Summary
Robust T cell responses have been associated with milder outcomes in many infections. T cells also establish long-term memory pools and, as they are predominantly directed toward epitopes encompassing conserved peptides, can respond to SARS-CoV-2 variants, including Omicron. Here, we discuss epitope-specific CD8+ and CD4+ T cell responses toward SARS-CoV-2 infection and vaccination, their subsequent persistence into long-term memory, and ongoing work to determine their role in limiting disease severity.
Kedzierska and Thomas review CD8+ and CD4+ T cell responses following SARS-CoV-2 infection and vaccination. They discuss epitope-specific T cell responses during acute disease, persistence into long-term memory, and cross-reactivity toward variants of concern.
Introduction
Two years since SARS-CoV-2 emerged in China, the virus has spread rapidly, causing the coronavirus disease 2019 (COVID-19) pandemic with >430 million infections and >5.9 million deaths. Infection with SARS-CoV-2 results in a spectrum of clinical presentations, ranging from asymptomatic to mild, severe, and fatal disease. Severe and fatal disease outcomes are predominantly associated with risk factors including age and pre-existing comorbidities. Last year the world transitioned into a new stage of the pandemic, in which COVID-19 vaccines became available and efforts have been focused on immunization of the global population. Progress has been made in terms of understanding immune responses to SARS-CoV-2 infection and COVID-19 vaccination. It is well-established that robust and broad immune responses precede patients’ recovery,1, 2, 3 while SARS-CoV-2-specific T cell and B cell responses generate long-lasting memory pools capable of recall following infection or vaccination. While antibodies produced by B cells, especially high titer neutralizing antibodies, can generate sterilizing immunity and prevent SARS-CoV-2 infection,4 it is hypothesized that T cells can limit disease severity, reduce its duration, and drive rapid recovery. While some have reported correlations between T cell responses and disease severity, these studies are limited compared to the extensive work demonstrating a protective role for antibodies. Indeed, a rigorous correlate of protection against severe disease based on T cell responses has not been demonstrated for SARS-CoV-2. The ongoing emergence of new variants with greater capacity for antibody escape highlights the need for an expanded understanding of protective roles played by T cells, tools to assess them in the broader population, and identification of those who may have poor T cell memory. Memory T cells may be of key importance when antibody levels wane or new variants of concern emerge that escape antibody responses. As T cells are generally directed at epitopes encompassing conserved viral regions, they can recognize emerging variants. We discuss T cell responses toward the pandemic SARS-CoV-2 infection and vaccination, focusing on conventional epitope-specific cytotoxic CD8+ T cells, helper CD4+ T cells, and T follicular helper (Tfh) cells during the acute phase and discuss their persistence into long-term memory.
T cell responses during acute SARS-CoV-2 infection
The importance of T cells in limiting disease severity and duration and driving recovery from SARS-CoV-2 infection has been shown in animal and human studies. Studies in non-human primates demonstrated that transfer of T cells offers a substantial level of protection from SARS-CoV-2 infection and leads to less severe disease outcome.5 T cell protection from coronaviruses and SARS-CoV-2 in mice has also been reported.6,7 In humans, early induction of interferon (IFN)-γ-secreting SARS-CoV-2-specific T cells was linked to mild disease in COVID-19 patients.8 Similarly, prominent CD8+ T cell responses directed against an immunodominant human leukocyte antigen (HLA)-B∗07:02-restricted N105-113 epitope (B7/N105) correlated with milder outcomes.9 The evidence for T cells driving recovery from COVID-19 also comes from immunosuppressed patients who lacked B cells/antibodies but recovered after eliciting T cell immunity.10 However, it is important to note that patients lacking B cells have higher mortality rates following SARS-CoV-2 infection.11
