Significance
The Mycobacterium tuberculosis (Mtb) ESX-3 type VII secretion system plays a critical role in iron acquisition. Infection of mice with highly attenuated Mtb deletion mutants lacking esxG or esxH, genes encoding key ESX-3 substrates, unexpectedly yielded suppressor mutants with restored capacity to grow in vivo and in vitro in the absence of iron supplementation. Whole-genome sequencing identified two mechanisms of suppression, the disruption of a transcriptional repressor that regulates expression of an ESX-3 paralogous region encoding EsxR and EsxS, and a massive 38- to 60-fold gene amplification of this same region. These data are significant because they reveal a previously unrecognized iron acquisition regulon and inform mechanisms of Mtb chromosome evolution.
Keywords: Mycobacterium tuberculosis, type VII secretion system, ESX-3, genetic accordion, TetR-family transcriptional regulator
Abstract
Mycobacterium tuberculosis (Mtb) possesses five type VII secretion systems (T7SS), virulence determinants that include the secretion apparatus and associated secretion substrates. Mtb strains deleted for the genes encoding substrates of the ESX-3 T7SS, esxG or esxH, require iron supplementation for in vitro growth and are highly attenuated in vivo. In a subset of infected mice, suppressor mutants of esxG or esxH deletions were isolated, which enabled growth to high titers or restored virulence. Suppression was conferred by mechanisms that cause overexpression of an ESX-3 paralogous region that lacks genes for the secretion apparatus but encodes EsxR and EsxS, apparent ESX-3 orphan substrates that functionally compensate for the lack of EsxG or EsxH. The mechanisms include the disruption of a transcriptional repressor and a massive 38- to 60-fold gene amplification. These data identify an iron acquisition regulon, provide insight into T7SS, and reveal a mechanism of Mtb chromosome evolution involving “accordion-type” amplification.
The major human pathogen, Mycobacterium tuberculosis (Mtb), causes tuberculosis in 10.4 million people annually, resulting in 1.4 million deaths (1). Significant gaps remain in our understanding of Mtb pathogenesis. Type VII secretion systems (T7SS) are an important, but incompletely understood class of Mtb virulence determinants that have been explored as targets for novel chemotherapies (2) and manipulated to generate attenuated vaccines (3–5). Mtb encodes five T7SS in genomic loci, designated esx-1 through esx-5 (6). Each of these loci encodes the secretion apparatus itself and associated substrates, including pairs of small helical proteins belonging to the WXG100 family, containing a Trp-Xaa-Gly (WXG) motif (7), as well as members of the proline-glutamic acid (PE) and proline-proline-glutamic acid (PPE) families (8). ESX-1 is the best studied of the Mtb T7SS. It modulates host signaling pathways, bacterial trafficking, and promotes virulence (9–13). However, the mechanisms by which individual protein substrates contribute to ESX functions remain incompletely understood.
The esx-3 locus encodes the secretion apparatus as well as the WXG100 pair EsxG/EsxH and a pair of PE/PPE protein family members, PE5/PPE4 (6). The esx-3 gene cluster (rv0282–rv0292) is regulated by the iron-dependent transcriptional repressor, IdeR (14), the zinc uptake regulator, Zur (15), the manganese transport regulator, MntR (16), and in Mycobacterium bovis may respond to oxidative stress through interactions between the CmtR regulator and Zur (17). Genetic studies implicate ESX-3 in mycobacterial utilization of iron bound to the siderophores mycobactin and carboxymycobactin (18–23). Whereas esx-3 was previously thought to be essential for growth, esx-3 deletion strains are in fact recoverable in medium supplemented with various iron complexes, whether the deletions encompass individual esx-3 genes or the full operon (22). In addition, evidence from studies of deletion mutants suggests that ESX-3 also plays iron-independent roles in Mtb pathogenesis. Notably, EsxH interacts with the host cell endosomal sorting complex required for transport (ESCRT) machinery, impairing the capacity of effector CD4+ T cells to target infected macrophages and clear Mtb infection (24). Additionally, EsxH is a T cell antigen that may serve as an immunological decoy. In mouse models, EsxH elicits robust CD8+ T cell responses, which fail to effectively recognize Mtb-infected macrophages (25).
Previous work demonstrated that Mtb strains bearing deletions of the entire esx-3 region or of the esxH gene alone are highly attenuated in their capacity to cause disease in immunocompetent mice (22). Here, we describe how a subset of immunocompetent mice infected with ΔesxG/ΔesxH mutants exhibited unexpectedly higher bacterial burdens or early mortality. The bacteria recovered from these mice no longer required iron supplementation in the form of hemin or mycobactin for in vitro growth. Whole-genome sequencing (WGS) of the in vivo isolates identified two general classes of suppressor mutations associated with these phenotypes. These include mutations in the putative transcriptional repressor protein Rv3058c, which is thought to regulate expression of a locus encoding several PE/PPE proteins and EsxR/EsxS, a WXG100 pair with high identity to EsxH/EsxG. The second class was a remarkable 38- to 60-fold gene amplification of esxR/esxS and flanking pe/ppe genes, involving a tandem duplication of a 2,440-bp region. These data implicate Rv3058c and the locus encoding EsxR/EsxS as a previously unrecognized iron regulon and point to a remarkable genetic plasticity of Mtb, which allows it to overcome impediments to in vivo growth and virulence.
Results
Isolation of ΔesxG and ΔesxH Phenotypic Revertants In Vitro and In Vivo.
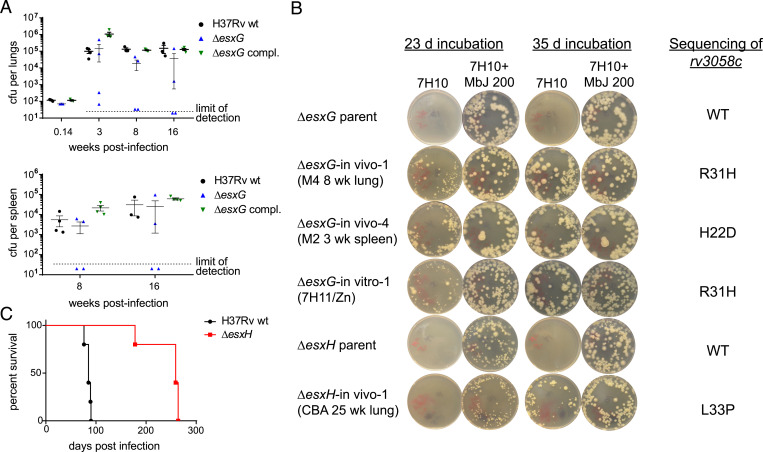
The in vitro iron-related growth phenotypes of Δesx-3, ΔesxH, and ΔesxG are similar in that none of the mutants grow effectively on standard 7H10 medium, and all require supplementation with iron complexes, such as mycobactin J or hemin, for growth (22). Furthermore, Mtb Δesx-3 region mutants and ΔesxH mutants are highly attenuated in vivo. Following aerosol infection of C57BL/6 mice, bacteria failed to proliferate in lungs, and dissemination to the spleen was not observed (22). Based on these findings, ΔesxG strains were expected to behave similarly after aerosol infection of C57BL/6 mice. Instead, we found that at 3 wk postinfection, two of four ΔesxG-infected mice had very low lung CFU levels (∼102 CFU), indicating the absence of growth, while the other two mice had bacterial burdens several logs higher, approximating those of mice infected with WT or ΔesxG-complemented strains (∼105 CFU) (Fig. 1A). A similar dichotomy was evident at the 8-wk and 16-wk harvests for the ΔesxG-infected mice. Dissemination to spleens was also observed in the ΔesxG-infected mice that exhibited elevated pulmonary bacterial burdens (Fig. 1A).
Fig. 1.
ΔesxG and ΔesxH recovered from mice form colonies on plain 7H10 and harbor point mutations in rv3058c. (A) C57BL/6 mice were infected by aerosol with H37Rv WT, the ΔesxG unmarked strain (mc27845), and ΔesxG complemented with an integrating vector expressing esxGH (mc27866). Lung CFU were determined at 24 h and at 3, 8, and 16 wk postinfection and spleen CFU were determined at 8 and 16 wk postinfection. n = 3 to 4 mice per group. Mean ± SEM are indicated. Dotted line denotes limit of detection. The H37Rv WT group CFU data were previously reported in comparison with ΔmbtB, ΔesxH, and Δpe5-ppe4 strains (22). The ΔesxG mutant and complemented strains tested in the present study were included in the same experiment but not previously reported. (B) The indicated isolates recovered after in vivo growth were plated onto 7H10 medium with and without 200 ng/mL mycobactin J, in comparison with parental strains. Results of sequencing analysis within the rv3058c gene as compared with the parental strain are shown at right. (C) CBA mice were intravenously infected with indicated strains and survival was monitored. n = 5 mice per group. The H37Rv WT group survival data were previously reported in comparison with ΔmbtB and Δpe5-ppe4 (22). The ΔesxH mutant tested in the present study was included in the same experiment but not previously reported.
Bacteria were recovered from the lungs and spleens of those ΔesxG-infected mice that exhibited high bacterial titers. These isolates were found to have acquired the capacity to grow on standard 7H10 medium in the absence of supplementation with mycobactin J (Fig. 1B). This growth phenotype contrasted with that of the ΔesxG and ΔesxH parental strains, which formed colonies on 7H10 supplemented with 200 ng/mL mycobactin J, but not on standard 7H10 medium, as previously reported (22) and as demonstrated in Fig. 1B. In a prior study, growth to high titers by ΔesxH was not observed in C57BL/6 mice following aerosol infection (22). In this study, CBA mice were chosen for infection by ΔesxH because these mice exhibit a greater susceptibility to infection with Mtb (26–30). Following intravenous infection of five CBA mice with ΔesxH, early morbidity was observed for one mouse, which succumbed ∼100 d prior to the remaining ΔesxH-infected mice (Fig. 1C). Notably, bacteria recovered from the lungs of this mouse had also acquired the capacity to grow on standard 7H10 medium lacking mycobactin J (Fig. 1B).
Phenotypic Revertants Harbor Mutations in Rv3058c, a Putative TetR-Family Regulator Implicated in esxR/esxS Expression.
Given that the ΔesxG and ΔesxH isolates exhibited altered growth properties following in vivo passage, we hypothesized that they acquired genetic differences resulting in suppressor phenotypes. Following in vitro culturing, complete genome sequences were determined for the ΔesxG parent, the ΔesxG strains with putative suppressor mutations, and our laboratory strain of H37Rv WT. Sequenced isolates included ΔesxG recovered from C57BL/6 mice at 8 wk and 16 wk postaerosol infection and are detailed in SI Appendix, Table S1.
