Significance
In metallurgy, annealing heats a metal or alloy to a predetermined temperature, holds this temperature for a certain time, and then decreases the metal to room temperature to change the physical and sometimes also the chemical properties of the material. Researchers introduce a similar concept to simulated annealing to predict minimum-energy conformations of biological macromolecules. In this work, we experimentally verify that annealing at a fast cooling rate can synchronize the 70S ribosome into a nonrotated state with minimum energy in our free-energy landscape analysis. Our results not only offer a facile yet robust approach to stabilizing proteins for high-resolution structural analysis but also contribute to understanding of protein folding and temperature adaptation.
Keywords: annealing, protein synchronization, ribosome, cryo-EM
Abstract
Researchers commonly anneal metals, alloys, and semiconductors to repair defects and improve microstructures via recrystallization. Theoretical studies indicate that simulated annealing on biological macromolecules helps predict the final structures with minimum free energy. Experimental validation of this homogenizing effect and further exploration of its applications are fascinating scientific questions that remain elusive. Here, we chose the apo-state 70S ribosome from Escherichia coli as a model, wherein the 30S subunit undergoes a thermally driven intersubunit rotation and exhibits substantial structural flexibility as well as distinct free energy. We experimentally demonstrate that annealing at a fast cooling rate enhances the 70S ribosome homogeneity and improves local resolution on the 30S subunit. After annealing, the 70S ribosome is in a nonrotated state with respect to corresponding intermediate structures in unannealed or heated ribosomes. Manifold-based analysis further indicates that the annealed 70S ribosome takes a narrow conformational distribution and exhibits a minimum-energy state in the free-energy landscape. Our experimental results offer a facile yet robust approach to enhance protein stability, which is ideal for high-resolution cryogenic electron microscopy. Beyond structure determination, annealing shows great potential for synchronizing proteins on a single-molecule level and can be extended to study protein folding and explore conformational and energy landscapes.
Annealing—common in metallurgy—heats a metal or alloy to a set temperature, holds this temperature, and then cools the metal to room temperature to improve the physical and sometimes also the chemical properties of the material (1, 2). During annealing, recrystallization repairs defects to afford a refined microstructure, which can relieve stress, soften the metal, increase the ductility, and improve the grain structure. One can also anneal organic/inorganic semiconductors via rearranging molecules, polymer chain segments, or entire polymer chains into active layer films. Optimizing the annealing temperature and time can either dramatically increase the crystal size from the submicrometer scale to several micrometers or yield different crystals (3–5).
Annealed materials tend to adopt homogenous states and readily assemble into either three-dimensional (3D) or two-dimensional (2D) crystals. One can readily visualize such regular packing via atomic force microscopy (AFM), X-ray diffraction (XRD), or electron microscopy (EM). Whether annealing exhibits similar effects on biological macromolecules, especially proteins, is a fascinating scientific question that remains unanswered. To date, annealing on proteins has mainly been limited to molecular dynamics simulations. Researchers have simulated annealing—such as with Monte Carlo algorithms—on proteins from a set of starting conformations, which has helped predict the final structures with minimum free energy (6–8). Distinct from metals and organic polymers, proteins and protein complexes are usually discrete entities that consist of chemically diverse subunits bound together in diverse geometries. This substantial structural heterogeneity hinders direct structural determination via AFM or XRD. In contrast, recent advancements in the resolution of cryo-EM offer great opportunities to obtain high-resolution protein structures at the single-molecule level (9–12). By using cryo-EM to compare detailed structures before/after annealing, one can obtain direct experimental evidence for annealing to affect protein conformations.
Many proteins, especially protein complexes, can, in principle, exhibit diverse conformations. Based on the Boltzmann distribution formula, each conformational state of a protein or protein complex is occupied with a statistical weight, , where is the free energy of the conformational state , is the Boltzmann constant, and is the absolute temperature. The Boltzmann distribution and the derived free energy are temperature dependent; an increase in temperature facilitates more rapid kinetics, and the speedup of the kinetics corresponds to more rapid crossing of the energy barrier between local minima. At room and physiological temperatures, conformations will usually differ from those at lower temperatures. This is the theoretical basis for protein conformational changes in accordance with temperature.
Results
Annealing Improves Local Resolution.
Here, we chose the apo-state 70S ribosome from Escherichia coli as a model, wherein the 30S subunit undergoes a thermally driven intersubunit rotation (13–18) and exhibits substantial structural flexibility as well as distinct free energy (19). We incubated purified apo-state 70S ribosome at 0 °C for 5 min, then immediately flash froze the ribosome for cryo-EM analysis, which presumably retained the same conformation as before vitrification (depicted as the unannealed state). We screened the collected 70S ribosome particles via discarding obvious junk and disassembled ribosomes through 2D and 3D classifications. Reconstruction from 200,000 randomly selected particles yielded a structure at a final resolution of 2.6 Å, based on gold-standard Fourier shell correlation (FSC; Fig. 1A and SI Appendix, Figs. S1–S3 and Table S1). Due to the lack of stabilizing factors, such as messenger (mRNA) and transfer RNA (tRNA), local resolution estimates on the unannealed 70S ribosome indicated variable resolutions across the entire density map over the 2.6- to 7.2-Å range (Fig. 1A). Relative to the 50S subunit, the 30S subunit—especially its head domain—was less well resolved, which is common in other apo-state ribosomes (20, 21).
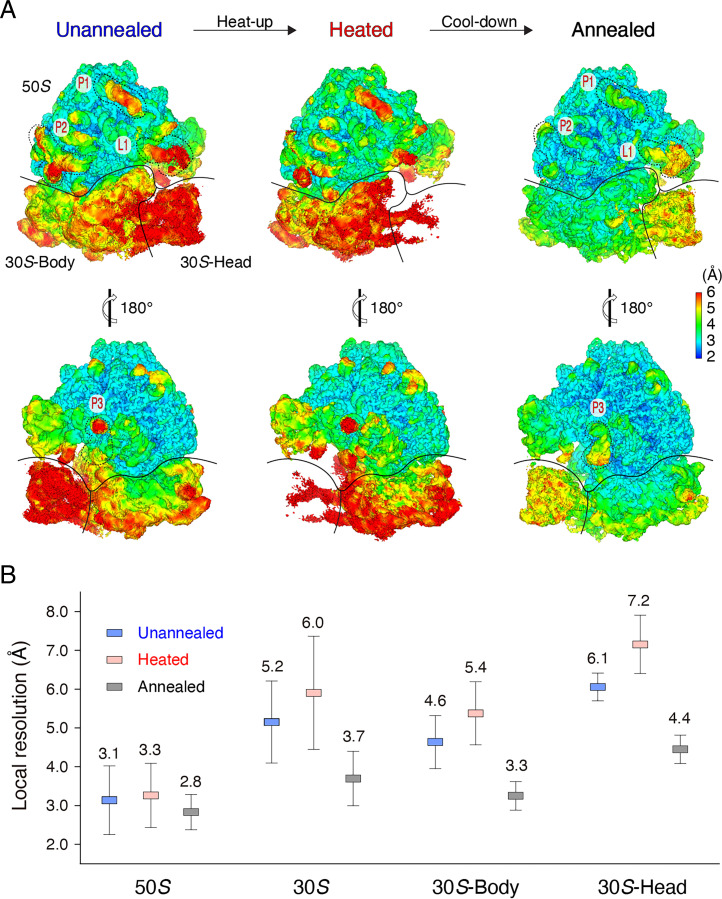
Fig. 1.
Annealing improves local resolution of the 70S ribosome. (A) Local resolution maps of the unannealed, heated, and annealed 70S ribosomes. The 50S subunit and the body and head domains of the 30S subunit are labeled. L1 stalk (L1) and other three subregions (P1, P2, and P3) on the periphery of the 50S subunit in the unannealed and annealed 70S ribosomes are marked for resolution comparison. (B) Local resolution comparison of the unannealed, heated, and annealed 70S ribosomes. The respective means and SDs were calculated from SI Appendix, Fig. S5.