SARS-CoV2-specific T cells are elicited during acute COVID-19. ICOS+PD-1+CD4+ Tfhs, activated CD38+HLA-DR+CD4+ T cells, and activated CD38+HLA-DR+CD8+ T cells emerge transiently in patients’ blood prior to recovery, suggesting involvement of T cells in resolution of COVID-19.3 Tfh cells are a specialized subset of CD4+ T cells playing key roles in the generation of antibodies and their affinity maturation and development of memory B cells and plasma cells. Moreover, activation of CXCR3+ circulating (c)Tfh1 cells in acute COVID-19 correlates with and predicts antibody levels at convalescence and neutralization titers in acute disease.2 In contrast, high levels (∼50%–90% in some ICU patients) of hyperactivated CD4+ and CD8+ T cells characterized by high and prolonged expression of CD38/HLA-DR correlate with COVID-19 severity.2
To define SARS-CoV-2-specific T cells, early studies used SARS-CoV-2 peptide “megapools” predicted to bind to common HLA-I and HLA-II alleles12 or overlapping peptides13 to stimulate peripheral blood mononuclear cells (PBMCs) from COVID-19 patients. Using CD69+CD137+ activation markers for CD8+ T cells and OX40+CD137+ for CD4+ T cells in an activation induced marker (AIM) assay, SARS-CoV2-reactive T cells directed toward peptides derived from S, M, N, ORFs were found in 70%–100% of COVID-19 patients in acute COVID-1914 and at convalescence.12 Furthermore, in vitro stimulation of PBMCs with SARS-CoV-2 overlapping peptides also led to clonal expansion of SARS-CoV-2-specific CD8+ and CD4+ T cells from COVID-19 patients and IFN-γ/TNF-α production, although CD4+ sets were generally numerically more prominent.15
SARS-CoV-2-reactive T cells were also detected by AIM assay in >20% of unexposed individuals,12, 13, 14 suggesting some level of cross-reactivity with T cells elicited by previously circulating common cold coronaviruses HCoV-OC43, HCoV-229E, HCoV-NL63, or HCoV-HKU116 or other pathogens. Cross-reactive T cells were mainly of a memory phenotype. Other studies have also provided evidence that SARS-CoV-2-exposed, but PCR-negative individuals had more prominent T cell responses in comparison to unexposed individuals,17, 18, 19 suggesting that pre-existing T cell responses are associated with protection from detectable SARS-CoV-2 infection.18,20 However, as high levels of cross-reactivity have not been observed by other assays (ELISpot, ICS, tetramers), this may suggest decreased specificity of the AIM assay relative to other assays. Furthermore, staining CD8+ T cells from pre-pandemic PBMCs with SARS-CoV-2-specific tetramers directly ex vivo across prominent epitopes with potential for cross-reactivity (B7/N105, B7/N257, A2/S269, and A24/S1208) identified a naive rather than memory phenotype of pre-existing precursor CD8+ T cell pools in children, adults, and elderly.9,15,21
Subsequent studies identified SARS-CoV-2-specific CD8+ T cell epitopes (peptides plus restricting Major Histocompatibility Complex [MHC]) using peptide-MHC-I tetramer binding.9,15,17,22 Identification of SARS-CoV-2 CD8+ T cell epitopes restricted by common human HLAs (e.g., A1/ORF1a1637, A2/S269, A3/N361, A24/S1208, B7/N105, and B40/N322) led to insights into CD8+ T cell origins, magnitude, phenotype, and immunodominance hierarchies directly ex vivo.21 The magnitude of SARS-CoV-2-specific CD8+ T cells ex vivo during acute COVID-19 or early convalescence varies broadly, across a range of 10−5 to as high as 10−1 in some individuals for certain epitopes.23 CD8+ T cells directed at the HLA-B∗07:02-restricted N105-113 (B7/N105+CD8+ T cells) are among the more immunodominant SARS-CoV-2 CD8+ T cell responses identified to date.9,21,22 The immunodominance of B7/N105+CD8+ T cells over other CD8+ T cell sets, including B7/N257+CD8+, A2/S269+CD8+, and A24/S1208+CD8+ T cells, was underpinned by high naive precursor frequencies in pre-pandemic children, adult, and elderly samples. Such high numbers of B7/N105+CD8+ naive precursors stemmed from highly diverse TCRαβ repertoire arisen from plasticity in TCRα-TCRβ pairing and lack of common TRAV, TRAJ, TRBV, or TRBJ gene segments.21 Importantly, robust B7/N105+CD8+ T cell responses characterized by expanded TCR clones with high functional avidity and anti-viral effector functions were found in patients with mild COVID-19 disease outcomes, while patients who recovered from severe disease had weaker B7/N105+CD8+ T cell responses,9 suggesting that B7/N105+CD8+ T cells contribute to SARS-CoV-2 control. Prominent B7/N105+CD8+ T cell populations responded to the ancestral strain and variants of concern, including the Delta strain. These findings support previous studies in animals and humans that TCR repertoire diversity plays a key role in selection of high-avidity CD8+ T cell responses important for protection from viral infections against both the wild-type virus and viral variants.
