Abstract
Viral infections are a common cause of morbidity worldwide. The emergence of Coronavirus Disease 2019 (COVID-19) has led to more attention to viral infections and finding novel therapeutics. The CRISPR-Cas9 system has been recently proposed as a potential therapeutic tool for the treatment of viral diseases. Here, we review the research progress in the use of CRISPR-Cas technology for treating viral infections, as well as the strategies for improving the delivery of this gene-editing tool in vivo. Key challenges that hinder the widespread clinical application of CRISPR-Cas9 technology are also discussed, and several possible directions for future research are proposed.
Keywords: CRISPR-Cas, Gene editing, Infection, Treatment, Virus
1. Background
Viral infections contribute to significant morbidity and mortality worldwide, and can potentially escalate into an epidemic or a pandemic that has serious socioeconomic implications [1], [2]. The evolution of drug-resistant viral strains, along with the continual emergence of new pathogenic viruses, has created a constant need for the design of novel antiviral therapeutic approaches [3], [4], [5]. Nevertheless, the development of therapeutic compounds based on drugs is challenging, as viruses often co-opt the host cellular machinery for their replication and transmission; thus, therapeutic agents that inhibit the replication of the virus are likely to interfere with the normal biological functions of the host [6]. Besides, the vastly different molecular characteristics among viruses have created significant obstacles for developing broad-spectrum antiviral therapeutic agents [7], [8]. For these reasons, the focus of more recent antiviral therapy development has shifted towards alternative strategies, such as with the use of RNA interference (RNAi)-based approach [9], [10]. However, these approaches have their inherent limitations, such as the inability to eradicate the cccDNA (covalently closed circular DNA) of hepatitis B virus (HBV); hence, cessation of RNAi treatment can cause HBV reactivation [11], [12].
In addition to structural proteins, the presence of nucleic acid is critical to assembling an infectious virion. Accordingly, the virus life cycle completion within the host cell depends on the efficient replication of the viral genome. Hence, it can be argued that one of the best antiviral methods may be the removal of viral genetic elements. However, this antiviral strategy of targeting the viral genome can face a major bottleneck involving the absence of a virus-specific gene degradation method. Some preclinical studies have recently examined the therapeutic activity of sequence-specific endonucleases to directly manipulate the viral genome, including clustered regulatory interspaced short palindromic repeat (CRISPR)-associated nucleases (Cas), transcription activator-like effectors nucleases (TALENS) and zinc finger nucleases (ZFNs) [13], [14]. In this regard, great attention has been attracted towards the CRISPR/Cas system because of simplicity, specificity and relative versatility [15], [16], [17], [18]. Six variants have been found for the CRISPR/Cas system [19], the most important of which is type II CRISPR/Cas9 system separated from Streptococcus pyogenes (Sp). The CRISPR/Cas9 system has been introduced as a promising approach to the knock-out of target genes and the knock-in of genetic material at specific sites in a variety of animal models like nonhuman primates, mice, Drosophila and zebrafish [20], [21]. This technique could enable the application of in vivo measures to treat some cancers and genetic defects in human beings. In recent decades, great attention has been paid towards the CRISPR/Cas9 rewiring strategies regarding DNA locus imaging, epigenetic modulation, targeting of human pathogens, gene activation, target gene inhibition and RNA tracking [22].
The recently emerged CRISPR-Cas have heralded a new era in the treatment of viral disorders. The nucleases were originally described as a mechanism of defense in archaea and bacteria, and have since been exploited as a genome-editing tool for precise manipulation of nucleic acid sequences in a variety of organisms [23]. The ability to modify the sequences of not only DNA but also RNA allows the nucleases to edit both the viral and the host genomes [24]. The nucleases are also capable of modifying multiple genes simultaneously at high-efficiency rates [25]. Moreover, unlike other types of sequence-specific endonucleases including the TALENs and zinc-finger nucleases (ZFNs), the use of the CRISPR-Cas system eliminates the need for engineering, selecting and validating the target site-specific protein pairs [26]. Rather, it uses small RNAs, which are easier to design and cheaper to produce, for its sequence-specific cleavage activity. These attributes, coupled with its high specificity and versatility, make the CRISPR-Cas system an ideal therapeutic option for viral diseases. Here, new advances in the use of CRISPR-Cas for treating viral disorders, with a special focus on human immunodeficiency virus (HIV), hepatitis B virus (HBV), herpesviruses, human papillomavirus (HPV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), were reviewed. We further discussed the use of vector systems to enhance the delivery of CRISPR-Cas. Finally, limitations of using CRISPR-Cas for antiviral therapy were illuminated, and possible directions for future research were proposed.
2. CRISPR-Cas systems
CRISPR-Cas is a prokaryotic adaptive immune system that plays a role in inactivating exogenous genetic elements. The CRISPR-Cas systems can be divided into two classes based on the effector protein used. Whereas Class 1 systems utilize multiple Cas effectors for their nucleic acid cleavage activities, Class 2 systems use a single multidomain effector, notably Cas9, Cas12 and Cas13, for the same purpose [27]. The type of effector protein used dictates a number of CRISPR-Cas features, including the target molecule, trans-cleavage ability, recognition site sequence, etc. As an example, Cas9-mediated cleavage targets double-stranded DNA (dsDNA), whereas Cas13-mediated cleavage targets single-stranded RNA molecules.
Among the numerous different CRISPR-Cas systems, CRISPR-Cas9 is undoubtedly the most well-characterized system. The CRISPR/Cas9 complex is composed of two key components: the Cas9 enzyme, which performs the endonuclease function; and a guide RNA (gRNA), which directs Cas9 to the target site. The Cas9 enzyme contains a HNH nuclease domain and a RuvC-like nuclease domain, each of which is in charge of cleaving one of the target DNA strands. The gRNA, on the other hand, comprises a short (~20 nt) CRISPR RNA (crRNA), which is the crucial element that determines the target specificity of CRISPR-Cas9, that is embedded within a longer trans-activating crRNA (tracrRNA). The CRISPR-Cas9-based genome editing technologies take advantage of the targeting specificity of the crRNA, such that the crRNA sequence is synthetically modified to mediate the cleavage of the desired sequence(s) by Cas9. The CRISPR-Cas9 system can cleave any dsDNA sequence in the genome, provided that it is located immediately adjacent to a protospacer adjacent motif (PAM), which serves as the site for Cas recognition [28]. The canonical sequence of the PAM is 5’-NGG-3’. Nonetheless, Cas also cleaves dsDNA sequence located next to other non-canonical PAMs, such as the 5’-NAG-3’ and 5’-NGA-3’ sequences, albeit to a much lower efficiency [28], [29].
Cleavage by Cas9 constitutes the first step in gene editing. The double-strand breaks (DSBs) induced by Cas9 need to be repaired by cellular DNA repair pathways, during which the gene modification is achieved. There are two major DNA repair pathways, namely the non-homologous end joining (NHEJ) and the homology directed repair (HDR) pathways. The latter pathway is preferred for precise gene editing, as the NHEJ repair pathway often results in the formation of indels and causes frameshifts to the genes of interest [30]. However, the choice of the repair pathway is dependent on a number of cellular regulatory factors, notably 53BP1 and BRCA1 [30]. High levels of 53BP1 have been associated with a preferential NHEJ repair pathway, whereas high levels of BRCA1 may promote HDR [30], [31]. A number of small-molecule drugs targeting the 53BP1 and BRCA1 pathways, including DNA ligase IV inhibitors, protein kinase catalytic subunit inhibitors, Rad51 agonists, Golgi apparatus inhibitors and β3-adrenergic receptor agonists, have been used to encourage HDR repair over NHEJ for Cas9-induced DSBs to facilitate precise gene editing [32], [33], [34], [35], [36].
As a precise genome modification tool, CRISPR-Cas9 holds great promise in molecular medicine. It has been widely used for the diagnosis of genetic and infectious diseases due to its ability to recognize and cleave specific sequences with high sensitivity, at a rapid rate. Several CRISPR-based diagnostics are commercially available for the clinical diagnosis of several viruses [37]. For example, DETECTR [38], SHERLOCK [39], FELUDA [40] and AIOD-CRISPR [41] kits have recently been developed for the detection of SARS-Cov-2 – of which the first two have received emergency use authorization from the Food and Drug Administration (FDA) [42]. Apart from diagnosis, CRISPR-Cas9 also finds its use in the treatment of diseases. In 2016, the CRISPR-Cas9 system was used for the first time in the clinic, where T cells with CRISPR-edited PD-1 were used to treat a lung cancer patient [43]. It has since been used in the treatment of pulmonary and gastrointestinal diseases, hematologic diseases, viral diseases, vector diseases, autoimmune and inflammatory diseases, and other cancers, where encouraging results have been obtained [44].
3. CRISPR-Cas for treatment of viral infections
Viruses are obligate parasites that infect and enter host prokaryotic or eukaryotic cells through their surface receptors [45], [46]. Their genome is composed of either ribonucleic acid (RNA) or deoxyribonucleic acid (DNA). A complete virus particle with all its structures is called a virion, which is a fully functional unit capable of infecting and delivering the viral DNA/RNA to the host cells. They can be good candidates to be targeted by CRISPR-based therapeutics due to their intracellular localization and cellular protein requirement for replication and survival [47], [48].
Viral infections may be classified into lytic, latent, or chronic. During lytic infection, the virus can actively replicate and produce offspring virions. Once the progeny viral genomes are packaged within the protein coats or capsids, they are released via cell lysis, a process which kills the host cell. Certain viruses may also induce latent infection, in which the virus enters a dormant condition and may stay inactive for years [49], [50]. Numerous human viruses, such as the herpes simplex virus 1 (HSV-1), Epstein-Barr virus (EBV), cytomegalovirus (CMV), human herpesvirus 6 (HHV-6) and HBV, may induce latent infections [51]. Finally, chronic infection is characterized by persistent low-level viral replication, which results in organ damage over time. Several major human viruses, including HBV and human immunodeficiency virus type 1 (HIV-1), exhibit characteristics of chronic infection [52], [53].
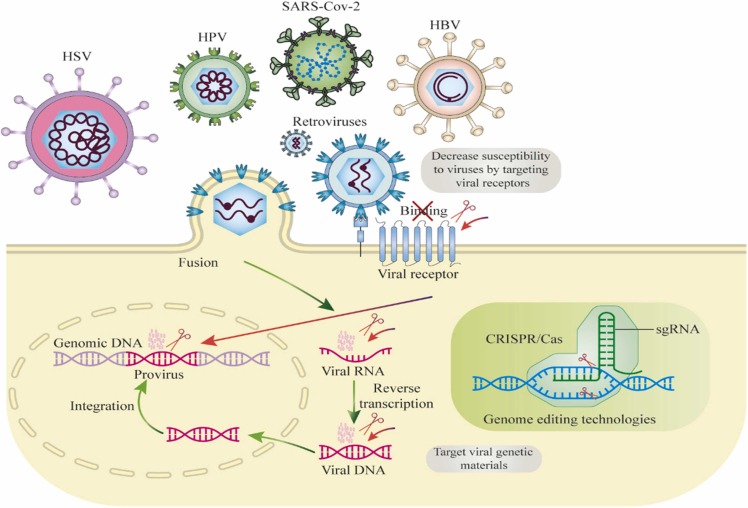
Eradication of virus in latent and chronic infections is difficult, as the viral genome is retained inside the host cell, either in the form of free-floating viral minichromosomes or incorporated into the host genome. Fortunately, CRISPR-based technologies allow specific targeting of the viral genome within the host cell, which abrogates the transcription and replication of the virus. For this reason, the CRISPR-Cas9 system has been widely used to target numerous types of human DNA viruses, including HSV-1, EBV, CMV, human papillomavirus (HPV), and JC polyomavirus [54], [55], [56], [57]. As seen in Table 1, the herpesviruses of HSV-1, HCMV and EBV were negatively manipulated by CRISPR/Cas9 in a study of Van Diemen and colleagues [43]. According to their efforts, the gRNAs were targeted to substantial genes of viruses (like EBNA 1and BARTs) to achieve efficient abrogation of HSV-1 and HCMV replication. Formation of infectious viral particles was completely abolished again by targeting HSV-1 concurrently with several gRNAs [43]. EBV was also completely cleared from latently infected tumor cells in human. Inefficient HCMV targeting via single gRNAs resulted in the choice of viral escape mutants following long-term replication [43]. Accordingly, co-targeting of various regions of the viral genome can be considered important to prevent the CRISPR/Cas9 digestion-resistant viruses with escape mutation. In the majority of cases, the DSBs induced by Cas9 are repaired via the NHEJ pathway, during which random nucleotide substitution, deletion and insertion disrupt the critical protein-coding regions and/or cis-regulatory elements of the viral genome, thus disabling the virus [58]. Apart from that, CRISPR-Cas complexes can be used to target RNA viruses. At least two RNA-targeting CRISPR-Cas systems have been developed for the treatment of viral infection. One such system uses the Cas9 protein, which, as mentioned above, does not target RNA by default [59]. However, when a distinct DNA oligomer carrying the PAM sequence is supplied to the system, the PAM oligomer can hybridize with the target RNA sequence to produce a double-stranded target required for Cas9-mediated cleavage [59]. The second CRISPR-Cas system utilizes Cas13 endonuclease, which has an affinity towards RNA [60], [61]. The CRISPR-Cas13 system has been suggested as a potential treatment strategy for RNA viruses [62]. Fig. 1 shows the mechanisms for viral genome editing to prevent or impede the virus-related infections.
Table 1.
Non-exhaustive list of the use of CRISPR-Cas systems for treating viral infections.
| Virus | CRISPR/Cas Nuclease used | Targets | Main findings | Ref |
|---|---|---|---|---|
| HIV | Cas9 | CCR5, LTR | Viral gene expression was reduced.Complete excision of the integrated proviral DNA. Complete virus inactivation. | [63], [64], [65], [66], [67], [68] |
| saCas9 | LTR, Gag | Viral gene expression was reduced and HIV-1 DNA was eradicated. | [17] | |
| Cas13a | LTR | Viral RNA in the capsid was destroyed. | [69] | |
| HBV | Cas9 | Multiple, HBsAg, S gene, X gene, cccDNA | HBV cccDNA and integrated HBV DNA were disrupted.cccDNA reservoirs were destroyed.HBV replication was disrupted. | [70], [71], [72], [73], [74], [75], [76], [77], [78] |
| EBV | Cas9 | BART, LMP1, LMP2A, Multiple | EBV infection and replication were inhibited.Latent virus infection was cleared.BART promoter was deleted. | [79], [80], [81] |
| HCMV | Cas9 | Multiple | Viral replication was impaired.Latent virus infection was cleared. | [80] |
| HSV-1 | Cas9 | Multiple, ICP0 | HSV-1 replication and infectivity were impaired.Latent virus infection was cleared. | [80], [82], [83] |
| HPV-6 HPV-11 | Cas9 | E7 | HPV-6 and − 11 E7 protein degradation. | [84] |
| HPV-16 | Cas9 | E7, E6, PD-1 | Deactivated HPV16 E6 and E7 and lead to inhibited cervical cancer cell growth.Downregulated PD-1 expression. | [85], [86], [87], [88], [89] |
| SaCs9 | E6, E7 | Suppressed HPV-positive anal cancer cell growth. | [90] | |
| HPV-18 | Cas9 | E6, E7 | Deactivated HPV-18 E7 E6 protein and suppressed cancer cell development. | [91], [92], [93] |
| SpCas9 | E7 | Downregulated HPV-18 E7 and suppressed carcinogenic potential of HPV. | [94] | |
| HPV-16 and − 18 | wild-type Cas9 and the highly specific FokI-dCas9 | E6 and E7 | Partial virus inactivation and triggered apoptosis of cancer cells. | [95] |
| HPV pseudo-type virus | HCas9 | E6 | HPV E6 protein degradation. | [96] |
| SARS-CoV-2 | Cas13 | Multiple | Inhibiting Coronavirus | [97] |
Fig. 1.
Mechanisms for genome editing to prevent or suppress the viral infections; the antiviral method mediated by genome editing is a facile design capable of rapidly setting for the control of infections caused by various viruses, thereby deleting latent infections and establishing related new resistance. The CRISPR-Cas system targets sequence-specific targeting of the viral genome (DNA or RNA) and/or host viral receptor (ligand–receptor interactions) as a way of protecting against viruses.
Current progress in the use of CRISPR-Cas systems for treating selected viral infections is summarized in Table 1 and is discussed in the following subsections.
3.1. Human immunodeficiency virus (HIV)
HIV is an RNA virus belonging to the Lentivirus genus of the Retroviridae family, which is evolutionarily derived from the same viruses found in chimpanzees [98]. The HIV genome is composed of two copies of single-stranded RNA molecules, which encode proteins required for its life cycle. The virus is known to attack the CD4+ T-cells and result in acquired immunodeficiency syndrome (AIDS), which exposes the infected person to secondary lethal infections and increases their risk of developing malignancies. HIV infection is a major global health concern with an increasing trend of incidence all over the world. There are two main types of HIV, namely HIV-1 and HIV-2, of which the former accounts for almost all cases of AIDS worldwide. Globally, approximately 40 million individuals are currently living with chronic HIV-1 [99].