To quantitate the resolution variation of different regions, we calculated the local resolution via averaging local resolution values within the selected regions (SI Appendix, Figs. S4–S6). Our analysis indicated that the 50S subunit exhibited an average local resolution of 3.1 Å, whereas the 30S subunit was much less resolved—only 5.2 Å. Furthermore, the 30S head domain was even less resolved—average resolution of 6.1 Å (Fig. 1B). The intersubunit ratcheting between the 50S and 30S subunits is the primary cause for poor resolvability; the intrasubunit swirling of the 30S subunit is secondary, which lessens the resolution of the head domain. For simplicity, we used the local resolution of the 30S subunit as a marker to monitor the effect of annealing on the 70S ribosome.
Full annealing consists of tandem heating and cooling steps. Cooling rate and annealing temperature are usually two critical elements for homogenization of metals, alloys, and semiconductors (22, 23). In metallurgy, the annealing temperature is usually 30 °C to 50 °C greater than the upper critical temperature of metals (24). In molecular dynamics simulations, researchers sometimes use extreme annealing temperatures—such as 127 °C—to help biological macromolecules escape energy barriers. To avoid heat-induced denaturation, we set the annealing temperatures of the 70S ribosome below the ∼72 °C melting temperature of bacterial ribosomes (25, 26). We carried out subsequent cooling on heated ribosomes in highly thermoconductive PCR tubes via either gradual cooling in the PCR machine or immediate immersion into an ice bath. We vitrified the annealed samples in a Vitrobot chamber preset at 4 °C and then subjected the samples to single-particle cryo-EM analysis in accordance with the same procedure used for unannealed ribosomes from the exact same number (200,000) of randomly selected particles (Fig. 2A).
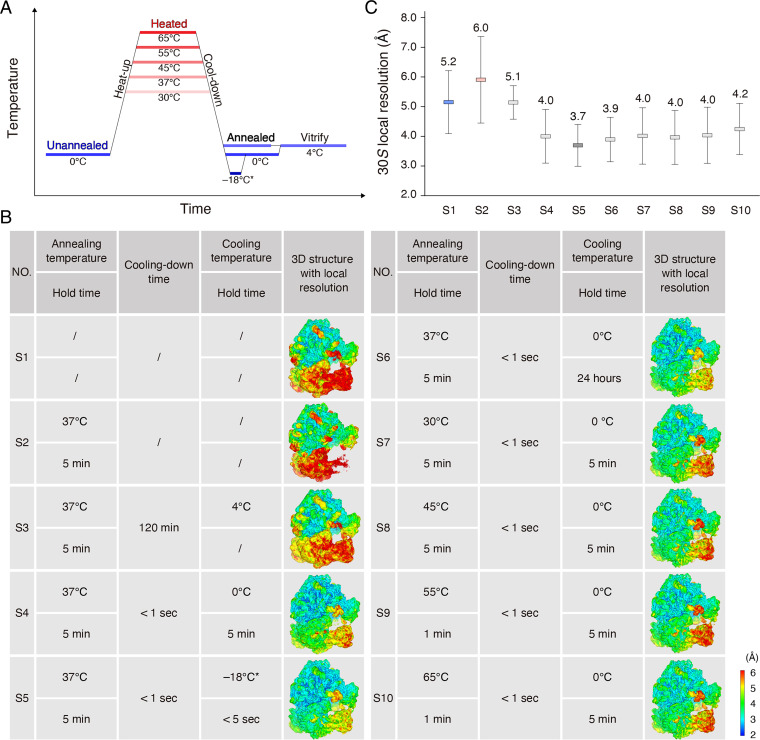
Fig. 2.
Systematic screening of annealing conditions. (A) Schematic for annealing of the 70S ribosome. The unannealed, heated, and annealed stages are labeled on the diagram. (B) Detailed annealing conditions of the 70S ribosomes and the respective 3D local resolution maps. We used sample names S1 through S10 in C and other figures. Regarding S5, we briefly (∼5 s) immersed the 70S ribosome into a mixture of salt, ice, and water precooled to −18 °C and then transferred the samples to an ice bath at 0 °C for 5 min before freezing. (C) Local resolution comparison of 30S subunits among various annealing conditions. The respective means and SDs were calculated from SI Appendix, Fig. S6.
We performed systematic screening of various combinations of cooling rates and annealing temperatures via comparing the local resolutions of the 30S subunit. When we gradually cooled the 70S ribosome over the course of 2 h from 37 °C to 0 °C, the average local resolution of the 30S subunit was 5.1 Å, and there was no obvious local resolution improvement with respect to the unannealed ribosome—5.2 Å (Fig. 2 B and C and SI Appendix, Fig. S6). Fast cooling via immersion into an ice bath improved the local resolution to a value of ∼4.0 Å on the 30S subunit (Fig. 2 B and C and SI Appendix, Fig. S6). A short-term immersion for ∼5 s into a mixture of salt, ice, and water (the measured temperature at −18 °C) prevented the 70S ribosome from freezing and yielded improved local resolution of the 30S subunit—3.7 Å; the 30S head was also dramatically improved to 4.4 Å (Figs. 1B and 2 B and C and SI Appendix, Fig. S6). Apparently, one can use fast instead of gradual cooling to improve the local resolution of the 30S subunit. As another critical parameter for annealing in metallurgy, annealing temperatures of 45 °C, 55 °C, and 65 °C did not further improve local resolutions of the 30S subunit, compared with 30 °C and 37 °C (Fig. 2 B and C and SI Appendix, Fig. S6). Thus, we considered fast cooling from 37 °C in a mixture of salt, ice, and water as ideal annealing conditions for improving the local resolution of the 70S ribosome.
To further understand the annealing process, we also investigated heating the 70S ribosome to an annealing temperature of 37 °C as an intermediate state by cryo-EM (Fig. 2A). Researchers recently developed temperature-resolved cryo-EM to study temperature-sensitive proteins, such as transient receptor potential vanilloid-3 (TRPV3) and ketol-acid reductoisomerase (KARI), at optimal temperatures (27, 28). We optimized sample preparation, such as by consistent high-temperature control and efficient prevention of ice contamination, and found these procedures useful for obtaining ideal cryo-EM grids (SI Appendix, Fig. S7 and Movie S1). We resolved 200,000 particles of the heated 70S ribosome at an FSC resolution of 2.7 Å, close to that of the unannealed ribosome. However, the average local resolution of the 30S subunit was 6.0 Å, substantially less than that of the unannealed and annealed structures (Figs. 1B and 2 B and C and SI Appendix, Fig. S6). The 30S head domain was only resolved at 7.2 Å, and the densities were almost indiscernible at the threshold level proper to the 50S subunit (Figs. 1 and 2 B and C and SI Appendix, Fig. S6). Thus, heating increased the kinetic energy of the 70S ribosome and imparted more structural flexibility to the 30S subunit, which, to some extent, hindered final reconstruction and afforded inferior resolution.
Annealing Stabilizes Flexible Regions.
The annealing-induced resolution improvement was not uniform across the entire 70S ribosome. Relative to a 1.5-Å resolution improvement of the 30S subunit, the well-resolved 50S subunit was only enhanced 0.3 Å (i.e., from a value of 3.1 Å to a value of 2.8 Å) after annealing (Fig. 1B). Thus, annealing was especially beneficial to low-resolution regions, which have greater structural flexibility. To further verify this deduction, we performed a comprehensive statistical analysis on the average local resolution of the same subregions between the unannealed and annealed 70S ribosomes. For example, annealing improved the average local resolutions of different regions at the level of ∼0.1, 0.6, 0.8, 1.2, and 2.0 Å; the corresponding regions in the unannealed ribosome had local resolution in the range of 2.5 to 3.0, 3.0 to 3.5, 4.0 to 4.5, 5.0 to 5.5, and 5.5 to 6.0 Å (Fig. 3A). An exponential curve fit well to the data, indicating that more flexibility in the unannealed 70S ribosome corresponded to more improvement in local resolution after annealing.