Conversely, CD8+ T cells directed at the HLA-A∗02:01-restricted S269-277 epitope (A2/S269), although immunodominant across known SARS-CoV-2-specific HLA-A∗02:01-restricted epitopes, are numerically subdominant when compared to B7/N105+CD8+ T cells.15,21 This stems from a biased TCR repertoire displaying common TRBV gene segments within A2/S269+CD8+ T cells (TRBV2/TRBV7-9/TRBV20-1), TRBJ (TRBJ2-2/TRBJ2-7), TRAV (TRAV12-1/TRAV12-2/TRAV14/DV4), and TRAJ (TRAJ43/TRAJ30). The molecular basis underlying biased TRAV12-mediated recognition of A2/S269 was defined by solving a ternary structure of TRAV12+ TCR complexed with A2/S269,24 which found that the TRAV12+ TCR docked atop HLA-A∗02:01, with both TRAV12 germline-encoded residues and amino acids derived from conserved CDR3α and CDR3β motifs playing a key role in A2/S269 recognition.
While SARS-CoV-2-specific CD8+ T cell epitopes restricted by predominant HLA-I are known, CD4+ T cell epitopes have been understudied, with fewer epitopes identified, in part due to the limitations in reagents for class-II multimers. As a result, rather than strictly mapped epitopes, reactive peptides have been identified that are strongly responsive across multiple individuals, including S816-830.25 Some of these regions may represent multiple overlapping epitopes. HLA-DRA-DRB1∗15:01S870-878 and HLA-DPA∗0103-DPB1∗04S167-180 are the most dominant SARS-CoV-2-specific CD4+ T cell epitopes described so far with known HLA restrictions in COVID-19 patients and vaccinees.26,27 S870-878 and S167-180 were identified using variations of “reverse epitope discovery,” wherein T cell receptors of interest are first isolated and then used to map the identity of specific epitopes. Overall, there is now enough evidence to show that T cells play a role in limiting COVID-19 severity and recovery from SARS-CoV-2 infection, especially in immunosuppressed individuals lacking B cells/antibodies. While COVID-19 patients with solid cancers had similar T cell characteristics to those of non-cancer patients, patients with hematologic malignancies displayed perturbed CD4+ T cells, but comparable highly activated CD38+HLA-DR+CD8+ T cell responses, correlating with IFN-γ in patients who survived, but not individuals with fatal disease outcomes.10
T cell responses toward COVID-19 vaccines
In 2020, global efforts focused on designing, developing, manufacturing, and evaluating COVID-19 vaccines. Clinical trials of first vaccines utilizing SARS-CoV-2 Spike by mRNA lipoparticles (Pfizer BNT162b2; Moderna VRC mRNA) or viral vectored vaccines (CanSino AdV5 COVID-19; Oxford/AstraZeneca ChAdOx) showed promising results for SARS-CoV-2-specific CD4+ and CD8+ T cells detected by IFN-γ secretion and ELISpot assays.28,29 Comparing to baseline, a ∼10-fold increase of IFN-γ-secreting T cells was found after vaccination,29 which was at levels of IFN-γ-producing T cells detected in COVID-19 patients. IFN-γ+CD4+ T cells numerically dominated over IFN-γ+CD8+ T cells after COVID-19 immunization, similar to what was reported for SARS-CoV-2 infection15 and non-human primates following COVID-19 vaccination (reviewed in Krammer, 202030).
Experiments using peptide-HLA multimers to detect SARS-CoV-2-specific CD4+ and CD8+ T cells provided exciting data that BNT162b2 mRNA and vectored ChAdOx COVID-19 vaccines elicit substantial CD4+ and CD8+ T cells, especially after the second dose. Indeed, BNT162b2 mRNA COVID-19 vaccines can induce prominent tetramer-specific CD8+ T cells toward Spike-derived epitopes.31,32 Combining 18 DNA-barcoded MHC-I tetramers for HLA-A∗01:01, HLA-A02∗01, HLA-A∗24:02, HLA-B∗15:01, and HLA-B∗40:02-derived epitopes, with scTCR-seq and scRNA-seq, tetramer-specific CD8+ T cell responses in COVID-19 patients and vaccinees were compared. CD8+ T cells directed toward Spike-derived epitopes had comparable magnitudes, phenotypes, and TCRαβ diversity after infection and immunization, demonstrating the robustness of SARS-CoV-2-specific CD8+ T cell pools induced by COVID-19 mRNA immunization. This is in contrast to vaccination with inactivated influenza vaccines, which do not elicit epitope-specific CD8+ T cell immunity.33 Although phenotypically similar SARS-CoV-2-specific CD8+ T cells were detected in both naive and recovered individuals after mRNA immunization, recovered individuals displayed CCR7−CD45RA+ effector phenotype after vaccination, indicating that each antigen exposure by vaccination/infection further matured the T cell compartment. With ongoing efforts to provide third and even fourth booster doses in response to Omicron, effects on memory T cell phenotypes should be carefully examined to ensure that they are retaining potency (Figure 1).
