Significance
Many forms of cardiomyopathy manifest with changes in sarcomeric structure, function, and energetics. We used small-angle X-ray diffraction and myosin super-relaxed (SRX) state approaches to investigate the mechanisms underlying the clinical phenotypes associated with HCM-related D166V (aspartate-to-valine) and DCM-linked D94A (aspartate-to-alanine) mutations in the cardiac myosin RLC (MYL2 gene). Modulation of myosin function through dysregulation of the SRX state was closely coupled with structural rearrangements and the Ca2+ dependence of force development in HCM–D166V mice. The DCM–D94A model favored the SRX state without altering structure/force–pCa relationships. Understanding the regulation of SRX ↔ DRX equilibrium in the normal heart and how it is changed in heart disease may advance future therapeutics of patients suffering from the mutated MYL2 gene.
Keywords: super-relaxed state of myosin, interfilament lattice spacing, equatorial intensity ratio, isometric force, transgenic RLC mice
Abstract
In this study, we investigated the role of the super-relaxed (SRX) state of myosin in the structure–function relationship of sarcomeres in the hearts of mouse models of cardiomyopathy-bearing mutations in the human ventricular regulatory light chain (RLC, MYL2 gene). Skinned papillary muscles from hypertrophic (HCM–D166V) and dilated (DCM–D94A) cardiomyopathy models were subjected to small-angle X-ray diffraction simultaneously with isometric force measurements to obtain the interfilament lattice spacing and equatorial intensity ratios (I11/I10) together with the force-pCa relationship over a full range of [Ca2+] and at a sarcomere length of 2.1 μm. In parallel, we studied the effect of mutations on the ATP-dependent myosin energetic states. Compared with wild-type (WT) and DCM–D94A mice, HCM–D166V significantly increased the Ca2+ sensitivity of force and left shifted the I11/I10-pCa relationship, indicating an apparent movement of HCM–D166V cross-bridges closer to actin-containing thin filaments, thereby allowing for their premature Ca2+ activation. The HCM–D166V model also disrupted the SRX state and promoted an SRX-to-DRX (super-relaxed to disordered relaxed) transition that correlated with an HCM-linked phenotype of hypercontractility. While this dysregulation of SRX ↔ DRX equilibrium was consistent with repositioning of myosin motors closer to the thin filaments and with increased force-pCa dependence for HCM–D166V, the DCM–D94A model favored the energy-conserving SRX state, but the structure/function–pCa data were similar to WT. Our results suggest that the mutation-induced redistribution of myosin energetic states is one of the key mechanisms contributing to the development of complex clinical phenotypes associated with human HCM–D166V and DCM–D94A mutations.
Inherited cardiomyopathies are a class of heart diseases caused by variations in multiple genetic loci, mostly originating from point mutations in sarcomeric proteins (1). The majority of hypertrophic cardiomyopathy (HCM)–causing mutations reside in human β-cardiac myosin (∼35%) and cardiac myosin-binding protein C (∼35%); however, according to recent genetic studies, mutations in the human cardiac regulatory light chain (RLC, MYL2 gene) are more common than previously reported, and they are often associated with adverse clinical outcomes (reviewed in refs. 2 and 3). In this study, we focused on the D166V (aspartate166 → valine)-RLC mutation shown to result in HCM (4) and the D94A (aspartate94 → alanine)–RLC mutation associated with dilated cardiomyopathy (DCM) in patients (5). Two “humanized” mouse models were explored, expressing human ventricular HCM–D166V and DCM–D94A MYL2 variants in mice (6, 7), and they served as model systems for investigating the mechanisms underlying human HCM and DCM.
The myosin RLC is a major subunit of striated-muscle myosin and a modulator of Ca2+ and tropomyosin–troponin-regulated cardiac muscle contraction (8). Together with the myosin essential light chain (ELC), the RLC binds to the lever arm domain of the myosin head, suggesting important roles for both light chains in maintaining the structural and functional integrity of the lever arm during the power stroke and sarcomere shortening (9, 10). Structurally, the RLC belongs to the EF-hand calcium-binding protein family and contains a homologous helix–loop–helix region composed of a 12-amino-acid Ca2+-binding loop flanked by two perpendicular α-helices (EF-hand motif) (8). The human cardiac RLC also contains a highly conserved N-terminal phosphorylatable serine (Ser-15), a target of cardiac myosin light chain kinase (11). Our previous investigations implied that both properties of the RLC, Ca2+ binding and Ser-15 phosphorylation, determine the role that RLC plays in cardiac muscle contraction in healthy and cardiomyopathic muscle (12, 13). The phosphorylation of cardiac RLC was shown to directly modulate the Ca2+-dependent force development and the kinetics of attachment and detachment of cycling myosin cross-bridges (14–16), and RLC phosphorylation-mediated sensitization to calcium was attributed to the switch between the OFF and ON conformations of myosin motors and a movement of myosin heads toward the thin filaments (17, 18).
Considering the importance of myosin RLC for the actin–myosin interaction and force production in the heart, it is not surprising that genetic mutations in MYL2 can result in structural and functional alterations and lead to cardiomyopathy in humans. To better understand the interplay between the mutation-induced molecular insult and heart dysfunction, we examined the effect of HCM–D166V and DCM–D94A mutations expressed in mouse models of HCM and DCM (6, 7) on sarcomeric structure, force production, and myosin energetic states using left ventricular (LV) papillary muscles (PM) from the hearts of mutant versus wild-type (WT) mice expressing a nonmutated human ventricular RLC. Importantly, we monitored the structural determinants of muscle contraction simultaneously with force development over the full range of Ca2+-activated force from relaxed through submaximal to maximum Ca2+ activation.
Small-angle X-ray diffraction is the technique of choice to obtain structural information concerning the sarcomere under physiological or disease-like conditions. The equatorial diffraction patterns yield two structural parameters, the interfilament lattice spacing (d10) that is proportional to the center-to-center distance between two adjacent thick and thin filaments (19) and the equatorial intensity ratio (I11/I10). I11/I10 is the ratio of the integrated intensity of the 1,1 equatorial reflections originating from the myosin and actin filaments to the 1,0 equatorial reflections arising from the myosin-containing thick filaments. These two parameters have been extensively used to characterize the normal sarcomere structure, but they also enable the assessment of the effects of various mutations in sarcomeric proteins on the proximity of myosin heads to actin, providing insights into the structural basis of disease phenotypes (18, 20, 21). I11/I10 reveals information about the proximity of myosin heads to the actin-containing thin filaments and is directly proportional to the number of attached cross-bridges in both cardiac (22) and skeletal (23) muscles. A relatively high value of I11/I10 indicates that myosin heads are more closely associated with the actin filaments, and a low value depicts myosin heads more closely associated with the thick filament backbone, apparently reflecting the formation of the “interacting-heads motif” (IHM) (22, 24). The IHM is thought to be, but not yet proven, the structural basis of the recently discovered super-relaxed (SRX) state of myosin (25, 26) that is considered central to modulating sarcomeric force production and energy utilization in cardiac muscle (27–29). The SRX is an energy-conserving state in which myosin cross-bridges cycle with a highly inhibited ATP turnover rate (25, 26). It has 10- to 100-fold slower ATPase than the already known non-actin–bound, disordered relaxed (DRX) state in which myosin heads protrude into the interfilament space but are restricted from binding to actin compared with SRX heads that are neatly ordered around the thick filament backbone (30). Structurally, the SRX state is associated with the IHM, comprised of the primary head–head interaction site of the “blocked” head and the converter domain of the “free” head, with both heads locked into the thick filament backbone, thus inhibiting ATP hydrolysis and withdrawing both heads from thin filament interaction and force production (1, 28, 31). This asymmetric IHM formation is presumed to define the SRX state of thick filaments in the heart (22) and could explain the structural origin of the slow (250 to 300 s) versus fast (<30 s) ATP turnover time in cardiac muscle (30, 31).
Under resting conditions, myosin heads can exist in a wide range of structural states and proximities to actin filaments, each associated with different rates of energy consumption (1). The relaxation phase of the cardiac cycle is critically important for normal heart function, and the disruption of IHM structures may contribute to energetically compromised myocardium and inefficient heart performance (24, 27, 29). The SRX conformation is supported by protein–protein interactions of myosin heads with the ELC and RLC that serve as scaffolding proteins, and mutations in these structural components of the IHM may lead to destabilization of SRX and unbalanced SRX ↔ DRX equilibrium. We thus tested whether the SRX state is affected by the two pathogenic variants of RLC and explored the mutation-specific redistribution of myosin energetic states in the hearts of HCM–D166V and DCM–D94A versus WT mice.
