Significance
A major contributor to heart valve disease is the excessive buildup of scar-like tissue in the valve, which can hinder the ability of the valve to open and close and can ultimately lead to heart failure. Controlling this scar tissue remodeling is very difficult due, in part, to a complex cellular regulation system and, in part, to large variabilities between different patients. We have built a computational model of the cell biochemical network that regulates valve remodeling, which enables virtual predictions of valve scarring given patient-specific biochemical levels. With this model, we ran personalized drug screens to predict each patient’s response to particular therapies, and follow-up cell culture experiments validated our predictions with over 80% accuracy.
Keywords: personalized medicine, signaling network, heart valve, myofibroblast, computational model
Abstract
Aortic valve stenosis (AVS) patients experience pathogenic valve leaflet stiffening due to excessive extracellular matrix (ECM) remodeling. Numerous microenvironmental cues influence pathogenic expression of ECM remodeling genes in tissue-resident valvular myofibroblasts, and the regulation of complex myofibroblast signaling networks depends on patient-specific extracellular factors. Here, we combined a manually curated myofibroblast signaling network with a data-driven transcription factor network to predict patient-specific myofibroblast gene expression signatures and drug responses. Using transcriptomic data from myofibroblasts cultured with AVS patient sera, we produced a large-scale, logic-gated differential equation model in which 11 biochemical and biomechanical signals were transduced via a network of 334 signaling and transcription reactions to accurately predict the expression of 27 fibrosis-related genes. Correlations were found between personalized model-predicted gene expression and AVS patient echocardiography data, suggesting links between fibrosis-related signaling and patient-specific AVS severity. Further, global network perturbation analyses revealed signaling molecules with the most influence over network-wide activity, including endothelin 1 (ET1), interleukin 6 (IL6), and transforming growth factor β (TGFβ), along with downstream mediators c-Jun N-terminal kinase (JNK), signal transducer and activator of transcription (STAT), and reactive oxygen species (ROS). Lastly, we performed virtual drug screening to identify patient-specific drug responses, which were experimentally validated via fibrotic gene expression measurements in valvular interstitial cells cultured with AVS patient sera and treated with or without bosentan—a clinically approved ET1 receptor inhibitor. In sum, our work advances the ability of computational approaches to provide a mechanistic basis for clinical decisions including patient stratification and personalized drug screening.
Fibrotic tissue remodeling is a significant driver of many pathologies, including heart failure, pulmonary fibrosis, wound healing, kidney disease, cancer, and others. But therapies to control fibrotic remodeling remain elusive due, in large part, to the complex intracellular signaling pathways and transcriptional networks that regulate the expression activities of the principal fibrosis-inducing cell type: the myofibroblast. For example, valvular remodeling is a major obstacle for the treatment of patients suffering from aortic valve stenosis (AVS). AVS is characterized by thickening of aortic valve leaflets via progressive extracellular matrix (ECM) accumulation and calcification, reducing blood flow through the valve and leading to compensatory cardiac hypertrophy, myocardial fibrosis, and, eventually, heart failure (1). Minimally invasive transcatheter aortic valve replacements (TAVR) serve as the current gold standard for treating severe AVS patients with high surgical risk (2). Unfortunately, interstitial fibrosis prior to surgery is not necessarily reversed after valve replacement, and high degrees of fibrosis are associated with reductions in long-term survival and cardiac function (3–5). Therefore, preventing valve fibrosis during initial AVS progression remains crucial for preserving heart function and preventing the late onset of heart failure. Identifying appropriate therapeutic compounds for this task remains a significant clinical need (6).
As mediators of ECM remodeling, valvular myofibroblasts play key roles during AVS progression, synthesizing matrix proteins, proteases such as matrix metalloproteinases (MMPs), and other regulatory matricellular proteins that orchestrate scar formation and increase tissue stiffness (7). Valvular interstitial cells (VICs) undergo activation to a myofibroblast phenotype in response to an influx of inflammatory cytokines, growth factors, and hormonal peptides, as well as changes in local biomechanical cues, including pathological stiffness and tensile loading (8–10). These extracellular cues are transduced through a complex network of regulation in which multiple intracellular signaling pathways interact via cross-talk and feedback mechanisms to dictate overall cell responses (11), thus further complicating efforts to understand how myofibroblasts respond within the valve microenvironment. While previous studies have shown that myofibroblasts can undergo deactivation in response to serum biomarkers after TAVR, macrophage-secreted inflammatory cues, and local mechanical stiffness (12–16), the complexities of valve myofibroblast signal transduction among a diverse microenvironment complicates efforts to control cell deactivation.
To account for the complex system of myofibroblast intracellular signaling, we employed a computational model in which environmental cues are transduced via a network of intracellular signaling and transcriptional interactions to predict changes in protein expression. Similar approaches have been used to model various cardiovascular and systemic diseases and successfully generate insight into influential mechanisms within this broad biological scope (17–21). In our current work, we adapted these approaches to model valve myofibroblast responses to patient-specific serum biomarker levels before and after TAVR by combining a network of myofibroblast signaling with a data-driven network of myofibroblast transcriptional regulation. We developed network reaction topology and fit reaction parameters using VIC RNA-sequencing data under various patient-matched serum conditions, then tested the correlations of personalized model predictions to clinically measured, patient-specific AVS severity. In addition, we conducted network perturbation analyses and patient-specific drug target screens, followed by in vitro validation experiments to test the model’s ability to predict variable drug responses across diverse patient sera contexts.
Results
Development of Myofibroblast Transcriptional Network.
Using a published dataset of valvular myofibroblast gene expression in response to pre- and post-TAVR patient sera (12), we developed a model of valve myofibroblast transcriptional regulation using established gene regulatory network inference algorithms. This dataset pairs proteomic signatures of patient sera collected prior to surgery and 1 mo postprocedure with RNA-sequencing signatures of porcine VICs treated with individual patient sera from both time points. The authors of this study found that altering protein composition after TAVR deactivated cultured VICs from a myofibroblast phenotype, suggesting that increases in inflammatory factors in circulating plasma can rescue resident cells from a profibrotic state. This study design provided a platform to unravel mechanisms of VIC deactivation through changes in patient proteomes after surgical replacement, and the use of computational networks can build on this insight in a predictive manner by prioritizing therapeutic strategies to rescue VIC activation before the need for valve replacement.
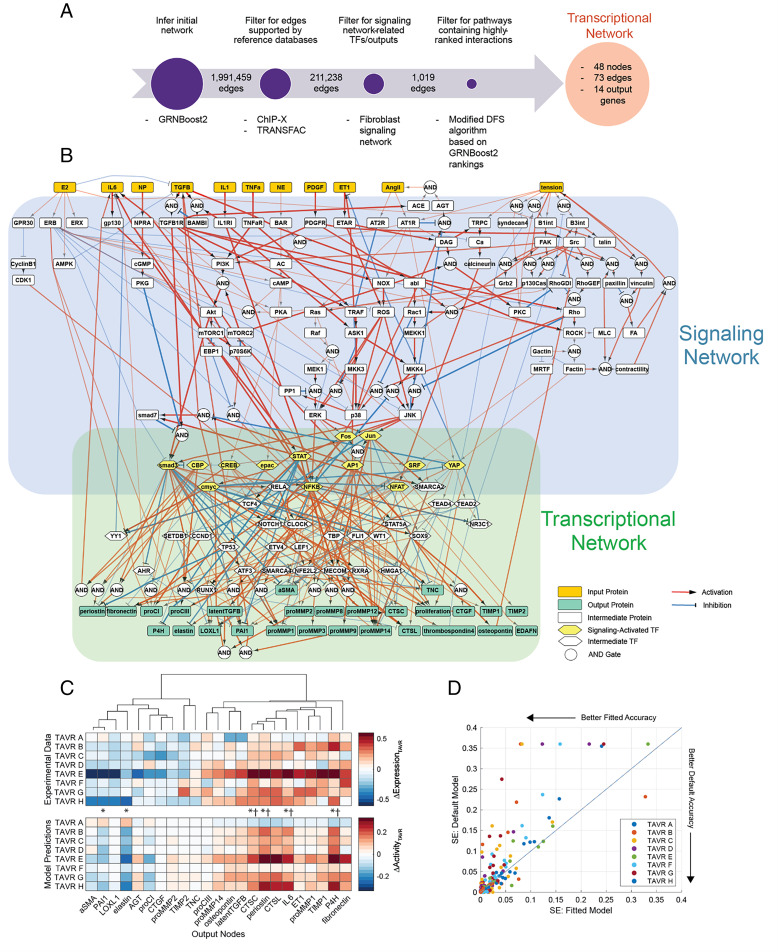
Upon initial topology inference and implementing several filtering steps to ensure sufficient experimental evidence and relation to fibrosis-related gene expression (Fig. 1A), our transcriptional network represents 10 transcription factors (TFs) activated by intracellular signaling, 27 intermediate TFs, and 18 target genes related to ECM turnover or autocrine feedback (SI Appendix, Fig. S1). This network inference strategy identified mechanisms mediating the expression of structural matrix proteins, matricellular proteins, and remodeling proteins related to collagen processing, cross-linking, and turnover, which is supported by substantial overlap with genes and pathways differentially expressed across normal, fibrotic, and calcific regions in aortic valve leaflet tissues obtained from AVS patients (22). Additionally, this data-driven approach identified several intermediate TFs shown to regulate valvular and ventricular myofibroblast behavior, including SOX9 (23, 24), CCND1 (25), ATF3 (26, 27), and NOTCH1 (28), suggesting agreement between inferred topology and previous experimental findings.