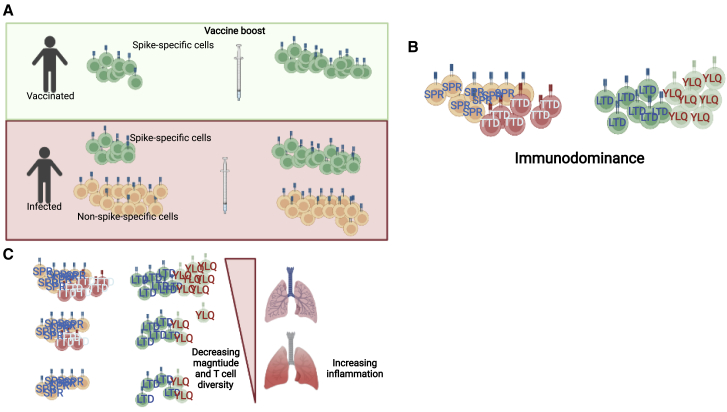
Figure 1.
Defining the features of protective T cell immunity
(A) Distinct antigenic histories generate differing levels of T cell memory targeting various viral targets. To date all approved vaccines have relied solely on Spike antigens, generating only Spike-specific memory. In most convalescent individuals, Spike responses are a significant minority of the repertoire but are further expanded by vaccine boosters.
(B) Immunodominance hierarchies have been defined for CD4+ and CD8+ T cell responses with some epitopes being targeted by up to 10% of CD4+ or CD8+ compartment. The consequences of differential epitope targeting remain to be defined.
(C) The spectrum of T cell specificity and magnitude generated in convalescent and vaccinated individuals may be driving variation in clinical outcomes, but rigorous correlates of protection of the T cell response have yet to be reported.
Created with BioRender.com.
For CD4+ T cells, the BNT162b2 vaccine induces robust tetramer-specific CD4+ T cell responses directed at the prominent DPB1∗04/S167-180 epitope in peripheral blood and lymph nodes.27 DPB1∗04/S167-180+ CD4+ T cells were detected at day 21 after the first vaccine dose, peaked at 7 days following the second dose, displayed a primarily CCR7−CD45RA− effector memory phenotype, and persisted for >200 days. Tetramer-specific Tfhs were detected in blood as circulating Tfh DPB1∗04/S167-180+CXCR5+PD-1+ cells and in lymph nodes at 30 days after the second BNT162b2 dose. Tfh cells subsequently persisted in lymph nodes at similar frequencies for >170 days. Both Tfh and Th1 responses positively correlated with neutralizing antibodies and CD8+ T cell responses generated after the boost.34 In general, CD4+ T cells developed more rapidly, while CD8+ T cells were induced gradually and varied across vaccinated individuals when detected by AIM assay, though it would be useful to assess CD4+ and CD8+ T cell kinetics with peptide-HLA (p-HLA) multimers. CD4+ T cell responses following mRNA vaccination were mainly of central memory T cells (TCM) and effector memory T cells (TEM) memory phenotypes. Comparison of immune dynamics with the UMAP approach showed different T cell trajectories within SARS-CoV-2-naive and recovered vaccinees.
Longevity of T cell responses following SARS-CoV-2 infection and vaccination
T cell responses appear to contract at slower rate than IgG antibodies during the first few months after SARS-CoV-2 infection.35 Studies of convalescent COVID-19 patients revealed durable T cell memory pools for up to 8 months.36, 37, 38 At 8 months, memory CD8+ T cells were predominantly directed toward peptides derived from Spike, Membrane, Nucleocapsid, and ORF3a. Similar to other viruses such as yellow fever, SARS-CoV-2-specific memory CD4+ and CD8+ T cell populations decreased within 3–6 months after disease onset.39 SARS-CoV-2 memory CD8+ T cells declined with a t1/2 of 190 days, while t1/2 was 64 days for CD4+ T cells. Durable cTfh memory was also detected at ≥6 months.39
Frequencies and phenotypes of SARS-CoV-2-specific T cells remain relatively stable in convalescent COVID-19 individuals. Immunodominant B7/N105-specific CD8+ T cells remained at comparable magnitudes ∼270 days post-disease onset when analyzed longitudinally.21 Tetramer-specific CD8+ T cells ex vivo were predominantly of TCM21 or stem cell memory (TSM)15 phenotype. Apart from slight decreases in TSM-like and increases in terminally differentiated effector memory T cells (TEMRA)-like populations, TCM-like phenotypes remained stable over time in convalescent individuals up to 270 days post-disease onset.