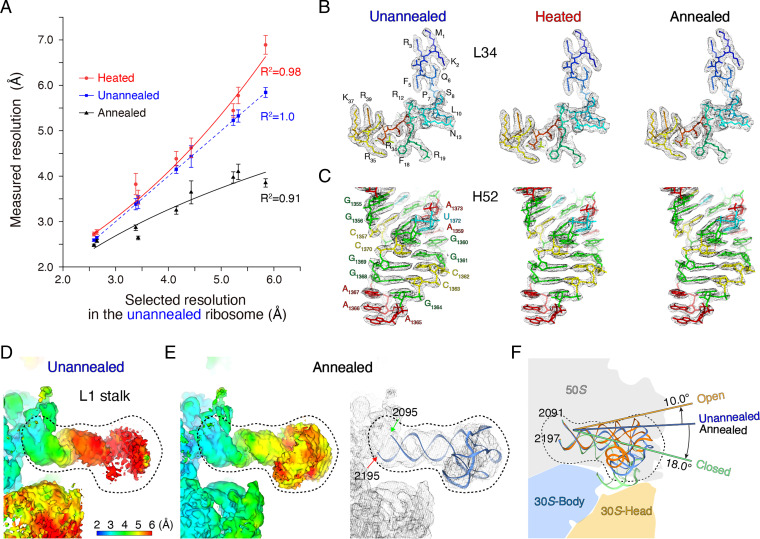
Fig. 3.
Annealing stabilizes flexible regions of the 70S ribosome. (A) The effects of heating and annealing on regions with different local resolutions. We selected regions with different local resolutions from 2.5 to 6.0 Å in the unannealed 70S ribosome and measured local resolutions in the corresponding regions in the heated and annealed 70S ribosomes. (B and C) Cryo-EM maps and fitted atomic models for L34 ribosomal protein and H52 helix in 23S rRNA under unannealed, heated, and annealed conditions. (D) Local resolution map of the L1 stalk in the unannealed ribosome. The position of the L1 stalk is shown in Fig. 1A. (E) Local resolution map of the L1 stalk in the annealed ribosome and fitted atomic model. (F) Intermediate states of the L1 stalk in the unannealed and annealed ribosomes, with respect to the known open and closed states. Atomic models of the L1 stalk are as follows: open (gold), intermediate (blue), and closed (green) states.
Rigid regions were well resolved in both unannealed and annealed 70S ribosomes, and most RNA fragments of 23S ribosomal RNA (rRNA) and ribosomal proteins in the 50S subunit were clearly resolved in the cryo-EM maps (Fig. 3 B and C). There were some flexible peripheral regions in the 50S subunit, which were poorly resolved in the unannealed ribosome. These regions were substantially more distinct after annealing (Fig. 1A). For instance, the L1 stalk, a mobile domain of the 50S subunit that shields the exit site of the deacylated tRNA, is usually invisible in X-ray crystallography and cryo-EM, especially in the absence of peptidyl–tRNA and aminoacyl–tRNA (29–31). After annealing, the L1 stalk was resolved at a resolution of 4.9 Å and adopted an orientation between the open and closed states, which researchers classify as an intermediate state during translation elongation (Fig. 3 D–F). In addition to the L1 stalk, regions from 1,034 to 1,121 and 1,437 to 1,553 in 23S rRNA are intrinsically flexible such that they interact with EF–G–GTP (during elongation and termination procedures) and EF–Tu–GTP–aminoacyl–tRNA (help coupling with the SecY complex). The resolution of these flexible regions was substantially improved: from a value of 4.9 Å to a value of 4.1 Å and from a value of 4.0 Å to a value of 2.9 Å, respectively, after annealing (SI Appendix, Fig. S8).
Compared with the unannealed ribosome, the overall structure of the heated ribosome was less well resolved. The resolution loss also occurred mainly on the flexible regions of the 70S ribosome. Similar local resolution comparisons between the unannealed and heated ribosomes indicated that more flexibility corresponded to a greater extent of resolution loss after heating (Fig. 3A). Briefly, the stable 50S subunit in the heated 70S ribosome had a similar resolution, at ∼3.3 Å, to the unannealed ribosome, whereas the flexible 30S subunit was substantially less well resolved, from 5.2 to 6.0 Å (Figs. 1B and 2B and SI Appendix, Fig. S8). Apparently, heating had a more substantial influence on the flexible rather than rigid regions, analogous to the effect of annealing.
Annealing Renders a Different State.
In metallurgy, recrystallization repairs defects and refines the microstructures after annealing. Thus, we tested whether the annealed 70S ribosome underwent conformational changes. To sample such changes before/after annealing, one requires accurate modeling—especially on the 30S subunit. We separately fit the 50S and 30S subunits from the crystallographic model with the Protein Data Bank Identification code (PDB ID 5MDZ) into the unannealed, heated, and annealed cryo-EM density maps via rigid body docking. Even though the fitting of the 30S head domain into the unannealed and heated 70S ribosomes was not as ideal as that of the annealed 70S ribosome, the well-resolved 30S body domains ensured docking accuracy among the unannealed, heated, and annealed 70S ribosomes (SI Appendix, Fig. S9).
We aligned the unannealed, heated, and annealed 70S ribosomes using the 50S subunit as a reference. We compared the orientations of 30S subunits via calculating the rotation angles relative to each other. The annealed 30S subunit rotated clockwise 7.4°—relative to the unannealed 30S subunit—exhibiting a distinct orientation. The motions of the 30S subunit after annealing resulted in shifts at the periphery of up to 12.0 Å (Fig. 4A). Apparently, annealing imparted a different orientation to the 30S subunit. The orientations of the 30S subunits between the unannealed and heated 70S ribosomes were quite close, with the maximal shift less than 1.8 Å (Fig. 4B). This further indicates that the annealing-induced orientation change occurs during cooling instead of heating.
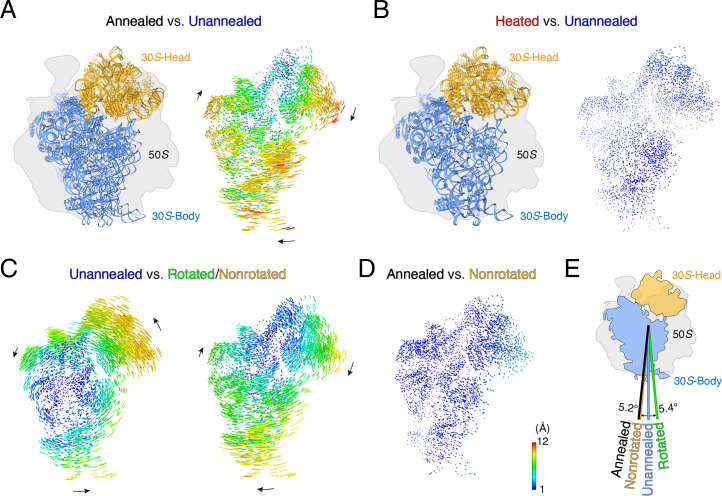
Fig. 4.
Annealing renders the 70S ribosome into a nonrotated state. (A) Rotational comparison of 30S subunits between the annealed and unannealed ribosomes. (Left) We aligned the 50S subunits in the annealed and unannealed ribosomes as a reference, and atomic models for the 30S subunits are shown. (Right) Difference vectors between phosphorous and Cα atoms in the 30S subunits, with arrows indicating the direction of the change. (B) Rotational comparison of 30S subunits between the heated and unannealed ribosomes. (C) Rotational comparison of the 30S subunit in the unannealed ribosome against the known fully rotated (PDB ID 4V9D, Left) and nonrotated (PDB ID 4V7C, Right) states. (D) Rotational comparison of the 30S subunit in the annealed ribosome against the nonrotated state. (E) Summary of rotation angles of the 30S subunit among the unannealed and annealed ribosomes and the rotated and nonrotated states.
