations in Tajima’s D and nucleotide diversity (Fig. 3). On broader spatial scales, the supergenes show distinct longitudinal clines on both sides of the Atlantic (21, 36, 37), consistent with a role in environmental adaptation. Combined, these results provide support for theoretical models that postulate a stabilizing role of intrapopulation diversity in species persistence (7–9, 28, 29).
Coastal and Offshore Ecotypes.
In line with previous findings, two genetically distinct ecotypes of Atlantic cod were identified within the North Sea–Skagerrak–Kattegat study system (Fig. 1). While the coastal ecotype was prevalent at inner-coast locations, as well as in the Kattegat and Öresund region, offshore cod dominate the North Sea and Skagerrak.
Oceanic modeling showed that coastal locations are generally more heterogeneous in terms of salinity stratification compared with offshore locations (SI Appendix, Fig. S2). The water column of the Kattegat–Öresund localities is also highly stratified due to mixing of high-saline water from the North Sea with low-saline water from the Baltic (23). While the adaptive component of the ecotypes’ spatial distribution is likely to be complex, a fitness advantage related to a stratified water column may contribute to explain the dominance of the coastal ecotype at both inner-coast locations and in the Kattegat–Öresund region (Fig. 1 A and B). It should also be noted that the severe declines both ecotypes have suffered in recent times (38–40) may have affected their relative frequencies within sampling locations, introducing bias in comparisons of ecotype distributions with environmental patterns and gradients.
Presence of coastal cod at the offshore Kattegat location, together with stable coexistence of both ecotypes, particularly at outer-coast locations, suggests that the present denotation of coastal and offshore cod might not represent a completely accurate distinction of the two ecotypes. The ecological factors influencing the spatial population structure both within fjords as well as at larger geographical scales, however, remain to be fully characterized. Based on this, we here choose to keep the current denotation, aware that future findings may motivate alterations.
Stabilizing Supergenes.
Selective processes acting on the three supergenes SG02, SG07, and SG12 were assessed using both SNP array and resequencing data. For supergene SG02 the II/II genotype for SG02 was absent in offshore cod from all the 16 sampling locations (Fig. 1B), indicating a fitness advantage of haplotype I in this ecotype. Directional selection acting on this supergene was also observed from the resequencing-derived AFS of offshore NS1 cod and coastal RIS cod (Fig. 2), while a pattern consistent with balancing selection was observed for coastal LIL cod. For supergene SG07, low frequencies of the I/I homozygote were observed in both ecotypes (Fig. 1B). For this supergene, a pattern consistent with balancing selection was observed for coastal LIL and RIS cod from the resequencing-derived AFS (Fig. 2), while directional selection acting on the supergene was observed for offshore NS1 cod. For supergene SG12, higher frequencies of the II/II homozygote was generally observed in coastal cod compared to offshore cod across the 16 sampling locations (Fig. 1B). For this supergene, signals of selection in the resequencing-derived AFS were less prominent, but coastal RIS cod showed some indication of directional selection (Fig. 2).
Variation in Tajima’s D and nucleotide diversity across locations (Fig. 3) were consistent with observations from the AFS (Fig. 2), showing a heterogeneous selective pressure acting on the supergenes within the study area. It should be noted that discrepancies in supergene genotype frequencies could potentially influence the selective patterns detected by the analyses conducted (e.g., deviation in AFS spectrum, nucleotide diversity, and Tajima’s D). For instance, the more even distribution of the allelic variants of the supergenes found in the genome-sequenced individuals from LIL, is likely the cause of the stronger signal of balancing selection in this population compared to RIS (SI Appendix, Table S3).
The term “balancing selection” is used to describe various mechanisms that may broadly be divided into two distinct selective modes in which two or more alleles contribute to population or species fitness: Selection for heterozygote individuals and temporally or spatially alternate selection for separate homozygotes or alleles (26). Both modes imply a role of each of at least two alleles in persistence of the populations or species in which they are maintained and may thus be considered “stabilizing” in the sense that the population or species is prevented from developing in one particular direction (28, 29). Variation in selective patterns observed across sampling locations (Figs. 2 and 3) show that the selective pressures acting on the three supergenes is heterogenous within the study area, supporting the presence of spatially and temporally alternating selective modes in the maintenance of the supergenes. Previous studies have shown “islands of divergence” consistent with supergene clines on both sides of the Atlantic Ocean (36, 41, 42). These findings are also in accordance with heterogeneity in the selective pressure acting on these supergenes across the species range. Trans-Atlantic segregation of SG02, SG07, and SG12 support stabilizing selection acting on the three supergenes (26) and are in line with a role of the supergenes in ecotype and species persistence over long time scales.