HIV infection is treated with highly active antiretroviral therapy (HAART), which significantly decreases HIV-1 replication in cells and suppresses plasma viral load to undetectable levels (<40 RNA copies/ml). Despite this, the treatment cannot completely eradicate the virus from the patient, as the viral DNA (provirus) is integrated into the genome of the host cells and continues to be replicated. This necessitates life-long drug administration to suppress the viral load [100]. Another key challenge to the effective treatment of HIV-1 infection is the emergence of antiretroviral drug resistance due to the accumulation of beneficial mutations that confer a selective advantage to the virus [101].
The development of genome editing tools including ZFNs, TALENs and CRISPR-Cas systems offers great promise for curing HIV infection. The CRISPR-Cas9 systems can efficiently suppress active HIV infection by introducing indel mutations to the target genes [102]. Among the important targets of these genome-editing technologies include C-C chemokine receptor type 5 (CCR5) and C-X-C chemokine receptor type 4 (CXCR4), which are co-receptors needed for entry of HIV-1 into CD4+ T cells [100], [103]. Defects in these co-receptors can make cells, especially immune cells, resistant to HIV [104], [105]. Disruption of genes encoding these co-receptors has also been shown to have no significant adverse effect on cell viability [106]. Preliminary findings from numerous clinical trials have also demonstrated the safety and efficacy of modifying these genes in human subjects [107]. Although all three aforementioned genome-editing tools have been widely used to induce mutations to CCR5 and CXCR4 [108], [109], CRISPR-Cas9 has the advantage of being able to induce mutations in a more specific and precise manner, and at higher efficiency [63], [64]. Interestingly, the CCR5-editing efficiency was found to be further improved when chemical alterations are introduced to the gRNA [110].
Elimination of the integrated proviral DNA represents another promising avenue for anti-HIV therapy. It was shown for the first time in 2013 that CRISPR/Cas9-targeting HIV-1 long terminal repeat (LTR) promoter U3 region, a sequence that contributes to the regulation of the HIV-1 gene expression, could eradicate the integrated HIV-1 DNA [65]. Besides, using Staphylococcus aureus Cas9 instead of the original Streptococcus pyrogens Cas9 used in typical genome engineering experiments can lead to a precise excision of a 9.7 kb fragment from the integrated proviral DNA in several organs, including liver, lung, kidney, and circulating lymphocytes, in vivo [17]. Importantly, apart from eradicating latent HIV infection, targeting proviral DNA using CRISPR-Cas9 has been shown to prevent new HIV infection [66], [111].
Besides, targeting genes associated with HIV-1 production, virion assembly and budding, and viral regulatory elements represent promising areas of focus for anti-HIV CRISPR-Cas9 therapy [112]. Unfortunately, numerous studies have reported the emergence of mutations at the gRNA-target site in the HIV-1 genome [18], [113], [114]. This has led to the generation of CRISPR-Cas9-resistant HIV-1 strains. To address this problem, a combinatorial CRISPR-Cas9 gene targeting strategy has been suggested, where two or more gRNAs are used to target the conserved HIV-1 regions. Studies utilizing this combinatorial CRISPR-Cas9 gene targeting strategy have reported successful inhibition of HIV-1 replication and overcome the problem of viral escape ( Fig. 2) [67], [68].
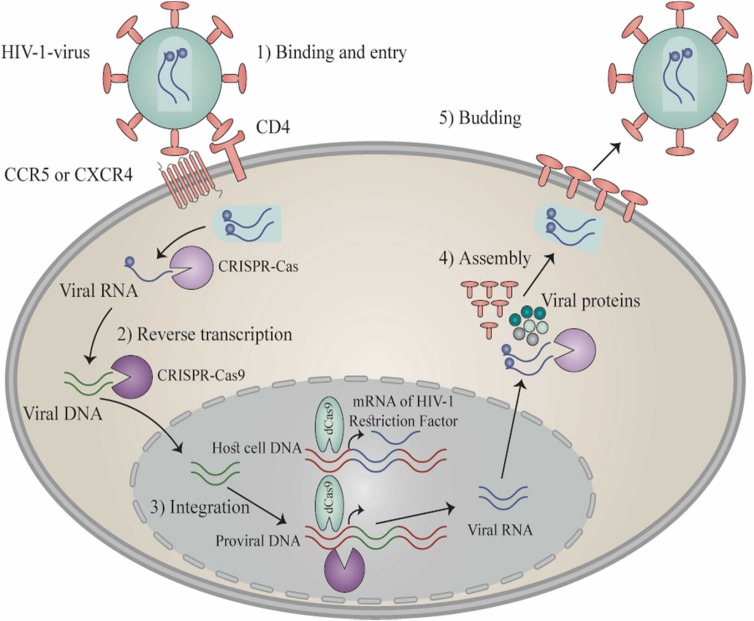
Fig. 2.
HIV-1 life cycle presents probable CRISPR-Cas9 targets. Five steps make up the life cycle of HIV-1 as follows: (a) Attachment and introduction: The penetration of HIV-1 into host cell occurs following the attachment of its gp120 to the surface receptor CD4 and subsequently to CXCR4 or CCR5 co-receptor, thereby leading to the fusion of cell membrane and HIV-1 and then the introduction of HIV-1 into host cells and the liberation of viral RNA. (b) Reverse transcription process: the viral RNA reverse transcription occurs in the host double-stranded DNA utilizing the reverse transcriptase. (c) Integration: After nuclear introduction of viral DNA, it is integrated into the genomic DNA of host cell exploiting the enzyme integrase. (d) Replication and assembly: After generation of viral RNA via the pro-viral DNA, the novel genomic RNA takes part for the synthesis of viral proteins that embrace the viral RNA to produce immature viral particles near the cell surface. (e) Budding: The novel particles are liberated outside to form viral protease capable of cleaving long protein chain leading to form viral maturation. Engineering processes convert the Cas9 to inactive Cas9 (dCas9) by inducing mutations in the two RuvC and HNH domains of nuclease. The subsequent dCas9 fusion using different agents into the DNA-binding domain specific to site can trigger latent virus through sgRNAs that target the LTR region of HIV-1.
Apart from CRISPR-Cas9, the use of CRISPR-Cas13a to target HIV-1 RNA is an alternative strategy that can be used to inhibit HIV infection. CRISPR-Cas13a targeting of HIV-1 RNA can reduce the expression of viral genes, as demonstrated by Yin et al. who used Leptotrichia buccalis (Lbu) Cas13a/crRNA for this purpose. It was found that CRISPR-Cas13a targeting of HIV-1 RNA resulted in an approximately 50% decrease in the expression of HIV-1 genes in HEK293T cells compared to the control group [69]. Promisingly, CRISPR-Cas13a targeted not only the newly synthesized viral RNA but also the viral RNA that entered the cells within the viral capsid, which suggests that gene editing using CRISPR-Cas13a provides a potential alternative for the control of HIV-1 infection [69].
The question is, what are the consequences of designing gRNAs that target non-conserved regions of the HIV-1 genome? In answer to this question, observations reported the presence of CD4 + T cells that are able to continuously express gRNA and Cas9. If non-conserved regions are targeted, there will be a significant barrier to infection in transient assays; however, all targeted infections reach high production levels of HIV-1 following a variable time. An escape was shown by targeting conserved regions in the genome of HIV-1 following a longer time. Based on the sequencing of escaped virus genome, multiple mutations through error-prone NHEJ repair route eliminated the region of PAM and the binding site of gRNA in HIV-1 genome [115]. There are several ways to overcome HIV-1 escape, such as concurrent targeting through a development of potent gRNAs to guide Cas9 on conserved regions [116], the use of Cas9 variants detecting various PAM formats [117], the application of CRISPR-like enzyme including Cpf1 that makes cut in the distal binding site [31], and NHEJ deletion via chemical agents like SCR7 [118], [119].
3.2. HTLV-1
The oncogenic human retrovirus of HTLV-1, Human T-cell leukemia virus type 1, causes CD4 + T-cell transformation, thereby developing numerous disorders like HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP) as neurodegenerative condition and adult T-cell leukemia/lymphoma (ATL) [120]. Although there are scientific progressions, some questions exist about diseases caused by HTLV-1, progression of illness and efficient therapies. Although some helper genes regarding the life cycle of virus are encoded by HTLV-1, Hbz and Tax are two important viral proteins implicated in HAM/TSP and ATL pathophysiology. The cellular immortalization and de novo infection are managed by Tax [121], but infected cell survival and proliferation by Hbz [122]. The comprehensive studies reported that these genes can be targeted to inhibit the survival and growth of cells infected with HTLV-1. The regions of long terminal repeat (LTR) in virus have role to combine host chromatin and viral genome, and also are promoters to express all genes of the virus. Hbz, Tax and LTR are the target regions are potentially effective in the treatment of new cases infected with HTLV-1, of asymptomatic carriers of the virus and of the subjects suffering from HAM/TSP and ATL. Two elements of the virus can be simultaneously impaired by designing gRNAs based on the over-lapping reading frames between 3’LTR and Tax as well as 3’LTR and Hbz [123]. Currently, the HIV-1 is the only for CRISPR editing in the retroviral proviruses [124]. Conversely, further concentrated gRNA targeting is presented by the HTLV-1 due to highly conserved genome of the virus with significant homogeneity in sequence within both various isolates of HTLV and the same host.
In a study by Baker et al. [125], the certain gRNAs were developed to target highly conserved regions of HTLV-1 long LTR and tax within 23 HTLV-1 genomes collected from various parts of the world. Based on their findings, the certain in vitro cleavage of HTLV-1 LTRs and tax was found in the gRNA/Cas9 ribonucleoproteins (RNPs). After delivering the RNPs via electroporation to two different cell lines infected with HTLV-1-, they exhibited effective HTLV-1 genome editing based on T7E1 mismatch detection test. In this work, the CIRCLE-SEQ was utilized to determine the specificity of HTLV-1-specific gRNA/Cas9 RNPs to detect probable off-target cleavage episodes. The HTLV-1 gRNAs were detected with the least off-target impact on the genomic DNA of human, which means promising tools to produce extremely specific CRISPR/Cas9 therapeutic agents versus HTLV-1. They could detect the pro-viral integration sites of HTLV-1 applying the data of CIRCLE-SEQ sequencing, which means a new approach to map the episodes of viral integration to genomic DNA [125].
3.3. Hepatitis B virus (HBV)
HBV is a non-cytopathic DNA virus belonging to the Hepadnaviridae family [126]. The HBV genome is circular in shape, partially double-stranded, and contains four major genes, namely S, C, X and P that respectively encodes the surface antigen, core protein, X protein and a DNA polymerase that possesses reverse transcriptase activity [127]. During infection, the partially double-stranded DNA is converted into an episomal cccDNA form, which serves as the template for generating its mRNAs and pre-genomic RNAs [128]. Nine major genotypes of HBV (A-I), along with a putative genotype (J), have been identified so far. These HBV genotypes show > 8% genome sequence divergence and have distinct epidemiology [129], [130]. Globally, more than 250 million people are living with chronic HBV, and roughly 800,000–900,000 people are dying from HBV-related liver disease each year [131]. Current antiviral therapies for HBV, such as the use of nucleoside analogue-based drugs [132], act by targeting different stages of the virus life cycle, disrupting the production of progeny virions, interfering with the critical molecular players in the viral pathogenesis, and also augmenting the host immune system to facilitate inhibition of the virus. These therapies are effective in suppressing the viral replication and diminishing the circulating surface antigen (HBsAg) of the virus. However, they are unable to fully eradicate the cccDNA and the integrated HBV DNA, which explains the common relapse of the disease after the cessation of the treatment [133], [134], [135]. Thus, contemporary therapeutic strategies for chronic hepatitis B have focused on targeting and eventually eradicating the HBV cccDNA and the integrated HBV genome.
The HBV cccDNA is double-stranded and contains evolutionarily conserved sequences. Therefore, it serves as a suitable target for CRISPR-Cas9 cleavage [77]. The possible use of CRISPR-Cas9 in targeting the HBV cccDNA and some promising results have been reported in some in vitro and in vivo investigations [70], [71], [72], [73], [74], [75], [76], [77], [78]. In the majority of studies, CRISPR-Cas9 was found to disrupt the HBV cccDNA and decrease the viral gene expression within the first week of transfection. The first use of CRISPR-Cas9 to target and destroy the HBV cccDNA was reported by Seeger and Sohn in 2014 [78]. CRISPR-Cas9 was revealed to induce on-target mutations, including single nucleotide indels and also large deletions with up to 2.3 kb length, in the cccDNA. This genetic modification successfully inactivated the episomal DNA in HBV-permissive HepG2/NTCP cells, which, in turn, inhibited the expression of HBV core antigen (HBcAg) and other viral proteins by up to tenfold [78]. In the same year, Lin et al. [70] demonstrated that CRISPR-Cas9 was capable of disrupting the intrahepatic HBV genome both in vitro and in vivo, which resulted in the decreased level of serum HBsAg in mouse models. This finding was corroborated by a number of subsequent studies, which showed that CRISPR-Cas9-mediated degradation of cccDNA suppressed HBV replication and disrupted its genome both in vitro and in vivo [71], [72], [73], [74]. Apart from cccDNA, several studies have demonstrated that CRISPR-Cas9 can disrupt the full-length integrated DNA of HBV [75], [76]. For example, Li et al. [76] reported the complete elimination of HBV infection in HepG2. A64 cells after the successful eradication of both cccDNA and the full-length integrated DNA using CRISPR-Cas9.
Despite the promising early results, some studies have reported a low efficiency of CRISPR-Cas9 targeting of the cccDNA [136]. The low efficiency could be attributed to epigenetic modifications, in particular DNA methylation, that reduced the accessibility of cccDNA for enzymatic excision [136]. Nonetheless, the efficiency could be improved by using two or more gRNAs to direct Cas9 to the target site. Besides, as mentioned above, a more precise genome editing can be achieved by suppressing the NHEJ repair pathway to allow the HDR repair mechanism to take place. With this in mind, Kostyushev et al. [77] suppressed the NHEJ pathway using the NU7026 inhibitor and observed an increased on-target deletion and inhibition of cccDNA following cleavage by Cas9.
In an in vitro study by Scott et al. [137]., multiple short gRNAs and SaCas9 were incorporated in ssAAVs, with efficient delivery and ability for development of HBV DNA mutations and inactivation of cccDNA [137]. In a recent report on a hydrodynamic in vivo model of HBV, the AAV vectors were successfully employed as a delivery vehicle for functional anti-viral SaCas9 and CRISPRs, resulting in decreased levels of HBV protein. Moreover, there was no important viral abrogation in an in vivo model of persistent HBV [138].
As Cas9-mediated DSBs expose the host genome to the risk of genomic damage, one study attempted the use of a modified version of the CRISPR-Cas system that did not involve DNA cleavage to achieve cccDNA editing [139]. This modified CRISPR-Cas editing was found to suppress HBV gene expression and achieve the permanent inactivation of the HBV genome, indicating its usefulness in treating HBV infection [139]. Finally, CRISPR-Cas9-mediated knockout of HBsAg in hepatocellular carcinoma cell lines was found to suppress the tumor cell proliferation in vitro and also attenuate the tumorigenicity of cancer cells in vivo [140]. Overall, these in vivo studies show the potential of anti-HBV CRISPR/Cas9 to disrupt HBV cccDNA in vivo mouse models. Nevertheless, additional studies are needed to assess whether CRISPR/Cas9 holds the therapeutic potential to limit productive HBV infections in vivo and is capable of eradicating multiple cccDNA copies present in infected hepatocytes.
3.4. Herpesviruses
Herpesviruses are a group of heterogeneous viruses belonging to the Herpesviridae family within the order Herpesvirales. To date, more than 100 species of herpesviruses have been identified, although only nine of the species are known to infect humans [141]. These include herpes simplex virus (HSV) types 1 and 2, which respectively cause oral cold sores and genital herpes; Epstein-Barr virus (EBV) or human herpesvirus-4 (HHV-4), which causes infectious mononucleosis and Burkitt lymphoma; varicella-zoster virus (VZV), which causes chickenpox in younger individuals and shingles in adults; Kaposi’s sarcoma-associated herpesvirus (KSHV) or HHV-8, which causes Kaposi’s sarcoma in AIDS patients; human cytomegalovirus (HCMV), which usually causes mild harmless symptoms; and HHV types 6 A, 6B and 7, which also cause mild symptoms [142]. Herpesviruses have a structure composed of the core particle (within which the genetic material is enclosed), the icosapentahedral capsid, the protein coat, and the bilayer envelope [143]. They possess a double-stranded linear DNA genome, which encodes for more than 20 structural proteins [144].
All herpesviruses have a latent period in their life cycle, during which the virus is “hidden”, no active virion is produced, and genes normally expressed during the lytic cycle are suppressed [141]. The ability to establish a latent infection represents a key challenge in human herpesvirus therapies. A number of nucleoside analogue-based antiviral drugs have been developed for the treatment of active herpesvirus infection by suppressing the virus replication, but they are not able to eradicate the latent infection. Fortunately, the development of the CRISPR-Cas9 technology provides a glimpse of hope for the eradication of latent herpesviruses, as the technology can disrupt the viral genome directly. The use of CRISPR-Cas9 for the treatment of selected herpesviruses is described in the subsections below.