Results
We aimed to investigate the impact of the two pathogenic MYL2 variants, HCM–D166V and DCM–D94A, on the Ca2+-dependent structure–function relationship and the effect of mutations in the RLC on myosin energetic states and the distribution of myosin cross-bridges between the SRX and DRX states. The X-ray experiments and force measurements were performed on skinned LVPM fibers of ∼5-mo-old male (M) and female (F) mice expressing ∼90% of human ventricular D166V RLC mutant (6), ∼50% D94A RLC mutant, and ∼90% of human nonmutated WT–RLC (UniProtKB P10916) (7). For SRX-pCa studies, LVPM were harvested from a larger group of animals that included 5- to 8-mo-old male and female HCM–D166V and DCM–D94A mice, and the data were compared with age-matched WT controls. All studies were carried out at room temperature (∼23 °C). Given the volume of structure/function–pCa data generated, SuperPlots (32) were used to clearly demonstrate the data points that were associated with statistically significant differences.
Assessment of X-ray Diffraction Patterns Concurrently with Force-pCa Traces in PM Fibers from HCM and DCM Models with Mutations in Myosin RLC.
To examine the spatial organization of the thick and thin filaments within the sarcomere, we employed low-angle X-ray diffraction of LVPM from mice, which reveals information concerning the distance between the thick and thin filaments (lattice spacing) and the relative degree of association of myosin heads with the actin-containing thin filaments. Lattice spacing, d10, and the equatorial intensity ratios, I11/I10, were determined in skinned LVPM of HCM–D166V, DCM–D94A, and WT mice at a sarcomere length of 2.1 μm. This structural information was obtained over a full range of Ca2+-activated force (from pCa 8 to 4) and correlated with simultaneously recorded force-pCa traces, allowing the structure–function analysis for all three genotypes.
HCM–D166V mutation and not DCM–D94A significantly affect equatorial intensity ratios and interfilament lattice spacing.
Measurements of X-ray diffraction patterns in skinned LVPM from mice at serial pCa solutions were assessed using 3M, 3F of WT; 2M, 2F of HCM–D166V; and 3M, 3F of DCM–D94A mice. The values obtained for several fibers per mouse were averaged and the data presented as mean ± SEM for N= number of mice. The significance (P < 0.05) was calculated by two-way ANOVA followed by Tukey’s multiple comparison test (Tables 1 and 2).
Table 1.
Assessment of X-ray diffraction patterns and I11/I10 ratios in PM fibers from WT, HCM–D166V, and DCM–D94A mice at serial pCa solutions
| pCa | WTI11/I10 | No. mice (M, F)No. fibers | HCM–D166VI11/I10 | No. mice (M, F) No. fibers | DCM–D94AI11/I10 | No. mice (M, F) No. fibers |
| 8 | 0.21 ± 0.01 | 6 (3, 3)14 | 0.39 ± 0.06* | 4 (2, 2)7 | 0.26 ± 0.03 | 6 (3, 3)14 |
| 7 | 0.26 ± 0.04 | 6 (3, 3)14 | 0.36 ± 0.06 | 4 (2, 2)7 | 0.25 ± 0.02 | 6 (3, 3)14 |
| 6 | 0.21 ± 0.02 | 6 (3, 3)14 | 0.36 ± 0.06 | 4 (2, 2)7 | 0.28 ± 0.04 | 6 (3, 3)13 |
| 5.4 | 0.27 ± 0.02 | 6 (3, 3)14 | 0.54 ± 0.05** | 3 (2, 1)6 | 0.30 ± 0.03+ | 6 (3, 3)13 |
| 5.2 | 0.42 ± 0.02 | 6 (3, 3)14 | 0.79 ± 0.07**** | 3 (2, 1)4 | 0.35 ± 0.03++++ | 6 (3, 3)13 |
| 4 | 0.64 ± 0.11 | 6 (3, 3)13 | 0.73 ± 0.11 | 3 (2, 1)3 | 0.58 ± 0.08 | 5 (2, 3)9 |
Data are mean ± SEM of N = number of mice with measurements for individual fibers (n) averaged per mouse. F, female; M, male. Significance was calculated by two-way ANOVA followed by Tukey’s multiple comparison test. *, **, ****P < 0.05, 0.01, and 0.0001 for mutant versus WT and +, ++++P < 0.05 and 0.0001 for DCM–D94A versus HCM–D166V.
Table 2.
Assessment of interfilament lattice spacings (d10) in PM fibers from WT, HCM–D166V, and DCM–D94A mice at serial Ca2+ concentrations
| pCa | WTd10 (nm) | No. mice (M, F) No. fibers | HCM–D166Vd10 (nm) | No. mice (M, F) No. fibers | DCM–D94Ad10 (nm) | No. mice (M, F) No. fibers |
| 8 | 41.58 ± 0.34 | 6 (3, 3)15 | 41.84 ± 0.45 | 4 (2, 2)7 | 41.18 ± 0.16 | 6 (3, 3)14 |
| 7 | 41.12 ± 0.30 | 6 (3, 3)14 | 42.07 ± 0.78 | 4 (2, 2)7 | 40.54 ± 0.26 | 6 (3, 3)14 |
| 6 | 40.60 ± 0.30 | 6 (3, 3)15 | 42.42 ± 0.39* | 4 (2, 2)7 | 40.69 ± 0.14+ | 6 (3, 3)14 |
| 5.4 | 41.10 ± 0.43 | 6 (3, 3)15 | 43.29 ± 0.81* | 4 (2, 2)7 | 41.72 ± 0.59 | 6 (3, 3)14 |
| 5.2 | 41.34 ± 0.36 | 6 (3, 3)15 | 42.86 ± 1.15 | 4 (2, 2)6 | 41.58 ± 0.31 | 6 (3, 3)13 |
| 4 | 41.19 ± 0.22 | 6 (3, 3)15 | 39.12 ± 0.9* | 4 (2, 2)6 | 41.41 ± 0.34+ | 6 (3, 3)12 |
Data are mean ± SEM of N = number of mice with measurements for individual fibers (n) averaged per mouse. F, female; M, male. Significance was calculated by two-way ANOVA followed by Tukey’s multiple comparison test. *P < 0.05 for mutant versus WT and +P < 0.05 for DCM–D94A versus HCM–D166V.
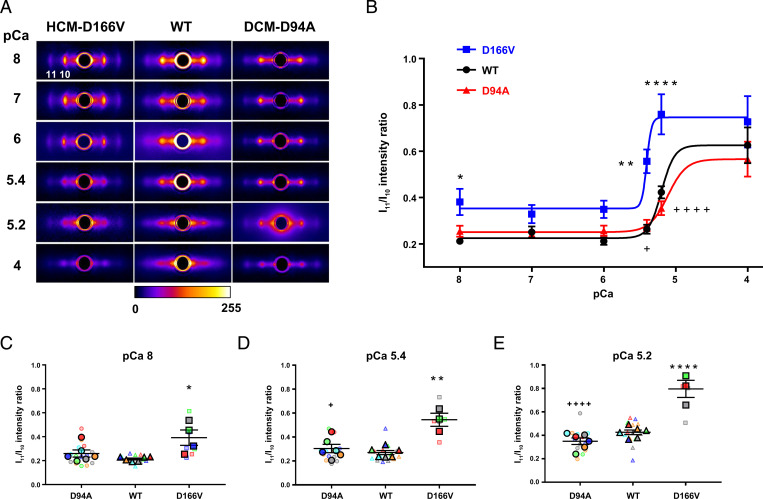
Fig. 1A shows representative equatorial X-ray diffraction patterns of LVPM fibers from all three genotypes at increasing calcium concentrations (from pCa 8 to 4). Because of the disordering of the myosin heads during activation, the absolute intensities of the 1,1 and 1,0 reflections were seen to be reduced at high calcium, but the ratio of the intensities I11/I10 increased with increasing [Ca2+]. The equatorial intensity ratios I11/I10 plotted as a function of [Ca2+] are presented in Fig. 1B. Among all three genotypes, the HCM–D166V model demonstrated the highest Ca2+ sensitivity of the I11/I10-pCa relationship, with pCa50 = 5.47 ± 0.15 compared with pCa50 = 5.20 ± 0.04 observed for WT and pCa50 = 5.11 ± 0.11 of DCM–D94A mice. The I11/I10 ratios determined at pCa 8, 5.4, and 5.2 are presented in the form of SuperPlots in Fig. 1 C–E, respectively. The values of I11/I10 per individual animal are color-coded and are superimposed with, respectively, color-coded measurements of I11/I10 obtained for multiple fibers. The results for pCa 8 and the submaximal calcium concentrations (pCa 5.4 and 5.2) indicated that more cross-bridges were formed in HCM–D166V samples compared with WT controls. No differences in I11/I10 were observed between the DCM–D94A model and WT, suggesting no changes in the redistribution of DCM–D94A heads between the vicinities of the thick and thin filaments.