Gene ontology overrepresentation of ion channel activity for the supergenes SG07 and SG12 suggests a link between the supergenes and salinity variation. Cod in the North Sea–Skagerrak–Kattegat system experience broad salinity ranges, and besides salinity tolerance and osmosis, ion channel activity might be related to traits such as sensory abilities or motion (43, 44). Stratification of the water column caused by mixing of water of different salinities may also relate to other environmental variables in addition to salinity. For example, both chemical and physical properties of the water may vary between layers of low-saline and high-saline water, confounding correlational patterns. Thus, correlations between supergenes and salinity gradients should be interpreted with caution, as fitness in stratified environments is likely to be highly complex. In an experimental setting, direct impacts on fitness in relation to both salinity and temperature has been observed for SG02 and SG07 (45). For SG07, larvae homozygous for haplotype II exhibited higher fitness than larvae homozygous for haplotype I at both intermediate and high temperatures yet showed lower fitness at low temperatures. This selective pattern in relation to temperature could be linked to the genomic signatures of balancing selection that we observe for this supergene. The clear climatic clines observed for the Atlantic cod supergenes on both sides of the Atlantic (21, 36, 37) also support a role in thermal adaptation.
Long-Term Pressure on Atlantic Cod in the North Sea System.
The North Sea–Skagerrak–Kattegat system was uninhabitable to marine life during the last glacial period due to glaciation, which also caused the sea levels to retract to as low as 120 m below present (46). Atlantic cod in the North Sea, Skagerrak, and Kattegat are thus colonizers that followed the retraction of ice sheets and rising sea levels through the late Pleistocene and early Holocene. To assess historical demographic developments of the Atlantic cod ecotypes in this region, we here used genomic inference from whole-genome sequencing data using two approaches, Markovian coalescent (32) and AFS diffusion approximation (34). While there are discrepancies between the two methods of inference in the exact timing of events, both approaches showed continuous declines in effective population size for both offshore and coastal cod through the past millennium. These results are in accordance with the well-documented expansion of North European cod fisheries since the Viking period (47–49).
To the Scandinavian Vikings, Atlantic cod fishing was essential, and their fisheries initialized an international trade that expanded through the medieval period (50). Vikings spread their fishing traditions to the British Isles, where zooarchaeological evidence supports large increases in cod catches at the transition between the Iron Age and the Viking Age (49). Excavated Atlantic cod bones from sites in northern Britain show that the average size of cod caught in the North Sea has been decreasing since the 11th century (47), supporting the results presented herein from genomic inference of historic population development (Fig. 4). Expansion of fisheries in Europe since the Viking period has included improved fishing vessels and gear, as well as a shift from limited inshore fisheries to large-scale exploitation of offshore resources (48). Together, these developments may provide an explanation for the continued pressure on cod through the past thousand years (Fig. 4). In the North Sea–Skagerrak–Kattegat system, several studies have shown severe declines in cod through the past century from abundance data (38–40), and phenotypic shifts and reduced phenotypic diversity have been observed on both sides of the Atlantic prior to cod stock collapses (51, 52). Currently, commercial fishing fleets have the largest impact on the offshore North Sea–Skagerrak cod (53, 54), while recreational fisheries may constitute the greatest cause of fishing mortality for the costal cod (54, 55).
Development of large-scale Atlantic cod fisheries in medieval Northern Europe was a direct result of the need to feed a steadily growing human population. These fisheries were targeted to the North Sea system in the temperate eastern Atlantic Ocean and is not likely to have affected cod homing to other regions. A recent study suggests that northeastern and northwestern Atlantic cod have not suffered marked declines in overall genomic diversity during the past century (56), through which it has been heavily exploited. However, historic and archeological data show industrialized exploitation of North Sea cod on longer time scales than for northern cod, and the trajectories of northern cod may thus not be comparable to those of temperate cod studied here. Generally, estimates from genomic inference for recent generations are considered less accurate than for more distant generations, and the declines inferred here may thus not be directly comparable to studies focusing on changes in genetic diversity over narrower timescales.
In the currently warming climate, there is a growing concern that cold-adapted species such as Atlantic cod may be unable to persist in areas of its current range. Still, the genetic component to temperature resilience of cod, and thus the potential for adaptive change, remains poorly characterized. Zooarchaeological excavations from “the Holocene warm period” (57), ∼9,000 to 6,000 y ago, show that Atlantic cod was abundant in coastal Scandinavia during a time where annual mean temperatures were generally 2 to 2.5 °C higher than present (58, 59). These findings suggest that Atlantic cod in the North Sea–Skagerrak–Kattegat system may be able to persist in a warming climate. A brief cooling period in the 15th century has previously been linked to an inferred bottleneck in Icelandic Atlantic cod (60). The study used genetic diversity in a mitochondrial gene as well as a gene positioned in a supergene on LG01 to infer historic effective population sizes. The LG01 supergene is linked to migratory behavior in northern cod but is only marginally polymorphic within the North Sea–Skagerrak–Kattegat system (61, 62). As reported here and elsewhere (63), Atlantic cod supergenes are under strong selective pressure and may be unsuited for inference of historic stock demography. Both our genomic inference and zooarchaeological records (47) suggest that cod declines in the North Sea–Skagerrak–Kattegat system were initiated at least half a millennium before the 15th century cooling period, supporting exploitation rather than climate as the main driver of declines. The Atlantic cod supergenes described herein provide a high potential for rapid adaptation given the high number of cosegregating genes they carry (64, 65). We have here shown that these supergenes are under strong and heterogenous selective pressure, which indicates a link to persistence through environmental fluctuations (7–9, 28, 29). Adaptive evolutionary change may allow species and populations to overcome declines through increase of favorable genetic variants (i.e., “evolutionary rescue”) (66), and for cod these supergenes may allow rapid adaptations necessary to persist through the environmental challenges we are currently facing despite the substantial long-term declines (Fig. 4).