Fig. 1.
Composite valve myofibroblast regulatory network accurately predicts fibrosis-related protein expression based on patient-specific biochemical stimuli. (A) Schematic of gene regulatory network inference and model pruning scheme resulting in a targeted myofibroblast transcriptional network. (B) Topology of combined network consisting of a curated network of myofibroblast signaling and an inferred transcriptional network derived from valve myofibroblast RNA sequencing. Biochemical and biomechanical input nodes, signaling intermediates, and cell-secreted output nodes are connected by directed edges representing activating and inhibiting reactions with reaction logic indicated by circular nodes (AND gates) or multiple edges (OR gates). Line widths represent average reaction weight parameters estimated via genetic algorithm. (C) Comparison of experimentally measured changes in output gene expression between pre- and post-TAVR conditions (ΔExpressionTAVR) with model-predicted changes in output protein activity between pre- and post-TAVR conditions (ΔActivityTAVR). Individual comparisons made for each patient (boxes) represent mean levels across an ensemble of estimated parameter sets, and statistical comparisons of activity levels between pre- and post-TAVR groups were conducted via two-tailed Student’s t tests. *P < 0.05, †P < 0.05 for model predictions and experimental values. (D) Comparison of squared errors (SE) of predicted patient ΔActivityTAVR levels between fitted model (x axis) and a default model (y axis). All SE values were calculated against experimental ΔExpressionTAVR levels.
Composite Myofibroblast Network Accurately Predicts Gene Expression.
Valve myofibroblast activation and expression of ECM components during AVS relies not only on regulatory mechanisms at the transcriptional level but also on changes in intracellular signaling that often involves cross-talk between multiple pathways. To provide a predictive solution that accounts for both levels of regulation simultaneously, we integrated the inferred transcriptional network above with our published network of myofibroblast signaling, incorporating signaling reactions and TF–target interactions into a single composite network for modeling via a system of ordinary differential equations (ODEs) (Fig. 1B). This composite network, consisting of 151 nodes and 334 edges in total, predicted the expression of 27 fibrosis-related proteins (output nodes) based on 11 canonical biochemical and mechanical stimuli (input nodes), providing a detailed footprint of valve myofibroblast responses to local environmental cues. This network was implemented as a system of logic-based ODEs as previously described (29), in which normalized node activity levels are approximated via Hill equations using normalized node and reaction parameters (see Materials and Methods for a full description).
We then contextualized this network to patient conditions before and after TAVR and maximized predictive capabilities of myofibroblast behavior during AVS progression. We implemented a genetic algorithm to estimate the relative weight parameters of all network reactions based on a published multiomic dataset consisting of proteomic profiling of pre- and post-TAVR patient sera and RNA sequencing of myofibroblasts cultured with patient-specific serum (12). An ensemble of parameter sets was estimated using serum proteomic and myofibroblast transcriptomic datasets as analogs for model inputs and outputs, respectively, and the resulting fitted network emphasized downstream signaling pathways from transforming growth factor-β (TGFβ), endothelin-1 (ET1), tumor necrosis factor-α (TNFα), and interleukins 1 and 6 (IL1 and IL6) as well as signal transducer and activator of transcription (STAT)- and nuclear factor kappa B (NFKB)-mediated transcriptional regulation (Fig. 1B).
We compared model predictions of myofibroblast-expressed proteins to measured gene expression levels by simulating cell responses to each patient’s pre- or post-TAVR serum conditions and measuring changes in output activity between pre- and post-TAVR conditions (ΔActivityTAVR). Trends in model predictions matched changes in gene expression experimentally observed in pre- and post-TAVR serum-treated myofibroblasts (ΔExpressionTAVR), with significant decreases in plasminogen activator inhibitor-1 (PAI1) and elastin activity along with significant increases in CTSC, CTSL, P4H, periostin, and IL6 activity mirroring significant changes in respective gene expression before and after TAVR (Fig. 1C). Across the ensemble of estimated parameter sets, the average mean-squared error (MSE) between ΔActivityTAVR and ΔExpressionTAVR levels for each patient and model output was 0.0329 ± 0.0531, reaching a level similar to other fitted mechanistic and logic-based networks (30, 31).
We additionally assessed the accuracy of our parameter estimation strategy against a default model without fitted reaction weight parameters (i.e., all reactions weight parameters set to one) by comparing ΔActivityTAVR levels for each model against the previous ΔExpressionTAVR levels. We found that parameter estimation improved predictive accuracy for individual outputs with respect to each patient serum, as fitted model ΔActivityTAVR levels for individual patients and outputs had lower squared errors against respective ΔExpressionTAVR levels compared to the default model (Fig. 1D). Moreover, comparisons of significantly altered output levels between pre- and post-TAVR conditions as predicted by the two models and as measured in myofibroblasts suggests a sizeable improvement in predictive accuracy. We found that, while the default model had a classification accuracy of 43.5% in predicting truly significant and nonsignificant changes in output activity compared to gene expression data, the fitted model improved the accuracy to 69.6%, thus predicting a higher proportion of truly significant and nonsignificant changes in output levels than in the default, unfitted model.
Model-Predicted ECM Expression Correlates with Clinical Measures of AVS Severity.
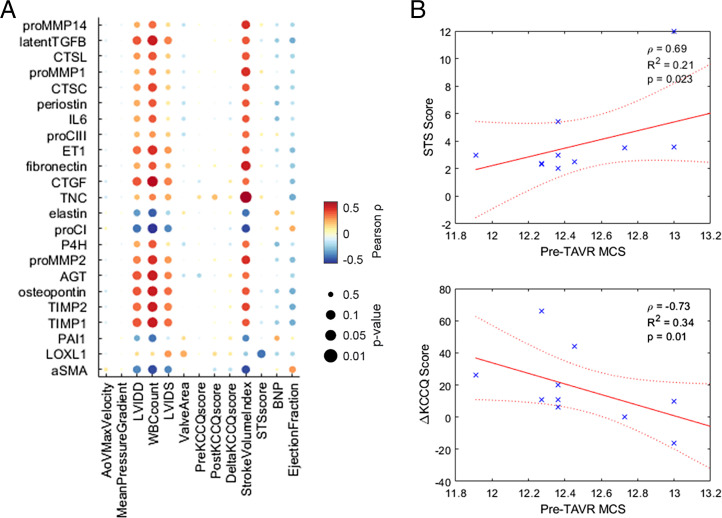
The relationship between valve fibrosis and reduced cardiac function during AVS is well known, but linking individual secreted proteins to changes in valve and heart function remains challenging because scarring involves a variety of matrix, protease, matricellular, and cross-linking proteins for overall changes in tissue architecture. Our myofibroblast network can account for many of the species involved in valve remodeling, so we hypothesized that comparing model-predicted trends in myofibroblast protein expression to patient-matched clinical measurements could identify strongly correlated proteins among a diverse protein expression profile. We performed a systematic correlational analysis between activity levels of 23 outputs predicted for each pre-TAVR patient serum and 13 clinical features measured prior to surgery, utilizing serum levels of the patient cohort used for model training as well as levels for four additional patients as a validation set (Fig. 2A). Several model-predicted outputs demonstrated significant positive correlations with left ventricle (LV) volume measurements (e.g., LV internal diameter at end diastole, LV internal diameter at end systole, and stroke volume index), including protease inhibitors TIMP1 and TIMP2, matricellular proteins osteopontin and tenascin C, and autocrine feedback signals latent TGFβ, CTGF, ET1, and angiotensinogen. These associations suggest that high levels of feedback signaling and matricellular protein expression persist for patients with worse valve function and greater severity of AVS, although the underlying mechanisms remain unclear.
Fig. 2.
Model-predicted protein expression correlates with clinical measures of disease severity. (A) Correlation analysis of pre-TAVR output node activity levels (rows) with clinical features measured before and after TAVR surgery (columns). All dots represent Pearson correlation coefficients and respective P values. (B) Notable correlations between aggregated MCS and clinical features. Spearman correlation coefficients (ρ) and P values were used to determine significance. Stars represent each patient's data point, solid line represents linear correlation fit, and dotted lines denote confidence intervals.