Following mRNA vaccination, durable CD4+ and CD8+ memory T cells are also generated. The contraction from the peak occurred within the first 3 months in the peripheral blood,40 in accordance with what has been shown for acute viral infections in humans and animal models. This was followed by establishment of relatively stable memory CD4+ T cell pools comprising mainly TCM and TEM phenotypes in SARS-CoV-2-recovered individuals.40 Furthermore, cTfh decreased rapidly within 6 months after mRNA vaccination, in agreement with studies showing transient appearance of cTfh cells in peripheral blood after influenza immunization, possibly reflecting their trafficking in and out of lymph nodes. As noted above, Tfh responses were more stable in lymph nodes, with high frequencies past 6 months after vaccination.27 Spike-specific Th1 cells were stable between 3 and 6 months after vaccination.
Overall, data on SARS-CoV-2-specific CD4+ and CD8+ T cells clearly demonstrate generation of long-term immunological epitope-specific memory T cell pools following BNT162b2 mRNA COVID-19 vaccination.
Cross-reactivity of SARS-CoV-2-specific T cells toward the variants of concern
Memory T cells established by infection, vaccination, or infection/vaccination can respond to SARS-CoV-2 and variants of concern.40 Viral peptides within dominant CD8+ and CD4+ T cell epitopes described in this commentary are conserved within the variants of concern and can respond to Delta and Omicron strains, as exemplified by B7/N105+CD8+ T cell responses.9 In the Omicron variant specifically, immunodominant T cell specificities identified in the Spike region (A2/S269-277-YLQPRTFLL; A24/S1208-1217-QYIKWPWYI; DPB4/S167-180-TFEYVSQPFLMDLE) and non-Spike regions (A1/ORF1a1637-1646-TTDPSFLGRY; A3/N361-369-KTFPPTEPK; B7/N105-113-SPRWYFYYL; B40/N322-331-MEVTPSGTWL) are conserved, suggesting that pre-existing memory T cells elicited by SARS-CoV-2 infection/vaccination can be recalled following infection with Omicron in a substantial proportion of world’s population. Recent studies demonstrate cross-reactivity of SARS-CoV-2-specific CD4+ and CD8+ T cells toward the SARS-CoV-2 Omicron variant using AIM assay, IFN-γ ICS, ELISpot, and/or proliferation approaches.41, 42, 43, 44, 45, 46, 47 By sequence analysis alone, many of the major epitopes previously characterized in both Spike and non-Spike proteins are completely conserved. The studies suggest that despite greatly reduced antibody responses, pre-existing T cell immunity can potentially provide some protection against Omicron. T cell responses induced by current Ad26.COV2.S and BNT162b2 SARS-CoV-2 vaccines were highly cross-reactive and durable against peptides from Omicron, similar to cross-reactivity observed against Delta and Beta. Generally, Omicron-specific CD8+ T cell responses constituted ∼70%–80% of CD8+ T cell responses induced toward the ancestral strain. Naranbhai and colleagues studied SARS-CoV-2 infected, vaccinated, infected plus vaccinated, and boosted individuals to reveal substantially preserved T cell responses toward Omicron-derived Spike and non-Spike peptides.46 Important findings were that the booster vaccination increased T cell responses to the Omicron variant. Surprisingly, 21% of individuals had >50% reduced T cell responses to Omicron-derived Spike peptides, similar to the findings by Keeton et al. showing that 15% of individuals lost CD8+ T cell reactivity to Omicron,43 which are consistent findings across the studies. Thus, while a large proportion of the world’s population has pre-existing T cells to Omicron, some individuals present non-conserved peptides and hence have limited T cell immunity to Omicron.
It is important to note that thus far, studies assessing responsiveness to variants primarily utilize peptide pools or multimer reagents to identify memory T cell populations. An understudied element of SARS-CoV-2 immunity is the extent to which antigen presentation varies among variants and whether processing and presentation of specific epitopes might be altered by mutations outside the minimal epitope. Furthermore, the virus, with its extensive array of innate immune modulation, could affect general features of the processing and presentation machinery and these mechanisms may differ across variants. Studies detecting T cell responses to infected cells directly will help address these considerations.
T cells in breakthrough infection
With the rapid spread of Omicron, even in previously vaccinated and boosted individuals, many questions need to be answered regarding T cell immunity. In particular, the extent to which CD4+ and CD8+ T cells provide protection and limit disease severity in humans has not been rigorously defined. Correlates of protection analyses in epidemiologically characterized populations are lacking, and such studies will have to be carefully integrated with detailed serological profiling to infer the causal contributions of T cells versus cross-reactive serological protection.48 Furthermore, the strong prior priming most individuals now have against Spike epitopes may bias post-infection memory T cell repertoires. The extent to which de novo non-Spike responses are recruited and expanded in breakthrough infections has not been quantified, and the potential for these cells to further limit future infections with new variants should be assessed. In future studies, features of protective T cell immunity following infection and vaccination need to be carefully defined (Figure 1). Well-curated, longitudinally tracked cohorts are necessary for these studies and should be a continued focus of the broad research community.