Ratcheting of the 30S subunit relative to the 50S subunit is the major rotation that facilitates tRNA translocation. Researchers thoroughly investigated the nonrotated (PDB ID 4V9D) and fully rotated (PDB ID 4V7C) states of the 30S subunit in the context of nascent peptide synthesis (32, 33). We aligned the unannealed, heated, and annealed ribosomes with both the nonrotated and fully rotated states, using the 50S subunits as a reference. In the unannealed 70S ribosome, the 30S subunit adopted an intermediate state between the nonrotated and fully rotated states. Specifically, the unannealed 30S subunit reached the fully rotated state via a 5.4° anticlockwise rotation and rotated clockwise 5.2° to remain in a nonrotated state (Fig. 4C). Distinct from the unannealed 30S subunit, the annealed 30S subunit adopted an orientation very close to the nonrotated state, with a rotation angle of 2.2° (Fig. 4D). The slight difference may be attributable to gentle swirling of the 30S head domain. Apparently, annealing switched the 30S subunit from an intermediate state to a nonrotated state (Fig. 4E), which is an appropriate state for a new ribosome cycle (34).
The 30S subunits under most annealing conditions adopted very similar orientations as the ideal annealing condition in the nonrotated states (SI Appendix, Fig. S10 A–F). Even when we extended the hold time after annealing to 24 h at 0 °C, we resolved the 30S subunit at an average resolution of 3.9 Å and the subunit remained in the nonrotated state, which indicated that the nonrotated state of the 30S subunit may be an energetically favorable state (Fig. 2 B and C and SI Appendix, Fig. S10B). The exception is that the orientation of the slowly cooled 30S subunit was similar to the unannealed and heated 30S subunits in the intermediate state (SI Appendix, Fig. S10G). Considering that the local resolutions of the unannealed, heated, and slowly cooled 30S subunits were substantially inferior compared with the annealed 30S subunits (Fig. 2 B and C), the different orientation of the 30S subunit imparted by annealing was relevant to the resolution improvement.
Annealing Minimizes the Free Energy.
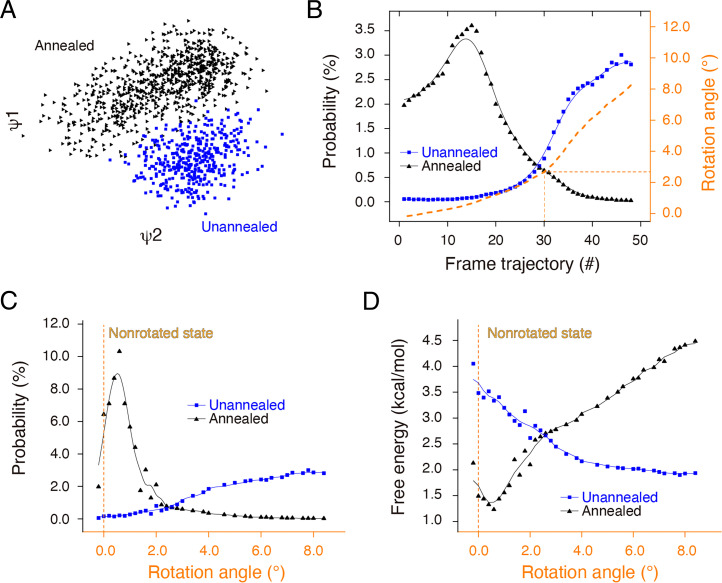
Cryo-EM 3D classification on the unannealed 70S ribosome indicated a variable orientation distribution of the 30S subunit around the intermediate state in each class (SI Appendix, Fig. S11A). As a control, most 3D classes from the annealed ribosome took a very similar orientation of the 30S subunit to the nonrotated state except one with low (∼1.1%) particle occupancy (SI Appendix, Fig. S11B). We further performed detailed free-energy landscape analysis to investigate the correspondence between the annealing-induced resolution improvement and continuous conformational change. For accurate comparison, we merged 300,000 particles from the unannealed and 300,000 particles from the annealed ribosomes with different labels into one dataset, and we subjected the merged dataset to a manifold-based analysis in the same conformational coordinate as described elsewhere (35). In the projection direction approximately orthogonal to the interface between the 50S and 30S subunits, the manifold indicated two clusters, wherein particles from the annealed and unannealed 70S ribosomes were well separated from each other (Fig. 5A and Movie S2). This result corresponds to the obvious orientation difference between the unannealed and annealed 70S ribosomes.
Fig. 5.
Annealing minimizes the free energy of the 70S ribosome. (A) Initial manifold snapshots of the 70S ribosome in one projection direction (points colored in accordance with the unannealed and annealed subsets). The projection direction is approximately orthogonal to the interface between the 50S and 30S subunits. (B) Particle distribution of unannealed and annealed ribosomes along the frame trajectory. We reconstructed the 3D structure at each frame and calculated the rotation angle of the 30S subunit with respect to the nonrotated state. (C) Particle distribution of the unannealed and annealed ribosomes along the rotation angle. We recalculated particle numbers in accordance with the rotation angle at intervals of 0.2° and used a moving average to smooth data variation. (D) Free-energy distribution of unannealed and annealed ribosomes along the rotation angle. The free energy for the unannealed and annealed ribosomes was calculated via the Boltzmann formula with the temperature at 273 K.
We classified the merged particles into 50 frames based on the orientation of the projections, and we separately calculated the particle number of each frame via the labels corresponding to either the unannealed or annealed ribosomes. The unannealed and annealed 70S ribosomes exhibit different peak positions along the frame trajectory. The annealed ribosome was mainly featured in the first 25 frames, whereas the unannealed ribosome started from frame 25 (Fig. 5B). Apparently, the unannealed and annealed ribosomes have distinct particle distributions along the frame trajectory. We reconstructed particles of each frame into a 3D structure with a resolution ranging from ∼5.0 to 8.0 Å, and we fit the respective atomic models for each frame via rigid body docking of the 50S and 30S subunits. As the entire body, we aligned the 70S ribosome from different frames based on the 50S subunit, and we calculated the rotation angles of the 30S subunit in different frames relative to the nonrotated state. The rotation angles from frames 1 through 30 were limited within an ultra-small range of 2.5°, a range that included more than 65% of the particles (the ratio to the total 300,000 particles) of the annealed ribosome. From frames 31 through 50, the rotation angles of the 30S subunit were ∼5.5°, a range that included ∼8% of the annealed particles (Fig. 5C). As the control, the unannealed ribosome included 3% of the particles in frames 1 through 30 and 46% in frames 31 through 50.
We recalculated the particle distribution of the unannealed and annealed ribosomes against the rotation angles at a step size of 0.2°, and we conducted a moving average to smooth data variations. In the new particle distribution curve, the annealed ribosomes exhibited a sharp peak, which was closer to the nonrotational state: a rotation angle of 0° (Fig. 5C). The sharp distribution indicated improved local resolution with stronger structural rigidity of the annealed ribosome. As the control, the unannealed ribosome exhibited no obvious peaks across the rotation angles from 0° to 8.4°. The wider distribution inevitably deteriorated the local resolution of reconstruction, which fits well with the lower resolution in the unannealed 30S subunit.
Based on the Boltzmann formula, we calculated the free energy of various conformations, including both unannealed and annealed ribosomes, against the rotation angles of the 30S subunit. Accordingly, the annealed ribosome exhibited a narrower and deeper well with a minimum energy (global minimum), whereas the unannealed ribosome exhibited a wider and shallower well (local minimum). Based on our experimental data, annealing led the 70S ribosome from a local minimum to a global minimum and dramatically improved the local resolution of the 30S subunit (Fig. 5D).
As the intermediate state of the annealing, we investigated the structural switch from a heated ribosome to an annealed ribosome via free-energy landscape analysis on the combined heated and annealed ribosomes. Similar to the combination of unannealed and annealed ribosomes, heated and annealed ribosomes also exhibited distinct particle distribution (SI Appendix, Fig. S12 A–D). As the control, when we analyzed the unannealed and heated ribosomes, the particle distribution was similar, and the heated ribosome had an even wider distribution (SI Appendix, Fig. S12E). These results indicate that the conformational change occurred during cooling.