In inference of historic effective population sizes, bias may be introduced through evolutionary influences such as selection or gene flow, as well inaccuracies in rates of mutation (67, 68). While the mutation rate utilized for inferring past effective population sizes herein is derived phylogenetically (69, 70), Markovian coalescent algorithms interpret this as a per-generation parameter. For the SMC++ software used here this effect should be attenuated since the algorithm also utilizes linkage information (32). Since SMC++ uses both AFS distributions and linkage between loci to infer past effective population size, the software is well suited for population genomic studies in comparison with other implementations that rely on deep sequencing of fewer individuals. Generally, a minimum of 8 individuals should be used in population studies for estimation of the AFS and genetic diversity derived parameters from high-density genetic markers such as SNP data from resequencing (71, 72). Here 19 coastal cod and 22 offshore cod were used to infer past effective population sizes, which should provide sufficient power and precision for the utilized approaches.
Early Onset of the Marine Anthropocene in Northern Europe?
Atlantic cod is a natural keystone predator in the ecosystems it inhabits. In areas of its range where stock collapses have been observed, detrimental ecosystem effects are reported (13–15). Because other species encompassing the same ecological role may mask the exploitation until they too suffer from depletion, substantial time lags may occur between the onset of overfishing and consequent ecological disruption.
Archeological excavations show that marine exploitation was crucial for early settlers on the Northern European shores after the last glacial period (59). Both marine mammals and fish were essential diet components in the earlier phases of human invasion, but later fish became predominant. Petroglyphs from the same period also testify to the cultural importance of both marine mammals and fish for early Northern Europeans. Fishing gear developed through the Mesolithic period (11,500 to 6,500 y before present) included fishhooks, leisters, fish traps, and fish nets (73). Based on archeological findings on the Swedish west coast it has been suggested that targeted fishing of Atlantic cod started as early as 9,000 y before present (74).
By whole-genome sequencing we here inferred the prehistoric abundances of Atlantic cod in the North Sea–Skagerrak–Kattegat system to be far beyond those observed today (Fig. 4). For such abundances to be supported, both suitable habitats and prey would need to have been proportionally available, and past stock sizes of Atlantic cod may therefore be interpreted as a proxy for prehistoric ecological state. The Anthropocene is characterized by human impacts that ultimately led to biological, climatic, and geochemical alterations, and our results indicate an early onset of the marine Anthropocene in Northern Europe.
Materials and Methods
Study Species and Experimental Setting.
Atlantic cod is a highly mobile fish distributed in coastal and shelf habitats on both sides of the North Atlantic. It is of high commercial value and has been subjected to substantial fisheries, both historically and contemporary. The Atlantic cod displays several life-history forms and exists as sedentary ecotypes that spawn inshore among fjords and skerries, and more dispersal-prone ecotypes that spawn offshore on banks. Partially coexisting coastal and offshore cod are observed on both sides of the Atlantic Ocean (20, 22, 35).
Within the marine environment coastal areas and the open ocean vary in several ecologically important aspects, such as temperature, salinity, habitat characteristics, and food composition (75, 76). The coastal zone often constitutes a highly heterogeneous seascape, scattered with skerries and islands. Representing the ultimate distributional margins of marine organisms, coastal areas are vital nursery habitats, feeding areas, and spawning grounds for a large number of species. In addition to great spatial variation, the coastal seascape is subject to substantial temporal variability both seasonally and annually. The Atlantic cod populations that now inhabit the North Sea–Skagerrak–Kattegat system are colonizers that followed the retraction of the Eurasian ice sheets and consequent rise of sea levels after the last glacial period. This recolonization of Scandinavia after the last glacial maximum has been postulated to drive “unexpected” genetic subdivision (77). Within this experimental setting, genetic differentiation between partially sympatric coastal and offshore cod is well documented (20, 22).
SNP Array Data.
Samples and SNP array genotypes.
Genotype data from a 10k Atlantic cod SNP array were retrieved from 752 specimens of Atlantic cod from a total of 16 sampling locations within the North Sea–Skagerrak–Kattegat system (Fig. 1B and SI Appendix, Table S1). Except from sampling locations NS2, SK1, SK2, and SK3, the SNP array data are previously described, as well as details of the genotyping and SNP array (20, 78). All utilized genotype data are available through Dryad (79). SNPs on linkage group 1 were excluded due to low variation within the study area (61). Genome-wide 8,253 SNPs gave informative and nonambiguous genotype calls across all sampling locations. Of these SNPs, 83 were positioned within supergene SG02, 187 were positioned within supergene SG07, and 199 were positioned within supergene SG12 (20). Both alleles of each of these three supergenes exist in both coastal and offshore cod, but the supergenes do not contribute to determine the ecotype, which is determined at the genome-wide level. When excluding chromosomes 2, 7, and 12, 6,683 SNPs remained.
