Significance
Photoacoustic (PA) imaging can provide information about deep-seated biological tissues. Unfortunately, signal intensity and resolution can be limited by background signals arising from endogenous chromophores. Photochromism, the use of agents that go from colorless to colored, has been suggested to be a promising avenue to overcome this limitation. However, a viable method that allows practical applications of this so-called background-suppressed PA imaging is still lacking. Here, we report the engineering of a phytochrome-based reporter protein (mDrBphP-PCMm/F469W) that displays high photoswitching contrast. Its expression in Escherichia coli enables point-specific imaging within the tumor region over several days and permits PA monitoring of deep-seated tumor tissues via reduction of background signals.
Keywords: tumor-targeted imaging, photoswitching, photoacoustic imaging, zero background, Escherichia coli
Abstract
Photoacoustic (PA) imaging offers promise for biomedical applications due to its ability to image deep within biological tissues while providing detailed molecular information; however, its detection sensitivity is limited by high background signals that arise from endogenous chromophores. Genetic reporter proteins with photoswitchable properties enable the removal of background signals through the subtraction of PA images for each light-absorbing form. Unfortunately, the application of photoswitchable chromoproteins for tumor-targeted imaging has been hampered by the lack of an effective targeted delivery scheme; that is, photoswitchable probes must be delivered in vivo with high targeting efficiency and specificity. To overcome this limitation, we have developed a tumor-targeting delivery system in which tumor-homing bacteria (Escherichia coli) are exploited as carriers to affect the point-specific delivery of genetically encoded photochromic probes to the tumor area. To improve the efficiency of the desired background suppression, we engineered a phytochrome-based reporter protein (mDrBphP-PCMm/F469W) that displays higher photoswitching contrast than those in the current state of the art. Photoacoustic computed tomography was applied to achieve good depth and resolution in the context of in vivo (mice) imaging. The present system effectively integrates a genetically encoded phytochrome-based reporter protein, PA imaging, and synthetic biology (GPS), to achieve essentially background-suppressed tumor-targeted PA monitoring in deep-seated tissues. The ability to image tumors at substantial depths may enable target-specific cancer diagnoses to be made with greater sensitivity, fidelity, and specificity.
Photoacoustic (PA), “light-in sound-out” imaging (1–6), is a promising imaging method that extends the penetration depth to the centimeter scale in vivo (7–10), a scope that is unattainable by conventional optical imaging technologies. In a clinical setting, this deep-tissue imaging capability could translate into diagnoses that are improved, relative to current imaging technologies. Recent efforts to improve image contrast and facilitate disease monitoring using PA have stimulated efforts to develop exogenous contrast agents that improve the signal-to-noise resolution (11–13). Unfortunately, the strong absorption from endogenous chromophores (e.g., hemoglobin and melanin) affects the sensitivity of PA. In fact, high background signals are routinely observed, which reduces the utility of the method. A common strategy used to overcome this latter limitation is to use multiwavelength imaging (i.e., PA spectroscopy) for the differentiation of each chromophore/contrast agent and deduction of its spatial distribution (14–18). However, this approach suffers from the drawback of wavelength-dependent optical attenuation in biological tissues. Optical fluence calibration between different wavelengths in deep tissues is currently a major challenge in PA spectroscopy. Furthermore, this spectral unmixing approach involves a large-scale inverse problem that presents a considerable computational burden, particularly for in vivo imaging which involves the presence of diverse chromospheres, including oxyhemoglobins, deoxyhemoglobins, lipids, water, and exogenously injected probes. Recently, reversibly photoswitchable chromoproteins (19–25), which are genetically encoded photoabsorbers manifesting two isomeric states with different absorption spectra, have shown great potential in addressing this challenge. Benefitting from this photochromic behavior, the subtraction of the PA images between the two isomeric states allows the overwhelming background signals to be effectively eliminated, preserving only the PA signals of the genetically encoded reporters. Such so-called background-suppressed PA imaging is expected to be particularly useful in the area of deep-seated tumor diagnosis.
In early proof-of-concept studies (19, 20, 26), photochromic proteins were demonstrated as allowing background signal subtraction; however, their limited targeting specificity has prevented their widespread use for in vivo applications. In previous approaches (19, 20), photochromic chromoproteins were implanted in vivo by injecting tumor cells expressing these reporter proteins directly into a specific location/organ. Although these demonstrations were successful, they rely on a delivery approach that is likely not practical in a clinical setting. For the promise of background-suppressed tumor-targeted imaging to be realized on a routine basis, systematic delivery systems with high targeting efficiency and specificity are needed, which we believe is the key to the wide application of genetically encoded probes.
Living microorganisms, such as Bacillus anaerobic bacteria and Calmette–Guérin, have shown recent promise as drug carriers for the tumor-targeted delivery of therapeutic agents (27–31). The hypoxic and immunosuppressive tumor microenvironment enables some bacterial strains to effect tumor-specific colonization (32–36). We therefore rationalized that this bacterial-based delivery strategy could be exploited to achieve the delivery of photoswitchable chromoproteins. The bacterial system offers two advantages. First, as microscopic robotic factories, bacterial vectors can be reprogrammed following genetic rules, facilitating their expression of genetically encoded proteins (e.g., photoswitchable chromoproteins). Second, recent studies have shown that bacteria selectively proliferate within tumor regions, particularly within necrotic tissues, without generating notable toxicity to normal organs and tissues (37). This is perhaps because bacteria in circulation and normal tissues are cleared within hours and days, respectively (32); in contrast, bacteria in tumor tissues continue to self-replicate to concentrations that often greatly exceed those present in the initial colony. This tendency not only reinforces tumor-targeting specificity but also facilitates PA detection sensitivity due to bacterial proliferation. In this regard, bacteria are expected to provide an attractive scheme for delivering genetically encoded photoswitchable chromoproteins into the microenvironment of tumor sites.
Here, we report the successful bacterial-based tumor delivery of a photochromic chromoprotein (mDrBphP-PCMm/F469W). As detailed below, the anaerobic nature of Escherichia coli enables the point-specific delivery of mDrBphP-PCMm/F469W at the tumor region. Subsequent PA imaging—photoacoustic computed tomography (PACT) (1, 38–40)—allows for tumor-targeting imaging in vivo. Elimination of background signals and the photomediated visualization of the PA signal corresponding to the tumor-targeting chromospheres afford unprecedented sensitivity for PA imaging. Moreover, the bacterial proliferation in tumor regions provided a stable PA signal during the imaging period over several days. After the PA imaging study has been performed, the E. coli could be largely removed by treating with an antibiotic (streptomycin). This system addresses the twin challenges of tumor-targeting and background suppression.
For ease of reference, we propose the term GPS-based imaging for this strategy. Here, G represents the genetically encoded contrast agent—an engineered photoswitchable chromoprotein (mDrBphP-PCMm/F469W), which display a higher photoswitching contrast compared to other chromoproteins reported to date (19–22), P indicates photoacoustic imaging—a custom-designed visualization tool to achieve deep-tissue tumor specific imaging, and S refers to synthetic biology—a bacterial vector (E. coli) reprogrammed to produce a photoswitchable chromoprotein and to deliver the protein specifically to the tumor region. The combination of attributes embodied in GPS imaging permits the visualization of deep-seated tumors via PA by effectively eliminating the background signals over relatively long measurement periods. We thus believe that the PA strategy reported here could help with the initial diagnosis and long-term monitoring of tumor-related disease.
Results
The Design and Evaluation of Genetically Encoded Photochromic Chromoproteins.