Discussion
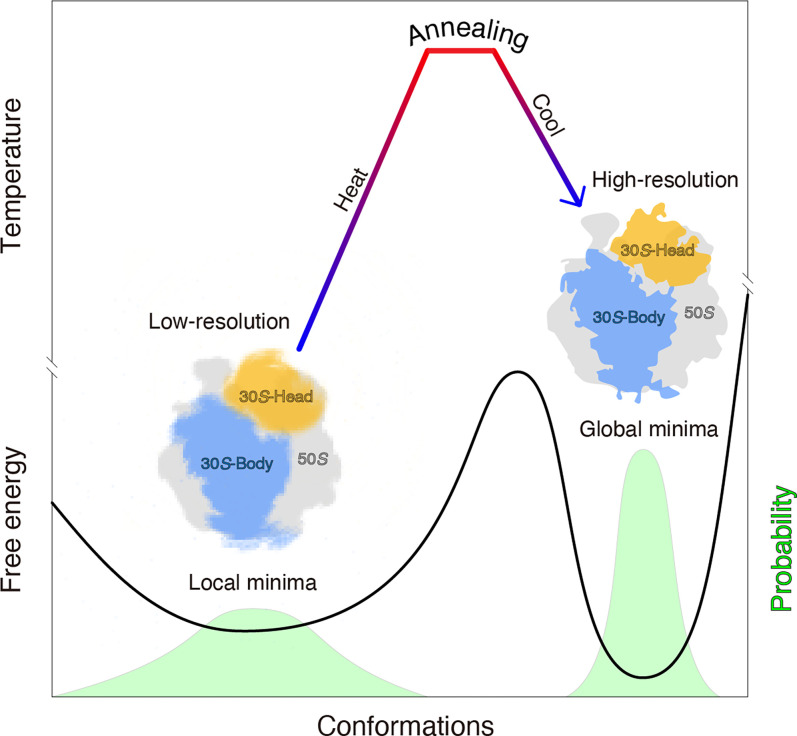
Not limited to metals, alloys, or semiconductors, we experimentally demonstrate that annealing can also synchronize the 70S ribosome into the minimum-energy state with a narrow conformational distribution (Fig. 6). Crystallization of ribosomes/nucleosomes features an annealing-like treatment, wherein researchers usually warm ribosomes/nucleosomes to between 37 °C and 55 °C and then decrease them to room temperature (19 °C) (36, 37). Strict annealing on the 70S ribosome for crystallization will be instructive to explore the physical and chemical effects of annealing on the 70S ribosome, as in metallurgy. Besides the 70S ribosome, annealing on other biological macromolecules, especially those with dynamic structures, will help verify the universality of the approach.
Fig. 6.
Model illustrating that annealing can synchronize a ribosome into a minimum-energy state with improved local resolution. Both the free-energy curve (solid line) and particle distribution probability (light green peaks) are shown.
Structural flexibility, albeit critical for protein function, hinders researchers’ ability to apply structural studies to elucidate function on a molecular level. Sustained efforts—such as mutations on key residues (38, 39), introduction of additional disulfide bonds (40, 41), addition of antibody/binding proteins (42–44), or cross-linking in a solution or within a glycerol/glucose gradient (45, 46)—have been useful for optimizing samples to increase structural stability. However, such efforts are time-consuming and can lack clear direction, and the final structures are confined to fixed states and sometimes are even distorted after additional manipulations. Annealing—a combination of appropriate heating and cooling—is nondestructive to proteins and is a facile yet robust approach for high-resolution cryo-EM. Intriguingly, annealing improves the local resolutions on both the 50S and 30S subunits compared with cross-linked 70S ribosomes via glutaraldehyde (SI Appendix, Figs. S13–S16).
Researchers have also attempted to improve local resolution via local refinement on flexible regions during cryo-EM image processing (47–50). We performed local refinements on the flexible 30S subunit of both unannealed and annealed ribosomes. The average local resolution of the 30S subunit of the unannealed ribosome was improved ∼1 Å to a value of 4.2 Å after local refinements (SI Appendix, Figs. S17 and S18). Distinct from the resolution improvement via annealing, local refinement in itself still resulted in an insufficient average resolution of 5.5 Å on the head domain of the 30S subunit (SI Appendix, Figs. S17 and S18). Evidently, annealing and local refinement improve local resolution via different mechanisms. Annealing can drive biological macromolecules into a minimum-energy state and globally increase the resolution across the entire map irrespective of the region size. As a control, local refinement worked on an algorithmic level and was only applicable to regions with reasonable sizes. When we applied local refinement to the annealed ribosome, both the body and head domains of the 30S subunit were improved to 2.9 and 3.9 Å, respectively (SI Appendix, Figs. S17 and S18). This indicates that annealing is compatible with local refinement on flexible regions and can further optimize local resolutions for detailed structural analysis.
One can use annealing to synchronize proteins into a minimum-energy state, which may benefit many single-molecule approaches such as optical tweezers and single-molecule fluorescence resonance energy transfer (51–53). One can also use annealing to investigate temperature adaptation and protein folding and facilitate algorithm development in molecular dynamics simulations. Thus, researchers should thoroughly investigate the mechanism of annealing and further optimize the annealing conditions for improved resolution.
Materials and Methods
Purification and Assembly of the Apo-state 70S Ribosome.
The 70S ribosomes were extracted from E. coli KC6/ΔsmpB/ΔssrA/S1::S1-8×His strain as previously described (54, 55). In brief, cells were lysed and the debris was removed via centrifugation. The supernatant was layered on the top of a sucrose cushion and centrifuged at 29,400 rpm using a 45Ti rotor (Beckman) for 16 h at 4 °C for the crude 70S ribosomes. The samples were further applied to a Butyl-650S column and eluted with a linear 1.5 to 0 M (NH4)2SO4 gradient in Tris⋅HCl (20 mM, pH 7.4) buffer containing 400 mM KCl, 10 mM Mg(OAc)2, and 6 mM 2-mercaptoethanol. The pooled fractions containing pure ribosomes were further passed through a 5-mL HisTrap Excel column (GE Healthcare Life Sciences) to remove residual ribosomes with ribosomal protein S1 bound. The S1-free 70S ribosome was loaded on a 10 to 40% (wt/vol) sucrose gradient using an SW32 rotor (Beckman) by centrifugation at 23,000 rpm for 14 h at 4 °C. The fractions containing 70S ribosomes were pooled and dialyzed against Hepes (5 mM, pH 7.5) buffer containing 50 mM KCl, 10 mM NH4Cl, 10 mM Mg(OAc)2, and 2 mM 2-mercaptoethanol. The dialyzed ribosomes were flash frozen in liquid nitrogen and stored under −80 °C.
The same batch of 70S ribosomes was used for cryo-EM sample preparation and image processing under various conditions.
Cryo-EM Sample Preparation.
The frozen 70S ribosomes were melted on ice and then diluted to a final concentration of 700 nM in Hepes buffer. To ensure fast heat exchange, no more than 20 µL of the diluted ribosomes were pipetted into one PCR tube. All the samples were subjected to various treatments as listed in Fig. 2B before vitrification.
For the annealed treatments (S4 through S10), the diluted ribosomes were placed on ice for 5 min and then transferred to a water bath, with preset temperatures ranging from 30 °C to 65 °C. After a 1- or 5-min hold time, the heated samples were immediately immersed into a mixture of salt, ice, and water (measured temperature: −18 °C) for ∼5 s and then to an ice bath (measured temperature: −0 °C) for 5 min (S5) or directly immersed into an ice bath for either 5 min (S4 and S7 through S10) or 24 h (S6). As a control, the unannealed ribosome (S1) was simply immersed in an ice bath for 5 min. For the slowly annealed sample (S3), the diluted ribosome in the PCR tube was heated to 37 °C for 5 min and then placed in a PCR machine (Bio-Rad). The temperature decreased gradually from an initial value of 37 °C to a final value of 4 °C over 2 h at a cooling rate of 0.275 °C/min.