While a true T cell-based correlate of protection has not been defined, several studies implicate T cells as protective based on associations with symptoms and outcomes.49,50 These studies identify robust T cell responses in subjects with asymptomatic disease or low symptom severity. In contrast, a number of studies have found that T cell responses in severely ill subjects show evidence of dysregulation, dysfunction, or deletion.51, 52, 53 These studies indicate that failure of T cells to elicit productive control may drive them toward pathological states that contribute to disease propagation without promoting viral clearance. Indeed, early dysregulated T cell responses may be causal in this phenotype, forming a vicious cycle whereby their initial failure to control infection drives further differentiation and functional profiles that inflict tissue damage without effectively eliminating viral reservoirs.
As noted, in contrast to antibody correlates for SARS-CoV-2 and other viruses,54, 55, 56, 57 no true T cell correlate of protection has been defined. To do so will require longitudinally monitored cohorts with known exposure data, where antibody and T cell responses can be integrated in models to capture their individual contributions. Ideally, these measures will include features of T magnitude, specificity, and possibly function, though each parameter increases the difficulty of assessment.
Concluding remarks
At 2 years of COVID-19 pandemic, remarkable progress has been made in terms of understanding immune responses to SARS-CoV-2 and developing and implementing vaccines. As we strive to immunize the global population, new variants emerge. Following SARS-CoV-2 infection and vaccination, long-lived CD4+ and CD8+ T cell memory pools are established, capable of being recalled following subsequent SARS-CoV-2 infection, even against highly drifted variants such as Omicron. So far, T cell responses are not greatly affected by the emergence of variants of concern, and thus can provide pre-existing immunity for the variants. However, as some of the most immunodominant SARS-CoV-2-specific CD8+ T cell responses (e.g., B7/N105+CD8+ T cells) are directed against epitopes encompassing peptides derived from non-Spike regions, extension of the current vaccines to non-Spike viral regions would increase the breadth of T cell responses toward future variants.
Acknowledgments
K.K. is supported by NHMRC Leadership Investigator Grant (1173871). P.G.T. and K.K. are supported by the Center for Influenza Vaccine Research for High-Risk Populations (CIVR-HRP) contract number 75N93019C00052 and U01AI144616-02S1. P.G.T. is supported by R01AI136514 and ALSAC at St. Jude.
Declaration of interests
P.G.T. serves on the scientific advisory boards of Immunoscape, Mirror Biologics, and Cytoagents and performs consulting for Johnson and Johnson. P.G.T. holds pending patents related to T cell receptor cloning and expression (US 2019/0040381, published February 7, 2019 and WO 2021/003114, published January 7, 2021) and COVID therapeutics (published November 11, 2021 as WO 2021/226174), along with an additional unpublished provisional application related to COVID diagnostics.
Contributor Information
Katherine Kedzierska, Email: kkedz@unimelb.edu.au.
Paul G. Thomas, Email: paul.thomas@stjude.org.
References
- 1.Juno J.A., Tan H.-X., Lee W.S., Reynaldi A., Kelly H.G., Wragg K., Esterbauer R., Kent H.E., Batten C.J., Mordant F.L., et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat. Med. 2020;26:1428–1434. doi: 10.1038/s41591-020-0995-0. [DOI] [PubMed] [Google Scholar]
- 2.Koutsakos M., Rowntree L.C., Hensen L., Chua B.Y., van de Sandt C.E., Habel J.R., Zhang W., Jia X., Kedzierski L., Ashhurst T.M., et al. Integrated immune dynamics define correlates of COVID-19 severity and antibody responses. Cell Rep. Med. 2021;2:100208. doi: 10.1016/j.xcrm.2021.100208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thevarajan I., Nguyen T.H.O., Koutsakos M., Druce J., Caly L., van de Sandt C.E., Jia X., Nicholson S., Catton M., Cowie B., et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat. Med. 2020;26:453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 5.McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao J., Zhao J., Perlman S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J. Virol. 