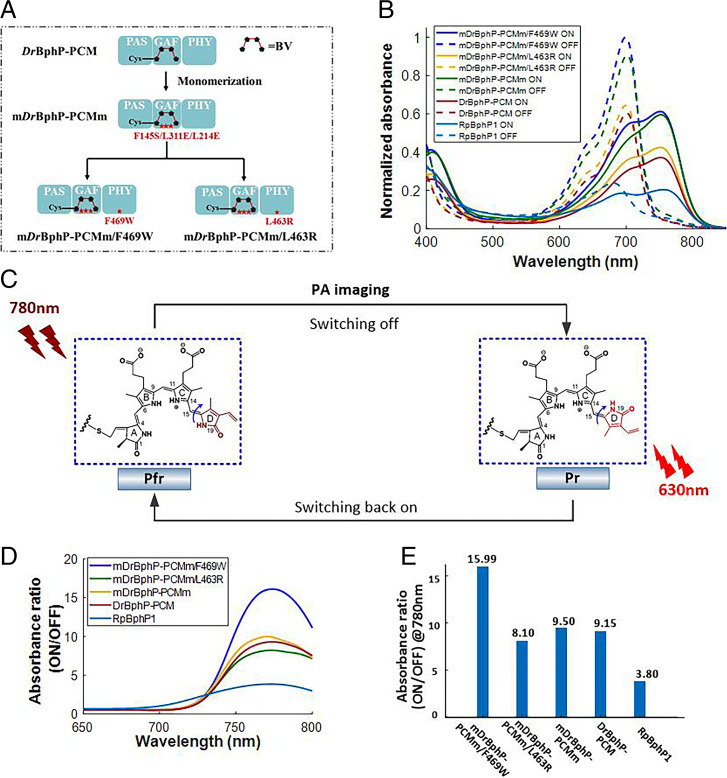
Phytochromes are light-sensitive proteins that originate from plants and bacteria (21, 22). In this work, we have engineered several photoswitchable chromoproteins based on bacterial phytochromes, named mDrBphP-PCMm, mDrBphP-PCMm/F469W, and mDrBphP-PCMm/L463R. The approach used to construct these genetically encoded chromoproteins is shown in Fig. 1A, and full detailed procedures leading to their generation are elaborated in SI Appendix, Supplemental Methods 1. Spectral and photoacoustic characterization studies were carried out for each chromoprotein (SI Appendix, Table S1, Fig. S1, and Supplemental Methods 3), the results of which are compared against the previously reported chromoproteins (DrBphP-PCM and RpBphP1) (19, 20). The evaluation of each chromoprotein enabled the identification of the best candidate for background-suppressed imaging. Notably, the three photoswitchable chromoproteins we engineered all exhibited a higher absorptivity than the previously reported chromoproteins (DrBphP-PCM and RpBphP1), a feature that was expected to translate into stronger PA signals. As shown in Fig. 1B, the two light-absorption states for these newly developed proteins are all between the red and near-infrared (NIR) regions. These spectra regions have relatively low optical attenuation in scattering media, thereby facilitating the photoconversion efficiency between the two states in deep tissues. As seen in Fig. 1C, the photoconversion arises from cis–trans photoisomerization of the D ring of the biliverdin (BV) chromophore (41) around the adjacent double bond (highlighted by the blue arrow). This photochemistry is the same for all of the genetic reporter proteins considered in this study. As noted in Fig. 1C, 780-nm light was used for both PA imaging and the photoconversion from the ON to the OFF state. Light irradiation using 630-nm light was then used to switch the proteins back to their respective ON states, thereby allowing repeated measurements.
Fig. 1.
(A) Evolution of our photoswitchable chromoproteins, including mDrBphP-PCMm, mDrBphP-PCMm/L463R, and mDrBphP-PCMm/F469W. Monomeric DrBphP-PCM (i.e., mDrBphP-PCMm) was constructed by introducing three mutations (F145S, L311E, and L314E). The mDrBphP-PCMm preserves three domains (PAS, GAF, and PHY) of DrBphP-PCM, the functions of which are elaborated in refs. 19 and 20. For the engineering of mDrBphP-PCMm/F469W and mDrBphP-PCMm/L463R, the mutations (F469W and L463R) were introduced into mDrBphP-PCMm to construct mDrBphP-PCMm/F469W and mDrBphP-PCMm/L463R, respectively. (B) The normalized absorption spectra of chromoproteins and their corresponding photoswitched isomers (mDrBphP-PCMm/F469W, mDrBphP-PCMm/L463R, mDrBphP-PCMm, DrBphP-PCM, and RpBphP1). (C) Basic schematic illustrating the proposed photoconversion process induced by light irradiation at 780 nm and 630 nm. Pr refers to the OFF state of photochromic bacteriophytochromes, while Pfr refers to the ON state. The phytochrome structures (i.e., BV) were adapted from ref. 41. (D) Absorbance ratio between the ON and OFF states of different photoswitchable chromoproteins as a function of wavelength. (E) Comparison of the absorbance ratios between the ON and OFF states at 780 nm. The results support the conclusion that mDrBphP-PCMm/F469W has the highest switching contrast among the different chromoproteins considered in this study.
Two selection criteria, namely, the molar absorption coefficient and the photoswitching contrast, were considered in evaluating various photochromic probes. The mDrBphP-PCMm/F469W shows the highest absorbance for both states, resulting in high imaging sensitivity in both the ON and OFF states, as mentioned earlier. The photoswitching contrast, on the other hand, is a key factor determining the efficiency of background suppression and imaging specificity. The photoswitching contrast is defined as the ratio of the absorption coefficient at the ON state to that at the OFF state. Thus, a higher photoswitching contrast indicates greater preservation of the intrinsic signal of the chromoproteins when the background signals are removed, thereby maximizing the image contrast after the PA image differential. As shown in Fig. 1D, mDrBphP-PCMm/F469W exhibits superior switching contrast in the NIR spectra region. For the wavelength used for PA imaging (780 nm), the photoswitching contrast is illustrated in Fig. 1E, in which mDrBphP-PCMm/F469W shows 1.75 times better switching contrast than that of DrBphP-PCM and 4.2 times better than RpBphP1. The PA signal ratios between the ON and OFF states follow a similar trend (SI Appendix, Fig. S1 D and E). On this basis, mDrBphP-PCMm/F469W was considered the most promising system for PA differential imaging.
Another advantage of mDrBphP-PCMm/F469W is that it remains a monomer (SI Appendix, Supplemental Methods 2 and Fig. S2A) as opposed to existing in dimeric form, as is true for DrBphP-PCM and RpBphP1; it is thus smaller than these latter constructs. This reduction in size means that there is a lower likelihood that mDrBphP-PCMm/F469W will interfere with the normal function of a protein partner to which it could be genetically fused. As a result, mDrBphP-PCMm/F469W was chosen in this study due to its high absorbance, high photoswitching contrast, and small size, attributes that we believe make it superior to DrBphP-PCM and RpBphP1 developed in previous studies (19, 20). In addition, mDrBphP-PCMm/F469W preserves the robust reversible photoswitching features of the photochromic bacteriophytochromes (SI Appendix, Fig. S2B). Multiple switching cycles did not trigger photoswitching fatigue or photobleaching. The large photoswitching contrast of mDrBphP-PCMm/F469W gives rise to a very dim fluorescence emission (SI Appendix, Fig. S3 and Supplemental Methods 4). This is because, in BV, the absorbed photon of light serves largely to drive cis–trans isomerization. This process competes with fluorescence emission.
