Significance
Diamond-based quantum sensing enables nanoscale measurements of biological systems with unprecedented sensitivity. Potential applications of this emerging technology range from the investigation of fundamental biological processes to the development of next-generation medical diagnostics devices. One of the main challenges faced by bioquantum sensing is the need to interface quantum sensors with biological target systems. Specifically, such an interface needs to maintain the highly fragile quantum states of our sensor and at the same time be able to fish intact biomolecules out of solution and immobilize them on our quantum sensor surface. Our work overcomes these challenges by combining tools from quantum engineering, single-molecule biophysics, and material processing.
Keywords: quantum sensing, NV center, diamond surface modification, biocompatible functionalization
Abstract
Quantum metrology enables some of the most precise measurements. In the life sciences, diamond-based quantum sensing has led to a new class of biophysical sensors and diagnostic devices that are being investigated as a platform for cancer screening and ultrasensitive immunoassays. However, a broader application in the life sciences based on nanoscale NMR spectroscopy has been hampered by the need to interface highly sensitive quantum bit (qubit) sensors with their biological targets. Here, we demonstrate an approach that combines quantum engineering with single-molecule biophysics to immobilize individual proteins and DNA molecules on the surface of a bulk diamond crystal that hosts coherent nitrogen vacancy qubit sensors. Our thin (sub–5 nm) functionalization architecture provides precise control over the biomolecule adsorption density and results in near-surface qubit coherence approaching 100 μs. The developed architecture remains chemically stable under physiological conditions for over 5 d, making our technique compatible with most biophysical and biomedical applications.
Recent developments in quantum engineering and diamond processing have brought us considerably closer to performing nanoscale NMR and electron paramagnetic resonance (EPR) spectroscopy of small ensembles and even individual biomolecules. Notably, these advances have enabled the detection of the nuclear spin noise from a single ubiquitin protein (1) and the probing of the EPR spectrum of an individual paramagnetic spin label conjugated to a protein (2) or DNA molecule (3). More recently, lock-in detection and signal reconstruction techniques (4, 5) have enabled one- and multidimensional NMR spectroscopy with 0.5-Hz spectral resolution (6–8). More advanced control sequences at cryogenic temperatures have further enabled mapping the precise location of up to 27 13C nuclear spins inside of diamond (9). Yet biologically meaningful spectroscopy on intact biomolecules remains elusive. One of the main outstanding challenges, which is required to perform nanoscale magnetic resonance spectroscopy of biomolecules, is the need to immobilize the target molecules within the 10- to 30-nm sensing range (2, 3, 7) of a highly coherent nitrogen vacancy (NV) qubit sensor. Immobilization is necessary because an untethered molecule would otherwise diffuse out of the detection volume within a few tens of microseconds.
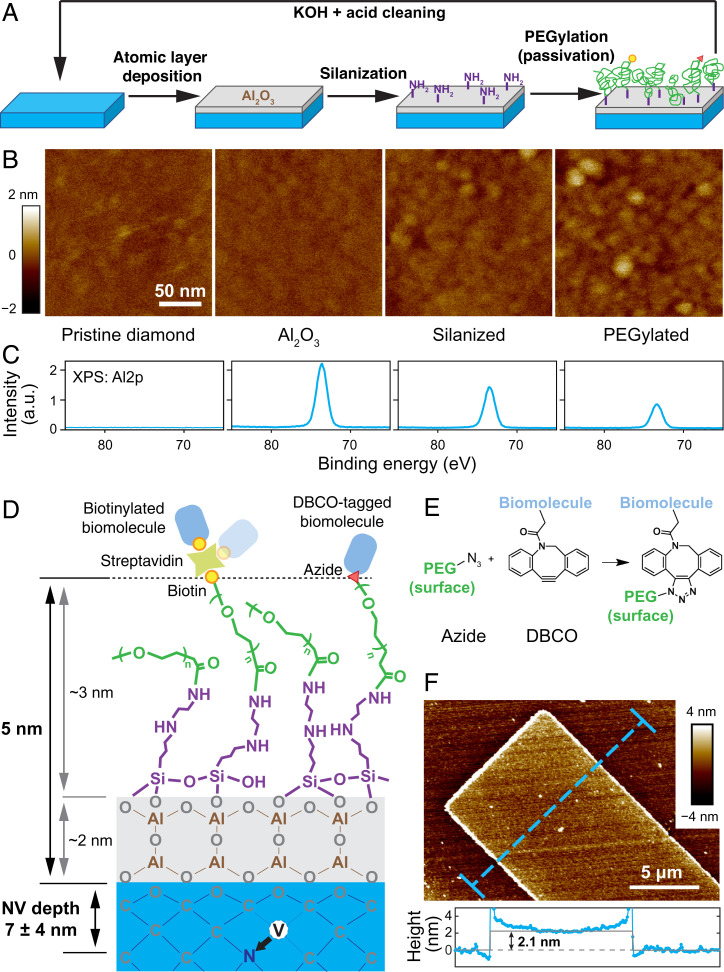
Various avenues to the functionalization of high-quality, single-crystalline diamond chips have been pursued over the last decade (10–12). However, none of the currently known approaches has led to the desired results of interfacing a coherent quantum sensor with target biomolecules. For example, hydrogen-terminated diamond surfaces can be chemically modified and form biologically stable surfaces (10, 13); but near-surface NV centers are generally charge-unstable under hydrogen termination (14), posing open challenges for NV sensing. On the other hand, oxygen-terminated diamond surfaces have been used to create charge stable NV– centers with exceptional coherence times within 10 nm from the diamond surface (15). However, perfectly arranged, ether-terminated diamond surfaces generally lack chemically functionalizable surface groups (such as carboxyl or hydroxyl groups), making it difficult to control immobilization density and surface passivation. Other platforms such as diamond nanocrystals can generally be functionalized (16, 17) because of their heterogeneous surface chemistry, but they do not possess the coherence times needed for nanoscale magnetic resonance spectroscopy. Our approach (Fig. 1A) overcomes these limitations by utilizing a 2-nm-thick Al2O3 layer deposited onto an oxygen-terminated diamond surface by atomic layer deposition (ALD). This Al2O3 “adhesion” layer is silanized by N-[3-(trimethoxysilyl)propyl]ethylenediamine to create an amine (–NH2) -terminated surface, which in turn is then grafted with a monolayer of heterobifunctional polyethylene glycol (PEG) via an N-hydroxysuccinimide (NHS) reaction, a process also referred as PEGylation. The PEG layer serves two purposes. First, it passivates the diamond surface to prevent nonspecific adsorption of biomolecules. Second, by adjusting the density of PEG molecules with functional groups (e.g., biotin or azide), we can control the immobilization density of proteins or DNA target molecules on the diamond surface. Furthermore, the small persistence length of the PEG linker (∼0.35 nm) allows the immobilized biomolecules to undergo rotational diffusion (18). This tumbling motion is the basis for motional averaging of the NMR spectra and helps to prevent immobilization of molecules in biologically inactive orientations.