Given the reversion of ΔesxG growth phenotypes in some mice, it was also of interest to attempt isolation of potential revertants in vitro. Large numbers of ΔesxG were plated onto 7H11 medium without additional iron supplements, resulting in the appearance of two large colonies, which were also subjected to WGS (SI Appendix, Table S1). Like the suppressor strains recovered from the in vivo studies, these in vitro suppressor strains also grew like WT in replating studies, exhibiting growth on 7H10 medium in the absence of mycobactin (Fig. 1B).
WGS confirmed the presence of the complete esxG coding sequence in the WT strain and the ∼180-bp deletion within the ΔesxG parent, as well as the various ΔesxG suppressor mutants (SI Appendix, Fig. S1). Sequencing data were further analyzed to identify the SNPs with the highest variant frequencies specific to the suppressor isolates, as these were thought most likely to represent the genetic changes responsible for the suppressor phenotypes. High-confidence SNPs with frequencies higher than 90% that were present in the putative ΔesxG suppressor mutants but absent from both H37Rv WT and the ΔesxG parent are summarized in SI Appendix, Table S2, with additional details in SI Appendix, Table S1 and Dataset S1.
One SNP mutation (SNP_1) (SI Appendix, Table S2) was shared by several in vivo suppressor strains that were isolated from organs of ΔesxG-infected mice with the highest bacterial titers, including a lung isolate obtained at 8 wk postinfection (ΔesxG in vivo-1), and two lung isolates obtained at 16 wk postinfection from the same mouse (ΔesxG in vivo-2 and -3) (SI Appendix, Table S1). SNP_1 is a missense mutation in the rv3058c gene, resulting in the substitution of histidine for arginine at position 31 (R31H) of the encoded protein. SNP_1 was also present in the large colonies isolated in vitro following plating of ΔesxG on 7H11 medium in the absence of mycobactin J (ΔesxG in vitro-1 and -2) (SI Appendix, Table S1).
Given that a mutation in rv3058c was associated with growth recovery of ΔesxG in vivo and in vitro, targeted PCR amplification and Sanger sequencing of the rv3058c gene was pursued in a variety of additional samples (SI Appendix, Tables S1 and S2). These included isolates from the lungs and spleens of ΔesxG-infected mice obtained at 8 and at 16 wk postinfection. This identified multiple isolates with the R31H change within rv3058c (SNP_1) (SI Appendix, Table S1). We also identified a different rv3058c mutation, SNP_2 resulting in aspartic acid for histidine at position 22 (H22D), in two isolates from the spleen of a ΔesxG-infected mouse (SI Appendix, Tables S1 and S2).
The rv3058c genes of multiple bacterial isolates from the ΔesxH-infected CBA mouse exhibiting early morbidity were also examined. These isolates (strains ΔesxH in vivo-1 to in vivo-8) (SI Appendix, Tables S1 and S2), recovered from lung, spleen, and liver, all shared SNP_3, resulting in the substitution of a proline for a leucine at position 33 (L33P) of Rv3058c. It should be noted that for mice in which multiple isolates were examined, including from different organs, only a single high-frequency SNP was identified per mouse (SI Appendix, Table S1). Additionally, representative isolates containing each rv3058c mutation that was identified, SNP_1, SNP_2, and SNP_3 were plated on media lacking mycobactin supplementation, and all exhibited restoration of growth (Fig. 1B).
Mutations in Rv3058c Map to a Putative DNA-Binding Domain and Disrupt DNA Binding.
The rv3058c gene is predicted to encode a member of the TetR family of transcriptional regulators, most of which function as transcriptional repressors, with the suppression of transcription relieved upon binding of a ligand (31, 32). TetR-family regulators contain conserved helix–turn–helix motifs at their N termini, which function in DNA binding, whereas the C terminus contains the ligand-binding pocket (31–33). As the suppressor mutations H22D, R31H, and L33P all mapped near the 5′ end of rv3058c, we hypothesized that they impair DNA binding. Both the Tuberculosis Database and published data, including chromatin immunoprecipitation-sequencing (ChIP-seq) analysis, predict three binding sites for Rv3058 (34–36): one in the intergenic region between rv3023c and pe29; another in the intergenic region between rv0834c and rv0835; and third, an intragenic site within the rv0869c gene. The two intergenic regions were used in EMSAs and reporter assays to characterize Rv3058c–DNA interactions, including the effects of the N-terminal amino acid substitutions on DNA binding.
Recombinant Rv3058c WT protein was expressed in Escherichia coli and purified (SI Appendix, Fig. S2A). The cy5-labeled DNA probes comprising the promoter regions upstream of rv0834c and pe29 were both found to be shifted markedly by the addition of recombinant purified Rv3058c WT protein (SI Appendix, Fig. S2 B, Left). After visualization of the cy5 signal, the gel was stained with Imperial blue protein stain (SI Appendix, Fig. S2 B, Right), revealing that Rv3058c WT protein comigrated with the shifted DNA probe, providing further evidence for their interaction. Using EMSA, we were unable to assess the DNA-binding properties of the Rv3058c R31H point mutant or of an H22D/R31H/L33P triple mutant because the proteins did not enter the gel efficiently, with Imperial blue staining revealing that most of the protein remained at the top of the wells.
For further analysis of Rv3058c interactions with predicted target promoters, a synthetic mammalian gene circuit was employed. It was comprised of a chimeric transcription factor expression plasmid and secreted embryonic alkaline phosphatase (SEAP) reporter plasmids (SI Appendix, Fig. S2C) and was adapted from an approach used to characterize the Mtb TetR-family member EthR2 (37). For the transcription factor plasmid, the coding sequence of Mtb rv3058c was fused to the herpes simplex VP16 transcription-activating domain in a mammalian expression vector, generating plasmid p3058c-VP16 (SI Appendix, Table S3). In this system, the Rv3058c portion of the chimera should bind to mycobacterial target promoters, and the VP16 domain serves as a transcriptional activator functional in mammalian cells. Constructs were made with WT Rv3058c as well as the R31H point mutant of Rv3058c, which was chosen due to its presence in multiple in vivo and in vitro suppressor isolates (SI Appendix, Table S3).
For the SEAP reporter plasmids, the ∼200-bp promoter regions of rv0834c and pe29 were inserted upstream of a minimal variant of the human cytomegalovirus-derived promoter (PhCMVmin) (SI Appendix, Fig. S2C and Table S3). The transcription factor plasmids (Rv3058c WT or R31H mutant) and SEAP reporter plasmids were cotransfected at various ratios into HEK 293T cells, and SEAP expression was quantified in cell culture supernatants 48 h posttransfection. Controls included individual plasmids, empty vectors (EV), and a positive control vector that constitutively expressed SEAP. Significantly higher levels of SEAP activity were detected with Rv3058c WT than with the R31H mutant for both the rv0834c and pe29 upstream sequences (SI Appendix, Fig. S2C). Western blots verified similar expression for the WT and mutant proteins. Therefore, the results support the ability of Rv3058c to interact with these promoter sequences and provide evidence that the R31H mutation impairs DNA-binding activity.
Deletion of rv3058c Phenocopies the Revertant In Vitro Growth Phenotypes.
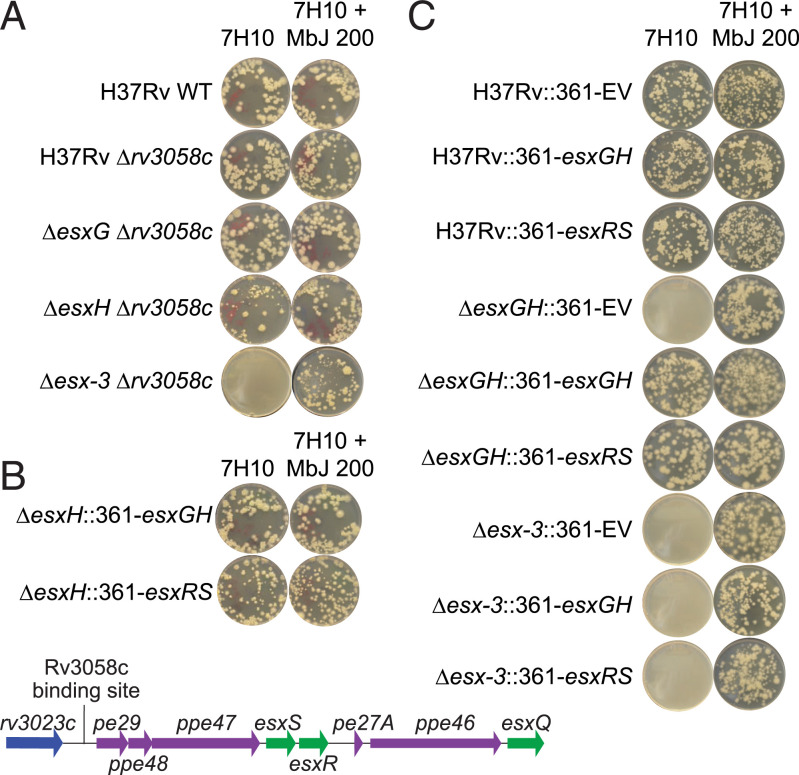
Given that missense mutations in Rv3058c were associated with impaired function of the protein, we hypothesized that a deletion mutant of rv3058c might phenocopy the suppressor mutants. Therefore, the rv3058c coding sequence was deleted from four genetic backgrounds (i.e., H37Rv WT, ΔesxG, ΔesxH, and Δesx-3) (SI Appendix, Table S4). The deletions were confirmed by three-primer PCR and further verified by WGS using Illumina MiSeq technology, which showed the absence of reads at the relevant loci (SI Appendix, Fig. S3). Growth of the mutant strains was assessed on 7H10 solid media prepared in the absence and presence of mycobactin supplementation. Similar to the ΔesxG and ΔesxH revertant strains (Fig. 1B), the ΔesxG Δrv3058c and ΔesxH Δrv3058c double-deletion mutants grew on 7H10 lacking additional iron supplements (Fig. 2A). However, the double Δesx-3 Δrv3058c mutant did not grow in the absence of mycobactin supplementation (Fig. 2A). These data indicate that the phenotypes of the ΔesxG and ΔesxH revertants are due to loss-of-function mutations in Rv3058c, but that rescue of the growth phenotype requires the presence of other components of the esx-3 locus, most likely those that comprise the secretion apparatus.
Fig. 2.