Genetic stratification.
Discrimination analysis of principal component (DAPC) was conducted with the Adegenet R package (80) to distinguish between the coastal and inshore Atlantic cod ecotypes based on 6,683 SNPs not situated in SG02, SG07, or SG12. In the DAPC, 600 principal components were retained and the minimum value for the Baysian information criterion was found for k = 2 clusters. Principal component analyses (PCA) was used to assign supergene genotypes to individual fish using 83 SNPs in SG02, 187 SNPs in SG07, and 199 SNPs in SG12. PCA revealed highly elevated divergence between three distinct clusters (k = 3; SI Appendix, Fig. S4), as previously observed (20). This trichotomous pattern is driven by the presence of two homozygotes (I/I and II/II) and one heterozygote (I/II) for the two haplotypes, or alleles, within each supergene. The nomination “haplotype I” represents the haplotype showing greater levels of diversity based on SNP array data (20). The utility of PCA for exploring variance structures and structural polymorphisms in genetic data has been widely appreciated (81, 82). The R software package (83) v3.4.1 was used for this and subsequent analyses. Deviations from Hardy–Weinberg expectations for the three supergenes were tested with the -hardy option in the vcftools (84) software package v0.1.13.
Gene Ontology Overrepresentation.
Gene models and predicted gene names with associated gene ontologies were retrieved for a total of 10,866 unique annotated genes within the gadMor2 assembly (24). In this assembly, supergenes SG02, SG07, and SG12 spanned a total of 157, 182, and 303 unique annotated genes, respectively. Overrepresented gene ontologies within the supergenes were identified with the R base package in the R software (83) v3.4.1.
Environmental Heterogeneity.
Coastal areas and estuaries are zones of transition where ocean and land processes interact. Discharge of freshwater from river outlets affects nutrient concentrations and primary production, as well as the thermodynamic properties of water. Salinity stratification may thus be a suitable proxy for environmental heterogeneity. Salinity data from six coastal cod and six offshore cod sampling sites were retrieved from a high-resolution coastal circulation model covering the Scandinavian estuary and the Norwegian coast (85). The ocean model applied was the Regional Ocean Modeling System (ROMS) (86) with a spatial resolution of 800 m. Predictions of daily salinity values at three depths (0, 5, and 10 m below surface) over a 23-y period (1996 to 2019) were used to investigate environmental heterogeneity.
Whole-Genome Resequencing Data.
Samples and DNA extraction.
A total of 41 Atlantic cod, 11 coastal cod from location LIL, 8 coastal cod from location RIS, and 22 offshore cod from location NS1 were chosen for resequencing. These samples have previously been included in analyses of the 10k SNP array data described above and elsewhere (20, 78). All those sequenced from LIL and RIS chosen for sequencing were of the coastal ecotype, while all sequenced specimens from NS1 were of the offshore ecotype. Genomic DNA was prepared for sequencing using the Truseq Library prep kit (Illumina) at the Norwegian Sequencing Centre (Oslo, Norway) and paired-end 150-bp sequencing was conducted with an Illumina HiSEq 2000 instrument. The sequencing data are available from the Sequence Read Archive (SRA) database with accession PRJNA689357 (87).
Quality control and SNP detection.
The Fastx software package (88) v0.0.14 was used to remove Illumina adapters and conduct quality filtering, ensuring that at least 60 bp of each read had a quality score above 20. Reads were aligned to the Atlantic cod reference genome gadMor2 (24) with the BWA software package (89) v0.7.5, before the alignments were indexed and sorted with the Samtools software package (90) v1.3.1. Variant detection was conducted with the Freebayes software (91) v1.1.0. Filtering was conducted with vcftools (84) v0.1.13. Genotypes with a quality score below 30 and read depth below 8 were excluded, as well as SNPs with more than 50% missing data. To avoid false positives in the highly repetitive Atlantic cod genome, SNPs showing a P value below 0.001 for deviations from Hardy–Weinberg proportions were also excluded. No filtering on minor-allele frequency or minor-allele count was conducted. Only biallelic SNPs were kept, leaving a total of 1,204,973 SNPs. Average read depth per individual was >11 times coverage across sites.
Allele-frequency spectra.
Allele-frequency spectra were constructed by calculating derived allele frequencies within the two coastal samples LIL and RIS, as well as in the offshore sample NS1, with the vcftools software from whole-genome resequencing data. The northeast Arctic cod was used as an outgroup (24) to construct unfolded spectra. Allele-frequency spectra were constructed separately for SNPs within the three supergenes SG02 (12,221 SNPs between 18,916,000 bp and 24,000,000 bp on chromosome 2), SG07 (30,937 SNPs between 13,641,000 bp and 22,999,792 bp on chromosome 7), and SG12 (27,916 SNPs between 676,900 bp and 13,758,590 bp on chromosome 12). For comparison with observed allele frequencies, an AFS developing neutrally under equilibrium expectations was estimated by xi = θ/i for i...i − n, where n is the number of chromosomes analyzed and θ is the population scaled mutation rate. Here, a θ of 0.005 was used. Tajima (25) showed that for low rates of mutation (θ→0) this corresponds to the expected proportion, rather than number, of mutations.