After treatments, all the unannealed and annealed samples in the PCR tubes were immediately placed in a Vitrobot chamber set at the minimum temperature of 4 °C and a humidity of 100% (Mark IV, Thermo Fisher Scientific). A gold-supported holey carbon grid was glow discharged and precooled in the Vitrobot chamber for 2 min, and 4 µL solution was applied onto the grid. The grid was immediately plunged into liquid ethane and stored under liquid nitrogen for future cryo-EM imaging.
Cryo-EM Sample Preparation with a Mist Umbrella.
In temperature-resolved cryo-EM, sudden exposure of high-temperature mist (∼37 °C) from a Vitrobot chamber to cooler ambient temperatures (∼20 °C) causes substantial ice contamination. To minimize ice contamination under various temperatures, especially at high temperatures, a mist umbrella was designed. The mist umbrella was made of a piece of round filter paper (40.0-mm diameter) with a precut square hole (9.0 mm × 3.6 mm) prepared via a homemade device (SI Appendix, Fig. S7). The peripheral belt of the mist umbrella was pretreated with low-adhesive glue. Before lifting the Vitrobot tweezer into the chamber, the mist umbrella was clamped to the tweezer via the precut hole. Over the course of lifting the grid to the chamber, the mist umbrella fell off the tweezer, yet glued to the bottom surface of the chamber. After blotting, the shutter of the Vitrobot chamber was immediately opened and the grid passed through the precut hole into liquid ethane. The high-temperature mist in the chamber was blocked by the mist umbrella, which prevented the exposure of high-temperature mist to the cool ambient. When the liquid nitrogen reservoir and cryogenic cup dropped away from the chamber, the tweezer reattached the mist umbrella to shield the grids (Movie S1). During grid transfer from liquid ethane to liquid nitrogen, one could readily remove the mist umbrella from the Vitrobot tweezer along the tear line.
The mist umbrella was used for the heated ribosome (S2) as an intermediate state. Diluted ribosomes were incubated in a water bath at 37 °C for 5 min and then transferred into the Vitrobot chamber at a temperature of 37 °C and a humidity of 100%. The grid was placed in the chamber for at least 100 s to ensure complete heat exchange, and then 5 µL solution was applied onto the grid via preheated tips. The grids were blotted with a 1-s blotting time and force level of 2, immediately plunged into liquid ethane, and stored under liquid nitrogen for future cryo-EM imaging.
Cryo-EM Data Collection.
Cryo-EM grids were examined in a low-dose mode on a Talos L120C transmission electron microscope (TEM; Thermo Fisher Scientific) for screening. Data collection on high-quality grids in all conditions was performed with the same Titan Krios G3i microscope (Thermo Fisher Scientific) equipped with a K3 BioQuantum direct electron detector (Gatan). Special care was taken to perform a coma-free alignment on the microscope.
Movies were collected via FEI EPU (Thermo Fisher Scientific) (56) automated data collection software at a total dose of 60 e−/Å2 fractionated over 50 frames in a defocus range of −1.0 to −1.5 µm. A superresolution mode was used with a final pixel size at 0.53 Å. The numbers of the collected movies under different conditions are listed in SI Appendix, Fig. S2B and Table S1.
Cryo-EM Data Processing and 3D Reconstruction.
For optimal structural comparison, the same image processing strategy was performed on ribosomes under all conditions with cryoSPARC version 3.1.0 (57); SI Appendix, Fig. S2 shows a detailed workflow. Specifically, raw movie stacks were aligned and summed in accordance with dose weighting with MotionCor2.1 (58). The contrast transfer function (CTF) parameters of the summed micrographs were determined with CTFFIND-4 (59). Micrographs with maximum resolution estimates of at least 5 Å were imported into cryoSPARC. Automatic particle picking was performed on the selected micrographs, and particle sets were created and subjected to reference-free 2D classifications. Obvious junk was excluded from the particle set. Six reference structures—including 2× 70S ribosome (PDB ID 5MDZ), 2× 50S subunit (derived from PDB ID 5MDZ), and 2× 30S subunit (derived from PDB ID 5MDZ)—were low-pass filtered to 40 Å and utilized for heterogeneous refinement. Only particles in the integral 70S ribosome with a sufficient signal on the 30S subunit were collected. A total of 200,000 particles were randomly selected from the new particle set for each condition, and per-particle refinement of CTF parameters was conducted. Particles with local CTF parameters were subjected to homogeneous refinement. All the final reconstructions were filtered and sharpened by a cryoSPARC postprocessing session, and the resolutions were determined by gold-standard FSC 0.143. For the unannealed (S1) or annealed (S5) 70S ribosomes, 200,000 particles were 3D classified into five classes for rotational comparison of 30S subunits between the 70S ribosome and their classified structures.
Model Building and Structural Analysis.
The crystal structure of the 70S ribosome, with an empty A site (PDB ID 5MDZ) (60), from E. coli was chosen as the initial template, and mRNA and fMet–tRNA were removed before fitting into the apo-state 70S ribosomes. The 50S and 30S subunits of the modified crystal structure were separately fit into the 70S ribosomes under various conditions via rigid-body docking, and the fits were ideal, except on the L1 stalk. A flexible fitting of the L1 stalk was performed with Rosetta 2018.33.60351 (61). The atomic model was further optimized for improved local density fitting with Coot 0.8.9.1 (62) and real-space refinement with PHENIX 1.17.1 (63).
Structures of the unannealed, heated, and annealed 70S ribosomes were aligned using their 50S subunits as a reference. The conformational change of the 30S subunits was analyzed in Pymol 2.4.0 as described elsewhere (16, 32). Difference vectors between the phosphorous and Cα atoms in the unannealed, heated, and annealed 30S subunits were calculated via the modified script (https://github.com/soothing35/cryoEM_annealling), and rotation angles between the 30S subunits were measured with the Pymol script “RotationAxis.py” (https://pymolwiki.org/index.php/RotationAxis).
Other structural analyses, such as structural superimposition, were fulfilled with UCSF Chimera 1.16 (64).
Local Refinement on the 30S Subunit.
Local refinement on the 30S subunit was also performed with cryoSPARC (57). Based on the docked atomic model, a mask for the 50S subunit was created and applied to subtract the corresponding densities in all of the particles. Local refinement was conducted on the 30S subunit density with rotations and shifts recentered every iteration. A new function in cryoSPARC termed “using pose/shift gaussian prior during alignment” was also used for improved reconstruction. The final reconstruction was filtered and sharpened by a cryoSPARC postprocessing session, and the resolutions were determined by gold-standard FSC 0.143.
Local Resolution Quantification.
To compare local resolutions of the 30S subunits and other subregions under various conditions, local resolution quantification was calculated via averaging values of the local resolution in selected regions (SI Appendix, Fig. S4). Specifically, local resolutions on the entire 70S ribosomes under various conditions were first estimated as per local resolution estimation with cryoSPARC (65). Then the regions within a certain local resolution range or within the same functional domain were selected as the targets. After atomic model building, a mask corresponding to the targeted region was created with the extension of two pixels, and the same mask strategy was applied to the targeted regions from other structures. The resulting subregions were adjusted to the same volume among different structures, and the values of the local resolution within the mask were read via our Python script (https://github.com/soothing35/cryoEM_annealling). The numbers of local resolution values in the range of 2.0 to 2.1 Å, 2.1 to 2.2 Å, and so on were counted in different subregions. The respective means and SDs were calculated for each subregion and compared with each other.
Free-Energy Landscape Analysis.
Free energy is inversely proportional to particle distribution, as described by the Boltzmann formula (66). The particle distribution of 70S ribosomes under various conditions was calculated with ManifoldEM (https://github.com/GMashayekhi/ManifoldEM_Matlab) as described elsewhere (35, 66). Specifically, 300,000 particles were randomly selected from screened particle sets for each condition to sample the entire conformational trajectory. Particles files, including both .star and .mrcs, were selected after autorefinement with RELION 3.0.8 (67). To ensure accurate alignment of structures from two different conditions, their particle files were merged together via appending one to the other with different labels, and the merged particle files were analyzed with ManifoldEM for further analysis.