Deletion of rv3058c or overexpression of esxS-esxR complements in vitro growth defects of esxG/H mutant strains. (A) Cultures of double mutants (ΔesxGΔrv3058c, ΔesxHΔrv3058c, and Δesx-3Δrv3058c) were plated in parallel onto media containing and lacking 200 ng/mL mycobactin J in comparison with the H37Rv WT strain. Deletion of rv3058c permits growth of ΔesxG and ΔesxH but not Δesx-3. (B) ΔesxH was transformed with pMV361 vector expressing esxGH or esxRS and selected on kanamycin; equivalent inocula were plated on 7H10 medium with and without mycobactin J. Both constructs rescued growth in the absence of mycobactin J. Below is a schematic illustrating the genomic organization of the esxRS locus; the location of the putative Rv3058c binding site upstream of pe29 is indicated. (C) H37Rv WT, ΔesxGH, and Δesx-3 were transformed with pMV361 EV or with pMV361 vector expressing esxGH or esxRS and selected on kanamycin. Equivalent numbers were plated on media with and without mycobactin J. Both the esxGH- and the esxRS-expressing constructs but not the EV rescued growth of ΔesxGH on standard 7H10 lacking mycobactin J. In contrast, growth of Δesx-3 was not rescued by any of the constructs.
Relevance of the Rv3058c-Regulated Genes esxR and esxS in the Revertant In Vitro Growth Phenotypes.
The WGS data, together with the reporter construct and EMSA results, led us to test whether a genetic locus or loci normally under negative regulation by Rv3058c might be derepressed in the double rv3058c/esxG or rv3058c/esxH deletion mutants, thereby compensating for the absence of EsxG and EsxH. The putative Rv3058c intergenic binding site found adjacent to pe29 (rv3022A) lies upstream of a locus encoding a pair of esx genes, esxS and esxR, which are flanked by pe-ppe genes (referred to here as the “esxRS locus,” defined as extending from pe29 [rv3022A] through esxQ [rv3017c]) (Fig. 2B). Because EsxR and EsxH exhibit substantial homology (84% amino acid identity), as do EsxS and EsxG (92% amino acid identity), this locus was considered to be a strong candidate for providing compensatory functions. Therefore, the effects of overexpression of esxS/esxR on the in vitro iron requirements of strains lacking esxG and/or esxH were investigated. Transformation of H37Rv ΔesxH or ΔesxGH with an integrating plasmid (pMV361) expressing esxG/esxH under control of the strong, constitutive hsp60 promoter (Phsp60) (38) restored the ability of the strains to grow on 7H10 medium in the absence of iron supplements (Fig. 2 B and C). A similar construct expressing esxS/esxR was equally effective in restoring growth, indicating the capacity of EsxS/EsxR to complement and substitute for EsxG/EsxH (Fig. 2 B and C). Although the ΔesxG/ΔesxH strains encode intact copies of esxS and esxR, the genes are presumably under negative regulation by Rv3058c at their native locus, which is bypassed by placing the genes under the control of Phsp60.
EsxG and EsxH are secreted via the ESX-3 secretion system, and deletion of the entire esx-3 region phenocopies the iron-dependent phenotypes of single and double esxG/esxH deletion mutants (22). To determine whether complementation with esxS-esxR requires an intact ESX-3 secretion system, plasmids expressing esxG/esxH or esxS/esxR were introduced into a Δesx-3 strain and plated on 7H10 medium lacking additional iron supplements (Fig. 2C). No growth was detected in the resulting strains, Δesx-3::361-esxGH and Δesx-3::361-esxRS, in the absence of iron supplementation, although each strain grew in the presence of mycobactin J. These data support the notion that both of these Esx heterodimeric pairs require an intact ESX-3 secretion system for their function (22, 39, 40).
Deletion of rv3058c or Overexpression of esxS/esxR Restores Virulence In Vivo.
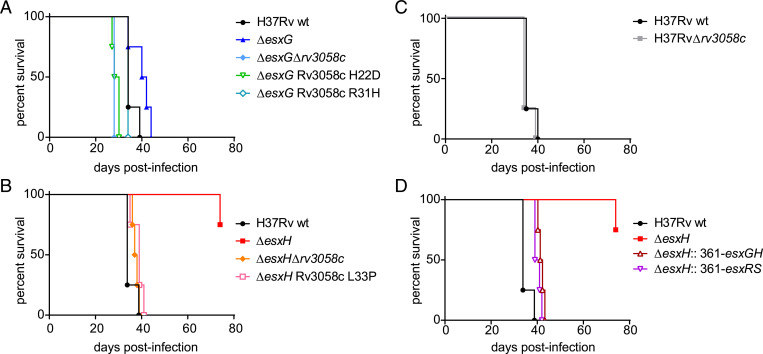
After confirming that deletion of rv3058c or overexpression of esxS/esxR corrected the in vitro iron-related growth defects of esxG and esxH deletion mutants, the effects of these genetic manipulations on virulence were examined by employing mouse models used previously to study in vivo behavior of the deletion mutants (22). In the first model, intravenous infection of severe combined immunodeficiency (SCID) mice, revealed that deletion of rv3058c reversed the attenuation of the ΔesxG and ΔesxH parental strains (an attenuation that was mild for the ΔesxG parent, and more marked for the ΔesxH parent), mimicking findings observed for the Rv3058c N-terminal point mutants tested in parallel (ΔesxG Rv3058c H22D, ΔesxG Rv3058c R31H and ΔesxH Rv3058c L33P) (Fig. 3 A and B). Deletion of rv3058c from the parental H37Rv WT strain did not alter the outcome of infection (Fig. 3C). Additionally, overexpression of either EsxG/H or EsxR/S in the ΔesxH strain restored virulence close to that of the WT strain (Fig. 3D).
Fig. 3.
Restoration of virulence of ΔesxG and ΔesxH strains by mutation of rv3058c or forced expression of esxRS. (A) Percent survival over time of SCID mice infected by the intravenous route with H37Rv WT, ΔesxG, ΔesxG Δrv3058c, or ΔesxG Rv3058c H22D or R31H point mutants. Similar experiments were performed but with H37Rv WT, ΔesxH, ΔesxH Δrv3058c, or ΔesxH Rv3058c L33P point mutant (B), with H37Rv WT or H37Rv Δrv3058c (C), and with H37Rv WT, ΔesxH, ΔesxH stably transformed with a plasmid that expresses esxG and esxH (ΔesxH::361-esxGH) or ΔesxH stably transformed with a plasmid that expresses esxS and esxR (ΔesxH:: 361-esxRS) (D). The H37Rv WT-infected group is identical for A–D, n = 4 mice per group.
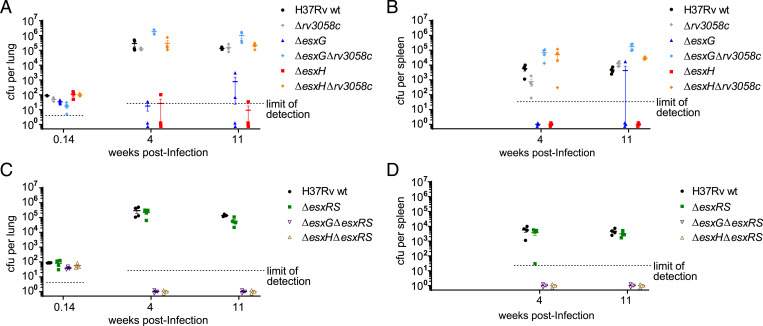
A second model used a low-dose aerosol infection of immunocompetent C57BL/6 mice, where bacterial load in lung and spleen was measured at 4 and 11 wk postinfection. Whereas ΔesxG and ΔesxH exhibited low bacterial burdens, at or below the limit of detection, deletion of rv3058c in either of these mutants restored growth to WT levels in both lung and spleen (Fig. 4 A and B). In contrast, deletion of rv3058 in the WT background did not appreciably alter bacterial burden (Fig. 4 A and B). Therefore, in this model, as in the SCID model described above, the absence of esxG or esxH is required to reveal a phenotype for loss of rv3058c. Combined deletion of genes esxR and esxS, presumed to be under the regulation of Rv3058c, in a WT background also did not alter bacterial burdens in lung or spleen (Fig. 4 C and D). Bacterial numbers for the ΔesxG/ΔesxR/ΔesxS and ΔesxH/ΔesxR/ΔesxS combined deletion mutants were below the limit of detection in both lung and spleen at the two time points examined (Fig. 4 C and D). Because the parental ΔesxG and ΔesxH strains already exhibit substantial growth defects in this C57BL/6 low-dose aerosol model (Fig. 4 A and B) (22), it is difficult to assess whether the combined deletions of esxR and esxS contribute any additional attenuating impact. However, no apparent outgrowth of suppressors was observed for either of these combined deletion mutants.
Fig. 4.
Deletion of rv3058c is sufficient to restore in vivo growth of the ΔesxG and ΔesxH strains in aerosol-infected C57BL/6 mice. (A) Lung titers at the indicated times postinfection from mice infected via the aerosol route with H37Rv WT, Δrv3058c, ΔesxG, ΔesxG Δrv3058c, ΔesxH, or ΔesxH Δrv3058c. (B) Spleen titers for the mice in A. (C) Lung titers for mice infected by the aerosol route with H37Rv WT, ΔesxRS, ΔesxG ΔesxRS, or ΔesxH ΔesxRS. (D) Spleen titers from the mice in C. For A–D, titers determined for individual mice are shown by symbols, while solid horizontal lines indicate means and error bars depict SEM. The limits of detection for the measurements are indicated by the dotted lines. n = 3 to 4 mice per group per time point for A–D.
Extensive Amplification of the esxRS Locus in a Mouse Isolate.