Diversity and neutrality statistics.
Tajima’s D (25) and nucleotide diversity (92) were calculated within sampling locations with Vcftools (84) v0.1.13 by a sliding window approach combining SNPs into 100,000-bp bins across each chromosome.
Historic effective population size.
Historic effective population sizes of coastal and offshore cod populations were inferred both by a diffusion approximation approach utilizing the site-frequency spectrum (34) and by a Markovian coalescent approach combining the efficiency of allele-frequency spectrum methodology with linkage disequilibrium information (32). The demographic inference was conducted excluding chromosomes 2, 7, and 12, as well as runs of homozygosity longer than 1 Mbp, as features such as centromeres and structural variants are known to bias demographic inference. A mutation rate of 0.6·10−8, determined from Atlantic cod phylogeny (69, 70), was used. Datasets were thinned to a minimum of 10 kbp distance between SNPs, resulting in a total of 42,788 SNPs when excluding chromosomes 2, 7, and 12. The distribution of interchromosomal linkage disequilibrium between these SNPs and the three supergenes did not show deviations from expectations (SI Appendix, Fig. S5). The northeast Atlantic cod (24) was used as an outgroup for defining unfolded allele-frequency spectrums as mentioned above.
The Markovian coalescent approach estimated historic effective population sizes without predefined demographic scenarios. The estimate option was used by combining genealogical information from all individuals from each population into a composite likelihood estimate.
The approximation diffusion approach implemented in the DADI python module (34) v2.0.1 was used to compare four predefined demographic models: 1) A standard neutral model of development (snm), 2) an exponential model of development (exp), 3) a two-epoch model of development (two_epoch), and 4) a three-epoch model development (three_epoch) (SI Appendix, Fig. S4 and Supplementary Notes). An optimization function that sequentially refined parameters (93) was used with four rounds (each with 10, 20, 30, and 40 replications), increasing maximum iterations (3, 5, 10, and 15) and decreasing fold in parameter generation (3, 2, 2, and 1), resulting in 100 replications. The model obtaining the best Akaike information criterion was deemed the most probable model (SI Appendix, Table S4). Ancestral population size (Ne) was calculated as Ne = θ/4 µL, where θ is the scaled population parameter as estimated by DADI, µ is the mutation rate, and L is the sequence length. Here, the ungapped sequence length of the gadMor2 assembly (646,375,971 bp) was multiplied by a factor of 0.04 to account for SNP thinning.
Supplementary Material
Acknowledgments
This work was supported by the Research Council of Norway (projects “CODFLICT,” “AQUAGENOME,” and “ECOGENOME”); the county of Aust-Agder Fylkeskommune; the European Regional Development Fund (Interreg IVa, “MarGen” and “Margen II” projects); and the Ministry of Trade, Industry, and Fisheries.
Footnotes
Reviewers: S.P., Stanford University; M.W., University of Auckland.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114904119/-/DCSupplemental.
Data Availability
Sequencing data have been deposited in the Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA689357) and genotype data have been deposited in the Dryad database (DOI: 10.5061/dryad.3bk3j9kj0).
References
- 1.Crutzen P. J., Geology of mankind. Nature 415, 23 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Wigginton N. S., Evidence of an Anthropocene epoch. Science 351, 134–136 (2016). [Google Scholar]
- 3.Stephens L., et al. , Archaeological assessment reveals Earth’s early transformation through land use. Science 365, 897–902 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Dirzo R., et al. , Defaunation in the Anthropocene. Science 345, 401–406 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Barnosky A. D., Koch P. L., Feranec R. S., Wing S. L., Shabel A. B., Assessing the causes of late Pleistocene extinctions on the continents. Science 306, 70–75 (2004). [DOI] [PubMed] [Google Scholar]
- 6.McCauley D. J., et al. , Marine defaunation: Animal loss in the global ocean. Science 347, 1255641 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Figge F., Bio-folio: Applying portfolio theory to biodiversity. Biodivers. Conserv. 13, 827–849 (2004). [Google Scholar]
- 8.Levene H., Genetic equilibrium when more than one ecological niche is available. Am. Nat. 87, 331–333 (1953). [Google Scholar]
- 9.Gillespie J. H., A general model to account for enzyme variation in natural populations. V. The SAS–CFF model. Theor. Popul. Biol. 14, 1–45 (1978). [DOI] [PubMed] [Google Scholar]
- 10.Eldredge N., Gould S., “Punctuated equilibria: An alternative to phyletic gradualism.” Models in Paleobiology, T. J. M. Schopf, Ed. (Freeman, Cooper Co., San Francisco, 1971) pp. 82–115. [Google Scholar]