Fig. 1.
Architecture and characterization of the diamond functionalization approach. (A) Schematic illustration of the functionalization process. A thin layer of Al2O3 (gray) was deposited to the pristine, oxygen-terminated diamond surfaces (blue), followed by silanization (purple) and PEGylation (green). Functional groups (biotin, yellow circle; azide, red triangle) allow for cross-linking with target biomolecules. AFM characterization of the surfaces (B) and XPS Al2p signal after each step of the functionalization (C). (D) Illustration of the overall chemical functionalization architecture (not to scale), with corresponding thicknesses. (E) Illustration of SPAAC reaction. (F) A lithographically fabricated Al2O3 pattern on the diamond surface by lift-off, with a thickness of ∼2.1 nm. The Al2O3 layer is uniform without the presence of pin holes. The elevated edges originate from lift-off combined with ALD deposition.
Diamond Surface Functionalization Architecture
Diamond-based sensing critically relies on minimizing the thickness of any functionalization layer while maintaining excellent surface morphology and surface coverage. We hence carefully characterized the surface at each step of our functionalization procedure. As confirmed by atomic force microscopy (AFM), thermal ALD enabled us to deposit a uniform, 2-nm-thick Al2O3 layer of excellent surface morphology (arithmetical mean deviation Ra = 459 pm) on an oxygen-terminated diamond surface (Ra = 446 pm) (Fig. 1 B and F). The changes in surface properties are corroborated by contact angle measurement (SI Appendix, Fig. S1). A slight increase of surface roughness can be observed after treatment with 10 mM KOH for 10 s, which serves the purpose of −OH activation for silanization (Ra = 841 pm) but also leads to hydrolysis of Al2O3 (SI Appendix, Fig. S2). A final surface roughness of Ra = 866 pm can be observed after PEGylation. X-ray photoelectron spectroscopy (XPS) further confirmed the presence of aluminum (especially the Al2p signal) after each surface treatment step, indicating that the Al2O3 layer remains stable during the processing (Fig. 1C). Angle-resolved XPS (ARXPS) allowed us to further estimate the thickness of the Al2O3 and PEG layer to be 2.0 ± 0.1 nm and 1.2 ± 0.2 nm (SI Appendix, Fig. S3). We note that ARXPS likely underestimated the true thickness of the PEG layer since ARXPS was performed under ultrahigh vacuum, which lead to a collapse of the PEG layer. Assuming a Gaussian chain model, we can estimate the thickness of the hydrated PEG layer to be 2.8 nm (SI Appendix, Fig. S4 and Table S1). From this, we estimate that the total thickness of the functionalization layer is on the order of 5 nm. Notably, shorter PEG can be employed to further reduce the overall thickness to 3 nm without impeding the fine control over the grafting density (vide infra), as demonstrated in SI Appendix, Fig. S5.
Single-Molecule Imaging and Bioconjugation
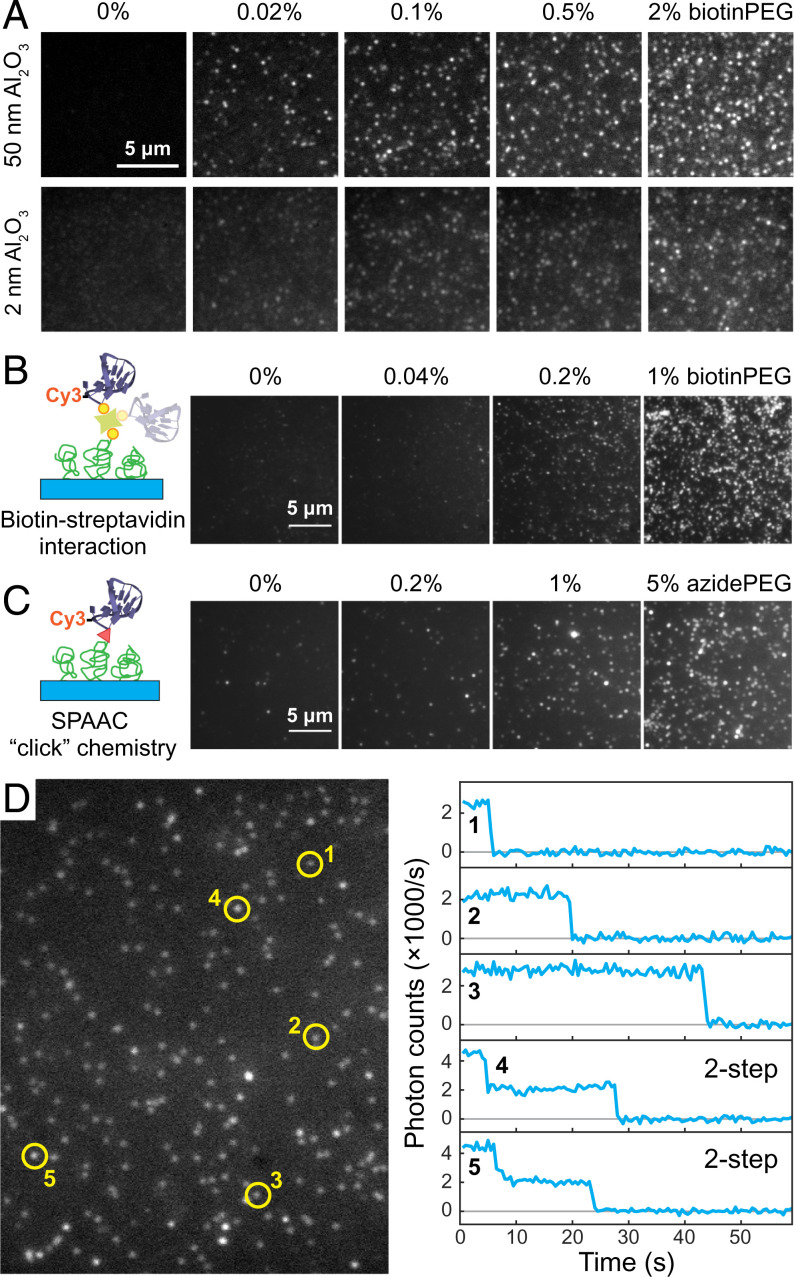
Next, we turned our attention to controlling and characterizing the adsorption density of proteins on a diamond surface. The density of binding sites can be controlled by adjusting the stoichiometric ratio of methyl-terminated PEG (mPEG) and functional PEG groups, for example, biotin-terminated PEG (biotinPEG) or azide-terminated PEG (azidePEG) for click chemistry (Fig. 1E). To investigate the effectiveness of our method, we characterized the adsorption density of Alexa 488 dye-labeled streptavidin (SA-488), our model system, by single-molecule fluorescence microscopy (19). Fig. 2A shows a series of fluorescence images for diamond samples with varying biotinPEG density that were incubated in 7 nM SA-488 for 20 min (for data of other concentrations, see SI Appendix, Fig. S6). The number of fluorescence spots (i.e., individual streptavidin molecules) shows a clear dependence of the adsorption density on the biotinPEG percentage. Importantly, for the diamonds coated solely with mPEG, we observed 4 × 10−3 SA-488 protein per square micrometer, whereas for 2% biotin we found roughly 0.5 SA-488 per square micrometer (SI Appendix, Fig. S7). This suggests that we can control the protein adsorption density over more than two orders of magnitude simply by changing the PEG composition. We note that for higher-biotinPEG densities individual SA-488 molecules were no longer optically resolvable. Furthermore, the immobilization of SA-488 was spatially homogeneous and highly reproducible, in sharp contrast to control experiments that skipped the silanization step, which resulted in a heterogeneous distribution of SA-488 on the diamond surface (SI Appendix, Fig. S8). Performing these titration series for two different Al2O3 layer thickness, we observed no qualitative difference in SA-488 adsorption density between a 2-nm and a 50-nm Al2O3 layer, indicating that working with an ultrathin Al2O3 layer does not negatively impact biofunctionalization. Interestingly, we did observe a reduction in the signal-to-noise ratio in our fluorescence microscopy images. At least partially, this observation can be explained by self-interference of an emitter at the diamond–Al2O3 interface (SI Appendix, Figs. S9 and S10).
Fig. 2.
Single-molecule characterization of fluorescently labeled biomolecules immobilized on diamond surfaces. (A) Fluorescence images of the immobilized SA-488 molecules for various biotinPEG percentages (0, 0.02, 0.1, 0.5, and 2%) and two different Al2O3 thickness (50 nm, imaged in buffer, and 2 nm, imaged in a refraction index = 1.42 Invitrogen Antifade medium). (B and C) Immobilization of a Cy3-ssDNA on diamond surfaces. This is achieved via either biotin–streptavidin interaction (B) or SPAAC (C). (D) A representative area of single-molecule fluorescence images of in B and the time traces of five selected fluorescence spots.
The developed diamond surface modification architecture can be readily combined with most well-established biochemical conjugation techniques. We demonstrate the versatility of our approach with two examples of bioconjugation: first, a molecular biological conjugation of target molecules to the diamond surface via biotin–streptavidin interaction and, second, a biochemical conjugation via “click chemistry.” For both examples, a Cy3 dye-labeled, 40-nt, single-stranded DNA (Cy3-ssDNA) served as a model molecule. In the first system (Fig. 2B), biotinylated Cy3-ssDNA was immobilized to the diamond surface mediated by streptavidin with no fluorescent label (nSA). Time-dependent fluorescence measurements show a single-step decay in the fluorescence signal for the majority of fluorescence spots, a hallmark for single-molecule measurements (Figs. 2D and SI Appendix, Fig. S11, also Movie S1) (20). In addition, we also observed two-step decay events, which can be explained by multiple ssDNA molecules binding to a single nSA homotetramer or coincidental colocalization of two nSA molecules. The second system exploited strain-promoted azide-alkyne cycloaddition (SPAAC), also known as “copper-free click chemistry” (Fig. 1E), for its reliable performance and fast kinetics (21). The diamond surfaces were prepared following the same procedure, except that biotinPEG was replaced by an azidePEG compound. Through SPAAC, the same Cy3-ssDNA engineered with a dibenzocyclooctyne (DBCO) label at its 5′ was successfully immobilized to diamond surfaces (Figs. 2C and SI Appendix, Fig. S11, also Movie S2). The large fluorescence spots, which include more than one fluorophore, were likely originated from the presence of aggregates in the DBCO-tagged Cy3-ssDNA sample.