We also aggregated individual output predictions into a single matrix content score (MCS) via a rank-based procedure as an analog for net matrix accumulation for each patient (see Materials and Methods for a full description). Notable correlations include pre-TAVR MCS values with pre-TAVR Society of Thoracic Surgeons risk scores (STS score), and pre- to post-TAVR changes in Kansas City Cardiomyopathy Questionnaire score (ΔKCCQ score), which show that simulated ECM production is predictive of declines in ventricular function, overall disease severity, and negative postsurgical outcomes (Fig. 2B).
Network Perturbation Analysis Reveals Influential Pathways Mediating Fibrosis during AVS.
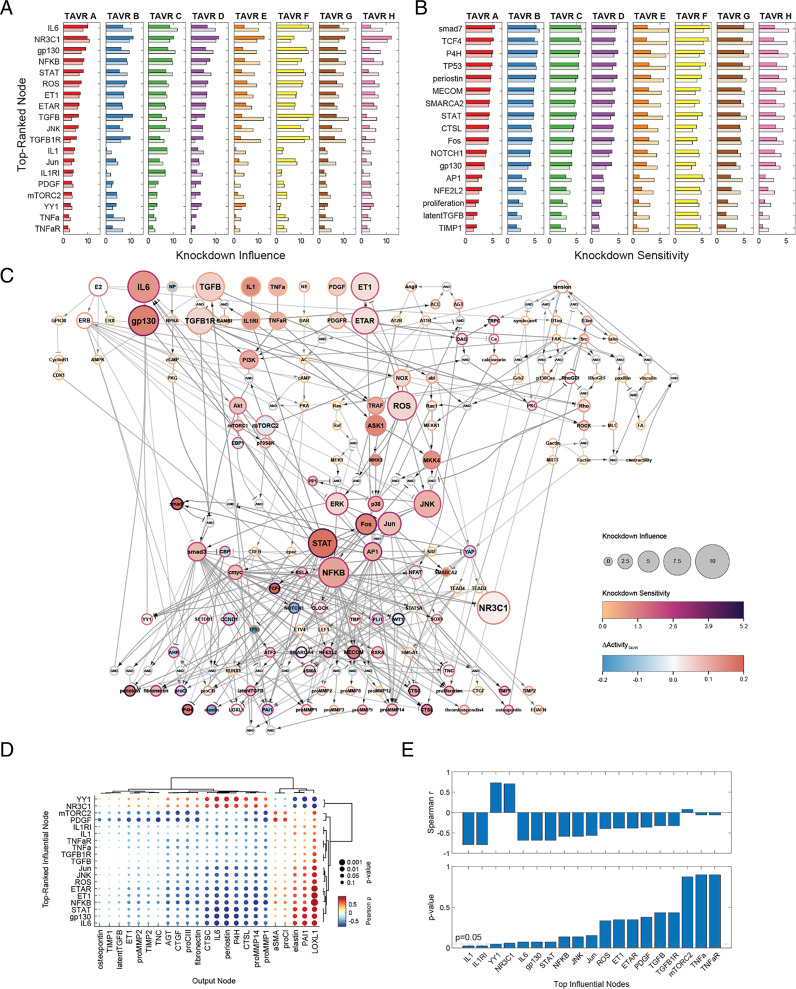
A key barrier to understanding myofibroblast activation during AVS is a lack of insight into the relative contributions of individual signaling molecules across the full, multipathway network. Thus, we performed comprehensive knockdown simulations of individual nodes for all individual patient serum levels, measuring changes in network activity with each knockdown (ΔActivityKD) to assess network-wide effects in a patient-specific manner. We calculated a knockdown influence level for each node as a metric for the total effects that one node has on all other nodes, and we calculated a knockdown sensitivity level for each node as a metric for the total change in activity for one node across the knockdown of all other nodes (see Materials and Methods for a full description). Selection of the top 10 scoring nodes in both metrics for each pre- and post-TAVR serum condition showed similar trends in node sensitivity and influence across patients, with a total of 19 nodes ranking in the top 10 knockdown influence and 17 nodes ranking in the top 10 knockdown sensitivity for at least one of the eight patients (Fig. 3 A and B, and see SI Appendix, Fig. S2A for network-wide sensitivity analysis).
Fig. 3.
Network perturbation analysis reveals influential pathways and differences in myofibroblast responses across patient conditions. (A and B) Comparison of top-ranked influential and sensitive nodes between patients and pre/post-TAVR conditions. Nodes shown reflect all unique nodes ranked in the top 10 based on knockdown influence (A) or knockdown sensitivity (B). Dark bars represent knockdown influence/sensitivity levels for each pre-TAVR patient condition, and light bars represent levels for each post-TAVR patient condition. (C) Topological representation of influential and sensitive pathways within the myofibroblast network. Knockdown influence/sensitivity levels and ΔActivityTAVR levels represent average values across all patient and pre- and post-TAVR conditions. (D) Correlational analysis between pre-TAVR knockdown influence levels for top-ranked nodes and ΔActivityTAVR levels for model outputs. All dots represent Pearson correlation coefficients and respective P values. (E) Correlational analysis between knockdown influence levels for top-ranked nodes and predicted ΔMCS levels between pre- and post-TAVR conditions.
Across all patient and serum conditions, several intracellular signaling pathways were consistently influential with respect to network-wide activity. Among the top influential pathways represented were IL6 signaling via gp130/STAT activation, canonical and noncanonical TGFβ signaling via smad3 and ROS, extracellular signal-regulated kinase (ERK) and JNK activation, and ET1 signaling via ROS/ERK/JNK activation along with minor influence via IL1, TNFα, and PDGF signaling (Fig. 3C). Nodes downstream from these pathways consistently scored highly in knockdown sensitivity, including targets of IL6 signaling such as smad7, TCF4, TP53, and NOTCH1 as well as targets of TGFβ/ET1 signaling such as AP1, NFKB, cmyc, and MECOM. These influential and sensitive species coincide with sizeable changes in activity between pre- and post-TAVR conditions, as average ΔActivityTAVR levels across all patients indicate up-regulation of IL6 pathway activation after TAVR and large increases or decreases in downstream activity, including connected TFs and target output nodes (Fig. 3C).
We next compared knockdown influence and sensitivity levels for top-ranked nodes across patient-specific conditions to assess differences both between individual patients and between pre/post-TAVR conditions. We found that influential nodes representing IL1 and TGFβ signaling pathways fluctuated between the patient cohort, as select patient conditions showed large network-wide changes in activity compared to others (Fig. 3A), and calculated coefficients of variation across patient conditions were largest for IL1- and TGFβ-associated nodes out of all top-ranked nodes (SI Appendix, Fig. S2B). While patient-specific differences in knockdown sensitivity levels were not as pronounced and coefficients of variation were generally lower (SI Appendix, Fig. S2C), overall knockdown sensitivity levels did vary by patient, with some patients showing little sensitivity across all top-ranked nodes compared to others (Fig. 3B). We also observed differences in influence and sensitivity from pre-TAVR to post-TAVR conditions across top-ranked nodes. Knockdown influence levels for IL6 signaling nodes increased for post-TAVR conditions compared to pre-TAVR conditions for seven of eight patients, which was consistent with increases in IL6-related activity during post-TAVR simulations.
As an additional exploratory analysis of influential pathway effects on myofibroblast behavior, we correlated pre-TAVR knockdown influence levels of top-ranked nodes with ΔActivityTAVR levels for all fibrosis-related outputs to discover relationships between pre-TAVR conditions and changes in cell behavior with surgical intervention. A total of 82 out of 437 correlations between pre-TAVR influence and output ΔActivityTAVR levels were significant, including 27 positive correlations and 55 negative correlations (Fig. 3D). Within these pairings, influence levels of IL6-associated nodes and ET1-associated nodes significantly correlated with nodes that ranked highly in knockdown sensitivity (Fig. 3D). Positive correlations with elastin, PAI1, and LOXL1 as well as negative correlations with multiple matrix proteases, periostin, and P4H suggest strong connections between pre-TAVR pathway knockdown and changes in protease, protease inhibitor, and collagen cross-linking protein expression over other ECM-related proteins. We additionally correlated pre-TAVR influence levels with changes in MCS values from pre-TAVR to post-TAVR (ΔMCS) as a metric for overall changes in myofibroblast-mediated matrix turnover, and we found that knockdown influence of the IL1- and ET1-associated nodes positively correlated with changes in matrix content for patients (Fig. 3E), suggesting that these pathways may contribute to overall changes in myofibroblast protein secretion within the larger network.