Expression of mDrBphP-PCMm/F469W in E. coli (MG1655).
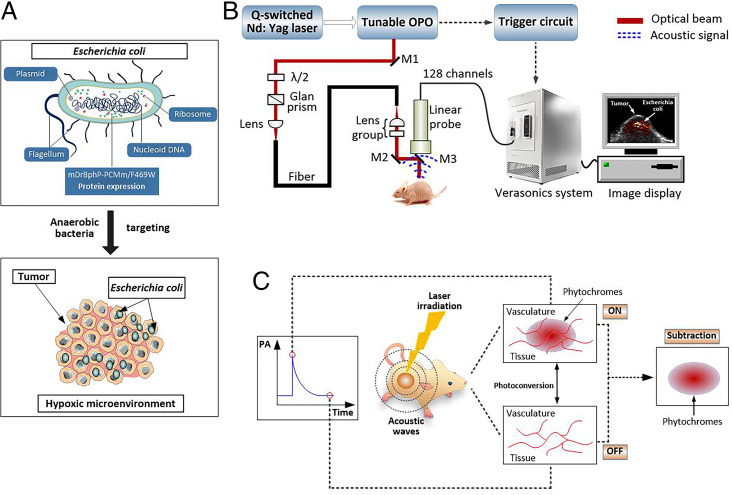
E. coli MG1655 are facultative anaerobic bacteria that have been used as drug delivery systems (42, 43). In this study, E. coli MG 1655 was reprogrammed to produce and deliver the photochromic PA imaging protein mDrBphP-PCMm/F469W into the tumor region. Among different bacteria, E. coli MG1655 was chosen for the following reasons: 1) Simple genetic manipulation of E. coli MG 1655 allows it to be used to express heterologous proteins; 2) the facultative anaerobic habits of E. coli MG1655 provide favorable niches for the specific colonization of hypoxic and necrotic regions within solid tumors (Fig. 2A). To introduce mDrBphP-PCMm/F469W within E. coli MG1655, a constitutive expression vector pJC-mDrBphP-PCMm/F469W-HO was constructed (see Materials and Methods), which is controlled by a constitutive synthetic prokaryotic promoter. By applying genetic rules, E. coli M1655 were reprogrammed (SI Appendix, Fig. S4 A and B) to construct mDrBphP-PCMm/F469W–expressing E. coli MG1655, simplified as E. coli (F469w) throughout the manuscript. Once isolated, these constructs were used in our phantom and in vivo PA imaging experiments. The expression results of E. coli (F469w) are shown in SI Appendix, Fig. S4 C and D.
Fig. 2.
(A) Schematic diagram of the present bacteria-based in vivo delivery system for photoswitchable chromoproteins. Owing to the unique hypoxic tumor microenvironment, E. coli (F469w) actively colonize in the tumor regions. (B) Experimental setup for the PACT system used in this study. M, mirror; λ/2, half-wave plate. (C) Cartoon illustration of the proposed background-suppressed PA molecular imaging. As detailed in the text, based on their unique photochromic features, the present genetically encoded probes allow background signals, including those from blood, skin, and melanin, to be eliminated by subtracting the PA signal produced by the red and NIR light-absorption states.
In Vitro PA Imaging of Bacteria.
A custom-built PACT system (Fig. 2B) was employed in this study. This PACT system provides 213-μm lateral resolution and 150-μm axial resolution (SI Appendix, Fig. S5). The full imaging procedures and instrument information are shown in detail in SI Appendix, Supplemental Methods 6. The PA performance of E. coli (F469w) was examined and analyzed (SI Appendix, Fig. S6). The results showed no obvious change in the PA signal after exposure to 2,000 laser pulses (over 1 min) at a wavelength of 700 nm (SI Appendix, Fig. S6D). This was taken as evidence that E. coli (F469w) displays good photostability.
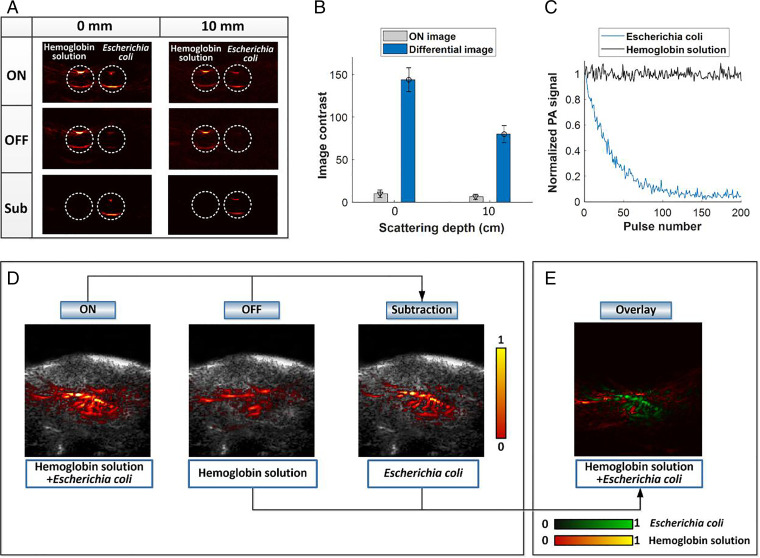
As an initial test, PA-based photochromic imaging was performed in transparent plastic tubes, and the PA signal of E. coli (F469w) was extracted by subtracting the ON- and OFF-state PA images under 780-nm irradiation (Fig. 2C). A transparent plastic tube was filled with a solution of E. coli (F469w) (5 × 107 colony-forming units [CFU]/mL). A second tube was filled with a hemoglobin solution. The latter solution, human hemoglobin (lyophilized powder) purchased from Sigma-Aldrich was dissolved in phosphate-buffered saline (PBS) to give a ca. 0.15 g/mL solution. These two solutions were subject to PA imaging using a clear medium (water) and a 10-mm-thick scattering medium (0.1% intralipid). Fig. 3A illustrates the PA images acquired before and after subjecting to 780-nm photoirradiation for both the E. coli solution and the hemoglobin solution (Fig. 3 A, Top and Middle), as well as the differential images between the two states (Fig. 3 A, Bottom). After PA image subtraction between the ON and OFF states, a global threshold was applied to all differential images, and values below the threshold were rejected. The threshold was set as 3 to 6 times the SD of the background noise outside the imaged region. These procedures were applied for all differential images in this study. The hemoglobin solution displayed high image contrast in the ON state, whereas, after subtraction (i.e., the differential image), the hemoglobin signals were effectively eliminated. In contrast, the PA signal corresponding to E. coli (F469w) remained. A similar imaging outcome was observed in the 10-mm-thick sample, leading us to conclude that PA imaging with essentially eliminated background signals at significant tissue depths (i.e., solid tumors) might be possible using the present mDrBphP-PCMm/F469W–based system. The image contrast was considerably enhanced after subtraction of the two states. The signal-to-noise ratio of the differential image showed a 14.5-fold increase compared with the original ON state (Fig. 3B) for water medium and a 12.7-fold enhancement for the scattering medium with a depth of 10 mm. The noise-equivalent bacteria concentration for E. coli (F469w) was estimated to be around 5 × 106 CFU/mL to ∼6 × 106 CFU/mL (SI Appendix, Fig. S4E) in the 10-mm-thick sample. Fig. 3C depicts the PA signal at 780 nm as a function of time (expressed in terms of the number of laser pulses used during laser irradiation) for both E. coli (F469w) and hemoglobin solution during the photoswitching process. To mimic the deep biological tissue environment, we performed tube tests in chicken breast tissues in which the photoswitching feature of E. coli was examined at different depths within the scattering medium (SI Appendix, Fig. S7). These studies revealed good photoswitching efficiency between the two states in deep tissue up to a centimeter scale.