2010;84:9318–9325. doi: 10.1128/JVI.01049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhuang Z., Lai X., Sun J., Chen Z., Zhang Z., Dai J., Liu D., Li Y., Li F., Wang Y., et al. Mapping and role of T cell response in SARS-CoV-2-infected mice. J. Exp. Med. 2021;218:e20202187. doi: 10.1084/jem.20202187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan M., Liu Y., Zhou R., Deng X., Li F., Liang K., Shi Y. Immunopathological characteristics of coronavirus disease 2019 cases in Guangzhou, China. Immunology. 2020;160:261–268. doi: 10.1111/imm.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng Y., Felce S.L., Dong D., Penkava F., Mentzer A.J., Yao X., Liu G., Yin Z., Chen J.-L., Lu Y., et al. COMBAT Consortium An immunodominant NP105-113-B∗07:02 cytotoxic T cell response controls viral replication and is associated with less severe COVID-19 disease. Nat. Immunol. 2022;23:50–61. doi: 10.1038/s41590-021-01084-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bange E.M., Han N.A., Wileyto P., Kim J.Y., Gouma S., Robinson J., Greenplate A.R., Hwee M.A., Porterfield F., Owoyemi O., et al. CD8+ T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat. Med. 2021;27:1280–1289. doi: 10.1038/s41591-021-01386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel N.J., D’Silva K.M., Hsu T.Y.-T., DiIorio M., Fu X., Cook C., Prisco L., Martin L., Vanni K.M.M., Zaccardelli A., et al. ACR Open Rheumatol; 2021. Coronavirus Disease 2019 Outcomes Among Recipients of Anti-CD20 Monoclonal Antibodies for Immune-Mediated Diseases: A Comparative Cohort Study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 14.Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van den Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M., et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5:eabd2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Habel J.R., Nguyen T.H.O., van de Sandt C.E., Juno J.A., Chaurasia P., Wragg K., Koutsakos M., Hensen L., Jia X., Chua B., et al. Suboptimal SARS-CoV-2-specific CD8+ T cell response associated with the prominent HLA-A∗02:01 phenotype. Proc. Natl. Acad. Sci. USA. 2020;117:24384–24391. doi: 10.1073/pnas.2015486117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.-B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., et al. Karolinska COVID-19 Study Group Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell. 2020;183:158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swadling L., Diniz M.O., Schmidt N.M., Amin O.E., Chandran A., Shaw E., Pade C., Gibbons J.M., Le Bert N., Tan A.T., et al. COVIDsortium Investigators Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature. 2022;601:110–117. doi: 10.1038/s41586-021-04186-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z., Yang X., Zhong J., Zhou Y., Tang Z., Zhou H., He J., Mei X., Tang Y., Lin B., et al. Exposure to SARS-CoV-2 generates T-cell memory in the absence of a detectable viral infection. Nat. Commun. 2021;12:1724. doi: 10.1038/s41467-021-22036-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kundu R., Narean J.S., Wang L., Fenn J., Pillay T., Fernandez N.D., Conibear E., Koycheva A., Davies M., Tolosa-Wright M., et al. Cross-reactive memory T cells associate with protection against SARS-CoV-2 infection in COVID-19 contacts. Nat. Commun. 2022;13:80. doi: 10.1038/s41467-021-27674-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen T.H.O., Rowntree L.C., Petersen J., Chua B.Y., Hensen L., Kedzierski L., van de Sandt C.E., Chaurasia P., Tan H.-X., Habel J.R., et al. CD8+ T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope display high naive precursor frequency and TCR promiscuity. Immunity. 2021;54:1066–1082.e5. doi: 10.1016/j.immuni.2021.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schulien I., Kemming J., Oberhardt V., Wild K., Seidel L.M., Killmer S., Sagar S., Daul F., Salvat Lago M., Decker A., et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8+ T cells. Nat. Med. 2021;27:78–85. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- 23.Gangaev A., Ketelaars S.L.C., Isaeva O.I., Patiwael S., Dopler A., Hoefakker K., De Biasi S., Gibellini L., Mussini C., Guaraldi G., et al. Identification and characterization of a SARS-CoV-2 specific CD8+ T cell response with immunodominant features. Nat. Commun. 2021;12:2593. doi: 10.1038/s41467-021-22811-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaurasia P., Nguyen T.H.O., Rowntree L.C., Juno J.A., Wheatley A.K., Kent S.J., Kedzierska K., Rossjohn J., Petersen J. Structural basis of biased T cell receptor recognition of an immunodominant HLA-A2 epitope of the SARS-CoV-2 spike protein. J. Biol. Chem. 2021;297:101065. doi: 10.1016/j.jbc.2021.101065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loyal L., Braun J., Henze L., Kruse B., Dingeldey M., Reimer U., Kern F., Schwarz T., Mangold M., Unger C., et al. Cross-reactive CD4+ T cells enhance SARS-CoV-2 immune responses upon infection and vaccination. Science. 