Fig. 1.
Assessment of X-ray diffraction patterns in PM fibers from ∼5-mo-old DCM–D94A and HCM–D166V hearts compared with WT mice at serial pCa solutions. (A) Representative fiber diffraction patterns of HCM–D166V (Left), WT controls (Middle), and DCM–D94A (Right) models at increasing calcium concentrations from pCa 8 to 4. The legend shows the mapping of false colors into detector intensity units (ADUs). For clarity, the images had a circular symmetric diffuse background subtracted using the “quadrant fold” routine in the MuscleX software package (https://musclex.readthedocs.io/en/latest/) developed at Biophysics Collaborative Access Team. (B) Equatorial intensity ratios I11/I10 plotted as a function of calcium. The values from several fibers per mouse were averaged and the data presented as mean ± SEM of N= number of mice (3M, 3F of WT; 2M, 2F of HCM–D166V; and 3M, 3F of DCM–D94A) with statistical significance calculated by two-way ANOVA followed by Tukey’s multiple comparison test. ****P < 0.0001 and **P < 0.01 for HCM–D166V versus WT and ++++P < 0.0001 and ++P < 0.01 for DCM–D94A versus HCM–D166V. All three groups of animals passed the Shapiro–Wilk normality test for N = no. animals per group. The pCa50 values of I11/I10–pCa relationship were 5.47 ± 0.15 (D166V), 5.11 ± 0.11 (D94A), and 5.20 ± 0.04 (WT). (C–E) SuperPlots of I11/I10 ratios for DCM-D94A, WT, and HCM-D166V mice at pCa 8 (C), pCa 5.4 (D), and pCa 5.2 (E). The data per animal (N) are pictured by large, color-coded symbols and the respective fiber measurements (n) by color-coded small symbols. Note the significantly increased I11/I10 ratios in HCM–D166V versus WT hearts at pCa 8 (*), pCa 5.4 (**), and pCa 5.2 (****) and between the HCM–D166V and DCM–D94A models at pCa 5.4 (+) and pCa 5.2 (++++).
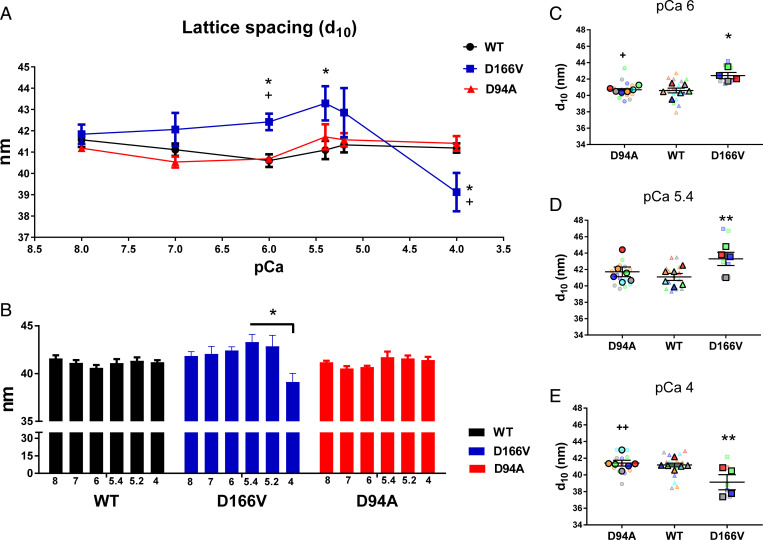
The plot of interfilament lattice spacings (d10) as a function of [Ca2+] for WT, HCM–D166V, and DCM–D94A mice is presented in Fig. 2A. Fig. 2B shows the comparison of d10 values within each genotype, and Fig. 2 C–E present the SuperPlots of d10 measurements for all mice at pCa 6, 5.4, and 4, respectively. At pCa 8 and 7, no significant differences in d10 were observed between all genotypes, indicating that the lattice spacing was relatively stable with respect to pathogenic RLC mutations under relaxed conditions. At intermediate Ca2+ concentrations (pCa 6 to 5.4), however, d10 values were significantly higher in HCM–D166V versus WT samples, and at pCa 6, they were also larger compared with DCM–D94A (Fig. 2 A, C, and D and Table 2). As shown in Fig. 2B, there were no significant differences in d10 as a function of calcium within WT or DCM–D94A groups, but significant differences were noted within the HCM–D166V model, and a decrease in d10 was observed for high calcium (pCa 4). Similarly, a decrease in d10 at pCa 4 for HCM–D166V mice was statistically significant compared with WT and DCM–D94A mice (Fig. 2 A and E), suggesting a less ordered sarcomere structure in HCM–D166V myocardium at high calcium. In agreement with this observation, the D166V mutation in myosin RLC was seen to largely decrease the level of maximal pCa 4 tension (6), indicating that susceptibility to damage with contraction might be a part of the HCM–D166V phenotype (33). However, since the normal operating range of physiological calcium concentrations in cardiac muscle is from pCa ∼7 to ∼5.8 (20, 34), working HCM–D166V hearts are not expected to experience significant deterioration of sarcomere ordering as observed at pCa 4. Taken together, while DCM–D94A did not show any differences in d10 compared with WT, the HCM–D166V model significantly differed from both DCM–D94A and WT, indicating that the calcium-dependent structural changes regulating the distance between the thick and thin filaments mostly occur in the myocardium of hypertrophic cardiomyopathy HCM–D166V mice.
Fig. 2.
Assessment of interfilament lattice spacings (d10) in PM fibers from ∼5-mo-old DCM–D94A and HCM–D166V models compared with WT mice. (A) Interfilament lattice spacings as a function of increasing calcium concentrations. The values of d10 measured for several fibers per mouse were averaged and the data presented as mean ± SEM of N = number of mice (3M, 3F of WT; 2M, 2F of HCM–D166V; and 3M, 3F of DCM–D94A) with statistical significance calculated by two-way ANOVA followed by Tukey’s multiple comparison test. **P < 0.01 and *P < 0.05 for HCM–D166V versus WT and ++P < 0.01 and +P < 0.05 for DCM–D94A versus HCM–D166V. All three groups of animals passed the Shapiro–Wilk normality test for N = no. animals per group. (B) The comparison of d10 values within each genotype. The statistical analysis was performed by one-way ANOVA followed by Tukey multiple comparison test. *P < 0.05 depicting significance in d10 between pCa 5.4 and 5.2 versus pCa 4 in the HCM–D166V model. (C–E) SuperPlots of d10 measured for DCM–D94A, WT, and HCM–D166V mice at pCa 6 (C), pCa 5.4 (D), and pCa 4 (E). The data per animal (N) are indicated by large symbols and the respective fiber measurements (n) by color-coded small symbols. Note the significantly increased d10 for HCM–D166V versus WT hearts at pCa 6 (*) and pCa 5.4 (**) and decreased d10 at pCa 4 (**). Significantly increased d10 was also observed in HCM–D166V versus DCM–D94A at pCa 6 (+) and decreased d10 in HCM–D166V versus DCM–D94A models at pCa 4 (++).
HCM–D166V mutation increases Ca2+ sensitivity of isometric force.