In ManifoldEM, merged particles were classified in accordance with the predetermined Euler angels, and particle images were deemed as dots in high-dimensional space. Clouds of dots in high-dimensional space were eigen-decomposed and embedded into low-dimensional space with a diffusion map algorithm, in which the kernel was defocus tolerant. The first five eigen-vectors were selected, and the conformational changes along each eigen-function were extracted into 2D movies by nonlinear Laplacian spectral analysis, also termed nonlinear singular value decomposition. Considering that the intersubunit rotation between the 50S and 30S subunits was considered to be the primary conformational change, movies with such conformational changes along each eigen-function were chosen manually. Particles involved in the conformational changes were further classified into 50 classes along the conformational trajectory.
For all 50 classes, 3D volumes were reconstructed via back-projection in RELION 3.0.8. Atomic models for the 50S and 30S subunits were separately docked in each volume via rigid-body docking. All structures were aligned to the nonrotated state (PDB ID 4V9D) with the 50S subunit as a reference. The rotation angle of the 30S subunit in each class was calculated with respect to the 30S subunit in the nonrotated state. Based on the labels of different conditions, particles in each class were separated. The particle distribution within the same condition was recalculated, and the respective free energy was calculated based on the Boltzmann distribution.
Cross-linking of 70S Ribosomes.
Glutaraldehyde (GA) was used to cross-link 70S ribosomes as described (68). To guarantee that GA cross-linking occurs on the interface between the 50S and 30S subunits, the effect of cross-linking was firstly estimated via checking the integrity of 70S ribosomes at low Mg2+ concentration via negative-stain EM. The frozen 70S ribosomes were melted on ice and then diluted to a final concentration of 700 nM in Hepes buffer. A small portion of ribosomes were diluted 20 times against Mg2+-free Hepes buffer for negative-stain EM to check the integrity of 70S ribosomes at 0.5 mM Mg2+ concentration. The other portion of 70S ribosomes were mixed with 2.5% (vol/vol) GA in a 2:1 volume ratio. The mixture was incubated under 0 °C for various cross-linking times from 10 to 65 min. At different time points, the mixture was diluted 20× with 0.5 mM Mg2+ in Hepes buffer and subjected to negative-stain EM. Via estimating the integrity of 70S ribosomes, 40 min is sufficient to cross-link the 30S and 50S subunits and is perfect to avoid over–cross-linking of the 70S ribosomes into clusters or aggregates.
The undiluted mixture of 70S ribosomes and GA after 40-min cross-linking was directly frozen into liquid ethane to fix the states and subjected to the same Titan Krios G3i microscope for cryo-EM data collection. After motion correction and CTF correction, 991,982 particles were picked up from 8,771 micrographs. Similar to the other annealing treatments, only particles in the integral 70S ribosome with sufficient signal on the 30S subunit were collected, and 200,000 particles were randomly selected for the final 3D refinement. The resolution was estimated at 2.63 Å based on gold-standard FSC 0.143.
Negative-Stain EM.
A total of 4 μL ribosome (∼35 nM) were applied to a glow-discharged EM grid covered by a thin layer of continuous carbon film and stained with 2% (wt/vol) uranyl acetate. Negatively stained grids were imaged on a Tecnai L120C TEM operating at 120 kV. Images were recorded at a magnification of ×73,000 and a defocus set to −2.0 µm, using a 4K × 4K Ceta CMOS camera (Thermo Fisher Scientific).
Supplementary Material
Acknowledgments
We thank Prof. Jianpeng Ma from Fudan University for discussions on simulated annealing. We thank Prof. Abbas Ourmazd and Dr. Ghoncheh Mashayekhi from University of Wisconsin-Milwaukee for help on ManifoldEM. We are grateful to Kang Li, Dianli Zhao, and Ceng Gao from the Cryo-EM Facility for Marine Biology at Qingdao National Laboratory for Marine Science and Technology for our cryo-EM data collection. This study was funded by the National Key R&D program of China Grants 2017YFA0504800 (Q.-T.S.), 2021YFF1200403 (Q.-T.S.), and 2018YFC1406700 (Q.-T.S.) and the National Natural Science Foundation of China, Grant 31870743 (Q.-T.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2111231119/-/DCSupplemental.
Data Availability
The cryo-EM density maps of ribosomes under all conditions (S1 through S10) were deposited in the Electron Microscopy Data Bank (EMDB) with accession IDs EMD-31266, EMD-31267, EMD-31268, EMD-31269, EMD-31270, EMD-31271, EMD-31272, EMD-31273, EMD-31274, and EMD-31275, as listed in SI Appendix, Fig. S2B. The locally refined cryo-EM maps of the 30S subunits from the unannealed (S1) and annealed (S5) ribosomes were also deposited in the EMDB with the respective accession IDs EMD-31277 and EMD-31279. Cryo-EM density maps and models for all 50 states in the free-energy landscape analysis are available on the Electron Microscopy Public Image Archive (EMPIAR-China) with the accession ID 100003. The codes for the local resolution analysis and modified ManifoldEM have been uploaded to GitHub: https://github.com/soothing35/cryoEM_annealling. All other study data are included in the article and/or supporting information.
References
- 1.Huang X., Sun H., Shen J., Cui K., Wang G., Different effects of annealing on microstructure evolution and SERS performance for Cu–Cr alloy film and bulk alloy. Materials (Basel) 12, 2990 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang C., et al. , Effects of annealing on microstructure and mechanical properties of metastable powder metallurgy CoCrFeNiMo0.2 high entropy alloy. Entropy (Basel) 21, 448 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang X., van Duren J. K. J., Janssen R. A. J., Michels M. A. J., Loos J., Morphology and thermal stability of the active layer in poly(p-phenylenevinylene)/methanofullerene plastic photovoltaic devices. Macromolecules 37, 2151–2158 (2004). [Google Scholar]
- 4.Zhu Z., et al. , Critical annealing temperature for stacking orientation of bilayer graphene. Small 14, e1802498 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Balagula R. M., et al. , Effects of thermal annealing on localization and strain in core/multishell GaAs/GaNAs/GaAs nanowires. Sci. Rep. 10, 8216 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheon S., Liang F., Folding small proteins via annealing stochastic approximation Monte Carlo. Biosystems 105, 243–249 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Schmitz M., Steger G., Description of RNA folding by “simulated annealing”. J. Mol. Biol. 255, 254–266 (1996). [DOI] [PubMed] [Google Scholar]
- 8.Chou K. C., Carlacci L., Simulated annealing approach to the study of protein structures. Protein Eng. 4, 661–667 (1991). [DOI] [PubMed] [Google Scholar]
- 9.Liao M., Cao E., Julius D., Cheng Y., Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504, 107–112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bai X. C., Fernandez I. S., McMullan G., Scheres S. H., Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. eLife 2, e00461 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakane T., et al. , Single-particle cryo-EM at atomic resolution. Nature 587, 152–156 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yip K. M., Fischer N., Paknia E., Chari A., Stark H., Atomic-resolution protein structure determination by cryo-EM. Nature 587, 157–161 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Cornish P. V., Ermolenko D. N., Noller H. F., Ha T., Spontaneous intersubunit rotation in single ribosomes. Mol. Cell 30, 578–588 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuwirth B. S., et al. , Structures of the bacterial ribosome at 3.5 A resolution. Science 310, 827–834 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Vila-Sanjurjo A., et al. , X-ray crystal structures of the WT and a hyper-accurate ribosome from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 100, 8682–8687 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W., Dunkle J. A., Cate J. H., Structures of the ribosome in intermediate states of ratcheting. Science 325, 1014–1017 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmed T., Shi J., Bhushan S., Unique localization of the plastid-specific ribosomal proteins in the chloroplast ribosome small subunit provides mechanistic insights into the chloroplastic translation. Nucleic Acids Res. 45, 8581–8595 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jha V., et al. , Structural basis of sequestration of the anti-Shine-Dalgarno sequence in the Bacteroidetes ribosome. Nucleic Acids Res. 49, 547–567 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer N., Konevega A. L., Wintermeyer W., Rodnina M. V., Stark H., Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature 466, 329–333 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Flygaard R. K., Boegholm N., Yusupov M., Jenner L. B., Cryo-EM structure of the hibernating Thermus thermophilus 100S ribosome reveals a protein-mediated dimerization mechanism. Nat. Commun. 9, 4179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy E. L., et al. , Cryo-electron microscopy structure of the 70S ribosome from Enterococcus faecalis. Sci. Rep. 10, 16301 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Divya P., et al. , Rapid and scalable wire-bar strategy for coating of TiO2 thin-films: Effect of post-annealing temperatures on structures and catalytic dye-degradation. Molecules 25, 1683 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhaskar A., Chang H. Y., Chang T. H., Cheng S. Y., Effect of microwave annealing temperatures on lead zirconate titanate thin films. Nanotechnology 18, 395704 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Singh R., “Heat treatment of steels” in Applied Welding Engineering, Singh R., Ed. (Butterworth-Heinemann, Boston, 2012), chap. 12, pp. 95–108. [Google Scholar]