3.4.1. Epstein-Barr virus (EBV)
In one of the earliest studies utilizing CRISPR-Cas9 for anti-EBV therapy, targeted deletion of the entire BART gene promoter was performed in epithelial cell lines latently infected with the virus [79]. The deletion was reported to be efficient and resulted in the loss of the gene expression, with no off-target modification. Another study demonstrated that targeting of the essential elements of EBV, including the nuclear antigen 1 (EBNA1) and origin of replication (OriP), resulted in the loss of the viral gene expression in Akata-Bx1 cells latently infected with the virus as well as the eradication of the virus from these cells [80]. Besides, CRISPR-Cas9-mediated knockout of the EBV’s latent membrane protein 1 (LMP1) gene in nasopharyngeal carcinoma (NPC) cell lines was found to suppress EBV replication and inhibit tumor cell growth through blockade of signaling activation [81]. These findings suggest that destruction of the EBV genome can suppress tumorigenicity in the virus-associated malignancies and may be considered a potential therapeutic strategy for the treatment of the diseases. Nevertheless, it is worthy of mention that similar to the HBV, the EBV genome contains copies of episomal cccDNA [145]. Targeting the linear EBV genome without considering the cccDNA can result in the incomplete eradication of the virus [79]. Hence, future investigation of the feasibility of using CRISPR-Cas9 for treating EBV infection should take into account the cccDNA of the virus.
3.4.2. Herpes simplex virus (HSV)
HSVs are highly contagious DNA viruses that cause oral and genital herpetic sores. There are two types of HSVs, namely HSV-1 and HSV-2. As in all herpesviruses, the HSV-1 is also able to develop a lytic infection because of exerting progeny and developing latent infection, which can persist throughout the host's lifetime [146]. HSV-1 is a ubiquitous virus living with over 50% of the US people, which can develop cold sores, keratitis and encephalitis [147], [148]. HSV-1 is a causative agent for mild or asymptomatic infections in immunocompetent or healthy people, as well as severe or life-threatening infections in infants with immature immunity or patients with immunodeficiency [146]. The HSV has highly prevalent, but no vaccine is available [149], [150]. However, acyclovir (ACV) is the first-line treatment for HSV-1 infection, with a history of about 50 years and subsequent analogs targeting the DNA polymerase of the virus. In a certain group of patients, such as those with immunodeficiency or those chronically treated with antiviral prophylaxis, there are repeated drug resistance episodes [151], [152], [153]. One of the earliest uses of CRISPR-Cas9 for HSV-1 therapy was reported by Bi et al., who replaced heterologous genes in the virus genome via HDR mechanisms and subsequently observed the inhibition of the viral replication [154]. Several other in vitro studies that followed found that CRISPR-Cas9 could modify the HSV-1 genome accurately and attenuate their virulence without causing off-target effects [82], [155], [156]. Besides, a study showed that gRNA-mediated targeting of HSV-1 ICP0 gene inhibited the virus replication in human oligodendroglioma cell line TC620 [83]. This observation was corroborated by the findings of other studies, which showed that CRISPR-Cas9 could target and destruct the HSV-1 genome at high efficacies [80], [157], [158]. Besides, similar results have been reported in HSV-2, where CRISPR-Cas9-mediated deletion of RL1 and LAT decreased neurovirulence, viral replication and plaque formation in Vero cells and also augmented immunity in virus-infected mice [159]. These observations suggest that CRISPR-Cas9 has the potential to be used clinically for HSV treatment in the future.
In a study by David Knipe et al., the CRISPR/Cas9 system was evaluated versus HSV-1 to detect the tricks of Cas9/gRNA to cleave the target lytic and latent viral genomes [160]. According to previous observations, the CRISPR/Cas9 editing system needs accessibility to target sites in the chromatin [161], [162]. Moreover, differential pathways of host cell are able to regulate chromatin assembly on the DNA of virus in epithelial cells (permissive) against nerves (non-permissive), helping the virus to develop the lytic versus latent infection [161]. In the latency, the chromatin of viral DNA is more compacted than replicating viral DNA, which results in less accessibility of Cas9/gRNA to the target sites of DNA [161], [162]. Accordingly, four main genes, including UL30, UL29, ICP4 and UL54, of virus are targeted by a complex of gRNAs [160]. Under an in vitro condition, a cleavage test based on SaCas9 was used to screen these gRNAs for detecting those capable of targeting lytic and quiescent genomes of HSV-1 in human cells. Staphylococcus aureus-derived SaCas9 is small and can be encoded by vector delivery systems based on AAV [163]. Numerous gRNAs constructed in the HSV-1-infected human foreskin fibroblasts could effectively decrease the replication of HSV-1 [160], in line with prior reports [164], [165] regarding the inhibition of lytic infection of HSV-1 by Cas9/gRNA system. Knipe et al. investigated the impact of generated gRNAs on the latent genome of virus using quiescent infection system of HSV-1 with replication-defective HSV-1 d109 virus. Based on their findings, heterochromatin loading in viral DNA [166] was the same as latent infection of mice [167], [168]. According to this model of quiescence infection, the reactivation of quiescent d109 genomes was decreased by anti- HSV-1 gRNAs, with two various gRNAs showing the additive impact [160]. Moreover, it was found that a significant level of indel mutations was triggered in the CRISPR/Cas9 in quiescent genomes of HSV-1. Therefore, the gRNAs targeting quiescent genomes of HSV-1 were detected and the indel mutations caused by SaCas9/gRNA resulted in loss-of-function mutations in related proteins [160]. Both replicating and non-replicating genomes were edited by CRISPR/Cas9, although it edited more effectively replicating genomes.
In a recent study by Yin et al. [169], the lentiviral particles carrying mRNA were applied to directly target the genomes of HSV-1, thereby concurrently delivering the mRNA of SpCas9 and the guide RNAs targeting viral gene (HELP or HSV-1-erasing lentiviral particle). In three various models of infection, the HELP could suppress the replication of HSV-1 and the incidence of herpetic stromal keratitis (HSK). The HELP could also remove the viral reservoir through retrograde transport to trigeminal ganglia from corneas. Further, the HELP suppressed viral replication in corneas derived from human with no off-target influences [169], highlighting the promising clinical application of HELP to manage refractory HSK. Together, these studies provide hope for potential future treatment of (latent) herpesvirus infections via anti-viral CRISPR/Cas9. Additional research, predominantly geared to in vivo studies, is needed.
3.4.3. Human cytomegalovirus (HCMV)
HCMV is a highly prevalent virus responsible for some congenital malformations like mental retardation and many disorders mainly among those suffering from some degree of immunodeficiency [170], [171]. Most HCMV-infected persons do not exhibit any sign or symptom throughout life. However, in a small number of patients, the clinical manifestation of HCMV infection can range from mild conditions such as rash and jaundice, to long-term health problems such as permanent neurological disorders. Currently available drugs for treating HCMV infection (e.g. ganciclovir, valganciclovir and foscarnet) are effective in attenuating the active infection and improving the clinical symptoms but are not useful for eliminating the latent infection [172]. The CRISPR-Cas9 system, as mentioned previously, enables the targeting of viruses in the latent phase; therefore, it serves as a potential therapeutic tool for eradicating latent HCMV. The use of CRISPR-Cas9 for manipulating the HCMV genome was reported by King and Munger, who explored the technology for the generation of recombinant HCMV [173]. It was demonstrated that CRISPR-Cas9 was capable of inducing precise mutagenesis at high efficiency [173]. Besides, Diemen et al. [80] disrupted seven essential genes of the HCMV genome using CRISPR-Cas9, and successfully impaired the replication of the virus. Nevertheless, virus escape was reported in the usage of single gRNAs [80]. Despite this, a more encouraging finding was reported in a more recent in vitro study, which demonstrated that targeting of the HCMV immediate-early genes not only decreased the viral gene expression and halted its virion production, but also inhibited viral replication and reactivation. Hence, genome editing using CRISPR-Cas9 may serve as a promising therapeutic strategy against the latent infection of the virus [174].
3.5. Human papillomavirus (HPV)
HPV is a part of Papillomaviridae family, which has DNA as its genomic material. It is the most prevalent sexually transmitted infection all over the world [175]. To date, more than 200 subtypes of HPV have been identified, which can be categorized into either low-risk or high-risk types. Low-risk HPVs cause genital warts, whereas high-risk HPVs cause lesions that can develop into various malignancies, notably cervical cancer, and multiple anogenital cancers (like penis, anus and vulva) and also head/neck cancers [176], [177], [178]. Among the many proteins encoded by the HPV genome, the E6 and E7 oncoproteins are the main driving force for malignant transformation. Hence, inactivation of the E6 and E7 oncogenes represents a possible strategy for inhibition of tumor cell proliferation. The CRISPR-Cas9 system allows precise disruption of these genes, so as to abolish their oncogenic functions [179]. Indeed, several in vitro and in vivo studies have investigated the applicability of CRISPR-Cas9 in the treatment of HPV-related cervical cancer. The findings indicate that CRISPR-Cas9-mediated knockdown of the HPV oncogenes not only induces apoptosis and suppresses tumorigenesis [85], [180], [181], but also improves radiosensitivity in cancer cells [181].
The HPV infections is progressed to invasive cancer via the regions of E6 and E7 integrated within the host cell with the aid of non-viral risk factors [182]. The high-risk HPV E6/E7 genes deactivated by CRISPR/Cas9 systems resulted in the restoration of pRb and p53 networks, thereby declining apoptosis and cell cycle arrest [87], [93], [95]. In a study by Jubair et al., the systemic Cas9/gRNAs in liposomes versus HPV16 E7 Caski cell and HeLa cell tumors removed the tumors through apoptosis in mice [95]. In line with this, lentiviral transduction of Cas9/gRNA versus HPV 18 E6/E7 genes caused an induction of cell death in HeLa cells after 10 days [93]. In a recent work by Inturi et al. [92], the HPV E6/E7 oncogenes were suppressed by the CRISPR/Cas9 in cervical cancer cell lines. Based on their findings, the knockout of E6/E7 exposed to CRISPR/Cas9 induced cellular senescence in the HeLa cells immortalized with HPV18. The HeLa cells inactivated with E6/E7 specifically displayed unique senescence indices such as large cell surface area, lamin B1 loss and high expression level of β-galactosidase. The bicistronic transcripts of E6 and E7, they are suppressed and the levels of pRb/p21 and p53/p21 are elevated by inactivating the HPV18 E6. HPV18 E7 knockout caused a reduced the expression of E6 by triggering the pRb/p21 pathway. Overall, cellular senescence was introduced to be alternative outcome of CRISPR/Cas9-inactivated HPV oncogene [92]. In a study by Gao et al. [183], the impact of CRISPR/Cas9 system was evaluated in the treatment of cervical carcinogenesis, in particular cervical precancerous lesions. The HPV16 E7 targeted CRISPR/Cas9, ZFN, TALEN plasmids were transfected in cervical pre-cancer/cancer cell lines. Based on their observations, when comparing with prior TALEN and ZFN systems, CRISPR/Cas9 had remarkable specificity and efficiency to impede colony generation and cell growth and to trigger apoptosis in cervical pre-cancer/cancer cell lines, especially in S12 cell line obtained from low-grade cervical lesion. During xenograft formation assays, the CRISPR/Cas9 could impede tumorigenesis in S12 cell line and influenced the expression of related protein in vivo. The cervical CRISPR/Cas9 therapy in K14-HPV16 transgenic mice model of HPV-driven spontaneous cervical carcinogenesis developed E7 gene mutations and restored E2F1, RB and CDK2 expression, which reversed the phenotype of cervical carcinogenesis. In their work, HPV16 E7-targeting CRISPR/Cas9 reverted cervical carcinogenesis associated with HPV under in vitro condition and also in K14-HPV16 transgenic mice, highlighting potent clinical application to treat cervical precancerous lesions [183].
In a study by Zhen et al., the performance of CRISPR/Cas9 to target HPV E6/E7 oncogenes was confirmed as promising sensitizer to cisplatin [184] and radiotherapy in the suppression of cervical carcinoma [185]. The E6 or E6/E7-targeting CRISPR/Cas9 in combination with cytotoxic factors may synergistically influence the restoration of pRB and/or TP53 performance, and may act as therapeutic option to eliminate the cervical cancer [185]. All these unknown points and the development of clinically CRISPR/Cas9-based combination therapies need further research. The basic pathway of synergy between therapies needs to be cleared to create proof-of-concept for combination therapies based on CRISPR/Cas9. Based on in vivo observations, the combination therapy is remarkably superior to both methods. Evaluation to confirm the absence of off-target impacts and minimal interferon induction are needed for the clinical use of combination therapies based on CRISPR/ Cas9. Though anti-viral CRISPR clinical trials are in preparation, it is still a long way to go before application in the clinic can become a reality. General challenges remain the efficient and specific targeted delivery of CRISPR/Cas9 to infected cells in vivo and the potential of the system to induce editing at undesired locations.
3.6. Merkel cell polyomavirus
Cyril Toker in 1972 introduced for the first time the Merkel cell carcinoma (MCC), as an aggressive skin cancer alongside immunodeficiency. Although rare, the diagnoses of MCC have tripled in the last twenty years. The introduction of a new human virus in 2008 called Merkel Cell polyomavirus (MCPyV) motivated researchers to investigate the virus as a major cause in 80% of MCC cases [186]. Evidence showed that MCPyV integrates into the tumor cell genome in the majority of MCCs. Tumor cells that are positive for this virus express two viral oncoproteins, including small (sT) and large (LT) tumor antigens (TAs) tumor antigens, which result in increased cell growth. Although immunotherapy has been a promising measure in MCC patients, not everyone has been able to respond to such therapies, which underscores the need to develop effective treatment strategies. In a recent work by Temblador et al. [187], the MCPyV TAs were inactivated by CRISPR/Cas9 editing in the MCPyV+ MCC cell lines of WAGA and MS-1. According to their observations, mutations in the frameshift of target sequence after repair of Cas9-induced DNA break resulted in a decrease in LT protein content, thereby disrupting cell proliferation, causing cell cycle arrest, and inducing apoptosis. There was no anti-proliferative activity in HEK293T cells (MCPyV−), which means no off-target performance of TAs-targeting sgRNAs capable of influencing the cell proliferation. The levels of cellular proteins responsible for the regulation of cell cycle did not change in WAGA cells, supporting the cell cycle. They found a central role for the MCPyV TAs in maintaining MCPyV+ MCC cells. There is a need for the evaluation of TAs impacts on cell cycle regulators to recognize the targets for new treatments of MCC. In total, they reported documents about obtaining a CRISPR/Cas9-based candidate for treatment of virus-positive MCC. They suggested that future experiments should focus on the development of a safe and efficient delivery system, especially in cases where the lesions are not directly reachable [187], [188], [189].
3.7. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)
SARS-CoV-2 is an RNA virus which is responsible for the ongoing Coronavirus Disease 2019 (COVID-19) pandemic. At the time of this writing, close to 240 million COVID-19 cases have been recorded worldwide, along with nearly 5 million deaths [190], [191]. One important lesson from this pandemic is that antiviral strategies capable of targeting new viruses for which no vaccines or medications are available are urgently needed. In line with this, a CRISPR-Cas-based technique, termed the prophylactic antiviral CRISPR in human cells (PAC-MAN), has been developed and evaluated for its potential to combat novel viruses, including SARS-CoV-2 [97]. The strategy involves the use of CRISPR-Cas13d to target highly conserved sequences of the SARS-CoV-2 genomes, which were identified using a bioinformatics workflow [97]. The approach was proven to be capable of cleaving SARS-CoV-2 fragments in human lung epithelial cells [97]. It was shown that the use of a six-crRNA panel allows targeting of 91% of the sequenced SARS-CoV-2, whereas a panel of 22-crRNAs was capable of targeting all sequenced coronaviruses [97]. It has also been proposed that the CRISPR-Cas13 machinery can be delivered into the airway epithelium with the use of a modified adeno-associated virus (AAV), i.e. a non-pathogenic human virus with an affinity for the airway epithelial cells, in order to improve the delivery efficiency [192]. Nonetheless, additional research is necessary to establish whether the approach is efficient against live SARS-CoV-2 and whether it is safe to be used in patients. Although a number of challenges need to be overcome before PAC-MAN can be applied clinically, the strategy shows great promise as a novel antiviral approach and has the potential to be quickly implemented in the event of future viral outbreaks.
4. Delivery strategies for CRISPR-Cas9-based therapeutics
Effective and safe delivery of genome editing agents to the intended target tissue is essential for the clinical application of CRISPR-Cas9-based therapeutics. Whilst carrier-free delivery of CRISPR-Cas9 agents has been demonstrated to be fairly effective in certain applications, there are still obvious potential limitations, including unwanted immunostimulation, difficulties in renal clearance, insufficient transport into the cytoplasm and nucleus, and unwanted degradation by cellular enzymes. To resolve these hurdles, several viral vectors for the delivery of CRISPR-Cas9 have been developed and used in preclinical contexts [193], [194]. These viral-based delivery systems offer numerous advantages, including a high transduction rate and transgene expression levels.