Stability under Physiological Conditions
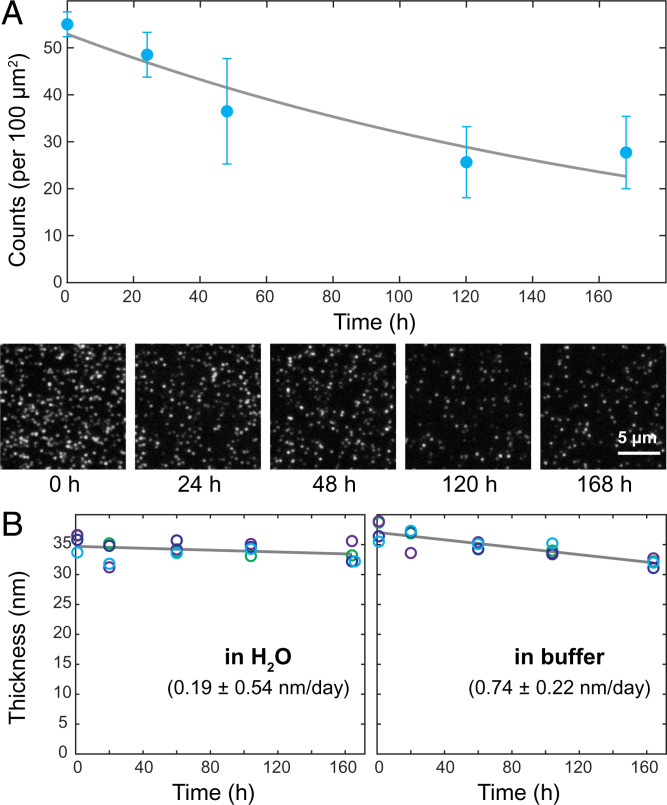
For any practical biophysical or diagnostics applications, it is important that the functionalization layer maintains good chemical stability without degrading over the course of a typical experiment. Oxides are known to hydrolyze over time when exposed to water, at a rate that depends on the film (e.g., composition, deposition method, and film quality) and solvent (e.g., pH and salinity) properties (22). Because the exact mechanism is difficult to predict, we experimentally tested the chemical stability of our functionalization architecture. We immobilized SA-488 to the diamond surface and monitored their fluorescence over the course of a week while storing the sample in a sodium phosphate buffer (pH 7.4, [NaH2PO4 + Na2HPO4] = 50 mM, [NaCl] = 100 mM) at 23.5 °C room temperature. Fig. 3A shows the observed fluorescence signal over time. The decrease in the number of SA-488 per field-of-view can be attributed to either a dissociation of the functionalization layer or photobleaching of the SA-488. This sets an upper limit for the functional layer dissociation rate to a half-life time of 5.7 d. In addition to these optical measurements, we also used AFM to monitor changes of the Al2O3 layer thickness as a function of submersion time in doubly deionized (DDI) water and sodium phosphate buffer. Fig. 3B shows the thickness of a lithographically patterned Al2O3 structure as a function of submersion time measured by AFM (SI Appendix, Fig. S12). We note that in water the dissociation is negligible (observed rate 0.19 ± 0.54 nm/d with a large uncertainty), which is in good agreement with the optical measurements. However, for the measurements in sodium phosphate buffer, we determined a dissociation rate of 0.74 ± 0.22 nm/d (and 1.02 ± 0.56 nm/d at 37 °C shown in SI Appendix, Fig. S13), which is slightly larger than what would be expected from the optical measurements in Fig. 3A and from the direct optical observation of the lithographic Al2O3 patterns in SI Appendix, Fig. S14. The observed optical and AFM results suggest that the Al2O3 layer on diamond undergoes a continuous dissolution process rather than a sudden detachment.
Fig. 3.
Time stability of the functionalization architecture in a physiological relevant environment. (A) Number of SA-488 molecules per 100-μm2 area (blue circles) detected by single-molecule fluorescence microscopy as a function of storage time in sodium phosphate buffer (pH 7.4, [NaH2PO4 + Na2HPO4] = 50 mM, [NaCl] = 100 mM) over a course of 1 wk. Each data point is based on three 2,800-μm2 field-of-view areas; error bars indicate one SD. Fit is an exponential decay. Representative single-molecule microscopy images on a 50-nm-thick Al2O3 layer are displayed at the bottom. (B) The overall thicknesses of the functional layer prepared on a 35-nm-thick, lithographically patterned Al2O3 structure in H2O (Left) and sodium phosphate buffer (Right) were tracked by AFM over a course of 1 wk at room temperature. Four unique sites (circles of the same color) were monitored for each sample, and the mean values were fitted to a linear model (gray), which allowed us to estimate the dissolution rates.
Qubit Coherence
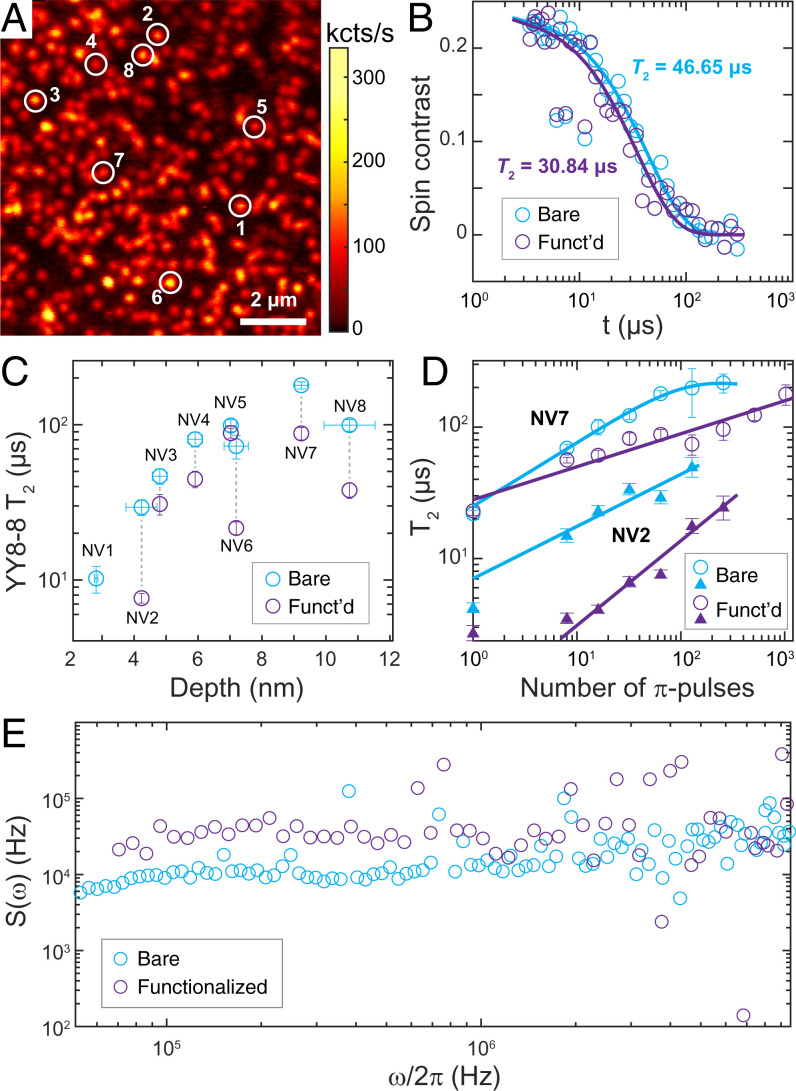
In parallel, we studied the impact of our functionalization architecture on the spin coherence (T2) of near-surface NV centers. Long coherence times are essential to NV-based quantum sensing because the sensitivity is generally proportional to (23). Fig. 4B shows an example of the coherence time of an NV center under (YY-8)N=8 dynamical decoupling (SI Appendix, Fig. S15 for pulse sequence diagram) before and after surface modification. YY-8 sequences were chosen for their robustness to pulse errors and the ability to suppress spurious signals from nearby nuclear spins (24–26). The observed coherence follows a stretched exponential exp[−(t/T2)n], with T2 = 47 μs before and T2 = 31 μs after the functionalization (the exponent n in Fig. 4 B and C ranges from 1 to 1.8). We further systematically investigated T2 and the longitudinal spin relaxation (T1) times of eight spatially resolved NV centers with depths ranging from 2.3 to 11 nm (Fig. 4C, and for NV position Fig. 4A), where the NV depths were determined by probing noise from the environmental 1H spins in the immersion oil following the method described in ref. 27. All investigated NV centers, with the exception of the shallowest NV (depth 2.3 nm), maintained their coherence after functionalization, with an observed characteristic increase in T2 as a function of NV depth (15). Overall, the T2 of these NVs upon surface functionalization is reduced by 49 ± 22% under (YY-8)N=8, or 15 ± 18% when using a spin-echo sequence (SI Appendix, Fig. S16). A careful investigation on spectral decomposition reveals a broadband noise spectrum across the frequency range of 0.05 to 10 MHz (Figs. 4B and SI Appendix, Fig. S17). We did not register a sizable reduction in T1 time (SI Appendix, Fig. S18) after surface treatment, suggesting that charge and magnetic field noise spectra have neglectable frequency components at 3 GHz. We note that the observed decrease in NV coherence cannot be explained by the presence of an 27Al nuclear spin bath (SI Appendix, Fig. S19) but can be attributed to a noisy environment introduced by paramagnetic defects in the Al2O3 layer (28).