- 11.Estes S., Arnold S. J., Resolving the paradox of stasis: Models with stabilizing selection explain evolutionary divergence on all timescales. Am. Nat. 169, 227–244 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Haller B. C., Hendry A. P., Solving the paradox of stasis: Squashed stabilizing selection and the limits of detection. Evolution 68, 483–500 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Frank K. T., Petrie B., Choi J. S., Leggett W. C., Trophic cascades in a formerly cod-dominated ecosystem. Science 308, 1621–1623 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Casini M., et al. , Trophic cascades promote threshold-like shifts in pelagic marine ecosystems. Proc. Natl. Acad. Sci. U.S.A. 106, 197–202 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson J. B. C., et al. , Historical overfishing and the recent collapse of coastal ecosystems. Science 293, 629–637 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Mills L. S., Soulé M. E., Doak D. F., The Keystone-Species Concept in Ecology and Conservation Management and policy must explicitly consider the complexity of interactions in natural systems. Bioscience 43, 219–224 (1993). [Google Scholar]
- 17.Hooper D. U., et al. , Effects of biodiversity on ecosystem functioning: A consensus of current knowledge. Ecol. Monogr. 75, 3–35 (2005). [Google Scholar]
- 18.Power M. E., et al. , Challenges in the Quest for Keystones Identifying keystone species is difficult—but essential to understanding how loss of species will affect ecosystems. Bioscience 46, 609–620 (1996). [Google Scholar]
- 19.Kirubakaran T. G., et al. , A nanopore based chromosome-level assembly representing Atlantic Cod from the Celtic Sea. G3 Genes Genomes Genet. 10, 2903–2910 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sodeland M., et al. , “Islands of divergence” in the Atlantic cod genome represent polymorphic chromosomal rearrangements. Genome Biol. Evol. 8, 1012–1022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berg P. R., et al. , Trans-oceanic genomic divergence of Atlantic cod ecotypes is associated with large inversions. Heredity 119, 418–428 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knutsen H., et al. , Stable coexistence of genetically divergent Atlantic cod ecotypes at multiple spatial scales. Evol. Appl. 11, 1527–1539 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersson L., Rydberg L., Trends in nutrient and oxygen conditions within the kattegat: Effects of local nutrient supply. Estuar. Coast. Shelf Sci. 26, 559–579 (1988). [Google Scholar]
- 24.Tørresen O. K., et al. , An improved genome assembly uncovers prolific tandem repeats in Atlantic cod. BMC Genomics 18, 95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tajima F., Evolutionary relationship of DNA sequences in finite populations. Genetics 105, 437–460 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Charlesworth D., Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet. 2, e64 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fijarczyk A., Babik W., Detecting balancing selection in genomes: Limits and prospects. Mol. Ecol. 24, 3529–3545 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Takahashi Y., Tanaka R., Yamamoto D., Noriyuki S., Kawata M., Balanced genetic diversity improves population fitness. Proc. Biol. Sci. 285, 20172045 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sides C. B., et al. , Revisiting Darwin’s hypothesis: Does greater intraspecific variability increase species’ ecological breadth? Am. J. Bot. 101, 56–62 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Castillo J. A., Agathos S. N., A genome-wide scan for genes under balancing selection in the plant pathogen Ralstonia solanacearum. BMC Evol. Biol. 19, 123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eckshtain-Levi N., Weisberg A. J., Vinatzer B. A., The population genetic test Tajima’s D identifies genes encoding pathogen-associated molecular patterns and other virulence-related genes in Ralstonia solanacearum. Mol. Plant Pathol. 19, 2187–2192 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terhorst J., Kamm J. A., Song Y. S., Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat. Genet. 49, 303–309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myers R. A., Mertz G., Fowlow P. S., Maximum population growth rates and recovery times for Atlantic cod, Gadus morhua. Oceanogr. Lit. Rev. 2, 401 (1998). [Google Scholar]
- 34.Gutenkunst R. N., Hernandez R. D., Williamson S. H., Bustamante C. D., Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5, e1000695 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruzzante D. E., Taggart C. T., Cook D., Goddard S., Genetic differentiation between inshore and offshore Atlantic cod (Gadus morhua) off Newfoundland: Microsatellite DNA variation and antifreeze level. Can. J. Fish. Aquat. Sci. 53, 634–645 (1996). [Google Scholar]