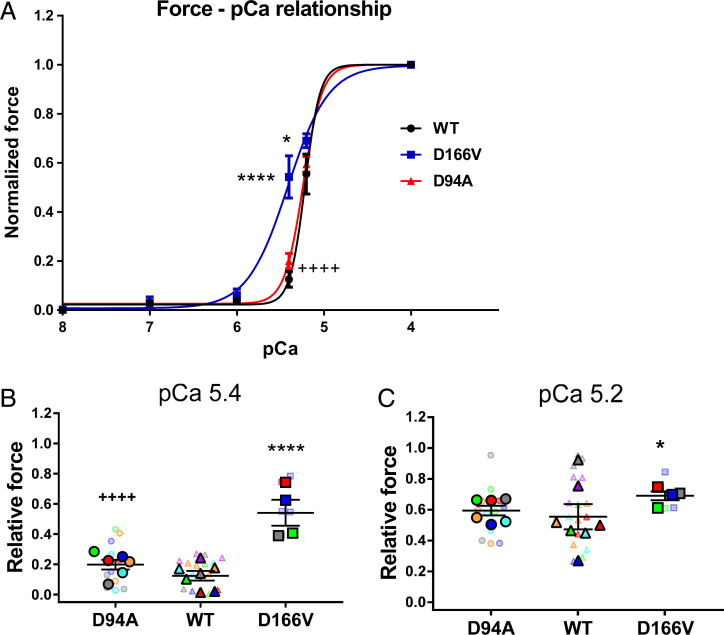
The force-pCa traces recorded simultaneously with I11/I10-pCa curves in PM fibers from WT, HCM–D166V, and DCM–D94A mice are plotted in Fig. 3. Fig. 3A presents normalized force-pCa relationships for all three genotypes, while absolute tensions are shown in SI Appendix, Table S1. The latter measurements are considered less reliable due to technical difficulties in accurately estimating the cross-sectional areas of large LVPM preparations (see Materials and Methods). Consistent with I11/I10-pCa data, the HCM–D166V model exhibited a leftward shift of the force-pCa relationship compared with WT and DCM–D94A mice (Fig. 3A and Table 3). The pCa50 value of force for HCM–D166V was 5.41 ± 0.04 compared with pCa50 = 5.22 ± 0.01 of WT and pCa50 = 5.24 ± 0.01 of DCM–D94A, indicating significantly increased Ca2+ sensitivity of force in the HCM–D166V model. Fig. 3 B and C show the SuperPlots of relative force for DCM–D94A, WT, and HCM–D166V mice at pCa 5.4 and 5.2, respectively. The data per animal (N) are pictured by large symbols and the respective fiber measurements (n) by color-coded small symbols. The values from several fibers per mouse were averaged and the data presented as mean ± SEM of N = number of mice (3M, 4F of WT; 2M, 2F of HCM–D166V; and 3M, 3F of DCM–D94A) with statistical significance calculated by two-way ANOVA followed by Tukey’s multiple comparison test. As observed in the X-ray experiments, the DCM–D94A model did not show any differences in force-pCa compared with WT controls (Fig. 3B and Table 3). The relative force in HCM–D166V fibers was significantly different from WT at pCa 5.4 and 5.2 (Fig. 3 B and C), confirming increased Ca2+ sensitivity of tension in the HCM model (Fig. 3A). The increased Ca2+ sensitivity of force was observed in HCM–D166V hearts previously and correlated with poor relaxation and diastolic dysfunction observed in transgenic D166V mice (33). Chronic sensitization of myofilaments to calcium as observed in HCM–D166V myocardium is an important finding of clinical significance since it can lead to cardiac arrhythmias and heart failure (35).
Fig. 3.
(A) The force-pCa dependence assessed in PM fibers from WT, HCM–D166V, and DCM–D94A mice. The force-pCa data were fitted to a four-parameter logistic equation yielding the pCa50 and nH (Hill coefficient) values (Table 3). Note that the HCM–D166V model demonstrates significantly increased calcium sensitivity of force (higher pCa50) compared with WT and DCM–D94A mice. All three groups of animals passed the Shapiro–Wilk normality test for N = no. animals/per group. (B and C) SuperPlots of relative force values at pCa 5.4 (B) and pCa 5.2 (C) for DCM–D94A, WT, and HCM–D166V mice. The data per animal (N) are pictured by large symbols and the respective fiber measurements (n) by color-coded small symbols. The values from several fibers per mouse were averaged and the data presented as mean ± SEM of N = number of mice (3M, 4F of WT; 2M, 2F of HCM–D166V; and 3M, 3F of DCM–D94A) with statistical significance calculated by two-way ANOVA followed by Tukey’s multiple comparison test. ****P < 0.0001 and *P < 0.01 for HCM–D166V versus WT and ++++P < 0.0001 for DCM–D94A versus HCM–D166V.
Table 3.
The force-pCa relationship assessed in PM fibers from WT, HCM–D166V, and DCM–D94A mice
| System | pCa50 | 95% CI | Hill coefficient (nH, slope) | 95% CI | No. mice (M, F)No. fibers |
| WT | 5.22 ± 0.01 | 5.18 to 5.25 | 5.03 ± 0.96 | 3.46 to 8.67 | 6 (3, 4)16 |
| HCM–D166V | 5.41 ± 0.04**** | 5.33 to 5.51 | 1.86 ± 0.36* | 1.31 to 2.77 | 4 (2, 2)6 |
| DCM–D94A | 5.24 ± 0.01++++ | 5.22 to 5.26 | 4.04 ± 0.39 | 3.29 to 4.92 | 6 (3, 3)13 |
Data are mean ± SEM of N = no. of mice. F, female; M, male. The statistical results were analyzed by two-way ANOVA with significance ****P < 0.0001, *P < 0.05 for HCM–D166V versus WT and ++++P < 0.0001, +P < 0.05 D94A for DCM–D94A versus HCM–D166V.
Altogether, the force-pCa data combined with I11/I10-pCa results suggest that the structural rearrangements observed in HCM–D166V sarcomeres and the apparent movement of D166V-mutated myosin cross-bridges closer to actin-containing thin filaments can explain the increased Ca2+ sensitivity of force generation in HCM–D166V hearts, providing insight into the structural basis of hypercontractile cross-bridge behavior and cardiac dysfunction in these mice.
The SRX State of Myosin in Mouse Models of HCM and DCM Versus WT Mice.
In resting cardiac muscle, myosin cross-bridges exist in two biochemical states: the SRX state, in which myosin heads hydrolyze ATP with a highly inhibited rate, and the DRX state, in which the heads cycle ATP with much higher rates. The SRX state has ∼10- to 100-fold slower ATP turnover rate compared with the DRX state. Structurally, the SRX heads are thought to bind closely to the thick filament backbone compared with the DRX heads that are disordered, free to diffuse into interfilament space and occupy a wide range of conformations with different proximities to the thin filaments (25, 30). During cardiac muscle contraction, the myosin heads undergo physiological transitions between the active and relaxed states, with the latter involving the SRX conformation to maximize the energy-saving state and the DRX conformation enabling cross-bridge formation with greater ATP consumption (31).
Structurally, the SRX state is associated with the formation and stability of the IHM in which the two myosin heads asymmetrically interact with one another while folding back onto the proximal tail (24, 28). Low-resolution structural models of the sequestered state suggest that HCM-associated mutations in myosin may weaken the IHM formation, thereby increasing the number of active heads and contributing to the phenotype of HCM-related hypercontractility (24, 36). The question that we asked next was whether the HCM–D166V and DCM–D94A mutations in myosin RLC affect the stability of the SRX energy-conserving state and whether this process is calcium dependent.
HCM-D166V and DCM-D94A mutations differently affect the SRX state of myosin.
Skinned LVPM fibers from mice were incubated in a rigor solution containing 250 μM mant-ATP, and decay fluorescence isotherms were collected during a rapid exchange of mant-ATP with 4 mM nonlabeled ATP and plotted as a function of time. Fluorescence decay curves were fitted to a double-exponential equation: F = 1 − P1 (1 − exp(−t/T1)) − P2 (1 − exp (−t/T2)), in which P1 and P2 (in %) are the amplitudes of fast and slow phases of fluorescence intensity decay, and T1 and T2 are their respective lifetimes (in seconds) (37, 38).