Fig. 4.
Impact of functionalization on NV electron spin coherence. (A) Confocal scan of near-surface NV centers (implantation energy 3 keV). The eight NV centers studied for their coherence times are marked by circles. (B) Typical time trace of coherence measured by a (YY-8)N=8 sequence before (blue) and after (purple) functionalization for NV center number 3 (depth 4.8 nm). T2 times are based on the fitted, stretched exponential decays (solid lines). (C) T2 measured by (YY-8)N=8 pulse sequence (total of 64 -pulses) plotted against NV depth before (blue) and after (purple) functionalization. Depth calibration was performed following ref 27. (D) T2 times as a function of number of -pulses for NV number 2 (depth 4.2 nm, triangles) and NV number 7 (depth 9.2 nm, open circles) before (blue) and after (purple) functionalization. Solid lines are fits based on Eqs. 1 and 2. (E) Spectral decomposition manifests a broadband noise spectrum across the frequency range of 0.05 to 10 MHz for NV number 7. All measurements were carried out at 1,750-G magnetic field strength.
Discussion
In conclusion, we developed a chemically stable, universal diamond surface functionalization architecture that can be combined with most of the established, biochemical conjugation techniques. While we demonstrated biotin–SA conjugation and SPAAC click chemistry, our functionalization approach can be readily extended to other conjugation techniques, such as maleimide reaction (29), Ni2+/His-tag interaction (30), and sortase-mediated enzymatic conjugation (31). Combined with single-molecule fluorescence imaging techniques, we have shown that this architecture allows for a precise control over the conjugation density of individual target proteins and DNA molecules. The observed NV coherence times of up to 100 μs are long enough to perform highly sensitive, state-of-the-art quantum sensing experiments on biological targets (6–8). Based on the demonstrated sensor–target distances and qubit coherence, we predict that the NMR signal of an individual 13C nuclear spin can be detected with integration times as short as 100 s (see Methods). The anticipated integration time can further be reduced by minimizing the overall thickness of the functionalization layer and increasing the NV coherence time. A decrease in the functionalization layer thickness can be achieved by the deposition of a sub–1-nm Al2O3 layer and the passivation with shorter PEG, whereas the coherence time can be increased through further material processing, such as optimization of Al2O3 growth parameters and additional annealing after Al2O3 deposition, as well as increasing the number of -pulses during dynamical decoupling (SI Appendix, Fig. S20). Finally, we note that our silanization technique could be extended to directly conjugate −OH-terminated diamond surfaces, eliminating the need for an Al2O3 adhesion layer. While creating a high-quality, −OH-terminated surface on (100) bulk diamond remains in an open-surface chemistry challenge, recent computational models (32) have suggested that near-surface NV centers can remain charge-stable in the presence of –OH termination, making this an interesting direction for future pursuits.
During the preparation of this manuscript, we became aware of recent work described in ref. 33, which applied NV sensing to probe the NMR signature of a self-assembled monolayer of organic molecules on an Al2O3-coated diamond sensors. Combining this (33) and other NV sensing techniques (6–8) with our molecular “pulldown” experiments will enable NMR and EPR spectroscopy of intact biomolecules in a relevant biological environment. Existing microfluidics platforms (34) can readily be combined with our diamond passivation and functionalization method, which will pave the way to label-free, high-throughput biosensing with applications in quality management in the pharmaceutical industry (35, 36), screening for targets in drug discovery (37), single-cell screening for metabolomics (38), proteomics (39), and detection of cancer markers (40). Furthermore, positioning individual biomolecules within the 10-nm sensing range of a single NV center brings us closer to performing EPR and NMR spectroscopy on individual-intact biomolecules. When combined with nanowire-assisted delivery platforms (41), such a technology could enable single-molecule magnetic resonance spectroscopy within the context of a cell. Magnetic resonance spectroscopy with single-molecule sensitivity could provide insights into receptor–ligand binding events (37), posttranslational protein modification [e.g., phosphorylation processes (42)], and the detection of subtle protein conformational changes in living cells (43), which can enhance our understanding of complex signaling pathways that are not accessible by current technologies.