Fig. 3.
(A) PA phantom imaging of E. coli (F469w) and hemoglobin solution at 780 nm in clear medium (water, Left) and in 10 mm-thick scattering medium (0.1% intralipid, Right). The ON and OFF states and the difference between them are shown in Top, Middle, and Bottom, respectively; Sub, subtraction. (B) Quantitative comparison of imaging contrast between the ON image and the differential image at different depths of scattering medium. The data were averaged over six measurements. Error bars indicate the standard error of the mean. (C) Normalized PA signal as a function of pulse number for both E. coli and hemoglobin solution, showing the switching-off process of E. coli and the steady state of the hemoglobin solution. The laser fluence was 18 mJ/cm2 at 780 nm, which is below the safety limit (20 mJ/cm2 at 780-nm wavelength) set by the American National Standards Institute. To convert the pulse number into time (seconds), the x axis should be multiplied by a factor of 0.033 since a 30-Hz pulse repetition frequency was applied. (D) Phantom images of E. coli (F469w) and hemoglobin solution at 780 nm in chicken breast tissue in the ON state (Left), the OFF state (Middle), and the difference between the two states (Right). The signals arising from E. coli are revealed in the differential image from which the background signals are eliminated. (E) Overlay image showing E. coli in green and the background hemoglobin signal in red. During the subtraction between the ON and OFF states, a global threshold (3 to 6 times SD of the background noise) was applied to all differential images.
To mimic the vascular environment in vivo within deep biological tissues, the hemoglobin solution was injected diffusively into 10-mm-thick chicken breast tissue samples. E. coli (F469w) were then injected into the same area. PA images in both the ON and OFF states were acquired. In the ON state (Fig. 3 D, Left), E. coli could not be identified, due to the high background signal arising from the hemoglobin solution. The differential image (Fig. 3 D, Right), in contrast, allows the distribution of E. coli to be seen clearly. The extracted signals of E. coli were then overlaid on the hemoglobin background. This allowed the individual spatial profiles of the two constituent chromophores to be distinguished (Fig. 3E). On the basis of these studies, we conclude that the signals of the present engineered E. coli can be successfully separated from background as modeled by hemoglobin in deep scattering tissues (∼10 mm).
In Vivo PA Imaging of E. coli Targeting Solid Tumors.
The putative tumor-selective colonization of E. coli (SI Appendix, Fig. S8) was then tested in the context of in vivo imaging. For these studies, E. coli (F469w) was delivered into female tumor‐bearing nude BALB/c mice via tail vein injection (Materials and Methods). A control group in which tumor-bearing mice were injected intravenously with stroke-physiological saline solution (SPSS) was also studied. Per our design expectations, E. coli acts both as a molecular probe to boost the detection sensitivity and specificity of the PA imaging and as a navigator to selectively colonize tumor tissues as a consequence of their anaerobic features.
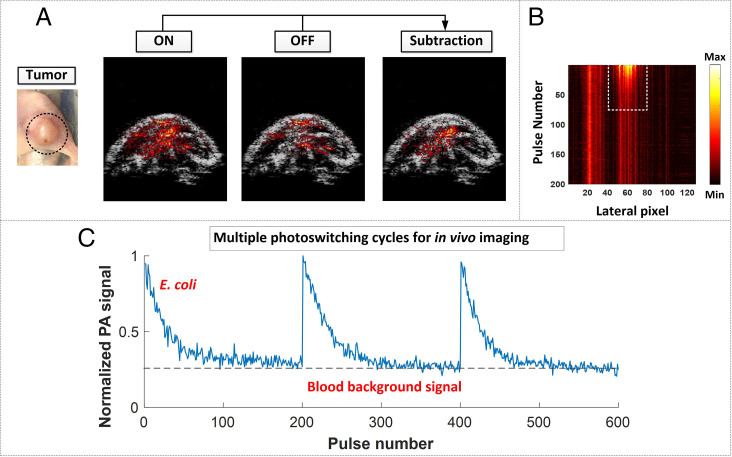
To visualize bacterial colonization, PA images of the tumor and adjacent regions were obtained by subtracting the OFF state from the original ON state (Fig. 4A). This subtraction ensures that the PA signals come from the E. coli that possess the photoswitching capability seen in the above model studies. Other background signals, such as tumor-associated vasculature, skin, and melanin, are thus subtracted out in the difference images. Fig. 4B shows the maximum amplitude projection (MAP) images of the same Bscan area of Fig. 4A plotted against time (i.e., laser pulse number), indicated in the y axis. It thus illustrates the variation in the PA signals seen during ON to OFF conversion triggered by illumination at 780 nm. The columns of the MAP image in which the PA signals decrease along the y axis reflect sites where E. coli accumulates, while the columns that exhibit steady-state PA signals represent the sites where endogenous chromospheres (such as hemoglobin and melanin) locate. In the E. coli accumulating region (dashed rectangle in Fig. 4B), the column showing the maximum PA signal was extracted, and its PA values were plotted as a function of pulse number to show, quantitatively, the PA signal variation seen during photoconversion (one cycle in Fig. 4C). The multiple ON/OFF cycles, triggered by the consecutive switching between 780-nm and 630-nm photoillumination, served to confirm the reversible photoswitching performance of E. coli, as well as the reproducible nature of our imaging results.
Fig. 4.
(A) PA imaging of mouse tumor region and adjacent tissues in the ON and OFF states as well as the subtracted image between them. The PA images are overlaid on ultrasound (US) images. A global threshold, calculated as 3 to 6 times SD of the background noise, was applied to the difference image. (B) MAP images of the colonization region plotted against time, that is, pulse number (y axis), showing the variation in the PA signal during the photoswitching process from the ON to the OFF state upon illumination at 780 nm. (C) Multiple switching cycles reflecting the consecutive application of 780- and 630-nm light and highlighting the reversible switching properties of E. coli (F469w). To convert the pulse number into time (seconds), the x axis should be multiplied by a factor of 0.033.