Targeted Virtual Drug Screen Predicts Stratified Patient Responses to Therapies.
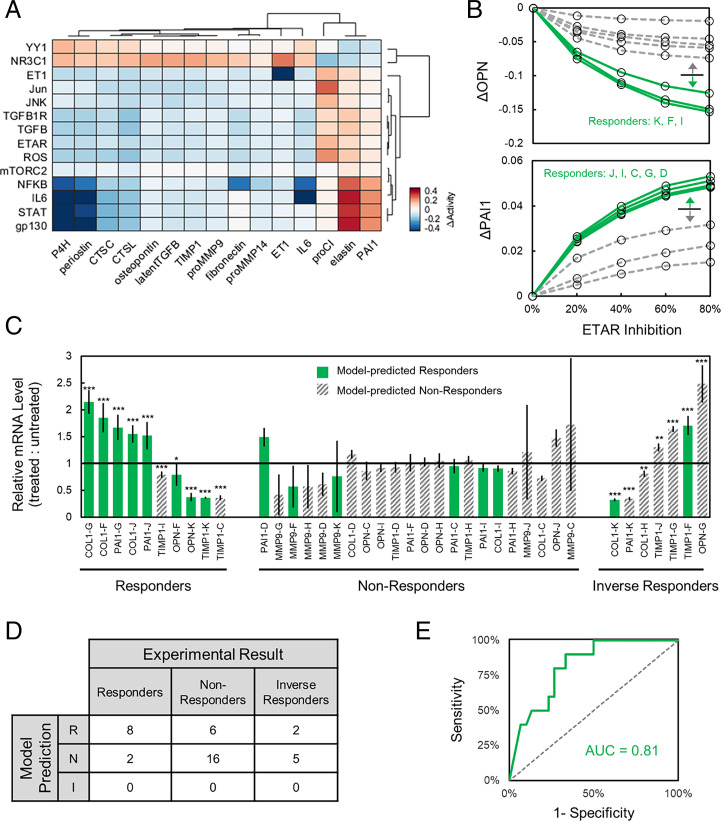
Our perturbation analyses above suggested that, while some signaling pathways tend to mediate valve myofibroblast protein expression over others, responses of individual patients to pathway perturbations can vary widely, with some patients experiencing little to no benefit for a given therapy. This variability is a common challenge for cardiovascular therapy development, so we investigated whether our network model could discern patient-specific responses to drugs by predicting personalized changes in matrix-related outputs. Inhibition of each node was performed for each patient’s pre-TAVR conditions in a dose–response manner by limiting the maximum activation of each node, and average changes in activity of output nodes (ΔActivityinhib) were measured for each patient relative to unperturbed levels. This knockdown analysis identified the 14 most influential nodes (Fig. 4A, rows) and the 15 most sensitive outputs (Fig. 4A, columns), on average, but also demonstrated wide distributions of patient responses to each perturbation. As an example, inhibiting endothelin-1 receptor (ETAR) produced either minimal, moderate, or substantial reductions in OPN production as well as minimal, moderate, or substantial increases in PAI1 production, depending upon the particular patient serum context (Fig. 4B). In addition, different patient backgrounds exhibited different response sensitivity, depending on the protein output of interest.
Fig. 4.
Virtual drug screen reveals stratified patient responses to inhibitor treatments. (A) Comparison of ΔActivityinhib levels for fibrosis-related model outputs upon knockdown of top-ranked influential nodes. Values represent average ΔActivityinhib levels across all patients for pre-TAVR conditions after 80% inhibition of each node relative to respective unperturbed conditions, and rows/columns were chosen for knockdowns that demonstrated an average 5% change in activity for at least five outputs and vice versa. (B) Simulated changes in OPN and PAI1 output activity for each patient with 0 to 80% inhibition of ETAR. Patients were designated as “Responder” or “Non-Responder” based on threshold classification of delta levels at 80% inhibition. (C) Relative in vitro gene expression levels of OPN, PAI1, COL1, MMP9, and TIMP1 across valve interstitial cell populations treated with patient-specific sera and ETAR inhibition with bosentan. Gene expression was measured via qPCR relative to RPL30 for cells dosed with individual patient sera for 24 h followed by 10 µM bosentan for 24 h, and statistical significance is shown as *P < 0.05, **P < 0.01, ***P < 0.001. Each experimental observation was designated as a responder (i.e., statistically significant effect of bosentan in the same direction as the model prediction), an inverse responder (i.e., statistically significant effect of bosentan in the opposite direction as the model prediction), or a nonresponder (i.e., statistically insignificant effect of bosentan in any direction). (D) Confusion matrix comparing model predictions of patient responder/nonresponder/inverse responder status compared to that determined via qPCR experimental results. (E) Receiver operating characteristic curve comparing demonstrated predictive accuracy of model predictions vs. in vitro patient-specific response status.
To experimentally validate the predictive accuracy of our model to capture patient-specific drug responses, we subjected porcine VIC populations to the ETAR inhibitor bosentan across eight patient-specific serum culture conditions (see Materials and Methods for a full description). Briefly, we cultured VICs on poly(ethylene glycol) hydrogels that served as precision biomaterials (32) and then exposed the cell-laden hydrogels to individual patient sera with or without 10 µM bosentan. Previous studies have demonstrated that VICs cultured on soft hydrogels retain a quiescent phenotype in the presence of serum, whereas the use of standard tissue culture plastic may activate VICs to a myofibroblast phenotype, due, in part, to the supraphysiological stiffness of tissue culture plastic (32, 33). After 24 h of drug treatment, we collected messenger RNA (mRNA) and measured OPN, PAI1, COL1, MMP9, and TIMP1 expression levels using RT-PCR (Fig. 4C). Patients were first classified as “responders” or “nonresponders” for each protein output, based on computationally predicted effects of ETAR 80% knockdown and a simple over–under threshold. Computational model-based classifications across patient backgrounds and genes were compared to experimental results for patient backgrounds and genes by designating each experimental result as a responder (i.e., statistically significant effect of bosentan in the same direction as the model prediction), an inverse responder (i.e., statistically significant effect of bosentan in the opposite direction from the model prediction), or a nonresponder (i.e., statistically insignificant effect of bosentan in any direction). The model-based stratification showed substantial predictive power, with 8 of 10 experimental responders correctly identified in the computational responder group, while only 6 of 16 nonresponders and only 2 of 7 inverse responders were incorrectly classified by the model as responders (Fig. 4 D and E). Varying the model stratification threshold produced a full receiver operating characteristic curve with an area under the curve of 0.81.
Discussion
Preventing valve fibrosis during AVS is crucial for long-term patient outcomes even after valve replacement interventions. Although previous studies have shown that alterations in serum protein levels after valve replacement can deactivate myofibroblasts (12, 13), the underlying mechanisms of this deactivation remain unclear due, in part, to the complexities of intracellular regulatory mechanisms. Using coupled serum proteomic and valve myofibroblast transcriptomic datasets for patients undergoing TAVR, we developed a strategy to integrate these patient-specific data into a computational model of cell signaling and transcriptional regulation to predict fibrosis-related protein expression in response to environmental signals. We found that this strategy improved the accuracy of patient-specific predictions over simulations based on model topology alone and resulted in a similar degree of accuracy against experimental gene expression data compared to other fitted network models (30, 31). Correlational analyses demonstrated strong trends between both individual matrix protein predictions and aggregated MCSs with clinical measures of disease severity and patient quality of life. Network-wide perturbation analyses identified influential signaling pathways and transcriptional regulators of network activity and output expression such as the IL6–gp130–STAT, TGFβ–ROS–JNK, and ET1–ROS–ERK/JNK signaling axes. We additionally demonstrated the capacity of the model to detect stratified patient responses to targeted inhibition in a virtual drug screen, showing that myofibroblasts respond substantially to simulated inhibitors for select patient conditions over others. Our results suggest that this strategy can accelerate the identification of antifibrotic therapies to slow AVS progression by accounting for individual patient conditions and generating insight into possible mechanisms governing cell behavior within a complex signaling context.
Identification of Transcriptional Regulators in Myofibroblast-Mediated Fibrosis.
While intracellular signaling processes remain a crucial step in transducing environmental cues into changes in cell protein expression and phenotype, regulation at a transcriptional level adds an additional layer of complexity and can confound understanding of cell behavior. Our construction and simulation of a myofibroblast transcriptional network derived from cell RNA-sequencing data suggest that this layer of regulation plays a highly influential role in myofibroblast behavior during AVS progression. Transcriptional regulators identified here via gene regulatory network inference have also been shown to mediate both VIC and cardiac fibroblast behavior in other cardiovascular pathologies. The TF SOX9 has been identified as a positive regulator of cardiac fibrosis and heart failure development in models of ischemia–reperfusion injury and myocardial infarction (MI), with ischemia causing increased SOX9 gene expression and spatial correlation with collagen gene expression and scar formation measured via histology (23). Fibroblast-specific deletion of SOX9 in an MI mouse model has also been shown to attenuate increases in αSMA expression, cell proliferation, scar formation, and functional measures. These data demonstrate a strong connection between this factor and fibrotic processes.