- 25.Pace B., Campbell L. L., Correlation of maximal growth temperature and ribosome heat stability. Proc. Natl. Acad. Sci. U.S.A. 57, 1110–1116 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cammarano P., et al. , Secondary structure features of ribosomal RNA species within intact ribosomal subunits and efficiency of RNA-protein interactions in thermoacidophilic (Caldariella acidophila, Bacillus acidocaldarius) and mesophilic (Escherichia coli) bacteria. Biochim. Biophys. Acta 740, 300–312 (1983). [DOI] [PubMed] [Google Scholar]
- 27.Chen C. Y., Chang Y. C., Lin B. L., Huang C. H., Tsai M. D., Temperature-resolved cryo-EM uncovers structural bases of temperature-dependent enzyme functions. J. Am. Chem. Soc. 141, 19983–19987 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Singh A. K., et al. , Structural basis of temperature sensation by the TRP channel TRPV3. Nat. Struct. Mol. Biol. 26, 994–998 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tishchenko S., et al. , High-resolution crystal structure of the isolated ribosomal L1 stalk. Acta Crystallogr. D Biol. Crystallogr. 68, 1051–1057 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Cornish P. V., et al. , Following movement of the L1 stalk between three functional states in single ribosomes. Proc. Natl. Acad. Sci. U.S.A. 106, 2571–2576 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fei J., Kosuri P., MacDougall D. D., Gonzalez R. L. Jr., Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol. Cell 30, 348–359 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Dunkle J. A., et al. , Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 332, 981–984 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brilot A. F., Korostelev A. A., Ermolenko D. N., Grigorieff N., Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc. Natl. Acad. Sci. U.S.A. 110, 20994–20999 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J., et al. , Coupling of mRNA structure rearrangement to ribosome movement during bypassing of non-coding regions. Cell 163, 1267–1280 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dashti A., et al. , Retrieving functional pathways of biomolecules from single-particle snapshots. Nat. Commun. 11, 4734 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luger K., Rechsteiner T. J., Richmond T. J., Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304, 3–19 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Flaus A., Luger K., Tan S., Richmond T. J., Mapping nucleosome position at single base-pair resolution by using site-directed hydroxyl radicals. Proc. Natl. Acad. Sci. U.S.A. 93, 1370–1375 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarkar C. A., et al. , Directed evolution of a G protein-coupled receptor for expression, stability, and binding selectivity. Proc. Natl. Acad. Sci. U.S.A. 105, 14808–14813 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lebon G., Bennett K., Jazayeri A., Tate C. G., Thermostabilisation of an agonist-bound conformation of the human adenosine A(2A) receptor. J. Mol. Biol. 409, 298–310 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu T., et al. , Enhancing protein stability with extended disulfide bonds. Proc. Natl. Acad. Sci. U.S.A. 113, 5910–5915 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zavodszky M., et al. , Disulfide bond effects on protein stability: Designed variants of Cucurbita maxima trypsin inhibitor-V. Protein Sci. 10, 149–160 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenbaum D. M., et al. , GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science 318, 1266–1273 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Cherezov V., et al. , High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science 318, 1258–1265 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rasmussen S. G., et al. , Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature 450, 383–387 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Stark H., GraFix: Stabilization of fragile macromolecular complexes for single particle cryo-EM. Methods Enzymol. 481, 109–126 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Kastner B., et al. , GraFix: Sample preparation for single-particle electron cryomicroscopy. Nat. Methods 5, 53–55 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Nakane T., Kimanius D., Lindahl E., Scheres S. H., Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. eLife 7, e36861 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Punjani A., Fleet D. J., 3D variability analysis: Resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol. 213, 107702 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Scheres S. H., Processing of structurally heterogeneous cryo-EM data in RELION. Methods Enzymol. 579, 125–157 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Chen B., Shen B., Frank J., Particle migration analysis in iterative classification of cryo-EM single-particle data. J. Struct. Biol. 188, 267–273 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lerner E., et al. , Toward dynamic structural biology: Two decades of single-molecule Förster resonance energy transfer. Science 359, eaan1133 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mazal H., Haran G., Single-molecule FRET methods to study the dynamics of proteins at work. Curr. Opin. Biomed. Eng. 12, 8–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sirinakis G., Ren Y., Gao Y., Xi Z., Zhang Y., Combined versatile high-resolution optical tweezers and single-molecule fluorescence microscopy. Rev. Sci. Instrum. 83, 093708 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin J., Gagnon M. G., Bulkley D., Steitz T. A., Conformational changes of elongation factor G on the ribosome during tRNA translocation. Cell 160, 219–227 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blaha G., et al. , Preparation of functional ribosomal complexes and effect of buffer conditions on tRNA positions observed by cryoelectron microscopy. Methods Enzymol. 317, 292–309 (2000). [DOI] [PubMed] [Google Scholar]
- 56.Thompson R. F., Iadanza M. G., Hesketh E. L., Rawson S., Ranson N. A., Collection, pre-processing and on-the-fly analysis of data for high-resolution, single-particle cryo-electron microscopy. Nat. Protoc. 14, 100–118 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.James N. R., Brown A., Gordiyenko Y., Ramakrishnan V., Translational termination without a stop codon. Science 354, 1437–1440 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lindert S., Meiler J., McCammon J. A., Iterative molecular dynamics-Rosetta protein structure refinement protocol to improve model quality. J. Chem. Theory Comput. 9, 3843–3847 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pettersen E. F., et al. , UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Kucukelbir A., Sigworth F. J., Tagare H. D., Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dashti A., et al. , Trajectories of the ribosome as a Brownian nanomachine. Proc. Natl. Acad. Sci. U.S.A. 111, 17492–17497 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zivanov J., et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Subramanian A. R., Glutaraldehyde fixation of ribosomes. Its use in the analysis of ribosome dissociation. Biochemistry 11, 2710–2714 (1972). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM density maps of ribosomes under all conditions (S1 through S10) were deposited in the Electron Microscopy Data Bank (EMDB) with accession IDs EMD-31266, EMD-31267, EMD-31268, EMD-31269, EMD-31270, EMD-31271, EMD-31272, EMD-31273, EMD-31274, and EMD-31275, as listed in SI Appendix, Fig. S2B. The locally refined cryo-EM maps of the 30S subunits from the unannealed (S1) and annealed (S5) ribosomes were also deposited in the EMDB with the respective accession IDs EMD-31277 and EMD-31279. Cryo-EM density maps and models for all 50 states in the free-energy landscape analysis are available on the Electron Microscopy Public Image Archive (EMPIAR-China) with the accession ID 100003. The codes for the local resolution analysis and modified ManifoldEM have been uploaded to GitHub: https://github.com/soothing35/cryoEM_annealling. All other study data are included in the article and/or supporting information.