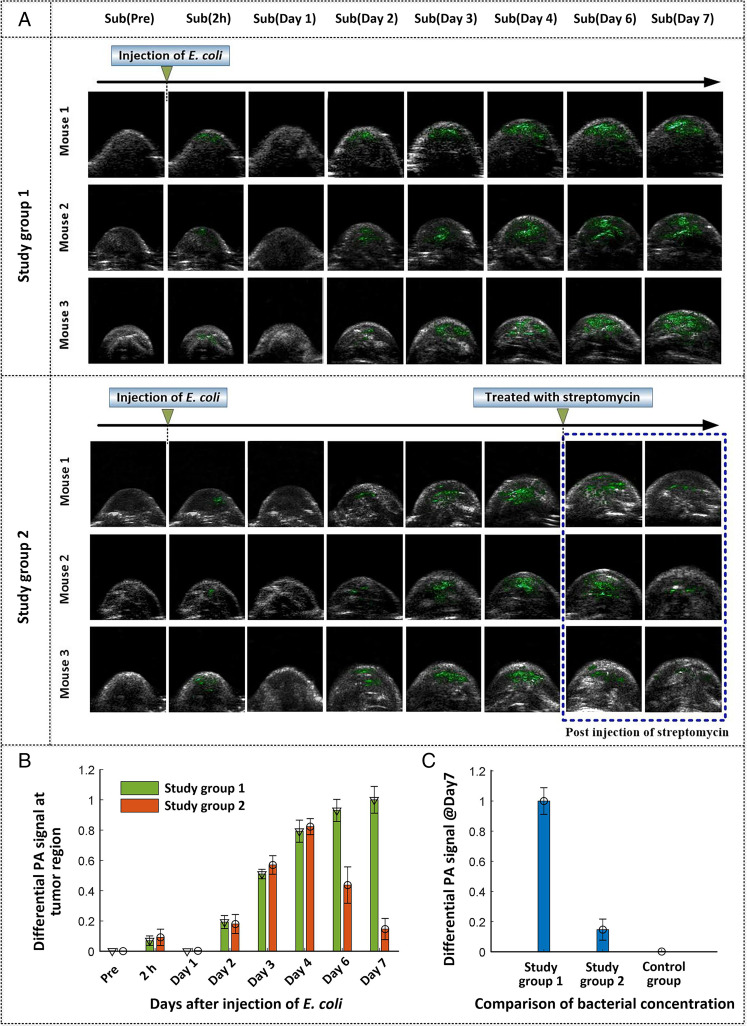
We performed PACT imaging of the tumor and adjacent regions of mice before injection and 2 h, and 1, 2, 3, 4, 6, and 7 d postinjection of E. coli (F469w) to evaluate the colonization of bacteria in tumor tissues as a function of time. The corresponding imaging results (Bscans) after subtraction of the OFF from the ON state are shown in Fig. 5A. Detailed results for the underlying ON and OFF states for each mouse can be found in SI Appendix, Fig. S9. The extracted signals of E. coli (F469w) are indicated in green in all of the images. We arranged two study groups in which mice were treated with and without streptomycin at day 4 postinjection of E. coli (F469w) (Fig. 5A). This latter study was undertaken to demonstrate that the E. coli used in the present approach could be largely eliminated from the tumor regions with drug intervention if desired. As shown in Fig. 5A, in all of the mice subject to study, slight signals ascribed to E. coli (F469w) were observed immediately, that is, 2 h after intravenous administration (SI Appendix, Figs. S10 and S11). This is likely due to the presence of bacteria in the blood circulatory system after intravenous injection. After approximately 12 h, signals ascribable to E. coli are essentially absent from the tumor or nearby regions (SI Appendix, Fig. S11), which we interpret in terms of considerable clearance of intravenously injected E. coli from blood circulation. Starting on day 2, E. coli signals reappeared in the tumor-associated region. The signal intensity increased on subsequent days, a result we take as evidence that the administered E. coli migrate to the cancerous tissues and start to proliferate. This selective colonization is consistent with the hypothesis that the unique hypoxic and necrotic regions of solid tumors offer a key niche for E. coli colonization. For the non–streptomycin-treated mice (study group 1), E. coli continued to proliferate during the ensuing days. For the streptomycin-treated mice (study group 2), the intensity of the difference-based PA signal decreased as a function of time poststreptomycin treatment and was largely absent by day 7, consistent with bacterial apoptosis after the injection of streptomycin. The effects of these two opposing dynamic processes (colonization vs. streptomycin-based eradication) are quantitatively reflected in Fig. 5B. It is worth noting that, in study group 1, the accumulation of E. coli starts to level off at about day 6, an observation that probably reflects the fact that the tumor region will not support infinite proliferation of the E. coli bacteria.
Fig. 5.
(A) Representative differential PA images of tumor‐bearing mice in two study groups at different time points before and after intravenous injection of E. coli (F469w), overlaid on US images. A global threshold, calculated as 3 to 6 times SD of the background noise, was applied to the difference image. (B) The relative PA signals of E. coli in the tumor region on different days after injection. The results provide support for the colonization of E. coli in tumor regions, as well as E. coli apoptosis following injection of streptomycin. (C) Statistical comparison of the E. coli signal for the control group and study groups at day 7 postinjection. Bars in B and C indicate the standard error of the mean (n = 3).
PA imaging was also performed on the control group wherein the mice were initially administered SPSS instead of E. coli (F469w). The difference PA images for the control group revealed no noticeable signal throughout the observation period (SI Appendix, Fig. S12). Fig. 5C provides a statistical comparison of the differential signal (E. coli signal) between the control group and two study groups on day 7 postinjection. The sharp contrast seen between the control group and study group 1 is taken as evidence that the presence of E. coli in the study groups originates from the intravenous injection instead of internal bacterial colonies that might be present in vivo. In contrast, the difference between study groups 1 and 2 provides support for the notion that the proliferated probes (E. coli (F469w)) in the tumor regions can be essentially eliminated from the colonized sites. We thus propose that the present E. coli–based photoswitchable chromoprotein targeting strategy can be implemented in a well-controlled manner with little of the PA probe persisting in the mouse tumor regions after drug (streptomycin) intervention.
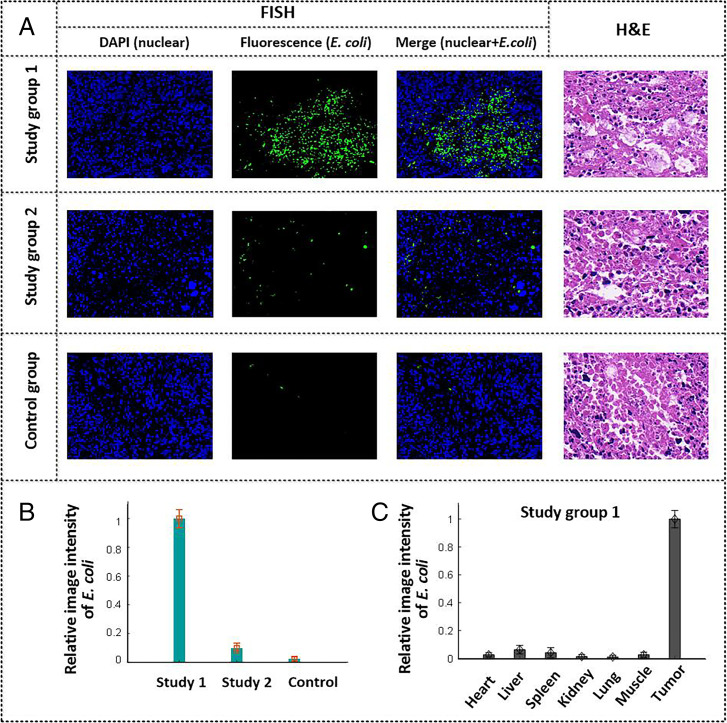
Following the PA imaging studies, the solid tumors were subject to histological examination to verify the colonization by E. coli. This was performed by employing fluorescence in situ hybridization (FISH), where a labeled nucleic acid strand was used to locate the DNA sequence of E. coli (SI Appendix, Supplemental Methods 8). The positive expression of E. coli was indicated by the presence of a fluorescence signal (green), while the cell nucleus was stained (blue) by DAPI. As shown in Fig. 6A (study group 1, FISH), labeled green regions were seen in the slices corresponding to tumor, as expected given the proposed presence of E. coli. Meanwhile, hematoxylin and eosin (H&E) staining of the adjacent slices of the same tumor samples was carried out to reveal the cell morphology and tissue structure of the cancerous region (Fig. 6 A, Right). It was inferred that the distribution of E. coli was heterogeneous (Fig. 5A and Fig. 6A, study group 1), which is likely due to the complexity and diversity (heterogeneity) of the immune features of the tumor microenvironment. Tumor heterogeneity in terms of immune cells and the hypoxic tumor microenvironment leads to an uneven spatial response to E. coli colonization in cancerous regions. The FISH results for study group 2 and the control group are provided in Fig. 6A. The E. coli signal in study group 2 was considerably lower than that of study group 1, presumably reflecting the streptomycin intervention. Note that very weak signals corresponding to E. coli are present in the control group. We assume that these latter signals may come from the internal bacterial colonies of E. coli inherent within the mice, since no E. coli (F469w) were administered to the control group. Fig. 6B provides statistical comparisons of the E. coli signal between the three groups. The FISH results illustrated in Fig. 6B are qualitatively consistent with the PA imaging outcomes shown in Fig. 5C.