NOTCH1 and ATF3 have also been implicated in cardiac fibrosis and myofibroblast activation, as overexpression of the NOTCH1 intracellular domain in rat cardiac fibroblasts antagonized TGFβ-induced smad3 activation and in vivo overexpression after MI reduced fibrotic area in rats (28). Further, cardiac fibroblast-specific overexpression of ATF3 attenuated scar formation and reduced the LV internal diameter in end systole and ejection fraction after MI (27). It should be noted that several regulators identified by our approach serve influential roles in osteogenic differentiation, such as NOTCH1 (34–36). Although the scope of our model does not directly reflect osteogenic phenotypes for valvular cells, this similarity in regulatory species suggests that pathways mediating myofibroblast and osteoblast phenotypes are not mutually exclusive. Our identification of transcriptional intermediates and connecting interactions suggests that the regulation of myofibroblast behavior involves numerous pathways simultaneously, and that computational approaches are advantageous for discerning mechanisms essential to cell function within a dense regulatory landscape.
Influential Signaling Axes and Regulatory Hubs Mediate Overall Myofibroblast Behavior.
Valve myofibroblasts respond to a variety of environmental stimuli, including inflammatory cytokines, growth factors, matrix stiffness, and hormonal peptides, all of which have been shown to alter cell behavior individually (7, 37). Within the context of AVS progression, however, the relative contribution of each environmental signal and its respective signaling pathway remains unclear. We performed a network-wide perturbation analysis to identify globally influential pathways with cells cultured in the presence of pre-TAVR and post-TAVR serum, and our simulations predicted that the transduction of IL6, TGFβ, and ET1 primarily influence activation levels across the network. Previous loss- and gain-of-function studies conducted both in vitro and in vivo have demonstrated the influence of these top nodes in fibrosis during AVS. Liu and Gotlieb (38) reported extensive changes in VIC phenotypes when exposed to exogenous TGFβ using an in vitro wound repair assay, as treated cells showed up-regulated αSMA expression, stress fiber formation, and wound closure in addition to attenuated apoptosis. Treatment with a TGFβ-neutralizing antibody showed attenuation across these categories, suggesting TGFβ as a major regulatory mechanism of VIC activation. A similar extent of behavioral change was observed by Jenke et al. (39) in a three-dimensional culture model of ovine VICs, as TGFβ treatment increased smad3/7 gene expression, latent TGFβ expression, cell proliferation via CCND1, and αSMA/COL1 expression. We additionally identified the glucocorticoid receptor NR3C1 as a top influencer of network-wide behavior, and, while no study has investigated glucocorticoid-mediated phenotypes in valvular cells, both a genetic loss-of-function study in mice and a gain-of-function study in dogs via the glucocorticoid prednisolone demonstrated that changes in NR3C1 activity promoted interstitial left ventricular fibrosis and up-regulated fibrotic gene expression (40, 41).
IL6 signaling and downstream activation of STAT has been previously linked to cardiac fibrosis across several cardiovascular pathologies with effects across multiple pathways (42). Dziemidowicz et al. (43) found that IL6−/− mice treated with isoproterenol developed interstitial ventricular fibrosis and increased phosphorylated levels of multiple signaling intermediates including ERK, Raf, and p38 compared to wild-type mice. In a similar finding, Hilfiker-Kleiner et al. (44) found decreases in ERK and Akt phosphorylation for mice carrying a point mutation in gp130 and subjected to MI compared to wild-type mice. Recent evidence of IL6 as a promoter of VIC osteogenic differentiation via the increased expression of RUNX2 and osteopontin suggests that this pathway may serve multiple phenotypic roles (13), and future targeted investigations of this behavior may explain possible context-dependent roles of IL6 signaling during AVS progression.
Noncanonical TGFβ signaling via NADPH oxidases (NOX) and ROS has also been previously revealed as an effective regulator of fibrosis in valvular and myocardial tissue, supporting the high influence that we found from the TGFβ–ROS–JNK axis in our computational network. Inhibition of NOX4 in cardiac fibroblasts has been shown to reduce TGFβ-stimulated intracellular superoxide production, which, in turn, mediates phosphorylation of smad2/3 and expression of αSMA, CTGF, and fibronectin, suggesting a largely profibrotic role of this pathway (45). The NOX–ROS signaling axis has also been shown to mediate changes across multiple pathways to alter fibrosis in mouse models of hypertension (46), and decreases in NOX4 expression and ROS generation following genetic knockout of SOD1 in mice mitral valves has been associated with changes in αSMA, CTGF, and MMP expression in mitral VICs (47), suggesting a similar systemic effect of this noncanonical mechanism on valvular myofibroblast behavior.
Our global analysis additionally suggests that select TFs may act as regulatory hubs for expression of ECM-related proteins and influence fibrotic behavior over others. We found that STAT, NFKB, and Jun influence large changes in network-wide activity, all of which have been well studied for their roles in pathological scar formation. In particular, we found STAT to be a primary influencer of myofibroblast behavior via the expression of matricellular proteins, cathepsin and MMP proteases, and cross-linking enzymes, and previous experimental studies have confirmed this central role of STAT in cardiac fibrosis and systemic sclerosis. Using IL-11 as a positive regulator of STAT3 after MI in mice, Obana et al. (48) demonstrated that STAT3 activation reduced LV scar formation and improved infarct wall thicknesses compared to untreated MI, demonstrating a primarily antifibrotic effect during ischemic injury. However, disruption of STAT3 phosphorylation via inhibition of EphrinB2 in a post-MI mouse model reduced αSMA protein expression, collagen I expression, and cell proliferation (49), and inhibition of STAT3 during bleomycin-induced skin fibrosis reduced gene expression of multiple ECM proteins (50), suggesting that STAT may exert both profibrotic and antifibrotic effects. In agreement with experimental evidence of IL6 signaling during AVS, the activation of STAT3 was also associated with osteogenic dedifferentiation in VICs via down-regulation of RUNX2 and BMP-2 (51), reinforcing the need to further investigate this intersection between fibrotic and osteogenic phenotypes.
Systems Models Relate Cell Phenotypes to Disease Severity and Predict Patient Responses.
Our model-centered approach provides unique advantages for relating cellular-level behavior to clinical manifestations of AVS through the prediction of protein expression based on patient-specific conditions. Our findings indicate that computational predictions of cell behavior correspond to multiple measures of AVS severity, including LV size and STS scores. Previous serum biomarker studies in AVS cohorts have shown individual biomarkers to be predictive of patient outcomes, such as N-terminal pro B-type natriuretic peptide (NT-proBNP) and C-reactive protein (52, 53), and, while useful for stratifying patient risk and informing surgical intervention, these biomarkers alone may not connect alterations in cellular function with overall disease progression. Our patient-specific predictions and correlation with pre-TAVR clinical measurements suggest that matricellular protein expression, autocrine feedback expression, and aggregation of all output proteins into a single ECM-related metric offer a predictive link between myofibroblast-mediated scar formation and reduced heart function. Virtual drug screens for individual patient conditions further demonstrated the ability of this model-based approach to discern responders versus nonresponders to possible therapies. Translating this approach into a clinical setting may improve overall responses to therapeutic intervention before and after surgery, via targeted drug and/or dose selection. While clinical validation of these trends is essential for further development and application of this approach, the framework developed here provides opportunities to further investigate fibrotic mechanisms of action for targeted patient cohorts or for other possible pathological scenarios.
Study Limitations.
Our study was primarily limited by the size of patient datasets used for model training and simulated scenarios. While this multiomic, patient-matched design formed an essential aspect of our model development strategy through the robust measurement of serum proteins and cell-specific gene expression, a lack of cohort size and diversity limited our predictive capabilities in terms of both model training and predicted trends in myofibroblast behavior. Additionally, the insufficient availability of select biochemical stimuli (e.g., estrogen), tissue biomechanical properties, and cell-secreted outputs limited our predictions related to these stimuli and associated regulatory pathways. In particular, the signaling network model was curated with substantial mechanotransduction pathways enabling the incorporation of local biomechanical signals such as tissue deformations and stiffness as an additional input which was not leveraged in these current simulations.