We first looked at the calcium dependence of fluorescence decay in WT–RLC mice. As indicated in Table 4, the ratio of P1 to P2 was ∼60:40, and no major changes in cross-bridge distribution between the two states were noted when different calcium concentrations were applied. This result is in accord with the report of Hooijman et al. (25) in which the SRX populations of myosin cross-bridges were present in active cardiac muscle but not active skeletal muscle, and the decay of fluorescence in cardiac fibers was not changed between chase with relaxing and activating solutions. The lack of a calcium effect observed for WT hearts was also true for the cardiomyopathy models, and no sensitivity to calcium in mant-ATP/ATP chase assays was noted in HCM–D166V and DCM–D94A mice (Table 4).
Table 4.
Summary of nucleotide exchange experiments in skinned PM fibers from DCM–D94A, HCM–D166V, and WT mouse modelsat serial pCa solutions
| DCM-D94A | WT | HCM-D166V | |||||||
| pCa | P1 (%) T1 (s) | P2 (%) T2 (s) | No. animals (F, M)No. fibers | P1 (%) T1 (s) | P2 (%) T2 (s) | No. animals (F, M) No. fibers | P1 (%) T1 (s) | P2 (%) T2 (s) | No. animals (F, M) No. fibers |
| 8 | 55 ± 3 | 42 ± 2 | 5 (3, 2)18 | 55 ± 4 | 42 ± 4 | 5 (3, 2)15 | 61 ± 3 | 35 ± 4 | 5 (4, 1)16 |
| 6.4 ± 1.6 | 138 ± 30 | 6.6 ± 1.1 | 213 ± 63 | 5.2 ± 1 | 110 ± 29 | ||||
| 7 | 55 ± 2 | 40 ± 31 | 5 (3, 2)22 | 58 ± 3 | 38 ± 3 | 5 (2, 3)15 | 62 ± 2 | 34 ± 2 | 5 (4, 1)22 |
| 5.9 ± 0.8 | 41 ± 27 | 6.8 ± 1.6 | 219 ± 50 | 5.3 ± 1 | 172 ± 44 | ||||
| 6 | 50 ± 2*,+ | 47 ± 2*, ++ | 5 (2, 3)20 | 60 ± 2 | 37 ± 2 | 5 (2, 3)15 | 61 ± 2 | 35 ± 2 | 5 (4, 1)15 |
| 6.3 ± 2.2 | 144 ± 66 | 8.3 ± 1.6 | 227 ± 46 | 4.5 ± 0.4 | 85 ± 18 | ||||
| 5 | 58 ± 3+ | 39 ± 3+ | 5 (2, 3)16 | 62 ± 4 | 35 ± 4 | 5 (2, 3)16 | 69 ± 3 | 27 ± 2 | 5 (4, 1)15 |
| 7.7 ± 1.2 | 136 ± 50 | 6.6 ± 1.7 | 207 ± 60 | 7.5 ± 0.9 | 202 ± 68 | ||||
| 4 | 56 ± 3+ | 42 ± 4+ | 5 (3, 2)20 | 60 ± 1 | 37 ± 1 | 5 (2, 3)19 | 66 ± 4 | 30 ± 4 | 5 (4, 1)15 |
| 6.8 ± 1.2 | 176 ± 63 | 9.9 ± 3.2 | 162 ± 56 | 5.1 ± 0.9 | 163 ± 56 | ||||
| Ave pCa | 55 ± 1*, ++++ | 42 ± 1*, ++++ | 6 (3, 3)96 | 59 ± 1 | 38 ± 1 | 6 (3, 3)80 | 64 ± 1* | 32 ± 1** | 5 (4, 1)83 |
| 6.6 ± 0.6 | 147 ± 21 | 7.6 ± 0.9 | 206 ± 23 | 5.6 ± 0.4 | 147 ± 21 | ||||
Data are mean ± SEM for N = no. animals. F, female; M, male. Significance was calculated by one-way (averaged pCa) or two-way ANOVA with Tukey’s multiple comparison test: *P < 0.05 and **P < 0.01 for mutant versus WT, +P < 0.05, ++P < 0.01, and ++++P < 0.0001 for DCM–D94A versus HCM–D166V.
The summary of nucleotide exchange experiments in skinned PM fibers from DCM–D94A, HCM–D166V, and WT mice is presented in Fig. 4. Fig. 4A depicts the genotype-dependent distribution of the fast (P1) and slow (P2) parameters derived from the two-exponential fit at serial calcium solutions, and the data are mean ± SEM for N = 5 to 6 animals per group and n = 15 to 22 fibers per each pCa condition. Significant differences in P1 versus P2 were observed between DCM–D94A and WT mice at pCa 6 and between the two pathogenic RLC mutants at pCa 6, 5, and 4. Considering no calcium dependence of P1 versus P2 distribution for any of the investigated mice, both amplitudes P1 and P2, determined in different calcium solutions, were averaged for each genotype and presented in Fig. 4B. The data points represent values averaged per animal at each pCa solution, and symbols of similar shapes indicate measurements performed at the same pCa for different genotypes. Pooling the data from different calcium concentrations revealed clear trends obscured by noise in the data at individual pCa values. As shown, both pathogenic models were statistically different from WT with the P1/P2 ratio = 55:42 for DCM–D94A and P1/P2 = 64:32 for HCM–D166V compared with P1/P2 = 59:38 observed for WT mice (Fig. 4B and Table 4).
Fig. 4.
The summary of nucleotide exchange experiments in skinned PM fibers from DCM–D94A, HCM–D166V, and WT mouse models. (A) The genotype-dependent distribution of the fast (P1) and slow (P2) parameters derived from the two-exponential fit at serial calcium solutions. Open symbols depict F mice, and closed symbols depict M mice. The data are mean ± SEM for N = 5 to 6 animals and n = 15 to 22 fibers per each pCa condition with significance calculated by two-way ANOVA with Tukey’s multiple comparison test: *P < 0.05 for DCM–D94A versus WT, +P < 0.05 and ++P < 0.01 for HCM–D166V versus DCM–D94A. Note the significant differences in P1 and P2 between DCM–D94A and WT mice at pCa 6 and between the two pathogenic RLC mutants at pCa 6, 5, and 4. (B) Comparison of P1 versus P2 in all three genotypes assessed at averaged calcium concentrations. Data points represent values averaged per animal at each pCa solution with one point = one animal at a certain pCa solution. Symbols of similar shapes between the genotypes indicate measurements performed at the same pCa. The data are mean ± SEM for N = no. animals with significance calculated with one-way ANOVA with Tukey’s multiple comparison test: *P < 0.05 and **P < 0.01 for mutant versus WT, +P < 0.05, ++P < 0.01, and ++++P < 0.0001 for DCM–D94A versus HCM–D166V. (C) The percent of myosin heads occupying the DRX versus SRX states was calculated using a correction factor of 0.44 for nonspecific mant-ATP binding (see Materials and Methods).
To relate the P1 and P2 values to the number of myosin heads occupying the DRX versus SRX states, we have determined the fraction of nonspecifically bound mant-ATP that contributes to the P1 parameter using a competition assay as described in Hooijman et al. (25). LVPM fibers from all three genotypes were incubated with a solution containing 250 µM mant-ATP and varying concentrations of ATP, and the fluorescence intensity was measured as a function of added [ATP] (SI Appendix). As shown in SI Appendix, Fig. S1, the fraction of nonspecifically bound mant-ATP = 0.44 ± 0.02 (SEM) (n = 10 fibers), and the population of myosin heads occupying the SRX state was calculated as P2/(1 − 0.44) (25).
Fig. 4C presents the distribution of myosin heads occupying the DRX versus SRX states in DCM–D94A, WT, and HCM–D166V mice in the form of a pie plot. The comparison of the two pathogenic models shows their different behavior in assuming the DRX or SRX conformations compared with WT mice. The DRX/SRX occupancy was statistically different between DCM–D94A (25:75) versus WT (33:67) versus HCM–D166V (43:57) (SI Appendix, Table S2). A disruption of the SRX conformation and a shift toward the DRX state was observed for HCM–D166V hearts, while the stabilization of the energy-conserving SRX state was noted in DCM–D94A (Fig. 4C). The HCM–D166V-dependent switch from the SRX to DRX state was consistent with the hypercontractility phenotype observed for this model (33) and other HCM-linked mutations in myosin (27, 29). The data are also in accord with the structure/function–pCa relationships observed in HCM–D166V fibers, including increased Ca2+ sensitivity of force (Fig. 3) and a left-shifted I11/I10-pCa relationship (Fig. 1B). The DCM–D94A model favored the energy-conserving SRX state without affecting the I11/I10 ratios or the force-pCa relationship.