Fig. 6.
(A) Representative images of FISH (Left, Center Left, and Center Right) and H&E-stained (Right) tumor slices for both the study groups and the control group at 7 d postinjection. (Magnification:x400.) (B) Statistical analysis of the E. coli distributions for the study and control groups at tumor regions based on the FISH images. (C) Quantitative comparison of E. coli distributions in different organs for study group 1 based on the FISH images. Bars in B and C indicate the mean and standard error of the fluorescence signals in the FISH images.
The major organs (heart, liver, spleen, lung, and kidney) and muscles of mice in the study group were also examined using the FISH technique. The organs were collected on day 7 postintravenous injection. The FISH results in SI Appendix, Fig. S13 reveal signals ascribable to the E. coli (F469w) within the solid tumors but no significant proliferation in normal tissues or organs. This difference is also reflected in the statistical analysis of the E. coli colonization in different organs and cancerous regions (Fig. 6C) based on the signal intensity in the FISH images. This sharp contrast is perhaps because, in normal organs, E. coli triggers the activation of antimicrobial defenses and is largely cleared by the host immune response; however, this is not the case in the immune-suppressed tumor microenvironment. Owing to the ineffective immune response, as well as the hypoxia microenvironment in tumor-associated regions, E. coli migrate to tumor sites and maintain positions of preferred low oxygen tension within the tumor region. This, in turn, leads to the superior tumor-targeting specificity seen for the E. coli (F469w).
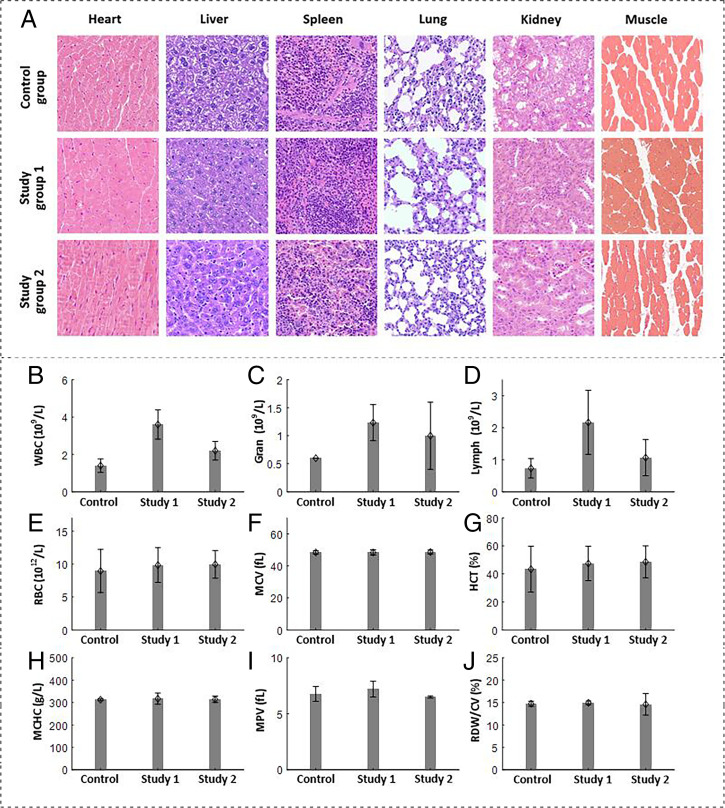
To evaluate the performance of E. coli (F469w) in the context of possible in vivo applications, H&E staining was also performed on different organs of the mice in both the study and control groups (Fig. 7A). No notable organ damage or inflammation was observed in study groups 1 and 2 as compared to the control group. Blood tests of the mice in the three groups were performed (Fig. 7 B–J). The numbers of white blood cells, neutrophilic granulocytes, and lymphocytes in study group 1 were higher than in the control group. This is likely due to bacterial infection of the mice in study group 1, which were intravenously injected with E. coli. In study group 2, where the mice were treated with streptomycin on day 4 postinjection of E. coli, the various blood panel parameters gradually returned to normal. For other blood parameters shown in Fig. 7 E–J, no significant discrepancies were observed between the three groups. Although the bacteria-based strategy may raise concerns of bacterial infection (32, 44, 45) in biomedical applications, the above results lead us to suggest that the proposed E. coli–based delivery approach detailed here can be conducted in a controllable manner when appropriate antibiotics are applied.
Fig. 7.
(A) Representative H&E-stained tissue images of major organs and muscle harvested from mice in the study groups and control group at 7 d after the intravenous injection of either E. coli or SPSS. (Magnification: ×400.) (B–J) Blood analysis of mice in the above three groups. Bars indicate the SD from the mean (n = 3). (B) Number of white blood cells (WBC). (C) Number of neutrophilic granulocytes (Gran), (D) Number of lymphocytes. (E) Number of red blood cells (RBC). (F) Mean corpuscular volume (MCV). (G) Hematocrit (HCT). (H) Mean corpuscular hemoglobin concentration (MCHC). (I) Mean platelet volume (MPV). (J) Red blood cell distribution width/coefficient of variation (RDW/CV).
Discussion
A major challenge associated with PA-based molecular imaging of tumors lies in the low specificity and sensitivity of the method, which has its origins in the overwhelming background signals. Owing to the photochromic change between two absorption states, genetically encoded photoswitchable chromoproteins show the potential to remove background signals. However, the in vivo applications of genetically encoded probes for tumor imaging have been hampered by the lack of a suitable delivery system with tumor-targeting efficiency and specificity. In this work, we developed an approach—a GPS localization-based targeting strategy wherein a bacterial vector (E. coli) is reprogrammed to express a photoswitchable chromoprotein and deliver the protein specifically to the tumor region. Owing to their anaerobic features, the E. coli (F469w) show tumor-preferential colonization ability; thus, genetically encoded probes can be delivered to the microenvironment of the tumor site. By employing the proposed GPS localization-based strategy, we were able to realize background-suppressed tumor-targeted deep PA-based tissue imaging (Fig. 5 and SI Appendix, Fig. S9).
The hypoxic tumor microenvironment initiates anaerobic bacterial migration to the tumor region, while the immunosuppressed tumor microenvironment allows the self-proliferation of E. coli (F469w) and accumulation of a PA-producing effect. Histological analyses conducted after PA imaging confirmed the accumulation of the targeting E. coli bacteria in tumor regions (Fig. 6A, study group 1). In contrast, E. coli in normal tissues/organs show rapid clearance due to host immune defense. This tumor-selective colonization and proliferation of E. coli overcomes the limitations of conventional nanoparticle (NP)-based molecular probes, which are easily diluted in vivo. NPs cannot self-proliferate, and their imaging sensitivity is limited by the initial concentration of molecular probes. In contrast, bacterial proliferation under immune-suppressed conditions enhances the targeting concentration, targeting period, and targeting specificity. In addition, the reporter proteins are genetically expressed during bacterial proliferation in tumor regions, which avoids NP disassembly in vivo that could occur for NP-based approaches. The E. coli (F469w) can be largely reduced by means of drug (streptomycin) intervention. Hence, this E. coli–based targeting strategy can be performed in a well-controlled manner.