Throughout our analysis, we did not find patient sex to be an influential factor in VIC responses to perturbation, as neither knockdown experiments in silico nor drug-induced gene expression in vitro showed substantial changes in behavior in cell behavior compared to those induced by knockdown or drug inhibition. While we did consider patient sex as a factor in our experimental design both through the incorporation of estrogen transduction in our network model and through the application of male and female sera to sex-matched VIC cultures, insufficient availability of both estrogenic biomarkers and female patients may be a factor in this discrepancy compared to previous studies demonstrating estrogen and sex dependence of fibrotic signaling pathways during AVS (54). Additionally, the young, healthy porcine VICs used for experimental validation may elicit differences in response compared to native, diseased VICs during AVS progression, and further experimental models representing aged and diseased cells are needed for full validation of our findings.
Because both fibrosis and calcification processes play key roles in the development of AVS, we acknowledge that our model of myofibroblast protein expression alone accounts for only one component mediating overall disease progression. Future adaptation of this modeling strategy toward osteogenic differentiation may provide additional insight into dominating mechanisms responsible for calcification and complement our current model of fibrosis-related gene expression for an overall picture of cell behavior. Because several influential pathways identified here also correspond to the expression of calcification-related proteins, implementing these same pathways along with those known to mediate severe AVS may be valuable for investigating differences in fibrotic versus osteogenic differentiation. Notably, recent transcriptomic and proteomic analyses across normal, fibrotic, and calcific disease stages of aortic valve leaflets from AVS patients have implicated important, stage-specific changes in the production levels of collagens, MMPs, TIMPs, proteoglycans, and most of the related signaling pathways included in our model (22).
Conclusion.
Our development of a cell regulatory network and analysis of patient-specific changes in valve myofibroblast protein expression demonstrated the translational possibilities of model-based approaches in identifying key mechanisms governing cell behavior within a larger signaling context. Predicted intracellular species and pathways influencing fibrosis-related gene expression provide targeted hypotheses for future clinical validation. Our findings of strong correlations between model-predicted protein expression and clinical measures of disease severity suggest that simulations of biological networks can be advantageous for relating cellular phenotypes to clinical manifestations of disease, and future exploration of the predictive capabilities of such models could prove useful for clinical translation. Our virtual drug screen of individual patient responses to pathway-inhibiting drugs further demonstrated the ability of this model-based approach to stratify patient cohorts based on individual serum biomarker levels. This strategy provides a framework for predicting cell behavior within a complex signaling context and enables further investigation into pathways mediating valvular fibrosis before and after surgical intervention.
Materials and Methods
Fibroblast Transcriptional Network Construction.
Gene regulatory network inference.
We employed a previously published method of gene regulatory network inference to derive a network of TF–target gene interactions based on a transcriptomic dataset of VIC populations after stimulation with patient sera. Bulk RNA-sequencing datasets recently published by our collaborators were used as a training set for network inference, and this dataset represents the gene expression of VIC populations stimulated with serum derived from a small cohort of patients undergoing a TAVR procedure (12). Briefly, patient blood samples were collected at the time of surgery and at the 1-mo follow-up visit (n = 4 female patient pairs and n = 8 male patient pairs, 24 samples total). Donors were 79.1 ± 8.4 y of age, on average, and all donors had a pre-TAVR aortic valve area of <1.5 cm2 (0.73 ± 0.24 cm2), suggesting either a moderate or severe stage of stenosis. The resulting serum samples were used to treat porcine VIC cultures seeded on soft poly(ethylene glycol) (PEG) hydrogels (Young’s modulus of 5.8 kPa) for 48 h prior to RNA sequencing, and counts per million for gene expression were used for subsequent network inference and model fitting methods. Of the total 24 samples collected, 16 samples were used for gene regulatory network inference and network parameter estimation (n = 4 female patient pairs and n = 4 male patient pairs), and 8 samples were retained for model validation only (n = 4 male patient pairs).
We utilized the GRNBoost2 machine learning algorithm to infer a network of TF–target interactions from the transcriptomic dataset above. This regression-based method is based on the GENIE3 (gene network inference with ensemble of trees) algorithm for predicting regulatory links between input genes and target genes via the construction of decision tree ensembles (54). Each ensemble of decision trees, which predicts the expression of a given target gene from the expression of all input genes, is used to determine the relative “importance” of each input gene in predicting the expression of the specified target gene. Decision tree ensembles are built for all genes across the transcriptome, and input–target gene links are aggregated to form a composite network of ranked interactions. The GENIE3 algorithm has been shown to outperform other methods in inferring gene regulatory networks, as part of the dialogue for reverse engineering assessments and methods (DREAM4) in silico multifactorial challenge (55), and it provides several advantages over other common inference algorithms: 1) Inference can be performed with minimal assumptions of network topology, 2) directed interactions (i.e., gene A activates gene B) can be inferred compared to correlation- and probability-based methods, and 3) nonlinear or combinatorial regulation can be derived compared to other regression-based methods (56). The GRNBoost2 implementation optimizes this approach using stochastic gradient boosting, which grows decision tree ensembles on a subset of observations and estimates the loss function on the remaining observations with each iteration. The algorithm implements an early stopping criterion if the loss function does not improve above a set threshold, thereby preventing unnecessary iterations of each decision tree and reducing overall computational time (57). The Arboreto library for python was used to apply this algorithm to the RNA-sequencing data described above, and the average computational time for network inference using a four-core computer was ∼10 min.
Network pruning.
Upon inference of the initial network, a three-step workflow was applied to filter the network for TF–target interactions that satisfy three requirements: 1) Interactions must be supported by experimental evidence, 2) interactions must relate to either literature-supported TFs (primary TFs) or fibrosis-related target genes in myofibroblasts, and 3) resulting pathways connecting primary TFs and target genes must have relatively strong links across all individual edges, as determined by interaction ranks assigned by the GRNBoost2 algorithm (Fig. 1A). The first step was performed by comparing inferred TF–target interactions with databases of known TF–target interactions aggregated from chromatin immunoprecipitation (ChIP) studies. Curated lists of known TF–target interactions from the ChIP-X Enrichment Analysis and transcription factor (TRANSFAC) databases were downloaded using the Harmonizome web interface (58), both of which were chosen to maximize coverage of TFs related to myofibroblast activation. Upon filtering the initial gene regulatory network for interactions contained in either database, the resulting network was filtered further for interactions containing TFs or fibrosis-related target genes in myofibroblasts. Lists of TFs and target genes were derived using our curated network model of myofibroblast signaling (59), which contains 11 primary TFs and 20 target genes coding for ECM-related proteins. For TFs that consist of several subunits (e.g., activator protein 1 [AP1] complex), both constituent genes were included for filtering. An additional eight target genes that were differentially expressed by patients between pre- and post-TAVR sera were also considered: CTSC, CTSL, COL4A5, LOXL1, P4HA1, P4HA3, LAMA4, and ELN. Proteins coded by these genes have been shown to alter matrix degradation and collagen processing, and alter material properties of cardiac tissue, and significant differences in expression within the RNA-sequencing dataset provide a rationale for exploring possible regulatory pathways affecting gene expression. After list construction, the database-filtered network above was filtered again for interactions containing genes in either list. After filtering, interactions contained only TF–TF interactions or TF–target interactions in which intermediate TFs not included in the primary TF list above regulate target gene expression (i.e., primary TF A activates secondary TF B, which activates target gene C).
After the second stage of filtering above, the final network topology was derived by ensuring that all resulting pathways between primary TFs and target genes contained interactions that ranked highly among possible regulatory links according to the GRNBoost2 algorithm. A modified depth-first search algorithm was implemented to find all possible pathways between each primary TF and target gene and check whether each interaction within that pathway met this requirement using the “importance” score for the interaction output by GRNBoost2. Individual interactions were only allowed if the importance score of each edge was greater than either a threshold of one or the 75th percentile of all interactions stemming from the same TF. This hybrid threshold was chosen to both limit interactions driven by noise, in which overall importance scores are low, and prevent premature exclusion of related interactions when all possible regulatory links may be ranked low relative to the entire network. By implementing this method, all interactions mediating expression of target genes were ensured to meet a threshold of confidence relative to neighboring interactions such that one TF within a pathway is not predictive of its downstream target. All network filtering steps were performed in a python environment using the numpy (60) and pandas (61) packages.
Composite Signaling/Transcriptional Network Implementation.
We combined the final transcriptional network with our previous myofibroblast signaling network describing intracellular mechanotransduction and chemotransduction (59) to form a composite network capable of predicting fibrosis-related protein expression in response to mechanical and biochemical stimuli. New TFs (model nodes) and/or transcriptional reactions (edges) were added to the cell signaling topology if they were not redundant to the original transcriptional reactions described by the signaling network. To account for potential differences in signaling between male/female cell signaling during fibrosis, we incorporated a curated set of intracellular reactions encompassing estrogen transduction found via a manual literature search. Reactions were included if at least two independent studies contained experimental evidence in either VICs or cardiac myofibroblasts, resulting in an additional seven nodes and 24 edges added to the network.