Discussion
Genetic cardiomyopathies are debilitating heart diseases affecting 1 in ∼200 of the general population (39), and they continue to be the leading cause of death worldwide (1). At present, there are no effective therapies, and multilaboratory efforts are directed toward developing phenotype-specific therapeutics that target the mechanisms involved in the etiology of the disease. An important tool in the investigation of the molecular, biochemical, and structural causes of cardiomyopathy phenotypes (e.g., HCM and DCM) has been the creation of transgenic mouse models with engineered point mutations in sarcomeric proteins that mimic those naturally occurring in humans. Previous studies in our laboratory have extensively characterized the two pathogenic RLC variants in transgenic mice expressing the HCM–D166V and DCM–D94A mutations as model systems for investigating the mechanisms underlying associated cardiomyopathies. Clinical population studies have shown that the D166V–RLC mutation can lead to malignant HCM (4), and the strong HCM phenotype was observed in transgenic HCM–D166V mice (6, 33). The D94A–RLC mutation was identified by exome sequencing in the family suffering from DCM (5), and a transgenic mouse model expressing the DCM–D94A mutation demonstrated a classic phenotype of DCM with largely reduced ejection fraction and systolic dysfunction (7). We have also reported on the profound effects of these two pathological RLC variants on the β-cardiac myosin motor function when reconstituted in porcine cardiac muscle preparations (5, 40), highlighting the importance of evaluating the impact of disease-causing mutations in a clinically relevant β-cardiac myosin background compared with the α-cardiac myosin present in transgenic mouse hearts (6, 7, 33). However, the validation of the in vitro data (e.g., force-pCa relationship) obtained in β-cardiac myosin reconstituted systems (5, 40) using HCM–D166V and DCM–D94A models (Fig. 3A) strongly argues for the value of mouse studies in elucidating the underlying mechanisms.
In this report, we investigated the impact of these two pathogenic MYL2 variants on the Ca2+-dependent structure–function relationships and the distribution of HCM–D166V and DCM–D94A cross-bridges between the SRX and DRX states. Our previous small-angle X-ray diffraction studies on HCM–D166V (33) and DCM–D94A (7) mice were restricted to the measurements of d10 and I11/I10 ratios for only relaxed muscle and, at most, for one submaximal contracting condition for DCM–D94A (7). These limited early measurements, however, built the scientific premise for the present investigation performed on skinned LVPM fibers from mice over the full range of force development from relaxed through submaximal to maximum activation (from pCa 8 to 4). Importantly, we report on X-ray structural data obtained simultaneously with force measurements for HCM–D166V, DCM–D94A, and WT controls that allowed a more comprehensive understanding of the structural basis of the calcium-dependent force development in cardiac muscle. Both the HCM–D166V and DCM–D94A mutations were previously shown to increase or decrease, respectively, the duration of force transients in the intact muscle (6, 7), thereby affecting the critical for normal heart function relaxation phase of muscle contraction. The effects of HCM–D166V and DCM–D94A mutations observed in this study could be due to changes in cross-bridge cycling; however, our experiments were not designed to directly address the question of cross-bridge kinetics but rather aimed to assess the effect of mutations on the stability of the SRX state of myosin and SRX ↔ DRX equilibrium.
The data showed a remarkable agreement between the I11/I10-pCa and force-pCa relationships in all investigated mice and between the structural and functional phenotypes and the myosin energetic states in the HCM–D166V model (Figs. 1–4). Consistent with previous muscle fiber mechanics results on HCM–D166V mice (6, 33), an increase in Ca2+ sensitivity of force-pCa was observed in this report. In parallel, the I11/I10–pCa dependence was leftward shifted in HCM–D166V versus WT mice, suggesting that the D166V-induced effect on lattice spacings and the movement of D166V heads toward the actin filaments was responsible for the observed increase in the Ca2+ sensitivity of force development. The structure–function data for the HCM–D166V model agree with the analysis of myosin energetic states that revealed the HCM–D166V-induced destabilization of the myosin energy-conserving SRX state and implicating a hypercontractile cross-bridge behavior (Fig. 4). The HCM clinical phenotype of hypercontractility is often associated with cardiac arrhythmias and heart failure (35), and the HCM–D166V model did show diastolic abnormalities in mice (33). The destabilization of the SRX state has been proposed to be a chief cause of HCM (20, 24), and therefore, dysregulation of the ordered resting configuration of myosin heads in HCM–D166V hearts observed in this report supports an adverse HCM phenotype of hypercontractility observed in humans and HCM–D166V mice (6, 33).
The DCM model, recapitulated in transgenic D94A–RLC mice (7), exhibited no changes in the Ca2+ sensitivity of either I11/I10–pCa or force-pCa relationships compared with WT mice. The DCM–D94A model favored an energy-conserving state characterized by a significant increase in the number of myosin heads occupying the SRX state (75%) compared with 67% of WT and 57% HCM–D166V (Fig. 4C). The result of a smaller proportion of heads occupying the DRX state in DCM–D94A versus WT fibers is in accord with shorter force transients measured in intact LVPM fibers from these mice (7), suggesting that fewer D94A heads are available to enter the force-producing phase and provide mechanical energy to support the pumping action of the heart. However, because of the lack of a consistent trend of lower I11/I10 ratio for DCM–D94A relative to WT (Table 1), one can speculate that the D94A mutation stabilizes an SRX state without entering a folded-back SRX–IHM structure as defined by quasi-atomic modeling studies (24, 28). Consistent with this notion, Anderson et al. demonstrated that a one-headed myosin (sS1) that lacks the S2 portion of the myosin rod was able to form the SRX-like state (∼10%), which increased to ∼40% on the addition of mavacamten (20). They hypothesized that myosin heads can occupy an SRX without adopting a folded IHM conformation that is further stabilized by intramolecular interactions similar to those existing in the IHM (20). Additional support comes from time-resolved fluorescence resonance energy transfer experiments showing that the SRX can exist in the absence of a conventional IHM, suggesting that the IHM is sufficient but not necessary to produce the SRX kinetic state (41). The molecular pathogenesis of DCM was recently assessed in the context of the myosin SRX state and IHM paradigm (24). Similar to the effects seen in this report, DCM-causing mutations in MYH7 were shown to have modest effects on IHM interactions but have substantially reduced myosin heavy chain (MHC) motor functions, particularly nucleotide binding, resulting in reduced ATP consumption and decreased contractility. The increased number of heads occupying the SRX state with reduced ATP consumption in DCM–D94A hearts (Fig. 4C) fully supports our previous observations of D94A-mediated diminished actin-activated myosin ATPase activity and a low ejection fraction in D94A mice in vivo (7). The complex mechanism involving changes in myosin motor function (steady-state and kinetics) seems to be a primary disease mechanism in DCM–D94A mice.
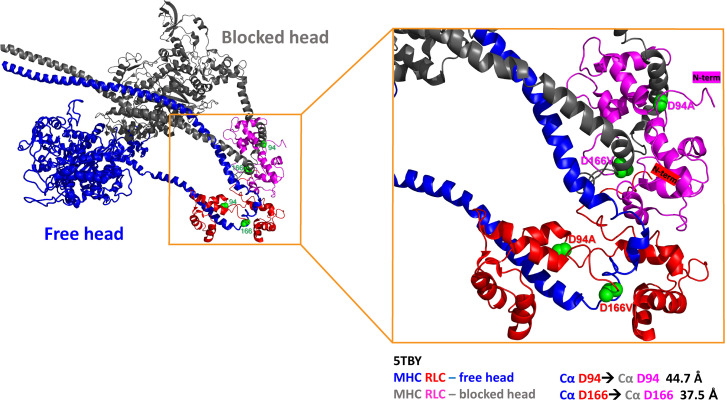
Fig. 5 illustrates the D94 and D166 residues of the RLC within the 5TBY structure of human β-cardiac heavy meromyosin IHM obtained by homology modeling by Alamo et al. (24). The authors defined various interaction sites between adjacent myosin heads and associated protein partners that can potentially be altered by disease-causing mutations in myosin. The asymmetrical sequestered state, or the IHM, is composed of two folded heads: the “blocked” head (BH) that is unable to interact with actin and the “free” head (FH) that can potentially be recruited to rapidly hydrolyze ATP in the DRX state, with both heads containing a pair of myosin ELC and RLC as scaffold proteins. As proposed by the authors, the interactions between myosin heads and myosin RLCs are important elements of the SRX conformation, which is further stabilized by intramolecular MHC–RLC and RLC–RLC interactions (24). Considering the RLC as an integral part of the IHM structure, one can speculate that both pathogenic variants differently affect these intramolecular interactions and discretely influence the SRX state of myosin. They are charge-changing mutations, prone to alter the local RLC structure and the binding of the RLC to the IQ motif of the myosin lever arm (5). This, in turn, may destabilize MHC–RLC interactions and alter the SRX conformation. It is difficult to conclude whether the mutations also destabilize the RLC–RLC interactions considering the long distances between the Cα atoms of the BH and the FH, 37.5 Å for D166 and 44.7 Å for D94, respectively (Fig. 5). Future structural studies would have to be performed to address this question.