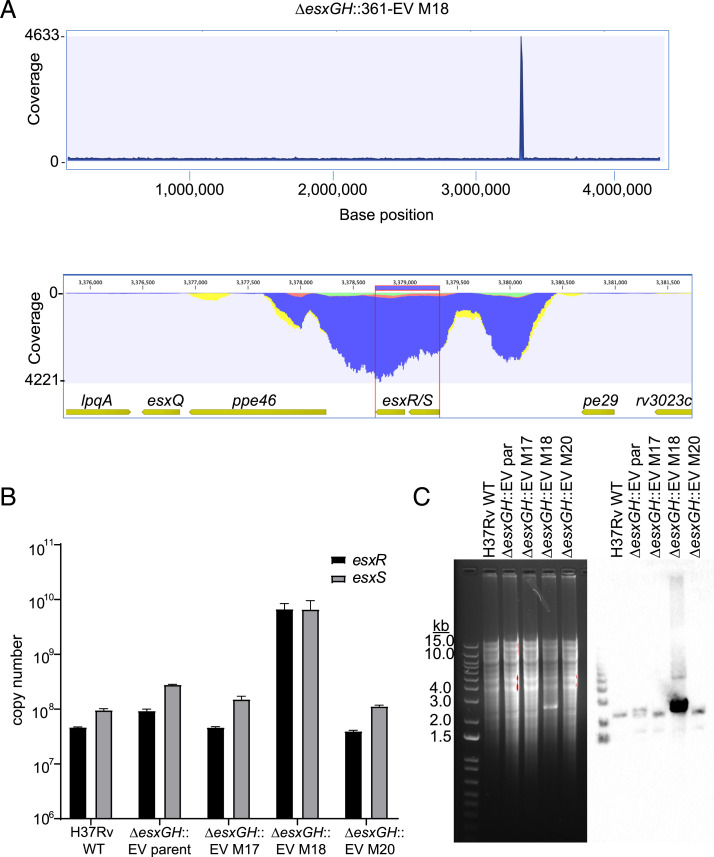
In a separate experiment, following low-dose aerosol infection of C57BL/6 mice with ΔesxGH::361-EV, which has the pMV361 EV integrated at the attB site and served as a control for the overexpression studies described above, a significant bacterial burden was detected at 1 mo postinfection. This result was surprising as the strain was expected to be growth-defective based on the esxG and esxH deletions. Genomic DNA prepared from bacteria isolated from lungs was subjected to WGS, and for one of the three isolates (ΔesxGH::361-EV M18, the ΔesxGH::361-EV isolate recovered from mouse #18) (SI Appendix, Table S1), sequencing coverage across the esxS-esxR genes and immediate flanking regions was ∼5,000-fold (Fig. 5A), as compared to only ∼30-fold for neighboring loci. This finding was confirmed by qPCR analysis of esxR and esxS copy numbers in H37Rv WT, the parental ΔesxGH::361-EV strain before passage through mice, and the three isolates recovered from mice (from mouse #17 [M17], #18 [M18], and #20 [M20]) (Fig. 5B). Again, ΔesxGH::361-EV M18 displayed close to 100-fold greater copy numbers for esxR and esxS compared with the other strains (Fig. 5B). The genomic DNA was also examined by Southern blot, using a 537-bp probe containing esxS-esxR coding sequences (Dataset S2), which revealed the expected ∼2,239-bp band for H37Rv WT (Fig. 5C). However, a tremendous increase in signal intensity was apparent for ΔesxGH::361-EV M18, so much so that a prominent band was visible on the ethidium-bromide stained gel. Southern blotting confirmed that the band contained esxS-esxR sequences. Neither the ΔesxGH::361-EV parent (before inoculation into animals) nor ΔesxGH::361-EV M17 and M20 exhibited the exceptional increase in band intensity observed for the ΔesxGH::361-EV M18 strain, although a doublet of two near-equal intensity bands was noted in the ΔesxGH::361-EV parental strain. The basis for this doublet is not clear.
Fig. 5.
Massive in vivo amplification of a portion of the esxRS locus in a ΔesxGH strain. (A) Whole-genome Illumina sequencing read coverage (read length 150 bp) for strain H37Rv ΔesxGH::361-EV M18. (Upper) Read coverage across the bacterial genome. (Lower) Zoom-in on an ∼3-kb region that includes the esxS-esxR genes and flanking sequences, demonstrating that this region corresponds to the sharp spike in coverage seen in the Upper panel. (B) qPCR was performed in order to determine the esxR and esxS copy numbers in equivalent amounts of genomic DNA from H37Rv WT, ΔesxGH::361-EV (the parental strain used to inoculate mice), and the three isolates recovered from mice #17, #18, and #20. The data represent the mean esxR and esxS copy numbers, determined in triplicate and calculated from standard curves. Error bars represent the SD. (C, Left) An ethidium bromide-stained agarose gel separating BamHI-digested genomic DNA from the indicated stains. (Right) The results of a Southern blot of the samples probed for esxS-esxR.
Illumina-based WGS using short reads, although well-suited for identifying SNPs and small insertions or deletions, has a limited ability to resolve large structural changes in the genome, such as gene duplications. Therefore, long-read Oxford Nanopore Technologies (ONT) MinION sequencing was employed to further characterize ΔesxGH::361-EV M18. For comparison, ΔesxGH::361-EV M20 was also sequenced as a representative in vivo isolate that lacked evidence of esxRS locus amplification. For ΔesxGH::361-EV M18, a total of 2435 ONT raw reads were extracted for assembly. The longest read was over 171 kb, and the average sequence depth was 113-fold for the shorter contig and 632-fold for the longer contig. The size of the stitched assembled genome was 4,583,652 bp.
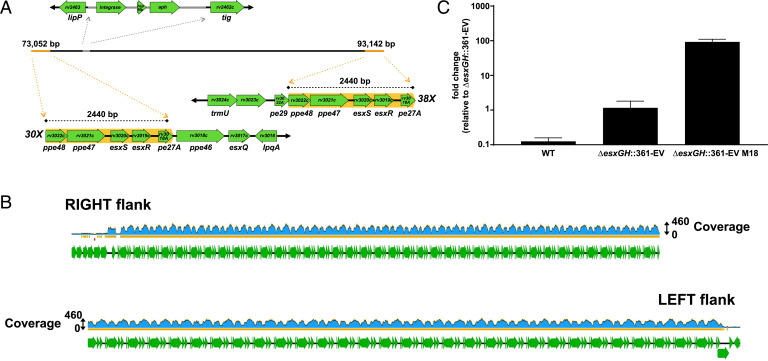
The sequencing of ΔesxGH::361-EV M18 revealed numerous tandem repeats, each 2,440 bp in length. Each repeat included the following gene sequence: ppe48-ppe47-esxS-esxR-pe27A (that is, rv3022c-rv3021c-rv3020c-rv3019c-rv3018A), with the nucleotide sequence of each repeat corresponding to that of the H37Rv parent strain (Fig. 6A). The longest repeat region captured was a single read greater than 132 kb in length, which consisted entirely of 54 tandem repeats. The sequencing also captured individual sequence “raw” reads consisting of 30 repeats adjacent to the chromosomal region downstream of esxRS and pe27A [i.e., genes ppe46 (rv3018c)–esxQ (rv3017c)–lpqA (rv3016)], and 38 repeats adjacent to the region upstream of ppe48 [i.e., genes trmU (rv3024c)–rv3023c–pe29 (rv3022A)] (Fig. 6 A and B). These reads established the amplification as occurring at the native locus within the bacterial chromosome, rather than as repeats scattered throughout the chromosome or as an extrachromosomal event. Nanopore sequencing of the ΔesxGH::361-EV M20 sample yielded a single large circular contig of ∼4.4 Mb, which represents the complete genome, with no evidence of tandem repeats (SI Appendix, Fig. S4). However, a fully closed genome for ΔesxGH::361-EV M18 (Fig. 6A) could not be assembled due to the tandem repeats, which exceeded the length of even the longest Nanopore reads. This suggests that there may be additional repeats not resolved by the Nanopore sequencing, a possibility supported by the estimated 100-fold greater copy number of esxR and esxS derived from qPCR.
Fig. 6.
Evidence for a massive in situ tandem amplification in ΔesxGH::361-EV M18. (A) Nanopore sequencing of ΔesxGH::361-EV M18 generated two contigs after Canu (v1.8) assembly, which were polished and stitched together to create a single linear contig on BioMatters Geneious R11 software by merging of overlapping sequences as described in SI Appendix, SI Materials and Methods. Annotation of the genome of ΔesxGH::361-EV M18 with reference genome H37Rv (NC_000962.3) revealed numerous tandem repeats of ∼2,440 bp in length at each end of the contig. The individual repeat units are highlighted with an orange background and flanking chromosomal sequences are also illustrated. As indicated, one end of the contig includes 30-fold repeats adjacent to neighboring genes ppe46, esxQ, and so forth, while the other end of the contig includes 38-fold repeats adjacent to the neighboring genes pe29, rv3023c, and so forth. A single copy of integrase-hsp60 promoter-aph from the pMV361 vector inserted between lipP (rv2463) and tig (rv2462c) as revealed by Nanopore sequencing and annotation is shown in gray on the assembly. (B) Mapping of the Illumina reads to the constructed ΔesxGH::361-EV M18 contig revealed a coverage as high as 460-fold along much of the tandem repeat region, versus 49-fold for the adjacent chromosomal loci on both the left and right flanks. This indicates that the number of repeats may actually be significantly larger than was captured by nanopore sequencing. (C) Expression of esxS in ΔesxGH::361-EV M18 as compared with the H37Rv WT and parental (ΔesxGH::361-EV) strains was quantified by qRT-PCR. Data are represented as fold-change of RNA levels in the indicated strains as compared with the ΔesxGH::361-EV strain. The data represent the mean and SD of three replicates.
To determine how the amplification impacts expression of the esxRS locus, we obtained total RNA from WT, the ΔesxGH parent of the amplification strain and the amplification strain itself, after growth in 7H9 media lacking additional iron supplements. We then performed real-time qRT-PCR for esxS and a housekeeping gene, sigA, to which esxS expression was normalized. EsxS expression increased approximately ninefold over WT levels in the ΔesxGH strain, possibly reflecting up-regulation due to iron starvation. Expression of esxS in ΔesxGH::361-EV M18, in contrast, was up-regulated ∼90-fold over its ΔesxGH parent (Fig. 6C). This confirms that expression is up-regulated by the amplification and the magnitude of the increase corresponds well to the number of repeats calculated by Nanopore sequencing and by qPCR of genomic DNA.
RNA Sequencing Provides Evidence that Derepression of esxRS Reverts the Iron-Starvation Phenotype of the ΔesxG Mutant.
To gain further insight into how derepression of esxRS can suppress ΔesxG phenotypes, we examined the transcriptomes of rv3058c deletion mutants in the H37Rv and ΔesxG backgrounds by RNA sequencing (RNA-seq). Following initial growth of all strains in 7H9 medium supplemented with 100 μM hemin to facilitate optimal growth of the ΔesxG mutants (22), bacteria were washed and inoculated into standard 7H9 medium without additional iron supplements, and triplicate samples were harvested for RNA isolation at day 3 (mid-log phase) and day 5 (late-log phase) postinoculation. Before harvesting, the OD600 values for the strains were in similar ranges (∼0.3 to 0.4 at day 3 and ∼0.7 to 1.3 at day 5).