The high detection sensitivity of mDrBphP-PCMm/F469W and the background-suppressed imaging it permits could prove useful in assisting intraoperative surgical navigation and for visualizing deep-seated nonpigmented tumor nodules. The bacteria models, on the other hand, could see application in investigations of bacterial infection, bacterial drug efficacy, and mechanisms of bacteria-based tumor inhibition.
We appreciate that certain issues need to be addressed to facilitate the clinical development of bacteria-based delivery systems. Bacteria vectors in the future need to be engineered to mitigate systemic toxicity and provide for improved biocompatibility. One strategy involves genetic modification to delete the virulence genes responsible for the pathogenicity. This is a strategy that is being pursued by a number of groups (46–48). Previous research showed that the removal of Salmonella msbB gene, which leads to the loss of a key component of lipopolysaccharide, helps diminish the toxicity of the Salmonella sp. by 10,000-fold (46). Similarly, a highly toxic strain of Clostridium novyi has been transformed into a substantially safer strain via genetic modification of a lethal exotoxin (47). Listeria monocytogenes have also been made safer (48) by removing prfA (the master virulence regulator gene). An alternative means of addressing the systemic toxicity of bacteria carriers is to employ probiotics, such as E. coli Nissle (EcN), wherein a nonpathogenic gram-negative strain is used as the active component of the microbial drug Mutaflor. EcN has a long track record of safety in humans (49, 50). Owing to its facultative anaerobic nature and compatibility with genetic engineering techniques (51) used to express heterologous proteins, EcN is attractive as a carrier for various probes.
Future clinical translation could also benefit from optimization of the tumor targeting. The present study exploits only the tumor hypoxic microenvironment as the target-localizing mechanism. Several studies (32, 52, 53) have shown that bacteria vectors can be made to express antibodies directed toward tumor-associated antigens, such as the colorectal cancer–associated carcinoembryonic antigen (54) and the lymphoma-associated antigen CD20 (52). Another approach would be to generate auxotrophic bacterial mutants (55). Such mutant strains are enriched in cancerous lesions, because the rapid metabolism within tumor regions provides sufficient levels of metabolites for these auxotrophic bacteria. In contrast, these mutant strains cannot replicate efficiently in normal tissue where the levels of nutrients (such as leucine and arginine) are too low (55). Improving targeting could also enhance the safety profile of the proposed E. coli–based delivery system, since it is expected to be reflected in a lower required bacterial injection dose (43).
Materials and Methods
Protein Expression in Bacteria.
For protein expression in E. coli, pJC-mDrBphP-PCMm/F469W-HO was transformed into E. coli MG1655, and a single transformant was transferred into a 100-mL flask containing 20 mL of super optimal broth (SOB) medium. The transformant was cultured in a shaker at a speed of 180 rpm/min for 24 h at 34 °C. Then, the bacteria were harvested by centrifugation at 5,000 rpm/min for 5 min.
In Vivo Delivery of Bacteria and Antibiotic.
For bacterial transfections, E. coli (F469W) were diluted in PBS to a concentration of 5 × 108 CFU/mL prior to tail vein injection. Then, 200-µL samples, which consisted of 1 × 108 E. coli, were delivered to the mice via intravenous injection using a syringe. The injection quantity of E. coli (∼108) was chosen as an empirical value here because higher quantities may lead to serious systemic bacterial infections, and lower quantities might reduce the PA detection sensitivity. After intravenous injection, PA monitoring of the mouse tumors and adjacent regions was made as a function of time (preinjection, 2 h postinjection, and from day 1 to day 7) to monitor both the targeting effect and the accumulation of E. coli at the tumor sites.
The Food and Drug Administration–approved antibiotic, streptomycin, was used for bacterial killing in the present study. Streptomycin was purchased from Acmec Biochemical Co., Ltd. and was initially dissolved in PBS to a concentration of 50 mg/mL, which was then stored at −4 °C until needed. We demonstrated the in vitro bacteria-killing effect at different concentrations (SI Appendix, Fig. S14). For in vivo bacterial infection treatment, the solution was diluted to a concentration of 150 µg/mL body blood weight and injected daily into the tail vein of the mice in study group 2 from day 4 to day 7 postinjection of the E. coli. Additional orthotopic injection of streptomycin was also applied specifically to the tumor region of mice in study group 2 to enhance the local bacteria-killing effect.
Expanded methods are given in SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by the National Key R&D Program of China (Grants 2020YFA0908800 and 2021YFE0202200), the National Natural Science Foundation of China (Grants 82122034, 92059108, 81927807, and 62105355), the Chinese Academy of Sciences (CAS; Awards YJKYYQ20190078, 2019352, and GJJSTD20210003), the CAS Key Laboratory of Health Informatics (Award 2011DP173015), the Guangdong Provincial Key Laboratory of Biomedical Optical Imaging (Award 2020B121201010), the Shenzhen Key Laboratory for Molecular Imaging (Award ZDSY20130401165820357), Shenzhen Science and Technology Innovation Funding (Awards JCYJ20210324101403010 and JCYJ20200109141222892), and the Robert A. Welch Foundation (Chair Funds F-0018).