Fig. 5.
The tertiary structure of human β-cardiac heavy meromyosin IHM (Protein Data Bank 5TBY) and PyMOL-derived distances between Cα atoms of D94 and D166 residues positioned on the RLC of the “blocked” head (MHC: gray, RLC: pink) and the “free” head (MHC: dark blue, RLC: red). The side chains of mutations are presented as green spheres, and the N-termini of RLCs are labeled with corresponding colors, pink for the RLC of BH and red for the RLC of FH.
As pictured in Fig. 5, the HCM–D166V mutation lies in the elbow of the MHC region that links the lever arm with the rod portion of myosin. The valine residue that replaces the aspartic acid in the HCM–D166V mutant introduces a loss of a negative charge in this region of the RLC backbone, which, in turn, may interfere with the myosin lever arm function to effectively execute the swinging motion and produce the power stroke while more cross-bridges enter the DRX–ON states. One can hypothesize that HCM–D166V disrupts the SRX state by facilitating the recruitment of the free head to rapidly hydrolyze ATP in the DRX state (31), while DCM–D94A favors the SRX–OFF conformation by weakening the interaction between the RLC with MHC (5) and diminishing the number of DRX heads (Fig. 4) that are readily available for binding to actin and producing force. The presence of the aspartic acid at both locations within the RLC molecule seems to be necessary for the structural stability of the lever arm to effectively execute the power stroke and muscle contraction.
Conclusions and Future Directions
HCM and DCM are complex diseases with many factors that contribute to myocardial dysfunction and the manifestation of clinical phenotypes. The HCM–D166V model promotes the SRX-to-DRX transition and increases the Ca2+ sensitivity of I11/I10 and force-pCa relationships, effects that are consistent with the hypercontractility phenotype, poor relaxation, and diastolic dysfunction in HCM–D166V hearts (33). The DCM–D94A mutation increases the number of heads occupying the energy-conserving sequestered state and reduces myosin motor function that, among other factors, may contribute to the development of hypocontractile phenotype, systolic dysfunction, and DCM in DCM–D94A mice (7). Understanding the mechanisms that foster the development of clinical phenotypes will propel new pharmacological treatments that may limit disease progression and adverse outcomes.
Materials and Methods
Detailed materials and methods are outlined in SI Appendix, Materials and Methods.
This study conforms to the Guide for the Care and Use of Laboratory Animals published by the US NIH (Publication No. 85–23, revised 2011) (42). All protocols were approved by the Institutional Animal Care and Use Committee at the University of Miami Miller School of Medicine (protocol No. 21–106 LF) and the Illinois Institute of Technology. The assurance number is No. A-3224–01, approved through November 30, 2023. The euthanasia of the mice was achieved through inhalation of CO2 followed by cervical dislocation.
Transgenic Mice.
The complementary DNA of human ventricular RLC (National Center for Biotechnology Information accession No. NP_000423.2), in the WT or D166V or D94A mutated form, was inserted downstream of the α-MHC promoter (clone 26 provided by Jeffrey Robbins, Cincinnati Children's Hospital Medical Center, Cincinnati, OH), producing transgenic lines expressing ∼90% of human nonmutated WT–RLC (UniProtKB P10916) (7, 43), ∼90% of human ventricular D166V RLC mutant (6), and ∼50% of human ventricular D94A RLC mutant (L1 and L2) (7) in the hearts of mice as described previously. Female and male DCM–D94A, HCM–D166V, and WT mice were used in the experiments.
Small-Angle X-Diffraction Study: Simultaneous Assessment of X-ray Diffraction Patterns and Isometric Force Measurements.
The equatorial X-ray diffraction patterns in LVPM fibers from HCM–D166V, DCM–D94A, and WT mice were acquired simultaneously with force-pCa traces over a wide range of calcium concentrations (from pCa 8 to 4) using the small-angle instrument at Biophysics Collaborative Access Team beamline 18ID at the Advanced Photon Source, Argonne National Laboratory, as described previously (7). The intensities of the 1,0 and 1,1 equatorial reflections were determined using nonlinear least square fits to one-dimensional projections of the integrated intensity along the equator assuming Gaussian peak shapes (44). The distances between the 1,0 and 1,1 equatorial X-ray reflections were used to calculate d10 lattice spacings using Bragg’s law. Isometric force was measured, and muscle length controlled as described previously (7).
SRX-pCa Measurements.
The LVPM fibers were isolated from HCM–D166V, DCM–D94A, and WT mice, chemically skinned, dissected to about 100 µm in diameter, and subjected to mant-ATP chase assays using the IonOptix instrumentation (IonOptix, LLC) as described previously (37, 38). The fibers were first incubated in a rigor buffer (120 mM K+-propionate [KPr], 5 mM MgPr, 2.5 mM K2HPO4, 2.5 mM KH2PO4, and 50 mM 3-(N-morpholino)propanesulfonic acid [MOPS], pH 6.8, with freshly added 2 mM DTT) containing 250 μM mant-ATP (Thermo Fisher Scientific) for several minutes until maximum fluorescence was reached. Fluorescence decay curves versus time were collected when mant-ATP was rapidly exchanged with 4 mM nonlabeled ATP applied in various pCa solutions containing 10−8 to 10−4 M [Ca2+], 5 mM free [Mg2+], total MgPr = 5.69 mM, 7 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid, 50 mM MOPS, pH 6.8, and ionic strength = 170 mM (adjusted with KPr). The sarcomere length of fibers was ∼2.1 ± 0.1 µm. Decay isotherms versus time (in seconds) were fitted to a two-state exponential equation using GraphPad 7 (GraphPad software) to derive the amplitudes of the fast (P1) and slow (P2) phases of fluorescence decay and their respective T1 and T2 lifetimes (in seconds) (37, 38). P1 and T1 represent the initial fast decay in fluorescence intensity, which comprises myosin in the DRX state and the release of nonspecifically bound mant-ATP, which is presumably also fast, while P2 and T2 are representative of the slow decrease in fluorescence intensity due to myosin in the SRX state (25).
Statistical Analyses.
The statistical analysis of the data was performed per N = no. hearts as independent biological samples, and the results from multiple fibers per single heart were averaged. Values are shown as means ± SEM of N = no. mice with statistically significant differences between multiple groups calculated using one-way or two-way ANOVA followed by Tukey’s multiple comparison test. Significance was defined as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Statistical analyses included the Shapiro–Wilk normality tests performed on all data sets of unequal sample size (45).