RNA-seq analysis comparing H37Rv Δrv3058c with H37Rv WT identified a total of seven genes that were up-regulated twofold or more in the Δrv3058c mutant at day 3 (SI Appendix, Table S5), consistent with Rv3058c acting as a repressor, similar to most other TetR-family members (31, 32). The sole down-regulated gene identified was rv3058c, which contained the targeted deletion; residual reads mapping to rv3058c were still detected in the mutant strain because the deletion spared the extreme 5′ and 3′ ends of the coding region (Dataset S3). Five of the seven up-regulated genes lie within the esxRS locus, including both esxR and esxS, and the ppe genes that flank them. Expression ratios of the genes in this region ranged from 2.28- to 3.50-fold greater for the mutant versus WT. The rv3022A gene at the 5′ end of this presumed operon did not meet criteria for induction, as expression was only ∼1.5-fold higher in the mutant than in WT (Dataset S3). The rv0834c gene (PE_PGRS14) was also up-regulated (3.21-fold) at day 3 in Δrv3058c, as was rv0989c (grcC2; 4.04-fold) (SI Appendix, Table S5). Notably, both rv0834c and rv3022A, which is at the start of the esxRS locus (Fig. 2B), were identified by ChIP-seq analysis as having proximal, upstream intergenic Rv3058c binding sites (35). Binding of purified recombinant Rv3058c at these sites by EMSA and reporter assays further confirmed these observations (SI Appendix, Fig. S2). These findings suggest that the enhanced expression observed in the deletion mutant is a direct result of the loss of repression by Rv3058c. Genes expressed at a higher level in Δrv3058c versus H37Rv at day 5 again included rv0989c, rv0834c, and esxS (Dataset S3). EsxR was just below the cutoff for fold-change in expression, at 1.965-fold higher in Δrv3058c than in WT.
When transcriptomes of ΔesxG Δrv3058c were compared with those of ΔesxG, the same pattern of gene-expression changes observed upon deletion of rv3058c in the H37Rv WT background was still apparent, although these were overlaid on a background of additional transcriptional changes, some metal-related, as described further below (Dataset S3). The seven genes up-regulated in the H37Rv Δrv3058c mutant at day 3 were also up-regulated approximately two- to fivefold in ΔesxG Δrv3058c as compared with the ΔesxG parent at this time point (SI Appendix, Table S5), and in fact were among the top 15 overexpressed genes when comparing the rv3058c deletion mutant to the parent. As noted above, five of these seven genes lie within the esxRS locus. Rv0989c, rv0834c, esxS, esxR, and ppe46 were also up-regulated at day 5 in ΔesxG Δrv3058c as compared with ΔesxG (Dataset S3). The common changes in both genetic backgrounds lend support to the transcriptional regulation of these genes by Rv3058c.
Additionally, several “metal-related” loci were down-regulated in ΔesxG Δrv3058c as compared with ΔesxG at both time points, including loci involved in synthesis of mycobactin siderophores and isonitrile lipopeptides (Dataset S3) (41–43). Also overexpressed in ΔesxG as compared with the double mutant at day 3 were rubA (rv3251c) and rubB (rv3250c) (Dataset S3), genes encoding rubredoxins, small iron-sulfur proteins that act as electron carriers and play roles in oxidative stress responses (44); the rubredoxins are induced in Mtb under iron starvation (45). Together, these changes suggest at least a partial reversion of the iron starvation phenotype in the ΔesxG Δrv3058c double mutant as compared with the ΔesxG parent.
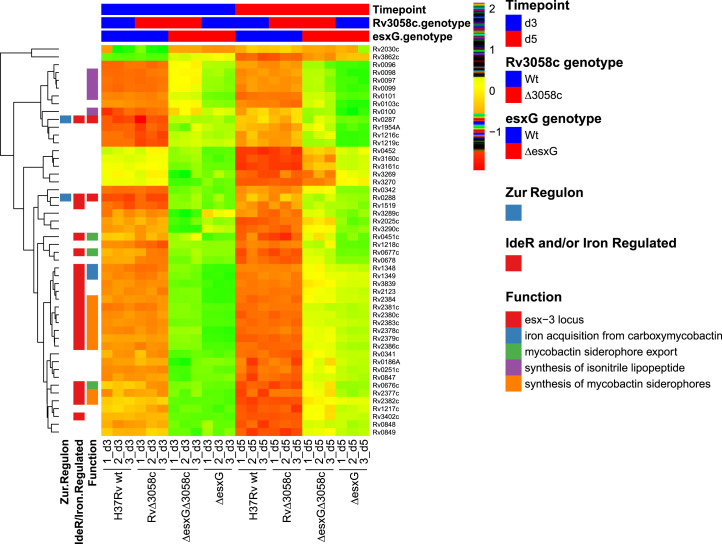
The global transcriptional impacts of esxG deletion and the extent to which these were modified by simultaneously deleting rv3058c were also assessed. A large number of differences were observed between the profiles of ΔesxG and H37Rv WT, many reflecting a state of iron deprivation for the mutant (Dataset S3). To investigate this further, the top 50 differentially expressed genes between ΔesxG and H37Rv WT at day 5 were identified, based on absolute log2 fold-change, and the relative expression levels of this gene set in H37Rv WT, Δrv3058c, ΔesxG, and ΔesxG Δrv3058c at days 3 and 5 were assessed (Fig. 7). The heat map revealed a metal starvation signature for ΔesxG, with up-regulation of numerous genes within the regulon of IdeR, as well as IdeR-independent, iron-repressed genes (14), and genes belonging to the Zur regulon (15). Similar transcriptional profiles were also observed upon down-regulation of the entire esx-3 locus in a conditional mutant (19). As illustrated by the heat map (Fig. 7), although ΔesxG Δrv3058c resembled ΔesxG more closely than WT in terms of transcription patterns, it was still apparent that deletion of rv3058c in the ΔesxG background partially reversed the iron-starvation signature, with the reversal being more apparent at day 5 than day 3 (Fig. 7). Principal component analysis also revealed the double mutant to lie between H37Rv WT and ΔesxG (SI Appendix, Fig. S5).
Fig. 7.
Transcriptomic analysis using RNA-seq reveals a metal starvation signature for the ΔesxG mutant grown in 7H9 medium, which is partially reversed by deleting rv3058c. The hierarchical clustering heatmap represents color-coded expression levels of the top 50 significant DEGs identified when comparing ΔesxG with H37Rv WT at day 5 of growth in 7H9 medium, based on the log2 fold-change values (false-discovery rate < 5%) as determined by RNA-seq analysis. Each row of the heat map represents a different gene as indicated to the right of the map and each column represents an individual sample as indicated below the map. The color and intensity of each cell are related to the normalized gene expression values, with green or red colors indicating up or down-regulation respectively and reordered according to the hierarchical clustering results. Several gene sets in functional categories relevant to this study are denoted by the colors in the legend on the right (Function). Genes that have previously been defined as being regulated by IdeR/iron or as belonging to the Zur regulon are indicated in the left-most columns. The groupings regarding time point, as well as rv3058c and esxG genotypes (WT or mutant) are indicated above the heat map.
Discussion
The ESX-3 T7SS of Mtb plays critical roles in both iron acquisition and virulence (18–23). In the present study, we made the fortuitous and surprising discovery of suppressor mutations that rescued Mtb esxG and esxH mutants, enabling them to multiply and cause disease in infected mice. Analysis of the suppressor mutants from the diseased mice revealed that the Mtb mutations fell into two distinct classes, both associated with the esxRS locus. One of these involved the unprecedented, nearly 100-fold amplification of the esxRS locus. The second and most frequently identified class of mutations resulted in introduction of single amino acid changes in the DNA-binding domain of the putative TetR-family transcriptional regulator (TFTR) Rv3058c. These mutations appear to suppress esxG or esxH deletion phenotypes by up-regulating the esxRS locus, which encodes EsxR and EsxS. EsxQ is also present in the esxRS locus and encodes a protein 65% identical to the N terminus of EsxH. Overall, the analyses performed further support the importance of ESX-3 in iron acquisition but also suggest that enhanced expression of the esxRS locus can compensate for the loss of the ESX-3 substrates EsxG and EsxH.
The first suppressor mutants recovered in vivo link the putative TFTR Rv3058c to the suppression of the ΔesxG and ΔesxH phenotypes and the regulation of esxRS expression. TFTRs possess N-terminal helix–turn–helix domains and C-terminal ligand-binding domains that regulate function. Although exceptions exist, in general, TFTRs act as repressors in the unliganded state (32, 46). Previous studies suggested a role for Rv3058c in regulation of the esxRS locus. Overexpression of rv3058c resulted in differential expression of 263 genes, including down-regulation of multiple genes within the esxRS locus (47). In this study, three independent nonsynonymous substitutions in rv3058c (H22D, R31H, and L33P) were identified in different suppressor strains. Based on the structures of TFTR family members (32, 33), these substitutions map to the putative N-terminal DNA-binding domain. A prior ChIP-seq study provided evidence for Rv3058c binding to the intergenic region between rv3023c and pe29, which is located upstream of the esxRS locus (35). Binding of recombinant purified WT Rv3058c to this site in EMSAs suggested that Rv3058c regulates at least some genes in this region, perhaps including esxR and esxS. WT and mutant Rv3058c were further assessed using a reporter gene assay that involved construction of a synthetic mammalian gene circuit to detect potential interaction of the Rv3058c protein with specific DNA sequences (37). An advantage of this approach, as opposed to testing in mycobacteria, is the expected absence of Rv3058c regulators and of competing mycobacterial transcription factors that might confound data interpretation. In this system, WT Rv3058c bound to the rv3023c/pe29 intergenic region, as evidenced by activation of the reporter gene, whereas the R31H mutant was significantly less capable of activating this promoter or another Rv3058c target promoter, suggesting a defect in binding. Based on these data, we propose that Rv3058c negatively regulates expression of the esxRS locus by binding to the rv3023c/pe29 intergenic region and that the point mutations disrupt binding, eliminating suppression of the locus. This interpretation is supported by the observation that the rv3058c deletion mutant phenocopied the suppressor mutants.
Overexpression of EsxR/S also phenocopied the rv3058c N-terminal point mutants and the deletion mutant with regard to complementing the iron supplementation requirements of the ΔesxG/H strains. Using a constitutive promoter that would not be repressed by Rv3058c, this overexpression was sufficient to restore growth in the absence of iron supplementation but only when the ESX-3 secretory apparatus was intact. This suggests that EsxR and EsxS serve as substrates for the ESX-3 secretion system, much like EsxG and EsxH. Given the capacity of Rv3058c to bind upstream of the esxRS locus, we hypothesize that Rv3058c and the esxRS locus can function as an iron-acquisition regulon. Central to this model is the role of Rv3058c in regulating iron responses. When the entire esx-3 region is intact, the function of the Rv3058c-EsxR/S regulon is presumably masked. However, impairment of ESX-3 function, as in the ΔesxG and ΔesxH strains, revealed the activity of the novel regulon. Consistent with the role of Rv3058c as a transcriptional repressor of the esxRS locus, RNA-seq revealed that seven differentially expressed genes (DEGs) were up-regulated in H37Rv Δrv3058c compared with H37Rv WT, including five genes from the esxRS locus. Comparison of ΔesxG and H37Rv WT transcriptomes revealed that many DEGs reflective of an iron-starvation state were up-regulated in ΔesxG, further supporting its importance in iron metabolism. The ΔesxG profile was characterized by up-regulation of the IdeR regulon, of IdeR-independent, iron-repressed genes (14), and of genes in the Zur regulon (15). When comparing this set of genes across the mutants, it was found that ΔesxG Δrv3058c resembled ΔesxG more closely than it resembled WT. Deletion of rv3058c in the ΔesxG background nonetheless resulted in a partial reversion of the iron-starvation signature and also up-regulated the esxRS locus. These findings lend additional support to the hypotheses that Rv3058c is a negative regulator of esxRS locus and that up-regulation of EsxR/S, possibly with contributions of the flanking pe-ppe genes, allows iron acquisition.