One of the most commonly used delivery platforms for CRISPR-Cas9 is the lentiviral vectors, which are derived from the HIV-1 [195]. The lentiviral vectors are replication-defective; they are produced by removing all unnecessary genes from the HIV-1 genome and segregating their cis-acting sequences from the trans-acting elements that are absolutely needed for viral particle production, infection, and integration into the host genome [196]. The third-generation lentiviral vector systems contain four plasmids: a transfer plasmid, which encodes the insert of interest; two separate packaging plasmids, one of which encodes Gag and Pol, and another encodes Rev; and an envelope plasmid, which encodes VSV-G. Lentiviral transfer vectors allow not just transgenes, but also short hairpin RNA (shRNAs) to be expressed in single units or in various combinations. Three more plasmids, namely pMDL, pRev and pVSVG, can be added to provide necessary packaging for the trans-acting components. Additionally, improved safety can be accomplished by introducing a deletion in the promoter-enhancer region located in the 3′ long terminal repeat (LTR), leading to the construction of self-inactivating vectors [197], [198], [199]. The lentiviral vectors have multiple strengths: they have the ability to integrate into the host genome in both dividing and non-dividing cells; possess broad cellular tropism; contain large packaging size for carrying the CRISPR-Cas9-gRNAs complex; allow easy assembly and modification; and ensure the stability of their payloads [200], [201]. For these reasons, lentiviral vectors have been used for delivering CRISPR-Cas9 components in many studies. In mice model with herpetic stromal keratitis (a condition caused by HSV-1 infection), for example, the introduction of SpCas9 mRNA and gRNA into the corneal tissue with the use of lentiviral particles was found to be effective in suppressing HSV replication and eradicating the latent HSV-1 [169].
Another frequently used virus-based gene delivery system is the adeno-associated virus (AAV) vectors. AAV is a non-enveloped, small (25 nm) virus with a linear single-stranded DNA genome of approximately 4.7 kb. There are higher than 200 molecularly manufactured or naturally occurring forms of AAV [202]. Of these, two AAV-based drugs have received the FDA approval for gene therapy: Glybera used to treat people who have a lack of lipoprotein lipase and Zolgensma used for children under the age of two who have spinal muscular atrophy (SMA) [203]. AAV vectors possess several characteristics that make them a suitable vehicle for delivering therapeutics: broad range of serotype specificity; capability to impact both dividing and non-dividing cells; non-pathogenicity; and low immunogenicity [204]. AAV8, an AAV vector, has been used for liver-specific delivery of reconstituted CRISPR-SaCas9, along with a liver-specific promoter [205]. The use of this vector was found to successfully enhance liver delivery and suppress HBV replication both in vitro and in vivo [205]. It should be noted, however, that the loading capacity of AAV vectors is modest (approximately 5 kb). For this reason, encapsulation of SpCas9 along with the efficient promoters into the vector may prove difficult. Fortunately, new Cas proteins that are remarkably smaller than the commonly used ones have been discovered recently [206], [207].
The third class of viral vectors that have great potential for CRISPR-Cas9 delivery is the adenoviral vectors (AdVs). These vectors are derived from adenoviruses, which are non-enveloped icosahedral viruses with linear DNA genomes of 30–40 kb that can be genetically modified with minimal effort [208], [209]. Typically, the antigen expression cassette is introduced into the E1 genomic region of the virus, rendering them incapable of replication. Furthermore, up to 7 kb expression cassettes can be accommodated by vectors in which nonessential E3 genes have been removed [210]. Schiwon and colleagues developed high-capacity adenoviral vectors (HCAdVs) that could efficiently deliver a multiplexed complete CRISPR-Cas9 system for the treatment of HBV. Specific delivery of this genome editing system contributed to a dramatic decrease in antigen production while enhancing the introduction of mutations in the HBV genome [211]. This allows a more potent degradation of the HBV genome, including the HBV cccDNA [211].
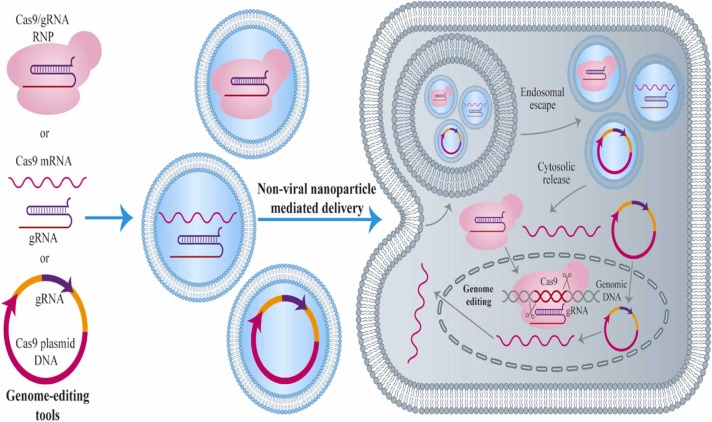
Although viral vectors serve as promising delivery vehicles for CRISPR-Cas systems, they have some inherent limitations. For example, the use of viral vectors increases the risk of tumorigenesis and may induce excessive immunogenicity [212], [213]. It is also difficult to insert new sequences to the viral vectors, and the scale-up process is difficult and time-consuming [212], [213]. On the other hand, non-viral vectors such as nanoparticles can overcome the drawbacks of viral-based delivery systems and have been shown to be highly efficient in delivering various therapeutic agents to the target tissue ( Fig. 3). However, no non-viral vectors have been used to deliver the CRISPR-Cas systems for the treatment of viral diseases so far. This represents an interesting avenue for future research.
Fig. 3.
CRISPR/Cas9 delivery system being plasmid protein, mRNA or DNA. The elements of this delivery system (being protein, mRNA or DNA) are delivered into cells to win a determined gene-editing performance. During delivery mediated by non-viral nano-particle, the encapsulation occurs for each element in nano-carriers to intracellularly uptake. Following the endocytosis, the nanoparticles inside the cell must move away from the endosome/ lysosome. To deliver the plasmid DNA, the DNA must be transported to the nucleus, and the pathway of native transcription of target cells needs be used for gene transcription to the mRNA, the mRNA transportation to the cytoplasm and then translation to the protein. The protein returns to the nucleus in which the CRISPR pathway influence the genomic DNA of the cell. To deliver the mRNA, it must be liberated in the cytosol to be translated to protein. Transient and instantaneous delivery of the protein results in the fastest initiation of gene editing, thus preventing the permanent integration of CRISPR genes into the host genome.
5. CRISPR/Cas9 gene editing mechanism and anti-viral vaccine production
Despite numerous works regarding gene performance, gene therapy and interaction of host with virus, the researchers tried to produce vaccines using the gene editing mechanism based on CRISPR/Cas9 because of its admirable features, including specificity, high efficiency, flexibility, versatility, simplicity and cost-effectiveness versus other viral genome editing pathways, thereby promising an effective approach to construct genetically engineered vaccines. Okoli et al. presented an overview on the advances in the employment of CRISPR/Cas9 in viral genome editing and the promotion of recombinant orthopoxvirus (OPXV) vaccines and vectors [214]. CRISPR/Cas9 genome-wide screening is an approach for detecting and ranking host limiting factors of H1N1 influenza virus in a production of cell-based vaccines, thereby potentially advancing vaccine production in the future [215].
Recent investigations focused on the use of CRISPR/Cas9 system regarding the development of animal anti-viral vaccine. Knock-in of DNA cassettes longer than 4 kb mediated by CRISPR/Cas9 system into PRV genome at 50% positive rate via NHEJ makes PRV a potent vector for vaccine development [216]. A study successfully employed CRISPR/Cas9 system to produce a vaccine through double-deletion of gE and TK genes of PRV [217]. HeN1 PRV strain inactivated by a triple gE/gI/TK gene was completely attenuated, which established immune protection versus parental PRV effort [218], [219]. According to NY strain (a PRV variant), a mutant deleted for triple gE/gI/TK was established via CRISPR/Cas9 gene-editing pathway and homologous DNA recombination, which was introduced as a next-generation vaccine to control novel PRV strains [220]. A non-essential gene of 8-DR was knocked out from the genome of virulent ASFV strain Georgia07 using CRISPR/Cas9 system in porcine macrophages, which was able to rescue the recombinant virus and was proposed as a platform for the development of the next generation vaccine to control African swine fever [218], [221].
CRISPR/Cas9 gene editing mechanism was introduced for the first in 2016 in avian herpesvirus mutagenesis in the live attenuated vaccine vector based on herpesvirus of turkeys (HVT) [222]. NHEJ and CRISPR/Cas9 system was recruited to insert a red fluorescent protein (RFP) into HVT UL45/UL46 region to generate a general donor plasmid. In the next step, the Cre-Lox system was applied to delete the inserted RFP gene and the gene VP2 from infectious bursal disease virus (IBDV) was inserted into the region UL45/UL46 to generate a recombinant vaccine of HVT/IBDV-VP2. After that, the expression cassettes of avian influenza virus (AIV)-H9N2 hemagglutinin (HA) and ILTV gD/gI genes were positioned in US2 and 065/066 regions of recombinant HVT/IBDV-VP2 virus, thereby promising a triple inserted HVT-VP2- gDgI-HA vaccine [223], [224]. In some studies, the HDR and CRISPR/Cas9 systems were recruited to position the expression cassette of AIV H7N9-HA into the same UL45/UL46 location in HVT genome, thereby developing a bivalent HVT-H7N9-HA vaccine [225]. In other works, the H9N2-HA gene from AIV BJ/15 strain was inserted into the UL45/UL46 site from HVT-BAC genome and residual HVT-BAC fragments were deleted by CRISPR/Cas9 mechanism for development of a recombinant vaccine of rHVT-H9N2-HA [226].
At the same time, this approach has been utilized to study other poultry herpesviruses. The HDR and CRISPR/Cas9 mechanisms have been recruited to insert HA gene of significantly pathogenic AIV H5N1, gE and pre-membrane (PrM) genes duck Tembusu virus (DTMUV) into the regions of US7/US8 and UL27/UL26 in the genome of DEV C-KCE strain for the construction of trivalent vaccine in controlling DTMUV, H5N1-AIV and DEV infections [223], [227]. The insertion of AIV H5N1-HA into the region of UL27/UL26 in the genome of DEV was carried out exploiting Cre-Lox and NHEJ-CRISPR/Cas9 systems for the development of a recombinant vaccine of DEV-AIV. Similarly, the genes of unique short 4 (US4) and thymidine kinase (TK) were omitted from the infections laryngotracheitis virus (ILTV) genome, and inserted fusion (F) gene of Newcastle disease virus (NDV) caused no complication on the replication of ILTV and the expression of F protein [228], [229]. For the first time, this study suppressed virulence factors and positioned heterologous genes into the genome of virus for production of multivalent recombinant vaccine. According to such documents, the CRISPR/Cas9 system can be employed for fast modification of genome and development of recombinant vaccine of avian herpesviruses.
A vaccine has been generated versus infections in dogs using CRISPR/Cas9 genome editing approach. The canine distemper virus (CDV) causes an infection in both domestic and wild carnivores, called canine distemper (CD). The CRISPR/Cas9 gene editing pathway was recently recruited to create an effective a recombinant virus consisting of CDV virus-like particles (VLPs), the matrix (M), F and H genes, capable of assembling CDVVLPs and presenting rapid seroconversion and greater antibody positivity when comparing with parental virus strain in minks and foxes [230]. For the first time, an effective and fast vaccine was developed applying the CRISPR/Cas9 pathway to prevent CD in foxes, dogs and minks. As seen in Fig. 4, the findings offered the CRISPR-Cas13 strategy as a therapeutic system for SARS-CoV-2.
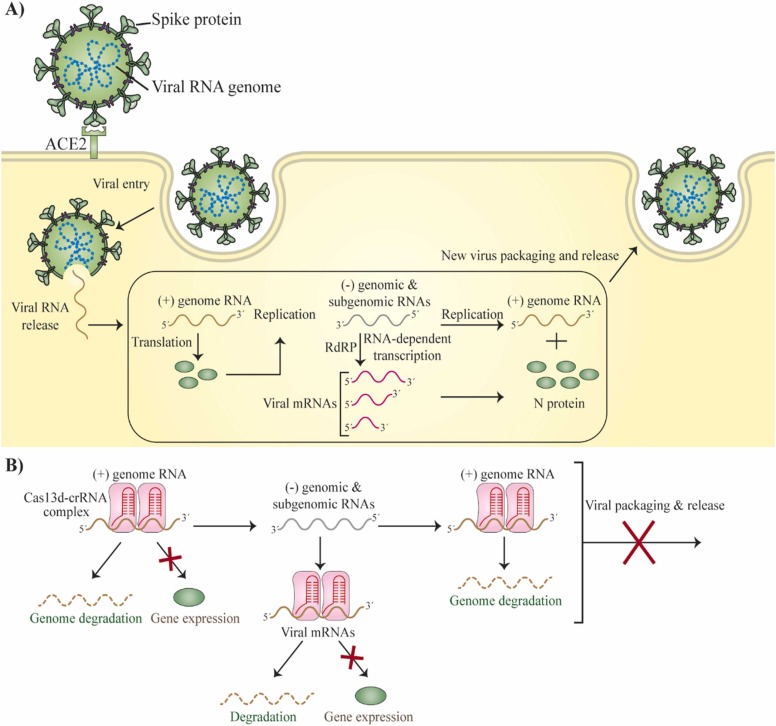
Fig. 4.
Hypothetical life cycle of SARS-CoV-2 and CRISPR-based pathway of PAC-MAN for RNA-guided degradation and suppression of viral RNA in CRISPR-Cas13-mediated suppression of coronavirus. (A) Hypothetical life cycle of SARS-CoV-2. After SARS-CoV-2 penetrates and the genome RNA is released, the positive-strand RNA genome is used as template to form genomic and sub-genomic negative-strand templates for the production of additional transcripts of viral genome and positive-stranded viral mRNAs. (B) The vial function and replication can be suppressed by Cas13d by directly targeting and cutting all viral positive-sense RNAs.
Highly infectious and wild strains of SARS-CoV-2 result from rapid evolution, which negatively affects human health. These continuous health concerns emphasize the availability of reliable diagnostic equipment with adjustable scales. In this regard, some promising CRISPR-based techniques include FELUDA, DETECTR and SHERLOCK for the diagnosis and management of SARS-CoV-2-induced COVID-19. There are several affordable portable detection kits that can be applied in hospitals and poor regions like small clinics, schools, airports and even at home. SARS-CoV-2 and its emerging strains can be detected by such CRISPR-supported diagnostic tools, thus providing one-hour results with high sensitivity, accuracy, and specificity when compared to standard PCR assay. Screening and early diagnosis of communities with COVID-19 at the individual level affects the community in impeding the spread of SARS-CoV-2 through immediate quarantine measures. Success has been reported for CRISPR-supported diagnostic tools, however, the use of these tools is restricted to the laboratory. There are needs for additional translational investigations to advance the laboratory achievements of these rapid diagnostic kits towards large-scale production and commercialization. The diagnosis of COVID-19 can be made anywhere by developing cost-effective, portable microfluidic cartridges based on CRISPR and lyophilized reagents. The kit needs to be used by trained and experienced staff in the field.
6. Potential challenges for CRISPR/Cas-based antiviral therapy
Promising applications are available for the CRISPR/Cas9 system for anti-virus purposes through manipulation. However, various obstacles have been reported to achieve these goals in human use. The safety of these strategies in vivo may be ensured by minimizing off-target performance. Incorrect deletion of a host gene with a high level of similarity with 20 bp seeding plus the PAM sequence at CRISPR/Cas9 target site can be highly destructive to the host cell. This off-target performance of the CRISPR/Cas9 system for human use can be precisely controlled through a various variant of Cas9 (Cas9nickase with decreased off-target features) resulting from the trigger of single-stranded DNA cutting rather than double-stranded [231], [232]. Three high gene-editing performance CRISPR/Cas systems with negligible off-target cleavage must be a much safer option for the clinical use of CRISPR/Cas9-supported antiviral approach [233], [234]. One major problem is to carry the components of CRISPR/Cas9 to any cells infected with virus, which must be bypassed for successful clinical use of this novel approach. This may be because the presence of cells infected with the remaining unmodified virus could be considered another repository of virus to release a novel virion into the previously modified healthy cells. One of the best delivery systems is AAV due to the high capability of viral titers with the potential to completely transmit all cells infected with virus to a patient [235]. Recombinant engineering of a capsid protein of AAV through tissue tropism for an infection site can lead to targeted delivery of AAV to specific tissues. One of the most important delivery strategies for CRISPR/Cas9 is a consistent record of safety and absence of AAV integration [235]. Based on previous findings, these AAV delivery methods were studied for feasibility in CRISPR/Cas9-supported antiviral investigations [90], [137], [138], [236], [237]. Viral vectors are extensively applied in CRISPR/Cas9 delivery under in vitro and in vivo conditions. Nevertheless, there are multiple bottlenecks for them, such as size limitations in the insertion of foreign gene, post-integration disruption of host gene and subsequent cancer development, difficulty in mass production, and strong immune behaviors. These limitations may be addressed by constructing non-viral vectors like lipid- or polymer-supported nanocarriers as an alternative delivery system for CRISPR/Cas9 [238], [239], [240]. Viruses can become resistant to the CRISPR/Cas9 system by producing an escape virus mutant, which must be addressed before it can be used in clinical applications. This episode can be prevented by carefully choosing the most essential and most conserved regions of the viral genome for gRNA design. The gRNAs must be carefully selected and optimally designed to effectively control viral infection via CRISPR/Cas9 resulting from variants of viruses with various sequences. The antiviral impact can be eliminated in many gRNA-supported monoplex strategies by selecting a mutant virus with a modified target site, which can no longer be separated by the CRISPR/Cas9 system. Accordingly, effective and successful effects were reported for multiplex strategies to suppress the production of a virus escape mutant and maintain long-term antiviral activity. The next issue is to more accurately validate the CRISPR/Cas9-supported host-targeting anti-virus strategy. A common example of an indirect use of the CRISPR/Cas9 technique is the removal of host factors essential for a particular stage of the virus life cycle. This host targeting through CRISPR/Cas9 has the advantage of greatly reduced viral resistance to direct modification of the virus genome. It is very difficult to choose a host dependency factor, which is unnecessary for a host and essential for a virus. We recommend a thorough analysis of the host's immune response to the CRISPR/Cas system to predict the side effects of this antiviral treatment. In practice, this CRISPR/Cas9-spported antiviral treatment should be cost-effective for patients.