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH (R01-HL143830 and R56-HL146133 to D.S.-C. and P41-GM103622 to T.C.I.). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110328119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Garfinkel A. C., Seidman J. G., Seidman C. E., Genetic pathogenesis of hypertrophic and dilated cardiomyopathy. Heart Fail. Clin. 14, 139–146 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sitbon Y. H., Yadav S., Kazmierczak K., Szczesna-Cordary D., Insights into myosin regulatory and essential light chains: A focus on their roles in cardiac and skeletal muscle function, development and disease. J. Muscle Res. Cell Motil. 41, 313–327 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yadav S., Sitbon Y. H., Kazmierczak K., Szczesna-Cordary D., Hereditary heart disease: Pathophysiology, clinical presentation, and animal models of HCM, RCM, and DCM associated with mutations in cardiac myosin light chains. Pflugers Arch. 471, 683–699 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richard P, et al. Hypertrophic cardiomyopathy: Distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation 107, 2227–2232 (2003), and erratum in Circulation 109, 3258 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Huang W., et al. , Novel familial dilated cardiomyopathy mutation in MYL2 affects the structure and function of myosin regulatory light chain. FEBS J. 282, 2379–2393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kerrick W. G. L., Kazmierczak K., Xu Y., Wang Y., Szczesna-Cordary D., Malignant familial hypertrophic cardiomyopathy D166V mutation in the ventricular myosin regulatory light chain causes profound effects in skinned and intact papillary muscle fibers from transgenic mice. FASEB J. 23, 855–865 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan C. C., et al. , Sarcomeric perturbations of myosin motors lead to dilated cardiomyopathy in genetically modified MYL2 mice. Proc. Natl. Acad. Sci. U.S.A. 115, E2338–E2347 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szczesna D., Regulatory light chains of striated muscle myosin. Structure, function and malfunction. Curr. Drug Targets Cardiovasc. Haematol. Disord. 3, 187–197 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Rayment I., et al. , Three-dimensional structure of myosin subfragment-1: A molecular motor. Science 261, 50–58 (1993). [DOI] [PubMed] [Google Scholar]
- 10.Geeves M. A., Stretching the lever-arm theory. Nature 415, 129–131 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Yadav S., Szczesna-Cordary D., Pseudophosphorylation of cardiac myosin regulatory light chain: A promising new tool for treatment of cardiomyopathy. Biophys. Rev. 9, 57–64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szczesna-Cordary D., Guzman G., Ng S. S., Zhao J., Familial hypertrophic cardiomyopathy-linked alterations in Ca2+ binding of human cardiac myosin regulatory light chain affect cardiac muscle contraction. J. Biol. Chem. 279, 3535–3542 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Szczesna D., et al. , Familial hypertrophic cardiomyopathy mutations in the regulatory light chains of myosin affect their structure, Ca2+ binding, and phosphorylation. J. Biol. Chem. 276, 7086–7092 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Chang A. N., et al. , Constitutive phosphorylation of cardiac myosin regulatory light chain in vivo. J. Biol. Chem. 290, 10703–10716 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olsson M. C., Patel J. R., Fitzsimons D. P., Walker J. W., Moss R. L., Basal myosin light chain phosphorylation is a determinant of Ca2+ sensitivity of force and activation dependence of the kinetics of myocardial force development. Am. J. Physiol. Heart Circ. Physiol. 287, H2712–H2718 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Toepfer C. N., West T. G., Ferenczi M. A., Revisiting Frank-Starling: Regulatory light chain phosphorylation alters the rate of force redevelopment (ktr ) in a length-dependent fashion. J. Physiol. 594, 5237–5254 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kampourakis T., Irving M., Phosphorylation of myosin regulatory light chain controls myosin head conformation in cardiac muscle. J. Mol. Cell. Cardiol. 85, 199–206 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colson B. A., et al. , Differential roles of regulatory light chain and myosin binding protein-C phosphorylations in the modulation of cardiac force development. J. Physiol. 588, 981–993 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Millman B. M., Irving T. C., Filament lattice of frog striated muscle. Radial forces, lattice stability, and filament compression in the A-band of relaxed and rigor muscle. Biophys. J. 54, 437–447 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson R. L., et al. , Deciphering the super relaxed state of human β-cardiac myosin and the mode of action of mavacamten from myosin molecules to muscle fibers. Proc. Natl. Acad. Sci. U.S.A. 115, E8143–E8152 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez-Martinez D., et al. , Structural and functional impact of troponin C-mediated Ca2+ sensitization on myofilament lattice spacing and cross-bridge mechanics in mouse cardiac muscle. J. Mol. Cell. Cardiol. 123, 26–37 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zoghbi M. E., Woodhead J. L., Moss R. L., Craig R., Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc. Natl. Acad. Sci. U.S.A. 105, 2386–2390 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma W., et al. , Myosin dynamics during relaxation in mouse soleus muscle and modulation by 2′-deoxy-ATP. J. Physiol. 598, 5165–5182 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alamo L., et al. , Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes. eLife 6, e24634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hooijman P., Stewart M. A., Cooke R., A new state of cardiac myosin with very slow ATP turnover: A potential cardioprotective mechanism in the heart. Biophys. J. 100, 1969–1976 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart M. A., Franks-Skiba K., Chen S., Cooke R., Myosin ATP turnover rate is a mechanism involved in thermogenesis in resting skeletal muscle fibers. Proc. Natl. Acad. Sci. U.S.A. 107, 430–435 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmid M., Toepfer C. N., Cardiac myosin super relaxation (SRX): A perspective on fundamental biology, human disease and therapeutics. Biol. Open 10, bio057646 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spudich J. A., Three perspectives on the molecular basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Pflugers Arch. 471, 701–717 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toepfer C. N., et al. , Myosin sequestration regulates sarcomere function, cardiomyocyte energetics, and metabolism, informing the pathogenesis of hypertrophic cardiomyopathy. Circulation 141, 828–842 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNamara J. W., Li A., Dos Remedios C. G., Cooke R., The role of super-relaxed myosin in skeletal and cardiac muscle. Biophys. Rev. 7, 5–14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alamo L., et al. , Conserved intramolecular interactions maintain myosin interacting-heads motifs explaining tarantula muscle super-relaxed state structural basis. J. Mol. Biol. 428, 1142–1164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lord S. J., Velle K. B., Mullins R. D., Fritz-Laylin L. K., SuperPlots: Communicating reproducibility and variability in cell biology. J. Cell Biol. 219, e202001064 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan C. C., et al. , Constitutive phosphorylation of cardiac myosin regulatory light chain prevents development of hypertrophic cardiomyopathy in mice. Proc. Natl. Acad. Sci. U.S.A. 112, E4138–E4146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spudich J. A., Hypertrophic and dilated cardiomyopathy: Four decades of basic research on muscle lead to potential therapeutic approaches to these devastating genetic diseases. Biophys. J. 106, 1236–1249 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huke S., Knollmann B. C., Increased myofilament Ca2+-sensitivity and arrhythmia susceptibility. J. Mol. Cell. Cardiol. 48, 824–833 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trivedi D. V., Adhikari A. S., Sarkar S. S., Ruppel K. M., Spudich J. A., Hypertrophic cardiomyopathy and the myosin mesa: Viewing an old disease in a new light. Biophys. Rev. 10, 27–48 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sitbon Y. H., et al. , Ablation of the N terminus of cardiac essential light chain promotes the super-relaxed state of myosin and counteracts hypercontractility in hypertrophic cardiomyopathy mutant mice. FEBS J. 287, 3989–4004 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yadav S., Kazmierczak K., Liang J., Sitbon Y. H., Szczesna-Cordary D., Phosphomimetic-mediated in vitro rescue of hypertrophic cardiomyopathy linked to R58Q mutation in myosin regulatory light chain. FEBS J. 286, 151–168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Semsarian C., Ingles J., Maron M. S., Maron B. J., New perspectives on the prevalence of hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 65, 1249–1254 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Muthu P., Liang J., Schmidt W., Moore J. R., Szczesna-Cordary D., In vitro rescue study of a malignant familial hypertrophic cardiomyopathy phenotype by pseudo-phosphorylation of myosin regulatory light chain. Arch. Biochem. Biophys. 552-553, 29–39 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chu S., Muretta J. M., Thomas D. D., Direct detection of the myosin super-relaxed state and interacting-heads motif in solution. J. Biol. Chem. 297, 101157 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.National Research Council, Guide for the Care and Use of Laboratory Animals ( National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
- 43.Wang Y., et al. , Prolonged Ca2+ and force transients in myosin RLC transgenic mouse fibers expressing malignant and benign FHC mutations. J. Mol. Biol. 361, 286–299 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Irving T. C., Millman B. M., Changes in thick filament structure during compression of the filament lattice in relaxed frog sartorius muscle. J. Muscle Res. Cell Motil. 10, 385–394 (1989). [DOI] [PubMed] [Google Scholar]
- 45.Ghasemi A., Zahediasl S., Normality tests for statistical analysis: A guide for non-statisticians. Int. J. Endocrinol. Metab. 10, 486–489 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.