It was previously demonstrated that deletions of the entire esx-3 region or of esxH alone are highly attenuating in vivo (22). Here, it was discovered that deletion of rv3058c in the ΔesxH or ΔesxG background served to restore virulence in both a SCID mouse intravenous infection model and a C57BL/6 mouse aerosol model. Similarly, the ΔesxH or ΔesxG isolates encoding N-terminal substitution mutations in Rv3058c, originally recovered from C57BL/6 mice infected by aerosol, exhibited enhanced virulence when compared with the parental deletion strains in the SCID mouse model. Therefore, virulence phenotypes correlate with in vitro growth. No obvious phenotype was apparent for the rv3058c deletion in the H37Rv WT background, demonstrating again that the role of Rv3058c in regulation of the esxRS locus is revealed by the absence of EsxG/H function, at least in the mouse models examined. Moreover, because the ΔesxG and ΔesxH parental strains already exhibit substantial growth defects, any further attenuation due to esxS-esxR deletion could not be discerned in these strains. The presence in numerous Mtb strains of the esxRS locus and of a specific transcription factor that interacts with cis-acting regulatory sequences upstream of esxRS nonetheless suggest that this iron-acquisition regulon also plays a role for Mtb in a WT background.
It should be noted that several strains of Mtb, as well as sequenced isolates of Mycobacterium microti, lack esxR and esxS, while retaining portions of the flanking pe-ppe sequences and esxQ (48, 49). The loss of these sequences has been proposed to result from homologous recombination between the highly homologous ppe genes, ppe46 (rv3018c) and ppe47/48 (rv3021c/rv3022c), located upstream and downstream of esxR and esxS (48). The impact on growth and virulence of these polymorphisms within the esxRS locus is unknown. It is of interest that deletion of pe18–ppe26, encoded within the esx-5 locus of Mycobacterium canettii, enhanced bacterial persistence in mice, demonstrating that loss of some esx region genes may facilitate pathogenesis (50). Related observations have been made for other mycobacteria as well. ESX-5–deficient Mycobacterium marinum displayed enhanced virulence in adult zebrafish (51). In the Mtb CDC1551 strain, deletion of the ppe38 locus interfered with the secretion of numerous ESX-5 substrates and resulted in enhanced virulence in a mouse model; this deleted configuration at the ppe38 locus is shared by Mtb strains from the Beijing lineage, which exhibit a hypervirulent phenotype (52, 53).
The most unexpected mechanism of suppression identified in this study was the massive, tandem amplification of a contiguous region of five genes from the esxRS locus: ppe48-ppe47-esxS-esxR-pe27A. This was identified in an in vivo isolate, suggesting that it restores virulence lost in ΔesxGH. Based on several methods of analysis, this region is estimated to be present in as many as 100 tandem copies within the bacterial chromosome. The use of Nanopore-based sequencing to resolve the amplification in detail highlights the utility of very long-read approaches for identifying and characterizing such events. Recent WGS studies of clinical isolates of Mtb have uncovered impressive strain diversity (54–58), and WGS has documented an abundance of in vivo SNP differences that occur in the context of antibiotic exposure, host immune pressures, or at distinct anatomic sites within an infected host (56, 59, 60). Of note, the majority of these studies using WGS have excluded repetitive regions, such as the pe/ppe gene families and transposable/mobile elements in their analyses (55), due to ambiguities in alignment and assembly imposed by the mapping of short reads. This omission may have failed to capture the full breadth of genetic variation in Mtb. Although chromosomal duplications have been reported in mycobacteria (61–67), we are not aware of previous reports of such extensive gene amplification in this genus, as described in this study. Recently, plasmids have been identified that harbor ESX loci, and it has been hypothesized that duplication and divergence of these plasmid loci followed by migration to the chromosome may have contributed to the complex evolutionary history of the ESX regions (68–71). The data presented here suggest that duplication and amplification of esx and pe/ppe genes can occur within the chromosome, apart from plasmid sequences. It should be noted that the strain in which the amplification occurred harbored at the attB site an integrated pMV361 EV that encodes integrase. Whether integrase contributed to the magnitude of the amplification remains to be determined.
The extensive amplification of the esxRS locus may represent a genetic accordion. Genetic accordions involve the amplification of genes whose products inefficiently counteract a given selective pressure (72–74), such that increased gene copy numbers increase the dose of the needed product. Additionally, the increased copy number can allow for rare mutations to occur to one copy of the amplified gene, which can then efficiently counteract the selective pressure. This can result in reduction of the amplification because high dosage is no longer needed and because there is a cost associated with maintaining the amplification. Examples of accordion phenomena have been described in a variety of biological systems, including bacteria and DNA viruses, and provide a means by which pathogens can coevolve as the host develops novel antimicrobial strategies (73–78). Belikova et al. (79) observed that gene duplication and amplification occur frequently in the Staphylococcus aureus chromosome, concentrated in loci containing repetitive DNA elements that facilitate recombination; findings were consistent with an “accordion” model with high copy-number variants favored under selective conditions. An especially affected locus encoding “conserved staphylococcal antigens 1” (csa1) displayed amplification up to 10 copies in clinical isolates, while introduction of an antibiotic resistance cassette within the locus resulted in amplifications up to 100- to 200-fold in the setting of antibiotic pressure (79). It will be of interest to determine if continued passage of the esxRS amplification strain under selective conditions in vitro or in vivo might yield strains with mutations that then allow contraction of the amplification.
Gene duplication, amplification, and divergence have been hypothesized to represent a major mechanism underlying the emergence of genes with novel functions (72, 80). Such phenomena appear to have played a role in the evolution of the PE/PPE families of proteins, as well as the emergence of the five ESX loci. In addition to the esx genes encoded within the ESX-1 to -5 regions (two per locus), at least 13 esx paralogs outside of these have been identified in H37Rv based on sequence homology, with an additional esx pair present in some isolates (6, 53, 81–84). Although most Esx proteins encoded apart from secretion systems lack well-defined functions, ESX-5 paralogs encoded within the ESX5a locus of Mtb and M. marinum facilitate the secretion of a subset of ESX-5 substrates (85). The EsxE-EsxF proteins, which lack homology with conserved ESX loci, mediate secretion of the tuberculosis necrotizing toxin (86). Similar to the present study, functional substitution by Esx paralogs has been reported in M. marinum, which possesses several duplications of esxB and esxA, which can functionally complement ESX-1–mediated activities (87).
The present study attributes function to the esxRS locus, which contains esxR, esxS, and esxQ, and is related to ESX-3. Specifically, we provide conclusive evidence that EsxR/S can assume the function of ESX-3 substrates in iron acquisition and virulence. Whether other functions of ESX-3 are also carried out by the esxRS region remains to be determined. For example, EsxH is an immunodominant antigen in the murine tuberculosis model where it stimulates potent T cell responses and may function as a decoy antigen; EsxH is also a target of human T cell responses and interacts with the host cell ESCRT machinery to modulate antigen presentation (24, 25, 88–92). Furthermore, a number of unique T cell epitopes have been identified in EsxR (89, 93); it remains to be determined whether EsxR has immunological functions similar to those reported for EsxH and whether such activities are relevant to the suppressor function of the esxRS locus.
In summary, our data illustrate how Mtb can take advantage of one apparent product of gene duplication in the esxRS locus to overcome, by at least two distinct genetic mechanisms, the severe attenuation conferred by mutations in EsxG and EsxH. The findings highlight the remarkable genetic plasticity that allows Mtb to overcome the loss of a key nutrient acquisition pathway. Beyond this, our work reveals the possibility that these genes can be amplified to high numbers using an accordion mechanism of amplification. Interestingly, despite its capabilities, PacBio sequencing has heretofore not revealed such amplifications. Our work clearly establishes that Mtb can accommodate such amplifications and future work will be required to elucidate the mechanisms.
Materials and Methods
Bacterial Strains and Culture Conditions.
Mycobacterial strains were grown in Middlebrook 7H9 medium (Difco) supplemented with 10% (vol/vol) OADC enrichment (0.5 g oleic acid, 50 g albumin, 20 g dextrose, 0.04 g catalase, 8.5 g sodium chloride in 1 L water), 0.2% (vol/vol) glycerol, and 0.05% (vol/vol) tyloxapol (Sigma). Supplemented media for culturing strains with mutations within the esx-3 locus also included 100 μM hemin (Sigma). Antibiotic selection media contained 50 to 75 µg/mL hygromycin B (Gold Biotechnology) or 25 µg/mL kanamycin as required. For cloning in E. coli, hygromycin was used at 150 µg/mL and kanamycin at 25 to 40 µg/mL
Bacterial Strain Construction.
Plasmids and mycobacterial strains are detailed in SI Appendix, Tables S3 and S4, respectively. H37Rv was the parental Mtb strain for all mutants used and generated in this study. The construction and unmarking of ΔesxG, ΔesxH, ΔesxGH, and Δesx-3 strains and the complementation of the unmarked ΔesxG strain have been described previously (22). The introduction of rv3058c deletions into H37Rv, ΔesxG, ΔesxH, and Δesx-3, and of esxS-esxR deletions into H37Rv, ΔesxG, and ΔesxH using specialized transduction (94, 95) is described in SI Appendix, SI Materials and Methods, as are the unmarking (removal of the sacB-hygromycin cassette) and complementation of the indicated strains.
Solid Media Growth Experiments.
Bacterial cultures grown to log phase were washed in PBS containing 0.05% tyloxapol (PBS-T), resuspended in PBS-T, and adjusted to equivalent OD600 values. Following serial dilution, equivalent inocula were plated in parallel onto 7H10 base medium containing 10% OADC and 0.5% glycerol, and either lacking or containing 200 ng/mL mycobactin J. Plates were incubated for ∼3 to 6 wk at 37 °C.