7. Conclusion and future perspectives
The CRISPR-Cas systems allow precise targeting and modification of both the viral and the host genomes, and offer a promising prospect for the therapy of viral diseases. However, before the approach can be translated into widespread clinical use, a number of key challenges need to be overcome. The most important consideration is to ensure that CRISPR therapeutics can be delivered specifically to their intended target site. This increases the efficiency of CRISPR-Cas-mediated targeting and at the same time avoids undesired effects of nonspecific targeting in other organs. Several viral vectors have been used as delivery systems for CRISPR-Cas therapeutics, including lentiviral vectors, AAV vectors and AdVs. However, each of these vectors has its own limitations. Non-viral vectors have not been used for the delivery of CRISPR-Cas, but they hold considerable promise to help mitigate the shortcomings of viral-based delivery systems. Therefore, it is imperative that future studies focus on investigating the feasibility of using non-viral vectors as a delivery vehicle for the CRISPR-Cas system.
Another challenge that needs to be addressed is the off-target genetic effects, in which DSBs occur at undesired genomic sites within the target cells. Many strategies have been proposed to decrease the off-target genetic effects, such as by using the Cas9-gRNA ribonucleoprotein (RNP) [241], employing RNA-targeting Cas systems, including RCas9 and Cas13 [242], modifying the Cas systems to improve DNA target sequence recognition [243], and altering the PAM preferences [244]. Interestingly it has been reported that by decreasing the length of gRNA from 20 bp to 17/18 bp, off-target effects can be reduced by 500-fold, without any adverse effects on the accuracy of Cas9 protein [245]. Modification of gRNA ribose-phosphate backbone with 2’-O-methyl-3’-phosphonoacetate was also found to significantly decrease the off-target events [246]. In addition to these strategies, using the natural inhibitors of CRISPR-Cas complexes, known as Anti CRISPR proteins, can reduce off-target cleavages [247]. However, these findings have not been extensively replicated. Hence, future studies should investigate whether the aforementioned strategies could reduce off-target genetic effects in the treatment of viral infections. Identification of additional strategies for optimizing the CRISPR-Cas system should also be encouraged.
In conclusion, the CRISPR-Cas systems are powerful tools that have a lot of potential as a new therapeutic technique for the treatment of viral infections. However, several key challenges must be overcome before this technology can be used in routine clinical settings. Further studies aimed at improving the safety and efficiency of the CRISPR-Cas therapy are also highly warranted.
CRediT authorship contribution statement
Sajad Najafi, Shing Cheng Tan, Shahin Aghamiri, Pourya Raee, Zahra Ebrahimi, Zahra Kargar Jahromi, Yazdan Rahmati, Javid Sadri Nahand, Vahid Jajarmi, Hamed Mirzaei contributed in data collection and manuscript drafting. Ahmad Piroozmand critically revised the manuscript. All authors approved the final version for submission.
Funding
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest statement
The authors declare no conflict of interest.
Acknowledgements
Not applicable.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.biopha.2022.112743.
Appendix A. Supplementary material
Supplementary material
.
Data Availability
Not applicable.
References
- 1.Qureshi A., Tantray V.G., Kirmani A.R., Ahangar A.G. A review on current status of antiviral siRNA. Rev. Med. Virol. 2018;28 doi: 10.1002/rmv.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu B., Guo H., Zhou P., Shi Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skegg D., Gluckman P., Boulton G., Hackmann H., Karim S.S.A., Piot P., Woopen C. Future scenarios for the COVID-19 pandemic. Lancet. 2021;397:777–778. doi: 10.1016/S0140-6736(21)00424-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamers R.L., Rinke de Wit T.F., Holmes C.B. HIV drug resistance in low-income and middle-income countries. Lancet HIV. 2018;5:e588–e596. doi: 10.1016/S2352-3018(18)30173-5. [DOI] [PubMed] [Google Scholar]
- 5.Tang L.S.Y., Covert E., Wilson E., Kottilil S. Chronic hepatitis b infection: a review. JAMA. 2018;319:1802–1813. doi: 10.1001/jama.2018.3795. [DOI] [PubMed] [Google Scholar]
- 6.Rotondo J.C., Mazziotta C., Lanzillotti C., Tognon M., Martini F. Epigenetic dysregulations in merkel cell polyomavirus-driven merkel cell carcinoma. Int J. Mol. Sci. 2021:22. doi: 10.3390/ijms222111464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mehta A., Michler T., Merkel O.M. siRNA therapeutics against respiratory viral infections—what have we learned for potential COVID-19 therapies? Adv. Healthc. Mater. 2021;10:2001650. doi: 10.1002/adhm.202001650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adalja A., Inglesby T. Broad-spectrum antiviral agents: a crucial pandemic tool. Expert Rev. Anti Infect. Ther. 2019;17:467–470. doi: 10.1080/14787210.2019.1635009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levanova A., Poranen M.M. RNA interference as a prospective tool for the control of human viral infections. Front. Microbiol. 2018:9. doi: 10.3389/fmicb.2018.02151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aghamiri S., Jafarpour A., Malekshahi Z.V., Mahmoudi Gomari M., Negahdari B. Targeting siRNA in colorectal cancer therapy: nanotechnology comes into view. J. Cell. Physiol. 2019;234:14818–14827. doi: 10.1002/jcp.28281. [DOI] [PubMed] [Google Scholar]
- 11.Aghamiri S., Jafarpour A., Gomari M.M., Ghorbani J., Rajabibazl M., Payandeh Z. siRNA nanotherapeutics: a promising strategy for anti-HBV therapy. IET Nanobiotechnol. 2019;13:457–463. doi: 10.1049/iet-nbt.2018.5286. [DOI] [Google Scholar]
- 12.Flisiak R., Jaroszewicz J., Łucejko M. siRNA drug development against hepatitis B virus infection. Expert Opin. Biol. Ther. 2018;18:609–617. doi: 10.1080/14712598.2018.1472231. [DOI] [PubMed] [Google Scholar]
- 13.Bella R., Kaminski R., Mancuso P., Young W.-B., Chen C., Sariyer R., Fischer T., Amini S., Ferrante P., Jacobson J.M. Removal of HIV DNA by CRISPR from patient blood engrafts in humanized mice. Mol. Ther. Nucleic Acids. 2018;12:275–282. doi: 10.1016/j.omtn.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carr A. Toxicity of antiretroviral therapy and implications for drug development. Nat. Rev. Drug Discov. 2003;2:624–634. doi: 10.1038/nrd1151. [DOI] [PubMed] [Google Scholar]
- 15.Liao H.K., Gu Y., Diaz A., Marlett J., Takahashi Y., Li M., Suzuki K., Xu R., Hishida T., Chang C.J., Esteban C.R., et al. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat. Commun. 2015;6:6413. doi: 10.1038/ncomms7413. [DOI] [PubMed] [Google Scholar]
- 16.Kaminski R., Chen Y., Fischer T., Tedaldi E., Napoli A., Zhang Y., Karn J., Hu W., Khalili K. Elimination of HIV-1 genomes from human T-lymphoid cells by CRISPR/Cas9 gene editing. Sci. Rep. 2016;6:22555. doi: 10.1038/srep22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaminski R., Bella R., Yin C., Otte J., Ferrante P., Gendelman H.E., Li H., Booze R., Gordon J., Hu W., Khalili K. Excision of HIV-1 DNA by gene editing: a proof-of-concept in vivo study. Gene Ther. 2016;23:690–695. doi: 10.1038/gt.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee C. CRISPR/Cas9-based antiviral strategy: current status and the potential challenge. Molecules. 2019:24. doi: 10.3390/molecules24071349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohanraju P., Makarova K.S., Zetsche B., Zhang F., Koonin E.V., Van der Oost J. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016:353. doi: 10.1126/science.aad5147. [DOI] [PubMed] [Google Scholar]
- 20.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sternberg S.H., Doudna J.A. Expanding the biologist’s toolkit with CRISPR-Cas9. Mol. Cell. 2015;58:568–574. doi: 10.1016/j.molcel.2015.02.032. [DOI] [PubMed] [Google Scholar]
- 22.Soppe J.A., Lebbink R.J. Antiviral goes viral: harnessing CRISPR/Cas9 to combat viruses in humans. Trends Microbiol. 2017;25:833–850. doi: 10.1016/j.tim.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 23.Mojica F.J., Rodriguez-Valera F. The discovery of CRISPR in archaea and bacteria. FEBS J. 2016;283:3162–3169. doi: 10.1111/febs.13766. [DOI] [PubMed] [Google Scholar]
- 24.Escalona-Noguero C., López-Valls M., Sot B. CRISPR/Cas technology as a promising weapon to combat viral infections. 2021;43 doi: 10.1002/bies.202000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou J., Shen B., Zhang W., Wang J., Yang J., Chen L., Zhang N., Zhu K., Xu J., Hu B., Leng Q., et al. One-step generation of different immunodeficient mice with multiple gene modifications by CRISPR/Cas9 mediated genome engineering. Int. J. Biochem. Cell Biol. 2014;46:49–55. doi: 10.1016/j.biocel.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 26.Janusz G., Pawlik A., widerska-Burek U., Polak J., Sulej J., Jarosz-Wilkołazka A., Paszczyński A. Laccase properties, physiological functions, and evolution. Int. J. Mol. Sci. 2020;21:966. doi: 10.3390/ijms21030966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makarova K.S., Wolf Y.I., Iranzo J., Shmakov S.A., Alkhnbashi O.S., Brouns S.J., Charpentier E., Cheng D., Haft D.H., Horvath P. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020;18:67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., Cradick T.J., et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y., Ge X., Yang F., Zhang L., Zheng J., Tan X., Jin Z.B., Qu J., Gu F. Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells. Sci. Rep. 2014;4:5405. doi: 10.1038/srep05405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang H., Ren S., Yu S., Pan H., Li T., Ge S., Zhang J., Xia N. Methods favoring homology-directed repair choice in response to CRISPR/Cas9 induced-double strand breaks. Int. J. Mol. Sci. 2020:21. doi: 10.3390/ijms21186461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirman Z., de Lange T. 53BP1: a DSB escort. Genes Dev. 2020;34:7–23. doi: 10.1101/gad.333237.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poonguzhali R., Basha S.K., Kumari V.S. Synthesis and characterization of chitosan-PVP-nanocellulose composites for in-vitro wound dressing application. Int. J. Biol. Macromol. 2017;105:111–120. doi: 10.1016/j.ijbiomac.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 33.Aksoy Y.A., Nguyen D.T., Chow S., Chung R.S., Guillemin G.J. Chemical reprogramming enhances homology-directed genome editing in zebrafish embryos. Commun. Biol. 2019;2:198. doi: 10.1038/s42003-019-0444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paulsen B.S., Mandal P.K. Ectopic expression of RAD52 and dn53BP1 improves homology-directed repair during CRISPR-Cas9 genome editing. Nat. Biomed. Eng. 2017;1:878–888. doi: 10.1038/s41551-017-0145-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Devkota S. The road less traveled: strategies to enhance the frequency of homology-directed repair (HDR) for increased efficiency of CRISPR/Cas-mediated transgenesis. BMB Rep. 2018;51:437–443. doi: 10.5483/BMBRep.2018.51.9.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurihara T., Kouyama-Suzuki E., Satoga M., Li X., Badawi M., Thiha, Baig D.N., Yanagawa T., Uemura T., Mori T., Tabuchi K. DNA repair protein RAD51 enhances the CRISPR/Cas9-mediated knock-in efficiency in brain neurons. Biochem. Biophys. Res. Commun. 2020;524:621–628. doi: 10.1016/j.bbrc.2020.01.132. [DOI] [PubMed] [Google Scholar]
- 37.Jolany vangah S., Katalani C., Boone H.A., Hajizade A., Sijercic A., Ahmadian G. CRISPR-based diagnosis of infectious and noninfectious diseases. Biol. Proced. Online. 2020;22:22. doi: 10.1186/s12575-020-00135-3. 10.1186/s12575-020-00135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patchsung M., Jantarug K., Pattama A., Aphicho K., Suraritdechachai S., Meesawat P., Sappakhaw K., Leelahakorn N., Ruenkam T., Wongsatit T., Athipanyasilp N., et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020;4:1140–1149. doi: 10.1038/s41551-020-00603-x. 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- 40.Azhar M., Phutela R., Ansari A.H., Sinha D., Sharma N., Kumar M., Aich M., Sharma S., Rauthan R., Singhal K., Lad H., et al. Rapid, field-deployable nucleobase detection and identification using FnCas9. bioRxiv. 2020 2020.2004.2007.028167. 10.1101/2020.04.07.028167. [Google Scholar]
- 41.Ding X., Yin K., Li Z., Liu C. All-in-one dual CRISPR-Cas12a (AIOD-CRISPR) assay: a case for rapid, ultrasensitive and visual detection of novel coronavirus SARS-CoV-2 and HIV virus. bioRxiv. 2020 doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rahman M.R., Hossain M.A., Mozibullah M., Mujib F.A., Afrose A., Shahed-Al-Mahmud M., Apu M.A.I. CRISPR is a useful biological tool for detecting nucleic acid of SARS-CoV-2 in human clinical samples. Biomed. Pharmacother. 2021;140 doi: 10.1016/j.biopha.2021.111772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cyranoski D. Chinese scientists to pioneer first human CRISPR trial. Nature. 2016;535:476–477. doi: 10.1038/nature.2016.20302. [DOI] [PubMed] [Google Scholar]
- 44.Rodríguez‑Rodríguez, D.R., Ramírez‑Solís, R., Garza‑Elizondo, M.A., Garza‑Rodríguez, M.D.L., and Barrera‑Saldaña, H.A Genome editing: a perspective on the application of CRISPR/Cas9 to study human diseases (review) Int. J. Mol. Med. 2019;43:1559–1574. doi: 10.3892/ijmm.2019.4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith A.E., Helenius A. How viruses enter animal cells. Science. 2004;304:237. doi: 10.1126/science.1094823. [DOI] [PubMed] [Google Scholar]