- 36.Kess T., et al. , Modular chromosome rearrangements reveal parallel and nonparallel adaptation in a marine fish. Ecol. Evol. 10, 638–653 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johansen T., et al. , Genomic analysis reveals neutral and adaptive patterns that challenge the current management regime for East Atlantic cod Gadus morhua L. Evol. Appl. 13, 2673–2688 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cook R. M., Sinclair A., Stefánsson G., Potential collapse of North Sea cod stocks. Nature 385, 521 (1997). [Google Scholar]
- 39.Jonsson P. R., Corell H., André C., Svedäng H., Moksnes P.-O., Recent decline in cod stocks in the North Sea–Skagerrak–Kattegat shifts the sources of larval supply. Fish. Oceanogr. 25, 210–228 (2016). [Google Scholar]
- 40.Barceló C., Ciannelli L., Olsen E. M., Johannessen T., Knutsen H., Eight decades of sampling reveal a contemporary novel fish assemblage in coastal nursery habitats. Glob. Change Biol. 22, 1155–1167 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Bradbury I. R., et al. , Genomic islands of divergence and their consequences for the resolution of spatial structure in an exploited marine fish. Evol. Appl. 6, 450–461 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hemmer-Hansen J., et al. , FishPopTrace Consortium, A genomic island linked to ecotype divergence in Atlantic cod. Mol. Ecol. 22, 2653–2667 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Hwang P.-P., Lee T.-H., Lin L.-Y., Ion regulation in fish gills: Recent progress in the cellular and molecular mechanisms. Am. J. Physiol. Regul. Integr. Comp. Physiol. 301, R28–R47 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Schulz D. J., Temporal S., Barry D. M., Garcia M. L., Mechanisms of voltage-gated ion channel regulation: From gene expression to localization. Cell. Mol. Life Sci. 65, 2215–2231 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oomen R. A., The Genomic Basis and Spatial Scale of Variation in Thermal Responses of Atlantic Cod (Gadus morhua) (Department of Biology, Dalhousie University, Canada, 2019). [Google Scholar]
- 46.Jelgersma S., “Sea-level changes in the North Sea basin” in The Quarternary History of the North Sea, vol. 2, E. Oele et al., Eds. (Univ. Ups Annum Quingentesium Celebrantis, Acta Univ. Upsaliensis, 1979), pp. 115–142. [Google Scholar]
- 47.Barrett J. H., An environmental (pre)history of European fishing: Past and future archaeological contributions to sustainable fisheries. J. Fish Biol. 94, 1033–1044 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Barrett J. H., et al. , Interpreting the expansion of sea fishing in medieval Europe using stable isotope analysis of archaeological cod bones. J. Archaeol. Sci. 38, 1516–1524 (2011). [Google Scholar]
- 49.Barrett J. H., Nicholson R. A., Cerón-Carrasco R., Archaeo-ichthyological Evidence for Long-term Socioeconomic Trends in Northern Scotland: 3500 BC to AD 1500. J. Archaeol. Sci. 26, 353–388 (1999). [Google Scholar]
- 50.Star B., et al. , Ancient DNA reveals the Arctic origin of Viking Age cod from Haithabu, Germany. Proc. Natl. Acad. Sci. U.S.A. 114, 9152–9157 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olsen E. M., Carlson S. M., Gjøsaeter J., Stenseth N. C., Nine decades of decreasing phenotypic variability in Atlantic cod. Ecol. Lett. 12, 622–631 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Olsen E. M., et al. , Maturation trends indicative of rapid evolution preceded the collapse of northern cod. Nature 428, 932–935 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Jorde P. E., et al. , Who is fishing on what stock: Population-of-origin of individual cod (Gadus morhua) in commercial and recreational fisheries. ICES J. Mar. Sci. 6, 2153–2162 (2018). [Google Scholar]
- 54.Kleiven A. R., et al. , Harvest pressure on coastal Atlantic cod (Gadus morhua) from recreational fishing relative to commercial fishing assessed from tag-recovery data. PLoS One 11, e0159220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fernández-Chacón A., Moland E., Espeland S. H., Kleiven A. R., Olsen E. M., Causes of mortality in depleted populations of Atlantic cod estimated from multi-event modelling of mark–recapture and recovery data. Can. J. Fish. Aquat. Sci. 74, 116–126 (2016). [Google Scholar]
- 56.Pinsky M. L., et al. , Genomic stability through time despite decades of exploitation in cod on both sides of the Atlantic. Proc. Natl. Acad. Sci. U.S.A. 118, e2025453118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Enghoff I. B., MacKenzie B. R., Nielsen E. E., The Danish fish fauna during the warm Atlantic period (ca. 7000–3900BC): Forerunner of future changes? Fish. Res. 87, 167–180 (2007). [Google Scholar]
- 58.Antonsson K., Holocene climate in central and southern Sweden: Quantitative reconstructions from fossil data (Uppsala, 2006) Acta Universitatis Upsaliensis.