The final network was implemented as a system of logic-based ODEs in which activity levels of all nodes were modeled by Hill equations. Logical NOT, AND, and OR gates were used for complex signaling interactions by applying the respective logical operations: for NOT gates, for AND gates, and for OR gates. The open source Netflux package for MATLAB was used to build this system of differential equations (28), and all simulations were conducted using MATLAB (Mathworks). All visualizations of network topology were constructed using Cytoscape (29, 62, 63).
Network Parameter Estimation.
Full details of our parameter estimation are included in SI Appendix, Supplementary Methods. To summarize, we first used k-means clustering based on a global sensitivity analysis to group closely connected reactions (thereby reducing the total number of parameter constraints). We then implemented a genetic fitting algorithm to optimize the reaction weight parameters (w) of all reactions in the composite network. Using serum protein levels for all patients as input concentrations and VIC gene expression levels as output concentrations, data for each input and output node were normalized and fit with the ga MATLAB function by minimizing the mean-squared error between the model-predicted and experimentally measured changes in pre-TAVR to post-TAVR expression levels.
Network Perturbation Analysis.
To identify influential signaling mechanisms across pre-TAVR and post-TAVR signaling contexts, a series of node knockdowns were simulated using normalized input levels from each patient serum sample. For each set of normalized input levels used during parameter estimation above, basal conditions (i.e., without any knockdown) were applied for 80 h, followed by knockdown of individual nodes using the Ymax parameter (Ymax,KD = 0.1*Ymax,basal) for 240 h. Steady-state activity levels of all nodes were measured with each knockdown, and changes in node activity (ΔActivityKD) were calculated as the difference between node activity after knockdown and basal node activity. Knockdown sensitivity of each node was calculated as the sum of absolute ΔActivityKD levels for the node across all knockdown simulations, and knockdown influence of each node was calculated as the sum of absolute ΔActivityKD levels for all other nodes in the network upon knockdown.
Patient Stratification Analysis.
Targeted simulations assessing stratified model responses to drug targets with patient-specific conditions were conducted using a series of dose–response simulations. For each node, a series of knockdown simulations was performed under each patient-specific condition. Following a simulation of basal conditions using each patient’s normalized input levels for 80 h, the Ymax parameter for each node was lowered to either 0.8, 0.6, 0.4, or 0.2 times the Ymax under basal conditions for 240 h (corresponding to 20 to 80% node inhibition). Steady-state activity levels of all nodes were measured with each dose and compared to steady-state levels prior to dosing. Patients were then classified as “responder” or “nonresponder” based on a simple over–under threshold, which was varied from zero to the maximum delta for each protein output to generate receiver operating characteristic curves.
In Vitro Validation Experiments.
Full details of our in vitro experiments and all statistical analyses are included in SI Appendix, Supplementary Methods. Briefly, PEG hydrogels were made and seeded with primary male or female porcine VICs as previously described (12, 64). VICs were seeded on PEG hydrogels at a density of 20,000 cells per square centimeter growth area in Media 199 supplemented with 1% serum (fetal bovine serum or human serum samples), 50 U/mL penicillin, 50 μg/mL streptomycin, and 1 μg/mL amphotericin B. In order to consider sex-specific VIC gene expression, VICs were sex matched to each human serum sample such that female sera were applied to female VICs and vice versa. RNA was extracted from cultured cells using a RNeasy Micro Kit (Qiagen) according to the manufacturer’s protocol. RNA quality was assessed via spectrophotometry (ND-1000, NanoDrop), and complementary DNA was synthesized using an iScript Synthesis kit (Bio-Rad) according to the manufacturer’s protocol. Relative mRNA expression was determined using SYBR Green reagents on an iCycler (Bio-Rad). Normalizations were performed using the RPL30 gene.
Supplementary Material
Acknowledgments
B.A.A. acknowledges funding from the NIH (Award R00HL148542) and the Burroughs Wellcome Fund Postdoctoral Enrichment Program; K.S.A. acknowledges funding from the NIH (Awards R01HL132353 and R01HL142935); and W.J.R. acknowledges funding from the NIH (Awards R01HL144927 and P20GM121342).
Footnotes
Reviewers: K.M., University of Wisconsin–Madison; and K.Y., Cincinnati Children's Hospital Medical Center.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2117323119/-/DCSupplemental.
Data Availability
The code generated during this study is available in the GitHub repository (https://github.com/jdroger/Fibroblast_Gene_Regulatory_Network). The published article includes all models generated or analyzed during this study. The experimental data generated from the in vitro VIC culture experiments and mRNA measurements are available in the Figshare repository (https://figshare.com/articles/dataset/Rogers_2022_NetworkModelingInVitroData_xlsx/19127951) (65).
References
- 1.Dweck M. R., Boon N. A., Newby D. E., Calcific aortic stenosis: A disease of the valve and the myocardium. J. Am. Coll. Cardiol. 60, 1854–1863 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Durko A. P., Osnabrugge R. L., Kappetein A. P., Long-term outlook for transcatheter aortic valve replacement. Trends Cardiovasc. Med. 28, 174–183 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Azevedo C. F., et al. , Prognostic significance of myocardial fibrosis quantification by histopathology and magnetic resonance imaging in patients with severe aortic valve disease. J. Am. Coll. Cardiol. 56, 278–287 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Milano A. D., et al. , Prognostic value of myocardial fibrosis in patients with severe aortic valve stenosis. J. Thorac. Cardiovasc. Surg. 144, 830–837 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Yarbrough W. M., Mukherjee R., Ikonomidis J. S., Zile M. R., Spinale F. G., Myocardial remodeling with aortic stenosis and after aortic valve replacement: Mechanisms and future prognostic implications. J. Thorac. Cardiovasc. Surg. 143, 656–664 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borer J. S., Sharma A., Drug therapy for heart valve diseases. Circulation 132, 1038–1045 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H., Leinwand L. A., Anseth K. S., Cardiac valve cells and their microenvironment—Insights from in vitro studies. Nat. Rev. Cardiol. 11, 715–727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yip C. Y. Y., Simmons C. A., The aortic valve microenvironment and its role in calcific aortic valve disease. Cardiovasc. Pathol. 20, 177–182 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Huang H. Y. S., Liao J., Sacks M. S., In-situ deformation of the aortic valve interstitial cell nucleus under diastolic loading. J. Biomech. Eng. 129, 880–889 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Rogers J., Holmes J., Saucerman J., Richardson W., Mechano-chemo signaling interactions modulate matrix production by cardiac fibroblasts. Matrix Biol.Plus 10, 100055 (2020). [DOI] [PMC free article] [PubMed]
- 11.Davis J., Molkentin J. D., Myofibroblasts: Trust your heart and let fate decide. J. Mol. Cell. Cardiol. 70, 9–18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aguado B. A., et al. , Transcatheter aortic valve replacements alter circulating serum factors to mediate myofibroblast deactivation. Sci. Transl. Med. 11, eaav3233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grim J. C., et al. , Secreted factors from proinflammatory macrophages promote an osteoblast-like phenotype in valvular interstitial cells. Arterioscler. Thromb. Vasc. Biol. 40, e296–e308 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kural M. H., Billiar K. L., Myofibroblast persistence with real-time changes in boundary stiffness. Acta Biomater. 32, 223–230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H., Haeger S. M., Kloxin A. M., Leinwand L. A., Anseth K. S., Redirecting valvular myofibroblasts into dormant fibroblasts through light-mediated reduction in substrate modulus. PLoS One 7, e39969 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kloxin A. M., Benton J. A., Anseth K. S., In situ elasticity modulation with dynamic substrates to direct cell phenotype. Biomaterials 31, 1–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeigler A. C., Richardson W. J., Holmes J. W., Saucerman J. J., A computational model of cardiac fibroblast signaling predicts context-dependent drivers of myofibroblast differentiation. J. Mol. Cell. Cardiol. 94, 72–81 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan P. M., Buchholz K. S., Omens J. H., McCulloch A. D., Saucerman J. J., Predictive model identifies key network regulators of cardiomyocyte mechano-signaling. PLOS Comput. Biol. 13, e1005854 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irons L., Humphrey J. D., Cell signaling model for arterial mechanobiology. PLOS Comput. Biol. 16, e1008161 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osmanbeyoglu H. U., Pelossof R., Bromberg J. F., Leslie C. S., Linking signaling pathways to transcriptional programs in breast cancer. Genome Res. 24, 1869–1880 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeigler A. C., Richardson W. J., Holmes J. W., Saucerman J. J., Computational modeling of cardiac fibroblasts and fibrosis. J. Mol. Cell. Cardiol. 