- 46.Nasir A., Romero-Severson E., Claverie J.-M. Investigating the concept and origin of viruses. Trends Microbiol. 2020;28:959–967. doi: 10.1016/j.tim.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Buhr H., Lebbink R.J. Harnessing CRISPR to combat human viral infections. Curr. Opin. Immunol. 2018;54:123–129. doi: 10.1016/j.coi.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 48.Escalona-Noguero C., López-Valls M., Sot B. CRISPR/Cas technology as a promising weapon to combat viral infections. BioEssays. 2021;43:2000315. doi: 10.1002/bies.202000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Traylen C.M., Patel H.R., Fondaw W., Mahatme S., Williams J.F., Walker L.R., Dyson O.F., Arce S., Akula S.M. Virus reactivation: a panoramic view in human infections. Future Virol. 2011;6:451–463. doi: 10.2217/fvl.11.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nehme Z., Pasquereau S., Herbein G. Control of viral infections by epigenetic-targeted therapy. Clin. Epigen. 2019;11:55. doi: 10.1186/s13148-019-0654-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Leo A., Calderon A., Lieberman P.M. Control of viral latency by episome maintenance proteins. Trends Microbiol. 2020;28:150–162. doi: 10.1016/j.tim.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fenwick C., Joo V., Jacquier P., Noto A., Banga R., Perreau M., Pantaleo G. T-cell exhaustion in HIV infection. Immunol. Rev. 2019;292:149–163. doi: 10.1111/imr.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsukuda S., Watashi K. Hepatitis B virus biology and life cycle. Antivir. Res. 2020;182 doi: 10.1016/j.antiviral.2020.104925. [DOI] [PubMed] [Google Scholar]
- 54.van Diemen F.R., Kruse E.M., Hooykaas M.J., Bruggeling C.E., Schürch A.C., van Ham P.M., Imhof S.M., Nijhuis M., Wiertz E.J., Lebbink R.J. CRISPR/Cas9-mediated genome editing of herpesviruses limits productive and latent infections. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsu J.L., Glaser S.L. Epstein–Barr virus-associated malignancies: epidemiologic patterns and etiologic implications. Crit. Rev. Oncol. Hematol. 2000;34:27–53. doi: 10.1016/S1040-8428(00)00046-9. [DOI] [PubMed] [Google Scholar]
- 56.Kennedy E.M., Kornepati A.V.R., Goldstein M., Bogerd H.P., Poling B.C., Whisnant A.W., Kastan M.B., Cullen B.R. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J. Virol. 2014;88:11965–11972. doi: 10.1128/JVI.01879-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chou Y.Y., Krupp A., Kaynor C., Gaudin R., Ma M., Cahir-McFarland E., Kirchhausen T. Inhibition of JCPyV infection mediated by targeted viral genome editing using CRISPR/Cas9. Sci. Rep. 2016;6:36921. doi: 10.1038/srep36921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hou P., Chen S., Wang S., Yu X., Chen Y., Jiang M., Zhuang K., Ho W., Hou W., Huang J., Guo D. Genome editing of CXCR4 by CRISPR/cas9 confers cells resistant to HIV-1 infection. Sci. Rep. 2015;5:15577. doi: 10.1038/srep15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O’Connell M.R., Oakes B.L., Sternberg S.H., East-Seletsky A., Kaplan M., Doudna J.A. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516:263–266. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim V.N. RNA-targeting CRISPR comes of age. Nat. Biotechnol. 2018;36:44–45. doi: 10.1038/nbt.4054. [DOI] [PubMed] [Google Scholar]
- 61.Abudayyeh O.O., Gootenberg J.S., Essletzbichler P., Han S., Joung J., Belanto J.J., Verdine V., Cox D.B.T., Kellner M.J., Regev A., Lander E.S., et al. RNA targeting with CRISPR–Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nguyen T.M., Zhang Y., Pandolfi P.P. Virus against virus: a potential treatment for 2019-nCov (SARS-CoV-2) and other RNA viruses. Cell Res. 2020;30:189–190. doi: 10.1038/s41422-020-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ye L., Wang J., Beyer A.I., Teque F., Cradick T.J., Qi Z., Chang J.C., Bao G., Muench M.O., Yu J., Levy J.A., et al. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Δ32 mutation confers resistance to HIV infection. Proc. Natl. Acad. Sci. 2014;111:9591–9596. doi: 10.1073/pnas.1407473111. 10.1073/pnas.1407473111%J Proceedings of the National Academy of Sciences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nerys-Junior A., Braga-Dias L.P., Pezzuto P., Cotta-de-Almeida V., Tanuri A. Comparison of the editing patterns and editing efficiencies of TALEN and CRISPR-Cas9 when targeting the human CCR5 gene. Genet. Mol. Biol. 2018;41:167–179. doi: 10.1590/1678-4685-gmb-2017-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ebina H., Misawa N., Kanemura Y., Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 2013;3:2510. doi: 10.1038/srep02510. 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hu W., Kaminski R., Yang F., Zhang Y., Cosentino L., Li F., Luo B., Alvarez-Carbonell D., Garcia-Mesa Y., Karn J., Mo X., et al. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc. Natl. Acad. Sci. USA. 2014;111:11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang G., Zhao N., Berkhout B., Das A.T. A combinatorial CRISPR-Cas9 attack on HIV-1 DNA extinguishes all infectious provirus in infected T cell cultures. Cell Rep. 2016;17:2819–2826. doi: 10.1016/j.celrep.2016.11.057. [DOI] [PubMed] [Google Scholar]
- 68.Lebbink R.J., de Jong D.C., Wolters F., Kruse E.M., van Ham P.M., Wiertz E.J., Nijhuis M. A combinational CRISPR/Cas9 gene-editing approach can halt HIV replication and prevent viral escape. Sci. Rep. 2017;7:41968. doi: 10.1038/srep41968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yin L., Zhao F., Sun H., Wang Z., Huang Y., Zhu W., Xu F., Mei S., Liu X., Zhang D., Wei L., et al. CRISPR-Cas13a inhibits HIV-1 infection. Mol. Ther. Nucleic Acids. 2020;21:147–155. doi: 10.1016/j.omtn.2020.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin S.-R., Yang H.-C., Kuo Y.-T., Liu C.-J., Yang T.-Y., Sung K.-C., Lin Y.-Y., Wang H.-Y., Wang C.-C., Shen Y.-C., Wu F.-Y., et al. The CRISPR/Cas9 system facilitates clearance of the intrahepatic HBV templates in vivo. Mol. Ther. Nucleic Acids. 2014;3 doi: 10.1038/mtna.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kennedy E.M., Bassit L.C., Mueller H., Kornepati A.V.R., Bogerd H.P., Nie T., Chatterjee P., Javanbakht H., Schinazi R.F., Cullen B.R. Suppression of hepatitis B virus DNA accumulation in chronically infected cells using a bacterial CRISPR/Cas RNA-guided DNA endonuclease. Virology. 2015;476:196–205. doi: 10.1016/j.virol.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhen S., Hua L., Liu Y.H., Gao L.C., Fu J., Wan D.Y., Dong L.H., Song H.F., Gao X. Harnessing the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated Cas9 system to disrupt the hepatitis B virus. Gene Ther. 2015;22:404–412. doi: 10.1038/gt.2015.2. [DOI] [PubMed] [Google Scholar]
- 73.Ramanan V., Shlomai A., Cox D.B.T., Schwartz R.E., Michailidis E., Bhatta A., Scott D.A., Zhang F., Rice C.M., Bhatia S.N. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci. Rep. 2015;5:10833. doi: 10.1038/srep10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang J., Xu Z.-W., Liu S., Zhang R.-Y., Ding S.-L., Xie X.-M., Long L., Chen X.-M., Zhuang H., Lu F.-M. Dual gRNAs guided CRISPR/Cas9 system inhibits hepatitis B virus replication. World J. Gastroenterol. 2015;21:9554–9565. doi: 10.3748/wjg.v21.i32.9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Karimova M., Beschorner N., Dammermann W., Chemnitz J., Indenbirken D., Bockmann J.-H., Grundhoff A., Lüth S., Buchholz F., Wiesch J.Sz, Hauber J. CRISPR/Cas9 nickase-mediated disruption of hepatitis B virus open reading frame S and X. Sci. Rep. 2015;5:13734. doi: 10.1038/srep13734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li H., Sheng C., Wang S., Yang L., Liang Y., Huang Y., Liu H., Li P., Yang C., Yang X., Jia L., et al. Removal of integrated hepatitis B virus DNA using CRISPR-Cas9. frontiers in cellular and infection. Microbiology. 2017:7. doi: 10.3389/fcimb.2017.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kostyushev D., Kostyusheva A., Brezgin S., Zarifyan D., Utkina A., Goptar I., Chulanov V. Suppressing the NHEJ pathway by DNA-PKcs inhibitor NU7026 prevents degradation of HBV cccDNA cleaved by CRISPR/Cas9. Sci. Rep. 2019;9:1847. doi: 10.1038/s41598-019-38526-6. 10.1038/s41598-019-38526-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seeger C., Sohn J.A. Targeting hepatitis B Virus With CRISPR/Cas9. Mol. Ther. Nucleic Acids. 2014;3 doi: 10.1038/mtna.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yuen K.-S., Chan C.-P., Wong N.-H.M., Ho C.-H., Ho T.-H., Lei T., Deng W., Tsao S.W., Chen H., Kok K.-H., Jin D.-Y. CRISPR/Cas9-mediated genome editing of Epstein–Barr virus in human cells. J. Gen. Virol. 2015;96:626–636. doi: 10.1099/jgv.0.000012. [DOI] [PubMed] [Google Scholar]
- 80.van Diemen F.R., Kruse E.M., Hooykaas M.J.G., Bruggeling C.E., Schürch A.C., van Ham P.M., Imhof S.M., Nijhuis M., Wiertz E.J.H.J., Lebbink R.J. CRISPR/Cas9-mediated genome editing of herpesviruses limits productive and latent infections. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005701. e1005701-e1005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huo H., Hu G. CRISPR/Cas9-mediated LMP1 knockout inhibits Epstein-Barr virus infection and nasopharyngeal carcinoma cell growth. Infect. Agents Cancer. 2019;14:30. doi: 10.1186/s13027-019-0246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lin C., Li H., Hao M., Xiong D., Luo Y., Huang C., Yuan Q., Zhang J., Xia N. Increasing the efficiency of CRISPR/Cas9-mediated precise genome editing of HSV-1 virus in human cells. Sci. Rep. 2016;6:34531. doi: 10.1038/srep34531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roehm P.C., Shekarabi M., Wollebo H.S., Bellizzi A., He L., Salkind J., Khalili K. Inhibition of HSV-1 replication by gene editing strategy. Sci. Rep. 2016;6:23146. doi: 10.1038/srep23146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu Y.-C., Cai Z.-M., Zhang X.-J. Reprogrammed CRISPR-Cas9 targeting the conserved regions of HPV6/11 E7 genes inhibits proliferation and induces apoptosis in E7-transformed keratinocytes. Asian J. Androl. 2016;18:475. doi: 10.4103/1008-682X.157399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhen S., Lu J., Liu Y.-H., Chen W., Li X. Synergistic antitumor effect on cervical cancer by rational combination of PD1 blockade and CRISPR-Cas9-mediated HPV knockout. Cancer Gene Ther. 2020;27:168–178. doi: 10.1038/s41417-019-0131-9. [DOI] [PubMed] [Google Scholar]
- 86.Zhen S., Hua L., Takahashi Y., Narita S., Liu Y.-H., Li Y. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9. Biochem. Biophys. Res. Commun. 2014;450:1422–1426. doi: 10.1016/j.bbrc.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 87.Hu Z., Yu L., Zhu D., Ding W., Wang X., Zhang C., Wang L., Jiang X., Shen H., He D. Disruption of HPV16-E7 by CRISPR/Cas system induces apoptosis and growth inhibition in HPV16 positive human cervical cancer cells. BioMed. Res. Int. 2014 doi: 10.1155/2014/612823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yu L., Wang X., Da Zhu W.D., Wang L., Zhang C., Jiang X., Shen H., Liao S., Ma D., Hu Z. Disruption of human papillomavirus 16 E6 gene by clustered regularly interspaced short palindromic repeat/Cas system in human cervical cancer cells. OncoTargets Ther. 2015;8:37. doi: 10.2147/OTT.S64092. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 89.Gao X., Jin Z., Tan X., Zhang C., Zou C., Zhang W., Ding J., Das B.C., Severinov K., Hitzeroth I.I. Hyperbranched poly (β-amino ester) based polyplex nanopaticles for delivery of CRISPR/Cas9 system and treatment of HPV infection associated cervical cancer. J. Control. Release. 2020;321:654–668. doi: 10.1016/j.jconrel.2020.02.045. [DOI] [PubMed] [Google Scholar]
- 90.Hsu D.S., Kornepati A.V., Glover W., Kennedy E.M., Cullen B.R. Targeting HPV16 DNA using CRISPR/Cas inhibits anal cancer growth in vivo. Future Virol. 2018;13:475–482. doi: 10.2217/fvl-2018-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yoshiba T., Saga Y., Urabe M., Uchibor R., Matsubara S., Fujiwara H., Mizukami H. CRISPR/Cas9–mediated cervical cancer treatment targeting human papillomavirus E6. Oncol. Lett. 2019;17:2197–2206. doi: 10.3892/ol.2018.9815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Inturi R., Jemth P. CRISPR/Cas9-based inactivation of human papillomavirus oncogenes E6 or E7 induces senescence in cervical cancer cells. Virology. 2021;562:92–102. doi: 10.1016/j.virol.2021.07.005. [DOI] [PubMed] [Google Scholar]
- 93.Kennedy E.M., Kornepati A.V., Goldstein M., Bogerd H.P., Poling B.C., Whisnant A.W., Kastan M.B., Cullen B.R. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J. Virol. 2014;88:11965–11972. doi: 10.1128/JVI.01879-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lao Y.H., Li M., Gao M.A., Shao D., Chi C.W., Huang D., Chakraborty S., Ho T.C., Jiang W., Wang H.X. HPV oncogene manipulation using nonvirally delivered CRISPR/Cas9 or Natronobacterium gregoryi Argonaute. Adv. Sci. 2018;5:1700540. doi: 10.1002/advs.201700540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jubair L., Fallaha S., McMillan N.A. Systemic delivery of CRISPR/Cas9 targeting HPV oncogenes is effective at eliminating established tumors. Mol. Ther. 2019;27:2091–2099. doi: 10.1016/j.ymthe.2019.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cheng Y.-x, Chen G.-t, Yang X., Wang Y.-q, Hong L. Effects of HPV pseudotype virus in cutting E6 gene selectively in SiHa cells. Curr. Med. Sci. 2018;38:212–221. doi: 10.1007/s11596-018-1868-3. [DOI] [PubMed] [Google Scholar]
- 97.Abbott T.R., Dhamdhere G., Liu Y., Lin X., Goudy L., Zeng L., Chemparathy A., Chmura S., Heaton N.S., Debs R., Pande T., et al. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell. 2020;181:865–876. doi: 10.1016/j.cell.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gao F., Bailes E., Robertson D.L., Chen Y., Rodenburg C.M., Michael S.F., Cummins L.B., Arthur L.O., Peeters M., Shaw G.M., Sharp P.M., et al. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 99.HIV/AIDS, J.U.N.P.o. (2019). Fact sheet—latest global and regional statistics on the status of the HIV epidemic. Geneva: Joint United Nations Programme on HIV/AIDS.