- 59.Persson P., Jonsson L., Riede F., Skar B., Ecology of Early Settlement in Northern Europe: Conditions for Subsistence and Survival (Equinox Publishing Limited, 2018). [Google Scholar]
- 60.Ólafsdóttir G. Á., Westfall K. M., Edvardsson R., Pálsson S., Historical DNA reveals the demographic history of Atlantic cod (Gadus morhua) in medieval and early modern Iceland. Proc. Biol. Sci. 281, 20132976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kirubakaran T. G., et al. , Two adjacent inversions maintain genomic differentiation between migratory and stationary ecotypes of Atlantic cod. Mol. Ecol. 25, 2130–2143 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Berg P. R., et al. , Three chromosomal rearrangements promote genomic divergence between migratory and stationary ecotypes of Atlantic cod. Sci. Rep. 6, 23246 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodríguez-Ramilo S. T., et al. , Strong selection pressures maintain divergence on genomic islands in Atlantic cod (Gadus morhua L.) populations. Genet. Sel. Evol. 51, 61 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rieseberg L. H., Chromosomal rearrangements and speciation. Trends Ecol. Evol. 16, 351–358 (2001). [DOI] [PubMed] [Google Scholar]
- 65.Noor M. A. F., Grams K. L., Bertucci L. A., Reiland J., Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. U.S.A. 98, 12084–12088 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carlson S. M., Cunningham C. J., Westley P. A. H., Evolutionary rescue in a changing world. Trends Ecol. Evol. 29, 521–530 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Beichman A. C., Phung T. N., Lohmueller K. E., Comparison of single genome and allele frequency data reveals discordant demographic histories. G3 (Bethesda) 7, 3605–3620 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scally A., Durbin R., Revising the human mutation rate: Implications for understanding human evolution. Nat. Rev. Genet. 13, 745–753 (2012). [DOI] [PubMed] [Google Scholar]
- 69.Tørresen O. K., et al. , Genomic architecture of haddock (Melanogrammus aeglefinus) shows expansions of innate immune genes and short tandem repeats. BMC Genomics 19, 240 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Malmstrøm M., et al. , Evolution of the immune system influences speciation rates in teleost fishes. Nat. Genet. 48, 1204–1210 (2016). [DOI] [PubMed] [Google Scholar]
- 71.Nazareno A. G., Bemmels J. B., Dick C. W., Lohmann L. G., Minimum sample sizes for population genomics: An empirical study from an Amazonian plant species. Mol. Ecol. Resour. 17, 1136–1147 (2017). [DOI] [PubMed] [Google Scholar]
- 72.Aguirre-Liguori J. A., Luna-Sánchez J. A., Gasca-Pineda J., Eguiarte L. E., Evaluation of the minimum sampling design for population genomic and microsatellite studies: An analysis based on wild maize. Front. Genet. 11, 870 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Clark J. G. D., The development of fishing in prehistoric Europe1. Antiqu. J. 28, 45–85 (1948). [Google Scholar]
- 74.Milner N., Craig O. E., Bailey G. N., University of York, Department of Archaeology, Eds., Shell Middens in Atlantic Europe (Oxbow Books, 2007). [Google Scholar]
- 75.Conover D. O., Clarke L. M., Munch S. B., Wagner G. N., Spatial and temporal scales of adaptive divergence in marine fishes and the implications for conservation. J. Fish Biol. 69, 21–47 (2006). [Google Scholar]
- 76.Sanford E., Kelly M. W., Local adaptation in marine invertebrates. Annu. Rev. Mar. Sci. 3, 509–535 (2011). [DOI] [PubMed] [Google Scholar]
- 77.Hewitt G., The genetic legacy of the Quaternary ice ages. Nature 405, 907–913 (2000). [DOI] [PubMed] [Google Scholar]
- 78.Barth J. M. I., et al. , Genome architecture enables local adaptation of Atlantic cod despite high connectivity. Mol. Ecol. 26, 4452–4466 (2017). [DOI] [PubMed] [Google Scholar]
- 79.Institute of Marine Research, Whole-genome re-sequencing North Sea and Skagerrak offshore and inshore Atlantic cod. Sequence Read Archive. https://www.ncbi.nlm.nih.gov/sra/PRJNA689357. Deposited 4 January 2021. [Google Scholar]
- 80.Jombart T., Ahmed I., adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sindi S. S., Raphael B. J., Identification and frequency estimation of inversion polymorphisms from haplotype data. J. Comput. Biol. 17, 517–531 (2010). [DOI] [PubMed] [Google Scholar]
- 82.Ma J., Amos C. I., Investigation of inversion polymorphisms in the human genome using principal components analysis. PLoS One 7, e40224 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.R Core Team, R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna, Austria, 2014).
- 84.Danecek P., et al. , 1000 Genomes Project Analysis Group, The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Asplin L., Albretsen J., Johnsen I. A., Sandvik A. D., The hydrodynamic foundation for salmon lice dispersion modeling along the Norwegian coast. Ocean Dyn. 70, 1151–1167 (2020). [Google Scholar]
- 86.Shchepetkin A. F., McWilliams J. C., The regional oceanic modeling system (ROMS): A split-explicit, free-surface, topography-following-coordinate oceanic model. Ocean Model. 9, 347–404 (2005). [Google Scholar]
- 87.M. Sodeland, Supporting data for “Stabilizing selection on Atlantic cod supergenes through a millennium of extensive exploitation.” Dryad. 10.5061/dryad.3bk3j9kj0. Deposited 26 January 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hannon G. J., FASTX-Toolkit (2010).
- 89.Li H., Durbin R., Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li H., et al. , 1000 Genome Project Data Processing Subgroup, The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). http://hannonlab.cshl.edu/fastx_toolkit/. Accessed 22 July 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Garrison E., Marth G., Haplotype-based variant detection from short-read sequencing. arXiv [Preprint] (2012). https://arxiv.org/abs/1207.3907. Accessed 26 October 2017.
- 92.Hudson R. R., Slatkin M., Maddison W. P., Estimation of levels of gene flow from DNA sequence data. Genetics 132, 583–589 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Portik D. M., et al. , Evaluating mechanisms of diversification in a Guineo-Congolian tropical forest frog using demographic model selection. Mol. Ecol. 26, 5245–5263 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited in the Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA689357) and genotype data have been deposited in the Dryad database (DOI: 10.5061/dryad.3bk3j9kj0).