WGS.
Genomic DNA was extracted from H37Rv WT and mutant strains by the hexadecyltrimethylammonium bromide-lysozyme method using established protocols (96) and WGS was conducted using Illumina and Nanopore MinION, as described in SI Appendix, SI Materials and Methods.
Protein Expression and Purification.
The expression and purification of Rv3058c WT, R31H, and H22D/R31H/L33P mutants is described in SI Appendix, SI Materials and Methods.
SEAP Reporter Assay.
Construction of transcriptional regulator-expressing plasmids encoding Rv3058c-VP16 chimeras and of plasmids containing the promoter regions from rv0834c and pe29 designed to assess the DNA-binding capacity of the chimeras is described in SI Appendix, SI Materials and Methods; plasmids used in this study are listed in SI Appendix, Table S3. The SI Appendix, SI Materials and Methods also provides details of the transfections and SEAP assays.
EMSA.
EMSA were performed to assess the DNA binding capacity of Rv3058c WT, using the ∼200-bp sequences upstream of rv0834c and pe29 as probes (Dataset S2). The EMSA reactions were prepared using the Invitrogen Molecular Probes SYBR SYPRO Electrophoretic Mobility-Shift Assay (EMSA) Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Details of probe preparation, binding reaction composition, and imaging are provided in SI Appendix, SI Materials and Methods.
qPCR of esxR and esxS in Amplification Strain.
An absolute quantitative real-time PCR assay using the Evagreen Dye and qPCR Master Mix kit (Biotium) was performed to determine esxR and esxS gene copy numbers in selected strains, as detailed in SI Appendix, SI Materials and Methods.
Southern Blot of esxRS Amplification Strain.
Genomic DNA was extracted using established protocols (96) from H37Rv WT, ΔesxGH transformed with pMV361 EV (ΔesxGH::361-EV), and three isolates of ΔesxGH::361-EV recovered from mice and Southern blotting performed as described in SI Appendix, SI Materials and Methods.
qRT-PCR Analysis of esxS Expression.
RNA was extracted from the relevant strains and subjected to qRT-PCR; relative esxS gene expression levels were determined using the 2−ΔΔCT method, with sigA serving as the housekeeping control gene. Further details are provided in SI Appendix, SI Materials and Methods.
Transcriptome Analysis.
Procedures for extraction of RNA from H37Rv WT, H37Rv Δrv3058c, ΔesxG, and ΔesxG Δrv3058c, for RNA-seq library preparation and sequencing and for bioinformatic analyses are provided in SI Appendix, SI Materials and Methods.
Mouse infections.
Mouse studies were performed in accordance with National Institutes of Health guidelines, and all work was approved by the Albert Einstein College of Medicine Institutional Animal Care and Use Committee. Details of aerosol and intravenous infection protocols are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Annie Dai, John Kim, Mei Chen, Laura Cole, and Jack Fischer for expert technical assistance; and Susan Howard for critical reading of the manuscript. This work was supported by NIH Grants U19AI109945 (to C.F.B.), P01AI120943 (to C.F.B., D.W.L., and G.K.A.), AI137344 (to J.C.), and AI026170 (to W.R.J.); Department of Defense Grant W81XWH2010099 (to J.M.T.); and funds to J.M.T. and C.F.B. from Georgia State University.
Footnotes
Reviewers: R.B., Institut Pasteur; K.D., Wadsworth Center, New York State Department of Health; and K.R., Cornell University.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112608119/-/DCSupplemental.
Data Availability
The raw reads reported in this paper have been deposited in the National Center for Biotechnology Information’s Sequence Read Archive (BioProject PRJNA784324) (97). All other study data are included in the article and/or supporting information.
References
- 1.World Health Organization, Global Tuberculosis Report 2016 (World Health Organization, Geneva, 2016). [Google Scholar]
- 2.Rybniker J., et al. , Anticytolytic screen identifies inhibitors of mycobacterial virulence protein secretion. Cell Host Microbe 16, 538–548 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Gröschel M. I., Sayes F., Simeone R., Majlessi L., Brosch R., ESX secretion systems: Mycobacterial evolution to counter host immunity. Nat. Rev. Microbiol. 14, 677–691 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Sweeney K. A., et al. , A recombinant Mycobacterium smegmatis induces potent bactericidal immunity against Mycobacterium tuberculosis. Nat. Med. 17, 1261–1268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tiwari S., et al. , BCG-prime and boost with Esx-5 secretion system deletion mutant leads to better protection against clinical strains of Mycobacterium tuberculosis. Vaccine 38, 7156–7165 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bitter W., et al. , Systematic genetic nomenclature for type VII secretion systems. PLoS Pathog. 5, e1000507 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pallen M. J., The ESAT-6/WXG100 superfamily—And a new Gram-positive secretion system? Trends Microbiol. 10, 209–212 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Abdallah A. M., et al. , Type VII secretion—Mycobacteria show the way. Nat. Rev. Microbiol. 5, 883–891 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Champion P. A., Cox J. S., Protein secretion systems in mycobacteria. Cell. Microbiol. 9, 1376–1384 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Stanley S. A., Cox J. S., Host-pathogen interactions during Mycobacterium tuberculosis infections. Curr. Top. Microbiol. Immunol. 374, 211–241 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Simeone R., Bottai D., Brosch R., ESX/type VII secretion systems and their role in host-pathogen interaction. Curr. Opin. Microbiol. 12, 4–10 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Stoop E. J., Bitter W., van der Sar A. M., Tubercle bacilli rely on a type VII army for pathogenicity. Trends Microbiol. 20, 477–484 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Bottai D., Gröschel M. I., Brosch R., Type VII secretion systems in Gram-positive bacteria. Curr. Top. Microbiol. Immunol. 404, 235–265 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez G. M., Voskuil M. I., Gold B., Schoolnik G. K., Smith I., ideR, An essential gene in Mycobacterium tuberculosis: Role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70, 3371–3381 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maciag A., et al. , Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J. Bacteriol. 189, 730–740 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pandey R., et al. , MntR(Rv2788): A transcriptional regulator that controls manganese homeostasis in Mycobacterium tuberculosis. Mol. Microbiol. 98, 1168–1183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X., et al. , A novel stress-inducible CmtR-ESX3-Zn2+ regulatory pathway essential for survival of Mycobacterium bovis under oxidative stress. J. Biol. Chem. 295, 17083–17099 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serafini A., Boldrin F., Palù G., Manganelli R., Characterization of a Mycobacterium tuberculosis ESX-3 conditional mutant: Essentiality and rescue by iron and zinc. J. Bacteriol. 191, 6340–6344 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serafini A., Pisu D., Palù G., Rodriguez G. M., Manganelli R., The ESX-3 secretion system is necessary for iron and zinc homeostasis in Mycobacterium tuberculosis. PLoS One 8, e78351 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siegrist M. S., et al. , Mycobacterial Esx-3 requires multiple components for iron acquisition. MBio 5, e01073-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siegrist M. S., et al. , Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc. Natl. Acad. Sci. U.S.A. 106, 18792–18797 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tufariello J. M., et al. , Separable roles for Mycobacterium tuberculosis ESX-3 effectors in iron acquisition and virulence. Proc. Natl. Acad. Sci. U.S.A. 113, E348–E357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L., et al. , Comprehensive analysis of iron utilization by Mycobacterium tuberculosis. PLoS Pathog. 16, e1008337 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Portal-Celhay C., et al. , Mycobacterium tuberculosis EsxH inhibits ESCRT-dependent CD4+ T-cell activation. Nat. Microbiol. 2, 16232 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J. D., et al. , Mycobacterium tuberculosis-specific CD4+ and CD8+ T cells differ in their capacity to recognize infected macrophages. PLoS Pathog. 14, e1007060 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beamer G. L., et al. , Interleukin-10 promotes Mycobacterium tuberculosis disease progression in CBA/J mice. J. Immunol. 181, 5545–5550 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller C., et al. , Genetically determined susceptibility to tuberculosis in mice causally involves accelerated and enhanced recruitment of granulocytes. Infect. Immun. 74, 4295–4309 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medina E., North R. J., Evidence inconsistent with a role for the Bcg gene (Nramp1) in resistance of mice to infection with virulent Mycobacterium tuberculosis. J. Exp. Med. 183, 1045–1051 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medina E., North R. J., Resistance ranking of some common inbred mouse strains to Mycobacterium tuberculosis and relationship to major histocompatibility complex haplotype and Nramp1 genotype. Immunology 93, 270–274 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner J., et al. , Immunological basis for reactivation of tuberculosis in mice. Infect. Immun. 69, 3264–3270 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balhana R. J., Singla A., Sikder M. H., Withers M., Kendall S. L., Global analyses of TetR family transcriptional regulators in mycobacteria indicates conservation across species and diversity in regulated functions. BMC Genomics 16, 479 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cuthbertson L., Nodwell J. R., The TetR family of regulators. Microbiol. Mol. Biol. Rev. 77, 440–475 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramos J. L., et al. , The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69, 326–356 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galagan J. E., et al. , TB database 2010: Overview and update. Tuberculosis (Edinb.) 90, 225–235 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Minch K. J., et al. , The DNA-binding network of Mycobacterium tuberculosis. Nat. Commun. 6, 5829 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reddy T. B., et al. , TB database: An integrated platform for tuberculosis research. Nucleic Acids Res. 37, D499–D508 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blondiaux N., et al. , Reversion of antibiotic resistance in Mycobacterium tuberculosis by spiroisoxazoline SMARt-420. Science 355, 1206–1211 (2017). [DOI] [PubMed] [Google Scholar]
- 38.O’Gaora P., “Expression of genes in mycobacteria” in Mycobacteria Protocols, Parish T., Stoker N. G., Eds. (Huaman Press, Totawa, NJ, 1998), pp. 261–273. [DOI] [PubMed] [Google Scholar]
- 39.Arbing M. A., et al. , The crystal structure of the Mycobacterium tuberculosis Rv3019c-Rv3020c ESX complex reveals a domain-swapped heterotetramer. Protein Sci. 19, 1692–1703 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ilghari D., et al. , Solution structure of the Mycobacterium tuberculosis EsxG·EsxH complex: Functional implications and comparisons with other M. tuberculosis Esx family complexes. J. Biol. Chem. 286, 29993–30002 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris N. C., et al. , Biosynthesis of isonitrile lipopeptides by conserved nonribosomal peptide synthetase gene clusters in Actinobacteria. Proc. Natl. Acad. Sci. U.S.A. 114, 7025–7030 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krithika R., et al. , A genetic locus required for iron acquisition in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 103, 2069–2074 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quadri L. E., Sello J., Keating T. A., Weinreb P. H., Walsh C. T.. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem. Biol. 5, 631–645 (1998). [DOI] [PubMed] [Google Scholar]