Materials and Methods
Functionalization.
Single-crystalline diamonds slabs (2 × 2 × 0.5 mm3, Element Six, electronic grade, Catalog No. 145-500-0385) were sonicated in acetone and isopropanol for 5 min each and dried with nitrogen gas before Al2O3 deposition. The deposition of Al2O3 layer was carried out in an Ultratech/Cambridge Savannah ALD System by alternatingly delivering trimethylaluminum and H2O gases at 200 °C, 20 cycles for 2-nm layer and 550 cycles for 50 nm Al2O3 thickness. The diamonds were then soaked in 10 mM KOH for 10 s before being rinsed with a copious amount of DDI water (Milli-Q), and dried in an oven set to 80 °C. Silanization was achieved using freshly prepared 3% N-[3-(trimethoxysilyl)propyl]ethylenediamine (CAS 1760-24-3, ACROS Organics, Catalog No. AC216531000) in anhydrous acetone (extra dry, ACROS Organics, Catalog No. AC326801000) at room temperature for 20 min. Upon completion, the surfaces were rinsed with acetone and DDI water and dried with nitrogen gas. For PEGylation, solutions of heterobifunctional PEG of various molecular weights (m.w.) were freshly prepared at ∼0.5 M concentration in 100-mM NaHCO3 buffer with a final pH between 8.0 to 8.5. Specifically, heterobifunctional PEG molecules mPEG-SVA (average m.w. 2,000 and 5,000) and biotinPEG-SVA (average m.w. 3,400 and 5,000) were purchased from Laysan Bio, azidoPEG-NHS (average m.w. 5,000, Catalog No. JKA5086) were purchased from Sigma-Aldrich, and mPEG9-NHS (m.w. 553.6, Catalog No. BP-22624) and biotinPEG8-NHS (m.w. 764.9 Catalog No. BP-22117) were purchased from BroadPharm. The diamonds were immersed in the PEG solutions and incubated for 1 to 2 h in the dark at room temperature, before being extensively washed with DDI water and dried with nitrogen gas.
Immobilization of Biomolecules.
To immobilized biomolecules, 3 μL of 7-nM SA-488 (in 50 mM pH 7.4 sodium phosphate buffer that also contained 100 mM NaCl) or 50 nM DBCO-tagged Cy3-ssDNA (in 50 mM pH 7.4 sodium phosphate buffer, 100 mM NaCl, which also contained 1 mM MgSO4) was carefully cast on the functionalized surface of each diamond and incubated at room temperature in a dark, moisturized environment for 20 min. For the streptavidin-mediated system, 20 nM biotin-tagged Cy3-labeled ssDNA was premixed with 40 nM nSA at 1: 1 volume ratio and incubated at room temperature for 20 min with mild agitation, which should predominantly result in 1-ssDNA: 1-streptavidin conjugates whose effective concentration was 10 nM. This solution was then applied to diamond surfaces in the same way. Upon completion, the diamond was gently rinsed with the same buffer and placed in an imaging dish for fluorescence microscopy study. The primary sequence of the 40-nt ssDNA is 5′-TTTTT TTTTT AGTCC GTGGT AGGGC AGGTT GGGGT GACTT-3′. The biotin-tag (or DBCO-tag), followed by a Cy3-label, is attached to the 5′ end of the ssDNA.
Single-Molecule Imaging.
Single-molecule fluorescence imaging was performed on a custom-built fluorescence microscope equipped with 488- and 532-nm lasers (Coherent Sapphire) and a 60× oil objective (Olympus UPLAPO60XOHR) in an inverted configuration with epi-illumination. Diamond samples were placed inside a buffer-containing dish with the functionalized side facing down and imaged through the glass coverslip bottom that was also passivated with mPEG to minimize nonspecific binding (SI Appendix, Fig. S21). For 488-nm excitation (SA-488), a ZET488/10x (Chroma) notch filter, a ZT488rdc-UF1 dichroic beamsplitter, and an ET525/50m emission filter were used. For 532-nm excitation (Cy3-ssDNA), a ZET532/10x notch filter, a ZT532rdc-UF1 dichroic beamsplitter, and an ET575/50m emission filter were used. Images were acquired by an Andor iXon Ultra 888 electron-multiplying charge-coupled device (EMCCD) camera (EMCCD cooled down to −60 °C) with 1 s exposure time and 200 (or 300) gain, or 500 × 120 ms for video.
Resetting Surface.
Diamonds were first soaked in 1 M KOH (typically overnight but can be shortened), which effectively removes Al2O3 at a rate of 3.6 nm/min (SI Appendix, Fig. S6). They were then immersed in NanoStrip (99% sulfuric acid + 1% H2O2) at room temperature for 5 min, rinsed extensively with DDI water, sonicated in acetone and 2-isopropanol for 5 min each, and finally dried with nitrogen gas. This cleaning procedure proved to be reliable to restore the diamond for single-molecule fluorescence imaging experiments. For spin coherent measurements, while the above-mentioned regenerating procedure may be sufficient, we in practice replaced the NanoStrip treatment by triacid cleaning, which uses a 1:1:1 mixture of nitric acid, perchloric acid, and sulfuric acid at boiling temperatures (44) to ensure minimal contamination.
NV Measurement Setup.
The electronic spin of individual NV centers was initialized and read out using a 520-nm green laser (Labs-Electronics, DLnsec). The spins were coherently manipulated by a microwave signal generator (Stanford Research Systems, SG 396) with a build-in in-phase and quadrature (IQ) modulator. The microwave pulse phase and length were controlled by an arbitrary waveform generator (Zurich Instrument, HDAWG8-ME) via IQ modulation. The modulated microwave was then amplified with a high-power amplifier (Mini-Circuits ZHL-16W-43+) and delivered via a coplanar waveguide to the diamond sample. A home-built confocal microscope was used to collect NV fluorescence, which was equipped with a dichroic beam splitter (Chroma, T610lpxr) to separate excitation and emission pathways. The emission was detected by a single-photon counter (Excelitas SPCM-AQRH-14) and processed by a time tagger (Swabian Instruments, Time Tagger 20). Confocal scanning was achieved by a piezo scanner (Mad City Lab, NANOM 350). External magnetic fields were provided by a neodymium-permanent magnet (K&J Magnet), which was mounted on a motorized translation stage with four degrees of freedom (Zaber technologies) for full control over the magnetic field alignment. The involved devices were synchronized and triggered by a transistor–transistor logic pulse generator (Swabian Instruments, Pulse Streamer 8/2).