93, 73–83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlotter F., et al. , Spatiotemporal multi-omics mapping generates a molecular atlas of the aortic valve and reveals networks driving disease. Circulation 138, 377–393 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lacraz G. P. A., et al. , Tomo-seq identifies SOX9 as a key regulator of cardiac fibrosis during ischemic injury. Circulation 136, 1396–1409 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Scharf G. M., et al. , Inactivation of Sox9 in fibroblasts reduces cardiac fibrosis and inflammation. JCI Insight 5, e126721 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenke A., et al. , Transforming growth factor-β1 promotes fibrosis but attenuates calcification of valvular tissue applied as a three-dimensional calcific aortic valve disease model. Am. J. Physiol. Heart Circ. Physiol. 319, H1123–H1141 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Sharma V., Dogra N., Saikia U. N., Khullar M., Transcriptional regulation of endothelial-to-mesenchymal transition in cardiac fibrosis: Role of myocardin-related transcription factor A and activating transcription factor 3. Can. J. Physiol. Pharmacol. 95, 1263–1270 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Li Y., et al. , Cardiac fibroblast-specific activating transcription factor 3 protects against heart failure by suppressing MAP2K3-p38 signaling. Circulation 135, 2041–2057 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou X. L., et al. , Notch signaling inhibits cardiac fibroblast to myofibroblast transformation by antagonizing TGF-β1/Smad3 signaling. J. Cell. Physiol. 234, 8834–8845 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Kraeutler M. J., Soltis A. R., Saucerman J. J., Modeling cardiac β-adrenergic signaling with normalized-Hill differential equations: Comparison with a biochemical model. BMC Syst. Biol. 4, 157 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schroer A. K., Ryzhova L. M., Merryman W. D., Network modeling approach to predict myofibroblast differentiation. Cell. Mol. Bioeng. 7, 446–459 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris M. K., Saez-Rodriguez J., Clarke D. C., Sorger P. K., Lauffenburger D. A., Training signaling pathway maps to biochemical data with constrained fuzzy logic: Quantitative analysis of liver cell responses to inflammatory stimuli. PLOS Comput. Biol. 7, e1001099 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aguado B. A., Grim J. C., Rosales A. M., Watson-Capps J. J., Anseth K. S., Engineering precision biomaterials for personalized medicine. Sci. Transl. Med. 10, eaam8645 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H., Tibbitt M. W., Langer S. J., Leinwand L. A., Anseth K. S., Hydrogels preserve native phenotypes of valvular fibroblasts through an elasticity-regulated PI3K/AKT pathway. Proc. Natl. Acad. Sci. U.S.A. 110, 19336–19341 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng Q., et al. , Notch1 promotes the pro-osteogenic response of human aortic valve interstitial cells via modulation of ERK1/2 and nuclear factor-κB activation. Arterioscler. Thromb. Vasc. Biol. 33, 1580–1590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J., et al. , Notch1 mutation leads to valvular calcification through enhanced myofibroblast mechanotransduction. Arterioscler. Thromb. Vasc. Biol. 35, 1597–1605 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menon V., Lincoln J., The genetic regulation of aortic valve development and calcific disease. Front. Cardiovasc. Med. 5, 162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rutkovskiy A., et al. , Valve interstitial cells: The key to understanding the pathophysiology of heart valve calcification. J. Am. Heart Assoc. 6, e006339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu A. C., Gotlieb A. I., Transforming growth factor-β regulates in vitro heart valve repair by activated valve interstitial cells. Am. J. Pathol. 173, 1275–1285 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jenke A., et al. , Transforming growth factor-β1 promotes fibrosis but attenuates calcification of valvular tissue applied as a three-dimensional calcific aortic valve disease model. Am. J. Physiol. Heart Circ. Physiol. 319, H1123–H1141 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Richardson R. V., et al. , Glucocorticoid receptor alters isovolumetric contraction and restrains cardiac fibrosis. J. Endocrinol. 232, 437–450 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka S., et al. , Long-term administration of prednisolone: Effects on the myocardial tissue of healthy beagle dogs. J. Vet. Med. Sci. 83, 84–93 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fischer P., Hilfiker-Kleiner D., Survival pathways in hypertrophy and heart failure: The gp130-STAT3 axis. Basic Res. Cardiol. 102, 279–297 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Dziemidowicz M., et al. , The role of interleukin-6 in intracellular signal transduction after chronic β-adrenergic stimulation in mouse myocardium. Arch. Med. Sci. 15, 1565–1575 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D. Hilfiker-Kleineret al., . Continuous glycoprotein-130-mediated signal transducer and activator of transcription-3 activation promotes inflammation, left ventricular rupture, and adverse outcome in subacute myocardial infarction. Circulation 122, 145–155 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Cucoranu I., et al. , NAD(P)H oxidase 4 mediates transforming growth factor-β1-induced differentiation of cardiac fibroblasts into myofibroblasts. Circ. Res. 97, 900–907 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Zhao Q. D., et al. , NADPH oxidase 4 induces cardiac fibrosis and hypertrophy through activating Akt/mTOR and NFκB signaling pathways. Circulation 131, 643–655 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hagler M. A., et al. , TGF-β signalling and reactive oxygen species drive fibrosis and matrix remodelling in myxomatous mitral valves. Cardiovasc. Res. 99, 175–184 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Obana M., et al. , Therapeutic activation of signal transducer and activator of transcription 3 by interleukin-11 ameliorates cardiac fibrosis after myocardial infarction. Circulation 121, 684–691 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Su S. A., et al. , EphrinB2 regulates cardiac fibrosis through modulating the interaction of Stat3 and TGF-β/Smad3 signaling. Circ. Res. 121, 617–627 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Chakraborty D., et al. , Activation of STAT3 integrates common profibrotic pathways to promote fibroblast activation and tissue fibrosis. Nat. Commun. 8, 1130 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deng X. S., Meng X., Song R., Fullerton D., Jaggers J., Rapamycin decreases the osteogenic response in aortic valve interstitial cells through the Stat3 pathway. Ann. Thorac. Surg. 102, 1229–1238 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Seoudy H., et al. , Periprocedural changes of NT-proBNP are associated with survival after transcatheter aortic valve implantation. J. Am. Heart Assoc. 8, e010876 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Imai K., et al. , C-reactive protein predicts severity, progression, and prognosis of asymptomatic aortic valve stenosis. Am. Heart J. 156, 713–718 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Sritharen Y., Enriquez-Sarano M., Schaff H. V., Casaclang-Verzosa G., Miller J. D., Pathophysiology of aortic valve stenosis: Is it both fibrocalcific and sex specific? Physiology (Bethesda) 32, 182–196 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huynh-Thu V. A., Irrthum A., Wehenkel L., Geurts P., Inferring regulatory networks from expression data using tree-based methods. PLoS One 5, e12776 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mercatelli D., Scalambra L., Triboli L., Ray F., Giorgi F. M., Gene regulatory network inference resources: A practical overview. Biochim. Biophys. Acta. Gene Regul. Mech. 1863, 194430 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Moerman T., et al. , GRNBoost2 and Arboreto: Efficient and scalable inference of gene regulatory networks. Bioinformatics 35, 2159–2161 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Rouillard A. D., et al. , The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016, baw100 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rogers J. D., Richardson W. J., Fibroblast mechanotransduction network predicts targets for mechano-adaptive infarct therapies. bioRxiv [Preprint] (2020). 10.1101/2020.08.13.250001 (Accessed 6 February 2022). [DOI] [PMC free article] [PubMed]
- 60.Harris C. R., et al. , Array programming with NumPy. Nature 585, 357–362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McKinney W., “Data structures for statistical computing in python” in Proceedings of the 9th Python in Science Conference, van der Walt S. J., Millman K. J., Eds. (SciPy, 2010), pp. 56–61.
- 62.Shannon P., et al. , Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeigler A. C., et al. , Computational model predicts paracrine and intracellular drivers of fibroblast phenotype after myocardial infarction. Matrix Biol. 91–92, 136–151 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mabry K. M., Lawrence R. L., Anseth K. S., Dynamic stiffening of poly(ethylene glycol)-based hydrogels to direct valvular interstitial cell phenotype in a three-dimensional environment. Biomaterials 49, 47–56 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.W. J. Richardson, Rogers+2022_NetworkModelingInVitroData.xlsx. Figshare. https://figshare.com/articles/dataset/Rogers_2022_NetworkModelingInVitroData_xlsx/19127951. Deposited 6 February 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code generated during this study is available in the GitHub repository (https://github.com/jdroger/Fibroblast_Gene_Regulatory_Network). The published article includes all models generated or analyzed during this study. The experimental data generated from the in vitro VIC culture experiments and mRNA measurements are available in the Figshare repository (https://figshare.com/articles/dataset/Rogers_2022_NetworkModelingInVitroData_xlsx/19127951) (65).