- 100.Lengauer T., Sing T. Bioinformatics-assisted anti-HIV therapy. Nat. Rev. Microbiol. 2006;4:790–797. doi: 10.1038/nrmicro1477. [DOI] [PubMed] [Google Scholar]
- 101.Günthard H.F., Calvez V., Paredes R., Pillay D., Shafer R.W., Wensing A.M., Jacobsen D.M., Richman D.D. Human immunodeficiency virus drug resistance: 2018 recommendations of the international antiviral society-USA panel. Clin. Infect. Dis. 2019;68:177–187. doi: 10.1093/cid/ciy463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yin L., Hu S., Mei S., Sun H., Xu F., Li J., Zhu W., Liu X., Zhao F., Zhang D., Cen S., et al. CRISPR/Cas9 inhibits multiple steps of HIV-1 infection. Hum. Gene Ther. 2018;29:1264–1276. doi: 10.1089/hum.2018.018. [DOI] [PubMed] [Google Scholar]
- 103.Lederman M.M., Penn-Nicholson A., Cho M., Mosier D. Biology of CCR5 and its role in HIV infection and treatment. JAMA. 2006;296:815–826. doi: 10.1001/jama.296.7.815. [DOI] [PubMed] [Google Scholar]
- 104.Wilen C.B., Wang J., Tilton J.C., Miller J.C., Kim K.A., Rebar E.J., Sherrill-Mix S.A., Patro S.C., Secreto A.J., Jordan A.P.O., Lee G., et al. Engineering HIV-resistant human CD4+ T cells with CXCR4-Specific zinc-finger nucleases. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hütter G., Nowak D., Mossner M., Ganepola S., Müssig A., Allers K., Schneider T., Hofmann J., Kücherer C., Blau O., Blau I.W., et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. New Engl. J. Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 106.Liu Z., Chen S., Jin X., Wang Q., Yang K., Li C., Xiao Q., Hou P., Liu S., Wu S., Hou W., et al. Genome editing of the HIV co-receptors CCR5 and CXCR4 by CRISPR-Cas9 protects CD4(+) T cells from HIV-1. infection. 2017;7:47. doi: 10.1186/s13578-017-0174-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ding J., Liu Y., Lai Y. Knowledge from London and Berlin: finding threads to a functional HIV cure. Front. Immunol. 2021:12. doi: 10.3389/fimmu.2021.688747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Allen A.G., Chung C.-H., Atkins A., Dampier W., Khalili K., Nonnemacher M.R., Wigdahl B. Gene editing of HIV-1 Co-receptors to prevent and/or cure virus infection. Front. Microbiol. 2018:9. doi: 10.3389/fmicb.2018.02940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hou P., Chen S., Wang S., Yu X., Chen Y., Jiang M., Zhuang K., Ho W., Hou W., Huang J., Guo D. Genome editing of CXCR4 by CRISPR/cas9 confers cells resistant to HIV-1 infection. Sci. Rep. 2015;5:15577. doi: 10.1038/srep15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hendel A., Bak R.O., Clark J.T., Kennedy A.B., Ryan D.E., Roy S., Steinfeld I., Lunstad B.D., Kaiser R.J., Wilkens A.B., Bacchetta R., et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015;33:985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ebina H., Misawa N., Kanemura Y., Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 2013;3:2510. doi: 10.1038/srep02510. 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Saayman S., Ali S.A., Morris K.V., Weinberg M.S. The therapeutic application of CRISPR/Cas9 technologies for HIV. Expert Opin. Biol. Ther. 2015;15:819–830. doi: 10.1517/14712598.2015.1036736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yoder K.E., Bundschuh R. Host double strand break repair generates HIV-1 strains resistant to CRISPR/Cas9. Sci. Rep. 2016;6:29530. doi: 10.1038/srep29530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang G., Zhao N., Berkhout B., Das A.T. CRISPR-Cas9 can inhibit HIV-1 replication but NHEJ repair facilitates virus escape. Mol. Ther. 2016;24:522–526. doi: 10.1038/mt.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang Z., Pan Q., Gendron P., Zhu W., Guo F., Cen S., Wainberg M.A., Liang C. CRISPR/Cas9-derived mutations both inhibit HIV-1 replication and accelerate viral escape. Cell Rep. 2016;15:481–489. doi: 10.1016/j.celrep.2016.03.042. [DOI] [PubMed] [Google Scholar]
- 116.Lebbink R.J., de Jong D.C., Wolters F., Kruse E.M., van Ham P.M., Wiertz E.J., Nijhuis M. A combinational CRISPR/Cas9 gene-editing approach can halt HIV replication and prevent viral escape. Sci. Rep. 2017;7:1–10. doi: 10.1038/srep41968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Anders C., Bargsten K., Jinek M. Structural plasticity of PAM recognition by engineered variants of the RNA-guided endonuclease Cas9. Mol. Cell. 2016;61:895–902. doi: 10.1016/j.molcel.2016.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xiong X., Chen M., Lim W.A., Zhao D., Qi L.S. CRISPR/Cas9 for human genome engineering and disease research. Annu. Rev. Genom. Hum. Genet. 2016;17:131–154. doi: 10.1146/annurev-genom-083115-022258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bayat H., Naderi F., Khan A.H., Memarnejadian A., Rahimpour A. The impact of CRISPR-Cas system on antiviral therapy. Adv. Pharm. Bull. 2018;8:591. doi: 10.15171/apb.2018.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tagaya Y., Gallo R.C. The exceptional oncogenicity of HTLV-1. Front. Microbiol. 2017;8:1425. doi: 10.3389/fmicb.2017.01425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Grassmann R., Aboud M., Jeang K.-T. Molecular mechanisms of cellular transformation by HTLV-1 Tax. Oncogene. 2005;24:5976–5985. doi: 10.1038/sj.onc.1208978. [DOI] [PubMed] [Google Scholar]
- 122.Arnold J., Zimmerman B., Li M., Lairmore M.D., Green P.L. Human T-cell leukemia virus type-1 antisense-encoded gene, Hbz, promotes T-lymphocyte proliferation. Blood J. Am. Soc. Hematol. 2008;112:3788–3797. doi: 10.1182/blood-2008-04-154286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ghezeldasht S.A., Shamsian S.A.A., Navashenaq J.G., Miri R., Ashrafi F., Mosavat A., Rezaee S.A. HTLV-1 oncovirus-host interactions: From entry to the manifestation of associated diseases. Rev. Med. Virol. 2021 doi: 10.1002/rmv.2235. [DOI] [PubMed] [Google Scholar]
- 124.Wang G., Zhao N., Berkhout B., Das A.T. CRISPR-Cas based antiviral strategies against HIV-1. Virus Res. 2018;244:321–332. doi: 10.1016/j.virusres.2017.07.020. [DOI] [PubMed] [Google Scholar]
- 125.Baker C., Chen Y., Sessions K., Howell A., Hayden M. CRISPR/Cas9 for HTLV-1: a novel therapeutic approach to the first discovered human retrovirus. J. Invest. Dermatol. 2019 ELSEVIER SCIENCE INC STE 800, 230 PARK AVE, NEW YORK, NY 10169 USA. [Google Scholar]
- 126.Lamontagne R.J., Bagga S., Bouchard M.J. Hepatitis B virus molecular biology and pathogenesis. Hepatoma Res. 2016;2:163–186. doi: 10.20517/2394-5079.2016.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McNaughton A.L., D’Arienzo V., Ansari M.A., Lumley S.F., Littlejohn M., Revill P., McKeating J.A., Matthews P.C. Insights from deep sequencing of the HBV Genome-Unique, Tiny, and Misunderstood. Gastroenterology. 2019;156:384–399. doi: 10.1053/j.gastro.2018.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Glebe D., König A. Molecular virology of hepatitis B virus and targets for antiviral intervention. Intervirology. 2014;57:134–140. doi: 10.1159/000360946. [DOI] [PubMed] [Google Scholar]
- 129.Lin C.L., Kao J.H. Hepatitis B virus genotypes and variants. Cold Spring Harb. Perspect. Med. 2015;5:a021436. doi: 10.1101/cshperspect.a021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kramvis A. Genotypes and genetic variability of hepatitis B virus. Intervirology. 2014;57:141–150. doi: 10.1159/000360947. [DOI] [PubMed] [Google Scholar]
- 131.Schweitzer A., Horn J., Mikolajczyk R.T., Krause G., Ott J.J. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet. 2015;386:1546–1555. doi: 10.1016/s0140-6736(15)61412-x. [DOI] [PubMed] [Google Scholar]
- 132.van Bömmel F., Berg T. Antiviral therapy of chronic hepatitis B. Intervirology. 2014;57:171–180. doi: 10.1159/000360945. [DOI] [PubMed] [Google Scholar]
- 133.Bang K.B., Kim H.J. Management of antiviral drug resistance in chronic hepatitis B. World J. Gastroenterol. WJG. 2014;20:11641. doi: 10.3748/wjg.v20.i33.11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Seeger C., Mason W.S. Molecular biology of hepatitis B virus infection. Virology. 2015;479–480:672–686. doi: 10.1016/j.virol.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fanning G.C., Zoulim F., Hou J., Bertoletti A. Therapeutic strategies for hepatitis B virus infection: towards a cure. Nat. Rev. Drug Discov. 2019;18:827–844. doi: 10.1038/s41573-019-0037-0. [DOI] [PubMed] [Google Scholar]
- 136.Kostyushev D., Brezgin S., Kostyusheva A., Zarifyan D., Goptar I., Chulanov V. Orthologous CRISPR/Cas9 systems for specific and efficient degradation of covalently closed circular DNA of hepatitis B virus. Cell. Mol. Life Sci. 2019;76:1779–1794. doi: 10.1007/s00018-019-03021-8. 10.1007/s00018-019-03021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Scott T., Moyo B., Nicholson S., Maepa M.B., Watashi K., Ely A., Weinberg M.S., Arbuthnot P. ssAAVs containing cassettes encoding SaCas9 and guides targeting hepatitis B virus inactivate replication of the virus in cultured cells. Sci. Rep. 2017;7:1–11. doi: 10.1038/s41598-017-07642-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liu Y., Zhao M., Gong M., Xu Y., Xie C., Deng H., Li X., Wu H., Wang Z. Inhibition of hepatitis B virus replication via HBV DNA cleavage by Cas9 from Staphylococcus aureus. Antivir. Res. 2018;152:58–67. doi: 10.1016/j.antiviral.2018.02.011. [DOI] [PubMed] [Google Scholar]
- 139.Chen L., Zhao J., Peng J., Li X., Deng X., Geng Z., Shen Z., Guo F., Zhang Q., Jin Y. Detection of SARS‐CoV‐2 in saliva and characterization of oral symptoms in COVID‐19 patients. Cell Prolif. 2020;53 doi: 10.1111/cpr.12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Song J., Zhang X., Ge Q., Yuan C., Chu L., Liang H.-f, Liao Z., Liu Q., Zhang Z., Zhang B. CRISPR/Cas9-mediated knockout of HBsAg inhibits proliferation and tumorigenicity of HBV-positive hepatocellular carcinoma cells. J. Cell. Biochem. 2018;119:8419–8431. doi: 10.1002/jcb.27050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mettenleiter T.C., Ehlers B., Müller T., Yoon K.-J., Teifke J.P. Herpesviruses. Dis. Swine. 2019:548–575. doi: 10.1002/9781119350927.ch35. [DOI] [Google Scholar]
- 142.Arvin A., Campadelli-Fiume G., Mocarski E., Moore P.S., Roizman B., Whitley R., Yamanishi K. Cambridge University Press; 2007. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. [PubMed] [Google Scholar]
- 143.Whitley R.J. In: Herpesviruses. In Medical Microbiology. Baron S., editor. University of Texas Medical Branch at Galveston Copyright © 1996, The University of Texas Medical Branch at Galveston; Galveston (TX: 1996. [Google Scholar]
- 144.Roizman B. Springer; US: 2013. The Herpesviruses. [Google Scholar]
- 145.Adams A., Lindahl T. Epstein-Barr virus genomes with properties of circular DNA molecules in carrier cells. Proc. Natl. Acad. Sci. USA. 1975;72:1477–1481. doi: 10.1073/pnas.72.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.P.E. Pellett B. Roizman Herpesviridae. Fields Virol. 2 2013 1802 1822.
- 147.Whitley R.J., Roizman B. Herpes simplex virus infections. Lancet. 2001;357:1513–1518. doi: 10.1016/S0140-6736(00)04638-9. [DOI] [PubMed] [Google Scholar]
- 148.Kennedy P.G., Steiner I. Recent issues in herpes simplex encephalitis. J. Neurovirol. 2013;19:346–350. doi: 10.1007/s13365-013-0178-6. [DOI] [PubMed] [Google Scholar]
- 149.Bolland S., Pierce S.K. Virology: Ups and downs in the search for a Herpes simplex virus vaccine. Elife. 2015;4 doi: 10.7554/eLife.06883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Awasthi S., Hook L.M., Pardi N., Wang F., Myles A., Cancro M.P., Cohen G.H., Weissman D., Friedman H.M. Nucleoside-modified mRNA encoding HSV-2 glycoproteins C, D, and E prevents clinical and subclinical genital herpes. Sci. Immunol. 2019:4. doi: 10.1126/sciimmunol.aaw7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Vadlapudi D., A., K, Vadlapatla R., Mitra K., A Update on emerging antivirals for the management of herpes simplex virus infections: a patenting perspective. Recent Pat. Anti Infect. Drug Discov. 2013;8:55–67. doi: 10.2174/1574891x11308010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Schaeffer H.J., Beauchamp L., de Miranda P., Elion G.B., Bauer D., Collins P. 9-(2-hydroxyethoxymethyl) guanine activity against viruses of the herpes group. Nature. 1978;272:583–585. doi: 10.1038/272583a0. [DOI] [PubMed] [Google Scholar]
- 153.Jiang Y.-C., Feng H., Lin Y.-C., Guo X.-R. New strategies against drug resistance to herpes simplex virus. Int. J. Oral. Sci. 2016;8:1–6. doi: 10.1038/ijos.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bi Y., Sun L., Gao D., Ding C., Li Z., Li Y., Cun W., Li Q. High-efficiency targeted editing of large viral genomes by RNA-guided nucleases. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Russell T.A., Stefanovic T., Tscharke D.C. Engineering herpes simplex viruses by infection-transfection methods including recombination site targeting by CRISPR/Cas9 nucleases. J. Virol. Methods. 2015;213:18–25. doi: 10.1016/j.jviromet.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 156.Xu X., Fan S., Zhou J., Zhang Y., Che Y., Cai H., Wang L., Guo L., Liu L., Li Q. The mutated tegument protein UL7 attenuates the virulence of herpes simplex virus 1 by reducing the modulation of α-4 gene transcription. Virol. J. 2016;13:152. doi: 10.1186/s12985-016-0600-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Karpov D.S., Karpov V.L., Klimova R.R., Demidova N.A., Kushch A.A. A plasmid-expressed CRISPR/Cas9 system suppresses replication of HSV type I in a vero cell culture. Mol. Biol. 2019;53:70–78. doi: 10.1134/S0026893319010059. [DOI] [PubMed] [Google Scholar]
- 158.Khodadad N., Fani M., Jamehdor S., Nahidsamiei R., Makvandi M., Kaboli S., Teimoori A., Thekkiniath J. A knockdown of the herpes simplex virus type-1 gene in all-in-one CRISPR vectors. Folia Histochem. Cytobiol. 2020;58:174–181. doi: 10.5603/FHC.a2020.0020. [DOI] [PubMed] [Google Scholar]
- 159.Liu L., Cheng J., Mou T., Zhang Y., Xu X., Zhang J., Li X., Feng X., Xu X., Liao Y., Fan S., et al. The mutation of the genes related to neurovirulence in HSV-2 produces an attenuated phenotype in mice. Viruses. 2020;12:770. doi: 10.3390/v12070770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Oh H.S., Neuhausser W.M., Eggan P., Angelova M., Kirchner R., Eggan K.C., Knipe D.M. Herpesviral lytic gene functions render the viral genome susceptible to novel editing by CRISPR/Cas9. Elife. 2019;8 doi: 10.7554/eLife.51662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Horlbeck M.A., Witkowsky L.B., Guglielmi B., Replogle J.M., Gilbert L.A., Villalta J.E., Torigoe S.E., Tjian R., Weissman J.S. Nucleosomes impede Cas9 access to DNA in vivo and in vitro. Elife. 2016;5 doi: 10.7554/eLife.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.van Diemen F.R., Kruse E.M., Hooykaas M.J., Bruggeling C.E., Schürch A.C., van Ham P.M., Imhof S.M., Nijhuis M., Wiertz E.J., Lebbink R.J. CRISPR/Cas9-mediated genome editing of herpesviruses limits productive and latent infections. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Roehm P.C., Shekarabi M., Wollebo H.S., Bellizzi A., He L., Salkind J., Khalili K. Inhibition of HSV-1 replication by gene editing strategy. Sci. Rep. 2016;6:1–11. doi: 10.1038/srep23146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ferenczy M.W., DeLuca N.A. Reversal of heterochromatic silencing of quiescent herpes simplex virus type 1 by ICP0. J. Virol. 2011;85:3424–3435. doi: 10.1128/JVI.02263-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang Q.-Y., Zhou C., Johnson K.E., Colgrove R.C., Coen D.M., Knipe D.M. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc. Natl. Acad. Sci. 2005;102:16055–16059. doi: 10.1073/pnas.0505850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cliffe A.R., Garber D.A., Knipe D.M. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J. Virol. 2009;83:8182–8190. doi: 10.1128/JVI.00712-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yin D., Ling S., Wang D., Dai Y., Jiang H., Zhou X., Paludan S.R., Hong J., Cai Y. Targeting herpes simplex virus with CRISPR–Cas9 cures herpetic stromal keratitis in mice. Nat. Biotechnol. 2021;39:567–577. doi: 10.1038/s41587-020-00781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.E. Mocarski T. Shenk R. Pass Fields virology. Knipe DM 2007 1960 2014.