Coherence Measurement.
The diamond sample used for NV spin coherence measurement was prepared following the procedure outlined in ref. 15. Briefly, a scaife-polished diamonds sample (Element Six, Catalog No. 145-500-0385) was etched with Ar/Cl2 followed by O2 plasma to remove the top few micrometers of material. We then intentionally graphitized the top few nanometers by annealing to 1,200 °C for 2 h in a vacuum tube furnace. The diamond was then implanted with 15N by Innovion Corporation (3 keV, 3 × 109/cm2, 0° tilt) and subsequently annealed to 800 °C in a vacuum tube furnace to form NV centers, followed by oxygen annealing at 460 °C. Coherence measurements before and after surface modification were performed at 1,750 G magnetic field strength. We use both YY-8 and spin-echo pulse sequences to measure the coherence and depth of NV centers.
The normalized signal (spin contrast) is defined by 2[F0(t) − F1(t)]/[F0(t) + F1(t)], which removes the common-mode noise [F0(t) refers to the detected fluorescence in |0⟩ and F1(t) to that in |−1⟩]. The coherence data in Fig. 4 are fitted to a stretched exponential function exp[−(t/T2)n]. T2 and number of π-pulse at which T2 saturates are fitted to either a saturation curve,
| [1] |
or to a power law,
| [2] |
where no saturation of T2 is observed (15). The final fitting parameters are given in Table S2.
Estimated Integration Time for Single–13C Spin Detection.
Based on the experimental parameters in Fig. 4B, we estimate the required integration time to detect a target nuclear spin. In NV sensing, the signal from an individual target spin is given by , where denotes the total measurement time, the phase accumulation time, the optical spin-readout time, () the average number of detected photons per readout window in ms = 0 (ms = 1), the spin contrast from Fig. 4B, the hyperfine coupling between the NV and the target spin, and the reduced Planck constant. The noise (SD) is given by . The NV center in Fig. 4B has a depth of 4.8 nm, with an additional 5 nm for the surface functionalization (i.e., 2 nm Al2O3 and the subsequent silanization and PEGylation using only mPEG). Taking into account that the diamond has a (1,0,0) cut, we estimate the average interaction strength between NV centers and 13C spins to be . Based on these parameters, we estimate the required integration time to be 2.8 h. These demanding integration times can further be reduced to 100 s by utilizing quantum logic sequences (e.g., 100 repetitions) (1).
Supplementary Material
Acknowledgments
We thank Dr. Nazar Delegan and Dr. Joseph Heremans for insightful discussions on diamond surface termination; Xinghan Guo, Dr. Noelia Bocchio, and Dr. Alexander High for help on ALD deposition and patterning; Dr. Jon Monserud, Sheela Waugh, Dr. Jason Cleveland, and Dr. Larry Gold for providing Cy3-ssDNA constructs and discussions on single-molecule imaging; and Uri Zvi, Zhendong Zhang, Dr. Joonhee Choi, and Hengyun Zhou for discussions and insights on quantum sensing. M.X., X.Y., and P.C.M. acknowledge financial support from the NSF Grant No. OMA-1936118 and OIA-2040520, and NSF QuBBE QLCI (NSF OMA- 2121044) and the Swiss National Science Foundation (SNSF) Grant No. 176875. X.Y. in addition acknowledges support from the US Department of Energy 1F-60579. N.Q. and A.T. acknowledge financial support from the SNSF Grant No. 183717. L.V.H.R. acknowledges support from the US Department of Defense through the National Defense Science and Engineering Graduate Fellowship Program. N.P.d.L. acknowledges support from the NSF Grant No. OMA-1936118 and DMR1752047. We acknowledge the use of the Pritzker Nanofabrication Facility at the University of Chicago (NSF ECCS-2025633), the University of Chicago Materials Research Science and Engineering Center (DMR-2011854), as well as the Imaging and Analysis Center at Princeton University (DMR-2011750).
Footnotes
Competing interest statement: The authors have filed a provisional patent application for the diamond functionalization process described in this manuscript.