Footnotes
Reviewers: S.H., Washington University in St. Louis; and J.Y., Duke University.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2121982119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Wang L. V., Hu S., Photoacoustic tomography: In vivo imaging from organelles to organs. Science 335, 1458–1462 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song H., Gonzales E., Soetikno B., Gong E., Wang L. V., Optical-resolution photoacoustic microscopy of ischemic stroke. Proc. SPIE 7899, 789906 (2011). [Google Scholar]
- 3.Wang X., et al. , Noninvasive laser-induced photoacoustic tomography for structural and functional in vivo imaging of the brain. Nat. Biotechnol. 21, 803–806 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Gao R., Xu Z., Ren Y., Song L., Liu C., Nonlinear mechanisms in photoacoustics—Powerful tools in photoacoustic imaging. Photoacoustics 22, 100243 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao R., et al. , Photoacoustic visualization of the fluence rate dependence of photodynamic therapy. Biomed. Opt. Express 11, 4203–4223 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu M., Wang L. V., Photoacoustic imaging in biomedicine. Rev. Sci. Instrum. 77, 305–598 (2006). [Google Scholar]
- 7.Jiang Y., Pu K., Advanced photoacoustic imaging applications of near-infrared absorbing organic nanoparticles. Small 13, 1700710 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Zhang C., et al. , Design and synthesis of a ratiometric photoacoustic probe for in situ imaging of zinc ions in deep tissue in vivo. Anal. Chem. 92, 6382–6390 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Attia A. B. E., et al. , A review of clinical photoacoustic imaging: Current and future trends. Photoacoustics 16, 100144 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao R., Xu Z., Song L., Liu C., Breaking acoustic limit of optical focusing using photoacoustic-guided wavefront shaping. Laser Photonics Rev. 15, 2000594 (2021). [Google Scholar]
- 11.Chen J., et al. , Expanded porphyrins: Functional photoacoustic imaging agents that operate in the NIR-II region. Chem. Sci. (Camb.) 12, 9916–9921 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren Y., et al. , Manganese(II) texaphyrin: A paramagnetic photoacoustic contrast agent activated by near-IR light. J. Am. Chem. Soc. 142, 16156–16160 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Fu Q., Zhu R., Song J., Yang H., Chen X., Photoacoustic imaging: Contrast agents and their biomedical applications. Adv. Mater. 31, e1805875 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Cox B., Laufer J. G., Arridge S. R., Beard P. C., Quantitative spectroscopic photoacoustic imaging: A review. J. Biomed. Opt. 17, 061202 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Chen Z., Yang S., Xing D., In vivo detection of hemoglobin oxygen saturation and carboxyhemoglobin saturation with multiwavelength photoacoustic microscopy. Opt. Lett. 37, 3414–3416 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Hussain A., Petersen W., Staley J., Hondebrink E., Steenbergen W., Quantitative blood oxygen saturation imaging using combined photoacoustics and acousto-optics. Opt. Lett. 41, 1720–1723 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Li M., Tang Y., Yao J., Photoacoustic tomography of blood oxygenation: A mini review. Photoacoustics 10, 65–73 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia J., et al. , Calibration-free quantification of absolute oxygen saturation based on the dynamics of photoacoustic signals. Opt. Lett. 38, 2800–2803 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao J., et al. , Multiscale photoacoustic tomography using reversibly switchable bacterial phytochrome as a near-infrared photochromic probe. Nat. Methods 13, 67–73 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li L., et al. , Small near-infrared photochromic protein for photoacoustic multi-contrast imaging and detection of protein interactions in vivo. Nat. Commun. 9, 2734 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chee R. K. W., Li Y., Zhang W., Campbell R. E., Zemp R. J., In vivo photoacoustic difference-spectra imaging of bacteria using photoswitchable chromoproteins. J. Biomed. Opt. 23, 1–11 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Märk J., et al. , Dual-wavelength 3D photoacoustic imaging of mammalian cells using a photoswitchable phytochrome reporter protein. Commun. Phys. 1, 3 (2018). [Google Scholar]
- 23.Mishra K., et al. , Multiplexed whole-animal imaging with reversibly switchable optoacoustic proteins. Sci. Adv. 6, eaaz6293 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vetschera P., et al. , Characterization of reversibly switchable fluorescent proteins in optoacoustic imaging. Anal. Chem. 90, 10527–10535 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Yang J., et al. , Focusing light inside live tissue using reversibly switchable bacterial phytochrome as a genetically encoded photochromic guide star. Sci. Adv. 5, eaay1211 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stiel A. C., et al. , High-contrast imaging of reversibly switchable fluorescent proteins via temporally unmixed multispectral optoacoustic tomography. Opt. Lett. 40, 367–370 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Felfoul O., et al. , Magneto-aerotactic bacteria deliver drug-containing nanoliposomes to tumour hypoxic regions. Nat. Nanotechnol. 11, 941–947 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quispe-Tintaya W., et al. , Nontoxic radioactive Listeriaat is a highly effective therapy against metastatic pancreatic cancer. Proc. Natl. Acad. Sci. U.S.A. 110, 8668–8673 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chowdhury S., et al. , Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat. Med. 25, 1057–1063 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chien T., Doshi A., Danino T., Advances in bacterial cancer therapies using synthetic biology. Curr. Opin. Syst. Biol. 5, 1–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Din M. O., et al. , Synchronized cycles of bacterial lysis for in vivo delivery. Nature 536, 81–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou S., Gravekamp C., Bermudes D., Liu K., Tumour-targeting bacteria engineered to fight cancer. Nat. Rev. Cancer 18, 727–743 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leschner S., et al. , Tumor invasion of Salmonella enterica serovar Typhimurium is accompanied by strong hemorrhage promoted by TNF-α. PLoS One 4, e6692 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang S.-N., et al. , Inhibition of tumor growth and metastasis by a combination of Escherichia coli-mediated cytolytic therapy and radiotherapy. Mol. Ther. 18, 635–642 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown J. M., Wilson W. R., Exploiting tumour hypoxia in cancer treatment. Nat. Rev. Cancer 4, 437–447 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Zheng J. H., et al. , Two-step enhanced cancer immunotherapy with engineered Salmonella typhimurium secreting heterologous flagellin. Sci. Transl. Med. 9, eaak9537 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Yi X., et al. , Bacteria-triggered tumor-specific thrombosis to enable potent photothermal immunotherapy of cancer. Sci. Adv. 6, eaba3546 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L. V., Yao J., A practical guide to photoacoustic tomography in the life sciences. Nat. Methods 13, 627–638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wray P., Lin L., Hu P., Wang L. V., Photoacoustic computed tomography of human extremities. J. Biomed. Opt. 24, 1–8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Na S., Wang L. V., Photoacoustic computed tomography for functional human brain imaging [Invited]. Biomed. Opt. Express 12, 4056–4083 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang Z., et al. , Near-infrared dyes, nanomaterials and proteins. Chin. Chem. Lett. 30, 1856–1882 (2019). [Google Scholar]
- 42.Kang S.-R., et al. , Imaging of tumor colonization by Escherichia coli using 18F-FDS PET. Theranostics 10, 4958–4966 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forbes N. S., Engineering the perfect (bacterial) cancer therapy. Nat. Rev. Cancer 10, 785–794 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bettegowda C., et al. , Imaging bacterial infections with radiolabeled 1-(2′-deoxy-2′-fluoro-β-D-arabinofuranosyl)-5-iodouracil. Proc. Natl. Acad. Sci. U.S.A. 102, 1145–1150 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diaz L. A. Jr., et al. , Imaging of musculoskeletal bacterial infections by [124I]FIAU-PET/CT. PLoS One 2, e1007 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Low K. B., et al. , Lipid A mutant Salmonella with suppressed virulence and TNFα induction retain tumor-targeting in vivo. Nat. Biotechnol. 17, 37–41 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Dang L. H., Bettegowda C., Huso D. L., Kinzler K. W., Vogelstein B., Combination bacteriolytic therapy for the treatment of experimental tumors. Proc. Natl. Acad. Sci. U.S.A. 98, 15155–15160 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freitag N. E., Rong L., Portnoy D. A., Regulation of the prfA transcriptional activator of Listeria monocytogenes: Multiple promoter elements contribute to intracellular growth and cell-to-cell spread. Infect. Immun. 61, 2537–2544 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Praveschotinunt P., et al. , Engineered E. coli Nissle 1917 for the delivery of matrix-tethered therapeutic domains to the gut. Nat. Commun. 10, 5580 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sonnenborn U., Escherichia coli strain Nissle 1917-from bench to bedside and back: History of a special Escherichia coli strain with probiotic properties. FEMS Microbiol. Lett. 363, fnw212 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Ou B., et al. , Genetic engineering of probiotic Escherichia coli Nissle 1917 for clinical application. Appl. Microbiol. Biotechnol. 100, 8693–8699 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Massa P. E., Paniccia A., Monegal A., de Marco A., Rescigno M., Salmonella engineered to express CD20-targeting antibodies and a drug-converting enzyme can eradicate human lymphomas. Blood 122, 705–714 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Piñero-Lambea C., et al. , Programming controlled adhesion of E. coli to target surfaces, cells, and tumors with synthetic adhesins. ACS Synth. Biol. 4, 463–473 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bereta M., et al. , Improving tumor targeting and therapeutic potential of Salmonella VNP20009 by displaying cell surface CEA-specific antibodies. Vaccine 25, 4183–4192 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toso J. F., et al. , Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J. Clin. Oncol. 20, 142–152 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.