- 171.Kirby T. Congenital cytomegalovirus-a neglected health problem. Lancet Infect. Dis. 2016;16:900–901. doi: 10.1016/s1473-3099(16)30226-2. [DOI] [PubMed] [Google Scholar]
- 172.Ahmed A. Antiviral treatment of cytomegalovirus infection. Infect. Disord. Drug Targets. 2011;11:475–503. doi: 10.2174/187152611797636640. [DOI] [PubMed] [Google Scholar]
- 173.King M.W., Munger J. Editing the human cytomegalovirus genome with the CRISPR/Cas9 system. Virology. 2019;529:186–194. doi: 10.1016/j.virol.2019.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Xiao J., Deng J., Zhang Q., Ma P., Lv L., Zhang Y., Li C., Zhang Y. Targeting human cytomegalovirus IE genes by CRISPR/Cas9 nuclease effectively inhibits viral replication and reactivation. Arch. Virol. 2020;165:1827–1835. doi: 10.1007/s00705-020-04687-3. 10.1007/s00705-020-04687-3. [DOI] [PubMed] [Google Scholar]
- 175.Tommasino M. The human papillomavirus family and its role in carcinogenesis. Semin Cancer Biol. 2014;26:13–21. doi: 10.1016/j.semcancer.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 176.Tan S.C., Ismail M.P., Duski D.R., Othman N.H., Ankathil R. Prevalence and type distribution of human papillomavirus (HPV) in Malaysian women with and without cervical cancer: an updated estimate. Biosci. Rep. 2018;38 doi: 10.1042/bsr20171268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Baghi H.B., Yousefi B., Oskouee M.A., Aghazadeh M. HPV vaccinations: a Middle Eastern and north African dilemma. Lancet Infect. Dis. 2017;17:18–19. doi: 10.1016/s1473-3099(16)30553-9. [DOI] [PubMed] [Google Scholar]
- 178.Moghoofei M., Keshavarz M., Ghorbani S., Babaei F., Nahand J.S., Tavakoli A., Mortazavi H.S., Marjani A., Mostafaei S., Monavari S.H. Association between human papillomavirus infection and prostate cancer: a global systematic review and meta-analysis. Asia Pac. J. Clin. Oncol. 2019;15:e59–e67. doi: 10.1111/ajco.13124. [DOI] [PubMed] [Google Scholar]
- 179.Aghamiri S., talaei S., Roshanzamiri S., Zandsalimi F., Fazeli E., Aliyu M., Kheiry Avarvand O., Ebrahimi Z., Keshavarz-Fathi M., Ghanbarian H. Delivery of genome editing tools: a promising strategy for HPV-related cervical malignancy therapy. Expert Opin. Drug Deliv. 2020;17:753–766. doi: 10.1080/17425247.2020.1747429. [DOI] [PubMed] [Google Scholar]
- 180.Gao X., Jin Z., Tan X., Zhang C., Zou C., Zhang W., Ding J., Das B.C., Severinov K., Hitzeroth I.I., Debata P.R., et al. Hyperbranched poly(β-amino ester) based polyplex nanopaticles for delivery of CRISPR/Cas9 system and treatment of HPV infection associated cervical cancer. J. Control. Release. 2020;321:654–668. doi: 10.1016/j.jconrel.2020.02.045. [DOI] [PubMed] [Google Scholar]
- 181.Pirouzfar M., Amiri F., Dianatpour M., Takhshid M.A. CRISPR/Cas9-mediated knockout of MLL5 enhances apoptotic effect of cisplatin in HeLa cells in vitro. EXCLI J. 2020;19:170–182. doi: 10.17179/excli2019-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Moody C.A., Laimins L.A. Human papillomavirus oncoproteins: pathways to transformation. Nat. Rev. Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 183.Gao C., Wu P., Yu L., Liu L., Liu H., Tan X., Wang L., Huang X., Wang H. The application of CRISPR/Cas9 system in cervical carcinogenesis. Cancer Gene Ther. 2021:1–9. doi: 10.1038/s41417-021-00366-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zhen S., Lu J.-J., Wang L.-J., Sun X.-M., Zhang J.-Q., Li X., Luo W.-J., Zhao L. In vitro and in vivo synergistic therapeutic effect of cisplatin with human papillomavirus16 E6/E7 CRISPR/Cas9 on cervical cancer cell line. Transl. Oncol. 2016;9:498–504. doi: 10.1016/j.tranon.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhen S., Li X. Oncogenic human papillomavirus: application of CRISPR/Cas9 therapeutic strategies for cervical cancer. Cell. Physiol. Biochem. 2017;44:2455–2466. doi: 10.1159/000486168. [DOI] [PubMed] [Google Scholar]
- 186.Chang Y., Moore P.S. Merkel cell carcinoma: a virus-induced human cancer. Annu Rev. Pathol. 2012;7:123–144. doi: 10.1146/annurev-pathol-011110-130227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Temblador A., Topalis D., Andrei G., Snoeck R. CRISPR/Cas9 editing of the polyomavirus tumor antigens inhibits merkel cell carcinoma growth in vitro. Cancers. 2019 doi: 10.3390/cancers11091260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Rui Y., Wilson D.R., Green J.J. Non-viral delivery to enable genome editing. Trends Biotechnol. 2019;37:281–293. doi: 10.1016/j.tibtech.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yin H., Xue W., Anderson D.G. CRISPR-Cas: a tool for cancer research and therapeutics. Nat. Rev. Clin. Oncol. 2019;16:281–295. doi: 10.1038/s41571-019-0166-8. [DOI] [PubMed] [Google Scholar]
- 190.Aghbash P.S., Eslami N., Shamekh A., Entezari-Maleki T., Baghi H.B. SARS-CoV-2 infection: the role of PD-1/PD-L1 and CTLA-4 axis. Life Sci. 2021;270 doi: 10.1016/j.lfs.2021.119124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Donyavi T., Bokharaei-Salim F., Baghi H.B., Khanaliha K., Alaei Janat-Makan M., Karimi B., Sadri Nahand J., Mirzaei H., Khatami A., Garshasbi S., Khoshmirsafa M., et al. Acute and post-acute phase of COVID-19: analyzing expression patterns of miRNA-29a-3p, 146a-3p, 155-5p, and let-7b-3p in PBMC. Int. Immunopharmacol. 2021;97 doi: 10.1016/j.intimp.2021.107641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Safari F., Afarid M., Rastegari B., Borhani-Haghighi A., Barekati-Mowahed M., Behzad-Behbahani A. CRISPR systems: novel approaches for detection and combating COVID-19. Virus Res. 2021;294 doi: 10.1016/j.virusres.2020.198282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Aghamiri S., Talaei S., Ghavidel A.A., Zandsalimi F., Masoumi S., Hafshejani N.H., Jajarmi V. Nanoparticles-mediated CRISPR/Cas9 delivery: Recent advances in cancer treatment. J. Drug Deliv. Sci. Technol. 2020;56 doi: 10.1016/j.jddst.2020.101533. [DOI] [Google Scholar]
- 194.Lundstrom K. Viral vectors in gene therapy. Diseases. 2018;6:42. doi: 10.3390/diseases6020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Johnson N.M., Alvarado A.F., Moffatt T.N., Edavettal J.M., Swaminathan T.A., Braun S.E. HIV-based lentiviral vectors: origin and sequence differences. Mol. Ther. Methods Clin. Dev. 2021;21:451–465. doi: 10.1016/j.omtm.2021.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Munis A.M. Gene therapy applications of non-human lentiviral vectors. Viruses. 2020:12. doi: 10.3390/v12101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zorzan M., Del Vecchio C., Vogiatzis S., Saccon E., Parolin C., Palù G., Calistri A., Mucignat-Caretta C. Targeting the regulatory subunit R2Alpha of protein kinase a in human glioblastoma through shRNA-expressing lentiviral vectors. Viruses. 2021;13 doi: 10.3390/v13071361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Tiscornia G., Singer O., Verma I.M. Production and purification of lentiviral vectors. Nat. Protoc. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- 199.Gándara C., Affleck V., Stoll E.A. Manufacture of third-generation lentivirus for preclinical use, with process development considerations for translation to good manufacturing practice. Hum. Gene Ther. Methods. 2018;29:1–15. doi: 10.1089/hgtb.2017.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lyu P., Wang L., Lu B. Virus-like particle mediated CRISPR/Cas9 delivery for efficient and safe genome editing. Life. 2020:10. doi: 10.3390/life10120366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ling S., Yang S., Hu X., Yin D., Dai Y., Qian X., Wang D., Pan X., Hong J., Sun X., Yang H., et al. Lentiviral delivery of co-packaged Cas9 mRNA and a Vegfa-targeting guide RNA prevents wet age-related macular degeneration in mice. Nat. Biomed. Eng. 2021;5:144–156. doi: 10.1038/s41551-020-00656-y. 10.1038/s41551-020-00656-y. [DOI] [PubMed] [Google Scholar]
- 202.Zhu D., Schieferecke A.J., Lopez P.A., Schaffer D.V. Adeno-associated virus vector for central nervous system gene therapy. Trends Mol. Med. 2021;27:524–537. doi: 10.1016/j.molmed.2021.03.010. [DOI] [PubMed] [Google Scholar]
- 203.Bulcha J.T., Wang Y., Ma H., Tai P.W.L., Gao G. Viral vector platforms within the gene therapy landscape. Signal Transduct. Target. Ther. 2021;6:53. doi: 10.1038/s41392-021-00487-6. 10.1038/s41392-021-00487-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Gupta V., Lourenço S.P., Hidalgo I.J. Development of gene therapy vectors: remaining challenges. J. Pharm. Sci. 2021;110:1915–1920. doi: 10.1016/j.xphs.2020.11.035. [DOI] [PubMed] [Google Scholar]
- 205.Yan K., Feng J., Liu X., Wang H., Li Q., Li J., Xu T., Sajid M., Ullah H., Zhou L., Zhou L., et al. Inhibition of hepatitis B virus by AAV8-derived CRISPR/SaCas9 expressed from liver-specific promoters. Front. Microbiol. 2021;12:956. doi: 10.3389/fmicb.2021.665184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Liu J.-J., Orlova N., Oakes B.L., Ma E., Spinner H.B., Baney K.L.M., Chuck J., Tan D., Knott G.J., Harrington L.B., Al-Shayeb B., et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature. 2019;566:218–223. doi: 10.1038/s41586-019-0908-x. 10.1038/s41586-019-0908-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Pausch, P., Al-Shayeb, B., Bisom-Rapp, E., Tsuchida, C.A., Li, Z., Cress, B.F., Knott, G.J., Jacobsen, S.E., Banfield, J.F., and Doudna, J.A. (2020). CRISPR-CasΦ from huge phages is a hypercompact genome editor. 369, 333–337. 10.1126/science.abb1400%J Science. [DOI] [PMC free article] [PubMed]
- 208.Gallardo J., Pérez-Illana M., Martín-González N., San Martín C. Adenovirus structure: what is new? Int. J. Mol. Sci. 2021;22:5240. doi: 10.3390/ijms22105240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Humphreys I.R., Sebastian S. Novel viral vectors in infectious diseases. Immunology. 2018;153:1–9. doi: 10.1111/imm.12829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Sasso E., D’Alise A.M., Zambrano N., Scarselli E., Folgori A., Nicosia A. New viral vectors for infectious diseases and cancer. Semin. Immunol. 2020;50 doi: 10.1016/j.smim.2020.101430. [DOI] [PubMed] [Google Scholar]
- 211.Schiwon M., Ehrke-Schulz E., Oswald A., Bergmann T., Michler T., Protzer U., Ehrhardt A. One-vector system for multiplexed CRISPR/Cas9 against hepatitis B virus cccDNA utilizing high-capacity adenoviral vectors. molecular therapy. Nucleic Acids. 2018;12:242–253. doi: 10.1016/j.omtn.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Vannucci L., Lai M., Chiuppesi F., Ceccherini-Nelli L., Pistello M. Viral vectors: a look back and ahead on gene transfer technology. New Microbiol. 2013;36:1–22. [PubMed] [Google Scholar]
- 213.Caffery B., Lee J.S., Alexander-Bryant A.A. Vectors for glioblastoma gene therapy: viral & non-viral delivery strategies. Nanomaterials. 2019:9. doi: 10.3390/nano9010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Okoli A., Okeke M.I., Tryland M., Moens U. CRISPR/Cas9—advancing orthopoxvirus genome editing for vaccine and vector development. Viruses. 2018;10:50. doi: 10.3390/v10010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sharon D.M., Nesdoly S., Yang H.J., Gélinas J.-F., Xia Y., Ansorge S., Kamen A.A. A pooled genome-wide screening strategy to identify and rank influenza host restriction factors in cell-based vaccine production platforms. Sci. Rep. 2020;10:1–15. doi: 10.1038/s41598-020-68934-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Xu A., Qin C., Lang Y., Wang M., Lin M., Li C., Zhang R., Tang J. A simple and rapid approach to manipulate pseudorabies virus genome by CRISPR/Cas9 system. Biotechnol. Lett. 2015;37:1265–1272. doi: 10.1007/s10529-015-1796-2. [DOI] [PubMed] [Google Scholar]
- 217.Liang X., Sun L., Yu T., Pan Y., Wang D., Hu X., Fu Z., He Q., Cao G. A CRISPR/Cas9 and Cre/Lox system-based express vaccine development strategy against re-emerging Pseudorabies virus. Sci. Rep. 2016;6:1–10. doi: 10.1038/srep19176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Teng M., Yao Y., Nair V., Luo J. Latest advances of virology research using CRISPR/Cas9-based gene-editing technology and its application to vaccine development. Viruses. 2021;13:779. doi: 10.3390/v13050779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Tang Y.-D., Liu J.-T., Wang T.-Y., An T.-Q., Sun M.-X., Wang S.-J., Fang Q.-Q., Hou L.-l, Tian Z.-J., Cai X.-H. Live attenuated pseudorabies virus developed using the CRISPR/Cas9 system. Virus Res. 2016;225:33–39. doi: 10.1016/j.virusres.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 220.Zhao Y., Wang L.-Q., Zheng H.-H., Yang Y.-R., Liu F., Zheng L.-L., Jin Y., Chen H.-Y. Construction and immunogenicity of a gE/gI/TK-deleted PRV based on porcine pseudorabies virus variant. Mol. Cell. Probes. 2020;53 doi: 10.1016/j.mcp.2020.101605. [DOI] [PubMed] [Google Scholar]
- 221.Borca M.V., Holinka L.G., Berggren K.A., Gladue D.P. CRISPR-Cas9, a tool to efficiently increase the development of recombinant African swine fever viruses. Sci. Rep. 2018;8:1–6. doi: 10.1038/s41598-018-21575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Yao Y., Bassett A., Nair V. Targeted editing of avian herpesvirus vaccine vector using CRISPR/Cas9 nucleases. J. Vaccin. Technol. 2016;1:1–7. [Google Scholar]
- 223.Tang N., Zhang Y., Sadigh Y., Moffat K., Shen Z., Nair V., Yao Y. Generation of a triple insert live avian herpesvirus vectored vaccine using crispr/cas9-based gene editing. Vaccines. 2020;8:97. doi: 10.3390/vaccines8010097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Tang N., Zhang Y., Pedrera M., Chang P., Baigent S., Moffat K., Shen Z., Nair V., Yao Y. A simple and rapid approach to develop recombinant avian herpesvirus vectored vaccines using CRISPR/Cas9 system. Vaccine. 2018;36:716–722. doi: 10.1016/j.vaccine.2017.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Chang P., Ameen F., Sealy J.E., Sadeyen J.-R., Bhat S., Li Y., Iqbal M. Application of hdr-crispr/cas9 and erythrocyte binding for rapid generation of recombinant turkey herpesvirus-vectored avian influenza virus vaccines. Vaccines. 2019;7:192. doi: 10.3390/vaccines7040192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Liu L., Wang T., Wang M., Tong Q., Sun Y., Pu J., Sun H., Liu J. Recombinant turkey herpesvirus expressing H9 hemagglutinin providing protection against H9N2 avian influenza. Virology. 2019;529:7–15. doi: 10.1016/j.virol.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 227.Zou Z., Huang K., Wei Y., Chen H., Liu Z., Jin M. Construction of a highly efficient CRISPR/Cas9-mediated duck enteritis virus-based vaccine against H5N1 avian influenza virus and duck Tembusu virus infection. Sci. Rep. 2017;7:1–12. doi: 10.1038/s41598-017-01554-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Atasoy M.O., Rohaim M.A., Munir M. Simultaneous deletion of virulence factors and insertion of antigens into the infectious laryngotracheitis virus using NHEJ-CRISPR/Cas9 and cre–lox system for construction of a stable vaccine vector. Vaccines. 2019;7:207. doi: 10.3390/vaccines7040207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Chang P., Yao Y., Tang N., Sadeyen J.-R., Sealy J., Clements A., Bhat S., Munir M., Bryant J.E., Iqbal M. The application of NHEJ-CRISPR/Cas9 and Cre-Lox system in the generation of bivalent duck enteritis virus vaccine against avian influenza virus. Viruses. 2018;10:81. doi: 10.3390/v10020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Gong Y., Chen T., Feng N., Meng X., Sun W., Wang T., Zhao Y., Yang S., Song X., Li W. A highly efficient recombinant canarypox virus-based vaccine against canine distemper virus constructed using the CRISPR/Cas9 gene editing method. Vet. Microbiol. 2020;251 doi: 10.1016/j.vetmic.2020.108920. [DOI] [PubMed] [Google Scholar]
- 231.Ran F.A., Hsu P.D., Lin C.-Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., Van Der Oost J., Regev A. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Kotterman M.A., Schaffer D.V. Engineering adeno-associated viruses for clinical gene therapy. Nat. Rev. Genet. 2014;15:445–451. doi: 10.1038/nrg3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Kaminski R., Bella R., Yin C., Otte J., Ferrante P., Gendelman H.E., Li H., Booze R., Gordon J., Hu W. Excision of HIV-1 DNA by gene editing: a proof-of-concept in vivo study. Gene Ther. 2016;23:690–695. doi: 10.1038/gt.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Yin C., Zhang T., Qu X., Zhang Y., Putatunda R., Xiao X., Li F., Xiao W., Zhao H., Dai S. In vivo excision of HIV-1 provirus by saCas9 and multiplex single-guide RNAs in animal models. Mol. Ther. 2017;25:1168–1186. doi: 10.1016/j.ymthe.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Khalili K., Kaminski R., Gordon J., Cosentino L., Hu W. Genome editing strategies: potential tools for eradicating HIV-1/AIDS. J. Neurovirol. 2015;21:310–321. doi: 10.1007/s13365-014-0308-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Li L., Hu S., Chen X. Non-viral delivery systems for CRISPR/Cas9-based genome editing: challenges and opportunities. Biomaterials. 2018;171:207–218. doi: 10.1016/j.biomaterials.2018.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Jiang C., Mei M., Li B., Zhu X., Zu W., Tian Y., Wang Q., Guo Y., Dong Y., Tan X. A non-viral CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA and pcsk9 in vivo. Cell Res. 2017;27:440–443. doi: 10.1038/cr.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.DeWitt M.A., Corn J.E., Carroll D. Genome editing via delivery of Cas9 ribonucleoprotein. Methods. 2017;121–122:9–15. doi: 10.1016/j.ymeth.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Konermann S., Lotfy P., Brideau N.J., Oki J., Shokhirev M.N., Hsu P.D. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173:665–676. doi: 10.1016/j.cell.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. J. Sci. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P.W., Li Z., Peterson R.T., Yeh J.-R.J., Aryee M.J., et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Lin Y., Cradick T.J., Brown M.T., Deshmukh H., Ranjan P., Sarode N., Wile B.M., Vertino P.M., Stewart F.J., Bao G. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42:7473–7485. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Ryan D.E., Taussig D., Steinfeld I., Phadnis S.M., Lunstad B.D., Singh M., Vuong X., Okochi K.D., McCaffrey R., Olesiak M., Roy S., et al. Improving CRISPR-Cas specificity with chemical modifications in single-guide RNAs. Nucleic Acids Res. 2018;46:792–803. doi: 10.1093/nar/gkx1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Shin J., Jiang F., Liu J.-J., Bray N.L., Rauch B.J., Baik S.H., Nogales E., Bondy-Denomy J., Corn J.E., Doudna J.A. Disabling Cas9 by an anti-CRISPR DNA mimic. J. Sci. Adv. 2017;3 doi: 10.1126/sciadv.1701620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Not applicable.