This article is a PNAS Direct Submission. F.J. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114186119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Lovchinsky I., et al. , Nuclear magnetic resonance detection and spectroscopy of single proteins using quantum logic. Science 351, 836–841 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Shi F., et al. , Protein imaging. Single-protein spin resonance spectroscopy under ambient conditions. Science 347, 1135–1138 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Shi F., et al. , Single-DNA electron spin resonance spectroscopy in aqueous solutions. Nat. Methods 15, 697–699 (2018). Correction in: Nat. Methods 15, 749 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Schmitt S., et al. , Submillihertz magnetic spectroscopy performed with a nanoscale quantum sensor. Science 356, 832–837 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Boss J. M., Cujia K. S., Zopes J., Degen C. L., Quantum sensing with arbitrary frequency resolution. Science 356, 837–840 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Glenn D. R., et al. , High-resolution magnetic resonance spectroscopy using a solid-state spin sensor. Nature 555, 351–354 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Aslam N., et al. , Nanoscale nuclear magnetic resonance with chemical resolution. Science 357, 67–71 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Smits J., et al. , Two-dimensional nuclear magnetic resonance spectroscopy with a microfluidic diamond quantum sensor. Sci. Adv. 5, eaaw7895 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abobeih M. H., et al. , Atomic-scale imaging of a 27-nuclear-spin cluster using a quantum sensor. Nature 576, 411–415 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Raymakers J., Haenen K., Maes W., Diamond surface functionalization: From gemstone to photoelectrochemical applications. J. Mater. Chem. C 7, 10134–10165 (2019). [Google Scholar]
- 11.Kawai S., et al. , Nitrogen-terminated diamond surface for nanoscale NMR by shallow nitrogen-vacancy centers. J. Phys. Chem. C 123, 3594–3604 (2019). [Google Scholar]
- 12.Li C., et al. , Systematic comparison of various oxidation treatments on diamond surface. Carbon 182, 725–734 (2021). [Google Scholar]
- 13.Nebel C. E., Rezek B., Shin D., Uetsuka H., Yang N., Diamond for bio-sensor applications. J. Phys. D Appl. Phys. 40, 6443 (2007). [Google Scholar]
- 14.Hauf M. V., et al. , Chemical control of the charge state of nitrogen-vacancy centers in diamond. Phys. Rev. B Condens. Matter 83, 081304 (2011). [Google Scholar]
- 15.Sangtawesin S., et al. , Origins of diamond surface noise probed by correlating single-spin measurements with surface spectroscopy. Phys. Rev. X 9, 031052 (2019). [Google Scholar]
- 16.Reina G., Zhao L., Bianco A., Komatsu N., Chemical functionalization of nanodiamonds: Opportunities and challenges ahead. Angew. Chem. Int. Ed. Engl. 58, 17918–17929 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Ryan R. G., et al. , Impact of surface functionalization on the quantum coherence of nitrogen-vacancy centers in nanodiamonds. ACS Appl. Mater. Interfaces 10, 13143–13149 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Dahan M., et al. , Ratiometric measurement and identification of single diffusing molecules. Chem. Phys. 247, 85–106 (1999). [Google Scholar]
- 19.Jain A., Liu R., Xiang Y. K., Ha T., Single-molecule pull-down for studying protein interactions. Nat. Protoc. 7, 445–452 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ha T., Tinnefeld P., Photophysics of fluorescent probes for single-molecule biophysics and super-resolution imaging. Annu. Rev. Phys. Chem. 63, 595–617 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dommerholt J., Rutjes F. P. J. T., van Delft F. L., Strain-promoted 1,3-dipolar cycloaddition of cycloalkynes and organic azides. Top. Curr. Chem. (Cham) 374, 16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang S.-K., et al. , Dissolution behaviors and applications of silicon oxides and nitrides in transient electronics. Adv. Funct. Mater. 24, 4427–4434 (2014). [Google Scholar]
- 23.Taylor J. M., et al. , High-sensitivity diamond magnetometer with nanoscale resolution. Nat. Phys. 4, 810–816 (2008). [Google Scholar]
- 24.Shu Z., et al. , Unambiguous nuclear spin detection using an engineered quantum sensing sequence. Phys. Rev. A 96, 051402 (2017). [Google Scholar]
- 25.Choi J., et al. , Robust dynamic Hamiltonian engineering of many-body spin systems. Phys. Rev. X 10, 031002 (2020). [Google Scholar]
- 26.Zhou H., et al. , Quantum metrology with strongly interacting spin systems. Phys. Rev. X 10, 031003 (2020). [Google Scholar]
- 27.Pham L. M., et al. , NMR technique for determining the depth of shallow nitrogen-vacancy centers in diamond. Phys. Rev. B 93, 045425 (2016). [Google Scholar]
- 28.de Graaf S. E., et al. , Direct identification of dilute surface spins on Al2O3: Origin of flux noise in quantum circuits. Phys. Rev. Lett. 118, 057703 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Zimmermann J. L., Nicolaus T., Neuert G., Blank K., Thiol-based, site-specific and covalent immobilization of biomolecules for single-molecule experiments. Nat. Protoc. 5, 975–985 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Zhen G., et al. , Nitrilotriacetic acid functionalized graft copolymers: A polymeric interface for selective and reversible binding of histidine-tagged proteins. Adv. Funct. Mater. 16, 243–251 (2006). [Google Scholar]
- 31.Srinivasan S., Hazra J. P., Singaraju G. S., Deb D., Rakshit S., ESCORTing proteins directly from whole cell-lysate for single-molecule studies. Anal. Biochem. 535, 35–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaviani M., et al. , Proper surface termination for luminescent near-surface NV centers in diamond. Nano Lett. 14, 4772–4777 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Liu K. S., et al. , Surface NMR using quantum sensors in diamond. Proc. Natl. Acad. Sci. U.S.A.119, e2111607119 (2022). [DOI] [PMC free article] [PubMed]
- 34.Andrich P., et al. , Engineered micro- and nanoscale diamonds as mobile probes for high-resolution sensing in fluid. Nano Lett. 14, 4959–4964 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Bingol K., et al. , Emerging new strategies for successful metabolite identification in metabolomics. Bioanalysis 8, 557–573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalfe A., Telfah A., Lambert J., Hergenröder R., Looking into living cell systems: Planar waveguide microfluidic NMR detector for in vitro metabolomics of tumor spheroids. Anal. Chem. 87, 7402–7410 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Meyer B., Peters T., NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew. Chem. Int. Ed. Engl. 42, 864–890 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Markley J. L., et al. , The future of NMR-based metabolomics. Curr. Opin. Biotechnol. 43, 34–40 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cleveland J. P., Holczer K., Vant-Hull B. P. J., “Proteomic assay using quantum sensors.” US Patent 16/686911 (2019).
- 40.Glenn D. R., et al. , Single-cell magnetic imaging using a quantum diamond microscope. Nat. Methods 12, 736–738 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shalek A. K., et al. , Vertical silicon nanowires as a universal platform for delivering biomolecules into living cells. Proc. Natl. Acad. Sci. U.S.A. 107, 1870–1875 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Selenko P., et al. , In situ observation of protein phosphorylation by high-resolution NMR spectroscopy. Nat. Struct. Mol. Biol. 15, 321–329 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Hänsel R., Luh L. M., Corbeski I., Trantirek L., Dötsch V., In-cell NMR and EPR spectroscopy of biomacromolecules. Angew. Chem. Int. Ed. Engl. 53, 10300–10314 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Brown K. J., Chartier E., Sweet E. M., Hopper D. A., Bassett L. C., Cleaning diamond surfaces using boiling acid treatment in a standard laboratory chemical hood. J. Chem. Health Saf. 26, 40–44 (2019). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.