- 44.Sushko T., et al. , A new twist of rubredoxin function in M. tuberculosis. Bioorg. Chem. 109, 104721 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Kurthkoti K., et al. , The capacity of Mycobacterium tuberculosis to survive iron starvation might enable it to persist in iron-deprived microenvironments of human granulomas. mBio 8, e01092-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nguyen Le Minh P., et al. , Ligand binding specificity of RutR, a member of the TetR family of transcription regulators in Escherichia coli. FEBS Open Bio 5, 76–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rustad T. R., et al. , Mapping and manipulating the Mycobacterium tuberculosis transcriptome using a transcription factor overexpression-derived regulatory network. Genome Biol. 15, 502 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marmiesse M., et al. , Macro-array and bioinformatic analyses reveal mycobacterial ‘core’ genes, variation in the ESAT-6 gene family and new phylogenetic markers for the Mycobacterium tuberculosis complex. Microbiology (Reading) 150, 483–496 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Orgeur M., et al. , Pathogenomic analyses of Mycobacterium microti, an ESX-1-deleted member of the Mycobacterium tuberculosis complex causing disease in various hosts. Microb. Genom. 7, 000505 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allen A. C., et al. , Parallel in vivo experimental evolution reveals that increased stress resistance was key for the emergence of persistent tuberculosis bacilli. Nat. Microbiol. 6, 1082–1093 (2021). [DOI] [PubMed] [Google Scholar]
- 51.Weerdenburg E. M., et al. , ESX-5-deficient Mycobacterium marinum is hypervirulent in adult zebrafish. Cell. Microbiol. 14, 728–739 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Ates L. S., et al. , Mutations in ppe38 block PE_PGRS secretion and increase virulence of Mycobacterium tuberculosis. Nat. Microbiol. 3, 181–188 (2018). [DOI] [PubMed] [Google Scholar]
- 53.McEvoy C. R., van Helden P. D., Warren R. M., Gey van Pittius N. C., Evidence for a rapid rate of molecular evolution at the hypervariable and immunogenic Mycobacterium tuberculosis PPE38 gene region. BMC Evol. Biol. 9, 237 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Rie A., Meehan C., The devil is in the diversity: Exploring within-person evolution of Mycobacterium tuberculosis. EBioMedicine 55, 102758 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ley S. D., de Vos M., Van Rie A., Warren R. M., Deciphering within-host microevolution of Mycobacterium tuberculosis through whole-genome sequencing: The phenotypic impact and way forward. Microbiol. Mol. Biol. Rev. 83, e00062-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nimmo C., et al. , Dynamics of within-host Mycobacterium tuberculosis diversity and heteroresistance during treatment. EBioMedicine 55, 102747 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ford C., et al. , Mycobacterium tuberculosis—Heterogeneity revealed through whole genome sequencing. Tuberculosis (Edinb.) 92, 194–201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Modlin S. J., et al. , Drivers and sites of diversity in the DNA adenine methylomes of 93 Mycobacterium tuberculosis complex clinical isolates. eLife 9, e58542 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lieberman T. D., et al. , Genomic diversity in autopsy samples reveals within-host dissemination of HIV-associated Mycobacterium tuberculosis. Nat. Med. 22, 1470–1474 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Q., et al. , Mycobacterium tuberculosis clinical isolates carry mutational signatures of host immune environments. Sci. Adv. 6, eaba4901 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brosch R., et al. , Genome plasticity of BCG and impact on vaccine efficacy. Proc. Natl. Acad. Sci. U.S.A. 104, 5596–5601 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Domenech P., Kolly G. S., Leon-Solis L., Fallow A., Reed M. B., Massive gene duplication event among clinical isolates of the Mycobacterium tuberculosis W/Beijing family. J. Bacteriol. 192, 4562–4570 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Galamba A., et al. , Disruption of adhC reveals a large duplication in the Mycobacterium smegmatis mc(2)155 genome. Microbiology (Reading) 147, 3281–3294 (2001). [DOI] [PubMed] [Google Scholar]
- 64.Leung A. S., et al. , Novel genome polymorphisms in BCG vaccine strains and impact on efficacy. BMC Genomics 9, 413 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang X. M., et al. , IS1096-mediated DNA rearrangements play a key role in genome evolution of Mycobacterium smegmatis. Tuberculosis (Edinb.) 88, 399–409 (2008). [DOI] [PubMed] [Google Scholar]
- 66.Weiner B., et al. , Independent large scale duplications in multiple M. tuberculosis lineages overlapping the same genomic region. PLoS One 7, e26038 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brosch R., et al. , Comparative genomics uncovers large tandem chromosomal duplications in Mycobacterium bovis BCG Pasteur. Yeast 17, 111–123 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ummels R., et al. , Identification of a novel conjugative plasmid in mycobacteria that requires both type IV and type VII secretion. MBio 5, e01744-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dumas E., et al. , Mycobacterial pan-genome analysis suggests important role of plasmids in the radiation of type VII secretion systems. Genome Biol. Evol. 8, 387–402 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Newton-Foot M., Warren R. M., Sampson S. L., van Helden P. D., Gey van Pittius N. C., The plasmid-mediated evolution of the mycobacterial ESX (type VII) secretion systems. BMC Evol. Biol. 16, 62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mortimer T. D., Weber A. M., Pepperell C. S., Evolutionary thrift: Mycobacteria repurpose plasmid diversity during adaptation of type VII secretion systems. Genome Biol. Evol. 9, 398–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bergthorsson U., Andersson D. I., Roth J. R., Ohno’s dilemma: Evolution of new genes under continuous selection. Proc. Natl. Acad. Sci. U.S.A. 104, 17004–17009 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Elde N. C., et al. , Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses. Cell 150, 831–841 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roth J. R., Andersson D. I., Poxvirus use a “gene accordion” to tune out host defenses. Cell 150, 671–672 (2012). [DOI] [PubMed] [Google Scholar]
- 75.Andersson D. I., Slechta E. S., Roth J. R., Evidence that gene amplification underlies adaptive mutability of the bacterial lac operon. Science 282, 1133–1135 (1998). [DOI] [PubMed] [Google Scholar]
- 76.Slechta E. S., et al. , Adaptive mutation: General mutagenesis is not a programmed response to stress but results from rare coamplification of dinB with lac. Proc. Natl. Acad. Sci. U.S.A. 100, 12847–12852 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pränting M., Andersson D. I., Escape from growth restriction in small colony variants of Salmonella typhimurium by gene amplification and mutation. Mol. Microbiol. 79, 305–315 (2011). [DOI] [PubMed] [Google Scholar]
- 78.Copley S. D., Toward a systems biology perspective on enzyme evolution. J. Biol. Chem. 287, 3–10 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Belikova D., Jochim A., Power J., Holden M. T. G., Heilbronner S., “Gene accordions” cause genotypic and phenotypic heterogeneity in clonal populations of Staphylococcus aureus. Nat. Commun. 11, 3526 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Francino M. P., An adaptive radiation model for the origin of new gene functions. Nat. Genet. 37, 573–577 (2005). [DOI] [PubMed] [Google Scholar]
- 81.Cole S. T., et al. , Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 (1998). [DOI] [PubMed] [Google Scholar]
- 82.Gey Van Pittius N. C., et al. , The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol. 2, RESEARCH0044 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tekaia F., et al. , Analysis of the proteome of Mycobacterium tuberculosis in silico. Tuber. Lung Dis. 79, 329–342 (1999). [DOI] [PubMed] [Google Scholar]
- 84.Uplekar S., Heym B., Friocourt V., Rougemont J., Cole S. T., Comparative genomics of Esx genes from clinical isolates of Mycobacterium tuberculosis provides evidence for gene conversion and epitope variation. Infect. Immun. 79, 4042–4049 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shah S., Cannon J. R., Fenselau C., Briken V., A duplicated ESAT-6 region of ESX-5 is involved in protein export and virulence of mycobacteria. Infect. Immun. 83, 4349–4361 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tak U., Dokland T., Niederweis M., Pore-forming Esx proteins mediate toxin secretion by Mycobacterium tuberculosis. Nat. Commun. 12, 394 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bosserman R. E., Thompson C. R., Nicholson K. R., Champion P. A., Esx paralogs are functionally equivalent to ESX-1 proteins but are dispensable for virulence in Mycobacterium marinum. J. Bacteriol. 200, e00726-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nunes-Alves C., et al. , Human and murine clonal CD8+ T cell expansions arise during tuberculosis because of TCR selection. PLoS Pathog. 11, e1004849 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Skjøt R. L., et al. , Epitope mapping of the immunodominant antigen TB10.4 and the two homologous proteins TB10.3 and TB12.9, which constitute a subfamily of the esat-6 gene family. Infect. Immun. 70, 5446–5453 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Skjøt R. L., et al. , Comparative evaluation of low-molecular-mass proteins from Mycobacterium tuberculosis identifies members of the ESAT-6 family as immunodominant T-cell antigens. Infect. Immun. 68, 214–220 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lewinsohn D. A., et al. , Comprehensive definition of human immunodominant CD8 antigens in tuberculosis. NPJ Vaccines 2, 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sutiwisesak R., et al. , A natural polymorphism of Mycobacterium tuberculosis in the esxH gene disrupts immunodomination by the TB10.4-specific CD8 T cell response. PLoS Pathog. 16, e1009000 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lindenstrøm T., Aagaard C., Christensen D., Agger E. M., Andersen P., High-frequency vaccine-induced CD8+ T cells specific for an epitope naturally processed during infection with Mycobacterium tuberculosis do not confer protection. Eur. J. Immunol. 44, 1699–1709 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bardarov S., et al. , Specialized transduction: An efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology (Reading) 148, 3007–3017 (2002). [DOI] [PubMed] [Google Scholar]
- 95.Jain P., et al. , Specialized transduction designed for precise high-throughput unmarked deletions in Mycobacterium tuberculosis. MBio 5, e01245-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Larsen M. H., Biermann K., Tandberg S., Hsu T., Jacobs W. R. J., Genetic manipulation of Mycobacterium tuberculosis. Curr. Protoc. Microbiol. 6, 10A.2.1–10A.2.21 (2007). [DOI] [PubMed] [Google Scholar]
- 97.Wang L., et al. , Mycobacterium tuberculosis massive gene amplification to overcome loss of ESX-3 secretion system substrates. National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA784324. Deposited 28 November 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw reads reported in this paper have been deposited in the National Center for Biotechnology Information’s Sequence Read Archive (BioProject PRJNA784324) (97). All other study data are included in the article and/or supporting information.