2021;374:h1823. doi: 10.1126/science.abh1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu X., Hosono Y., Nagae M., Ishizuka S., Ishikawa E., Motooka D., Ozaki Y., Sax N., Maeda Y., Kato Y., et al. Identification of conserved SARS-CoV-2 spike epitopes that expand public cTfh clonotypes in mild COVID-19 patients. J. Exp. Med. 2021;218:e20211327. doi: 10.1084/jem.20211327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mudd P.A., Minervina A.A., Pogorelyy M.V., Turner J.S., Kim W., Kalaidina E., Petersen J., Schmitz A.J., Lei T., Haile A., et al. SARS-CoV-2 mRNA vaccination elicits a robust and persistent T follicular helper cell response in humans. Cell. 2021;185:603–613.e15. doi: 10.1016/j.cell.2021.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sahin U., Muik A., Vogler I., Derhovanessian E., Kranz L.M., Vormehr M., Quandt J., Bidmon N., Ulges A., Baum A., et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature. 2021;595:572–577. doi: 10.1038/s41586-021-03653-6. [DOI] [PubMed] [Google Scholar]
- 29.Zhu F.-C., Li Y.-H., Guan X.-H., Hou L.-H., Wang W.-J., Li J.-X., Wu S.-P., Wang B.-S., Wang Z., Wang L., et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020;395:1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 31.Minervina A.A., Pogorelyy M.V., Kirk A.M., Allen E.K., Allison K.J., Lin C.-Y., Brice D.C., Zhu X., Vegesana K., Wu G., et al. Convergent epitope-specific T cell responses after SARS-CoV-2 infection and vaccination. medRxiv. 2021 doi: 10.1101/2021.07.12.21260227. Preprint at. [DOI] [Google Scholar]
- 32.Oberhardt V., Luxenburger H., Kemming J., Schulien I., Ciminski K., Giese S., Csernalabics B., Lang-Meli J., Janowska I., Staniek J., et al. Rapid and stable mobilization of CD8+ T cells by SARS-CoV-2 mRNA vaccine. Nature. 2021;597:268–273. doi: 10.1038/s41586-021-03841-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koutsakos M., Wheatley A.K., Loh L., Clemens E.B., Sant S., Nüssing S., Fox A., Chung A.W., Laurie K.L., Hurt A.C., et al. Circulating TFH cells, serological memory, and tissue compartmentalization shape human influenza-specific B cell immunity. Sci. Transl. Med. 2018;10:eaan8405. doi: 10.1126/scitranslmed.aan8405. [DOI] [PubMed] [Google Scholar]
- 34.Painter M.M., Mathew D., Goel R.R., Apostolidis S.A., Pattekar A., Kuthuru O., Baxter A.E., Herati R.S., Oldridge D.A., Gouma S., et al. Rapid induction of antigen-specific CD4+ T cells is associated with coordinated humoral and cellular immunity to SARS-CoV-2 mRNA vaccination. Immunity. 2021;54:2133–2142.e3. doi: 10.1016/j.immuni.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cromer D., Juno J.A., Khoury D., Reynaldi A., Wheatley A.K., Kent S.J., Davenport M.P. Prospects for durable immune control of SARS-CoV-2 and prevention of reinfection. Nat. Rev. Immunol. 2021;21:395–404. doi: 10.1038/s41577-021-00550-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Payne R.P., Longet S., Austin J.A., Skelly D.T., Dejnirattisai W., Adele S., Meardon N., Faustini S., Al-Taei S., Moore S.C., et al. PITCH Consortium Immunogenicity of standard and extended dosing intervals of BNT162b2 mRNA vaccine. Cell. 2021;184:5699–5714.e11. doi: 10.1016/j.cell.2021.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodda L.B., Netland J., Shehata L., Pruner K.B., Morawski P.A., Thouvenel C.D., Takehara K.K., Eggenberger J., Hemann E.A., Waterman H.R., et al. Functional SARS-CoV-2-Specific Immune Memory Persists after Mild COVID-19. Cell. 2021;184:169–183.e17. doi: 10.1016/j.cell.2020.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuo J., Dowell A.C., Pearce H., Verma K., Long H.M., Begum J., Aiano F., Amin-Chowdhury Z., Hoschler K., Brooks T., et al. Robust SARS-CoV-2-specific T cell immunity is maintained at 6 months following primary infection. Nat. Immunol. 2021;22:620–626. doi: 10.1038/s41590-021-00902-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goel R.R., Painter M.M., Apostolidis S.A., Mathew D., Meng W., Rosenfeld A.M., Lundgreen K.A., Reynaldi A., Khoury D.S., Pattekar A., et al. UPenn COVID Processing Unit‡ mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science. 2021;374:abm0829. doi: 10.1126/science.abm0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao Y., Cai C., Grifoni A., Müller T.R., Niessl J., Olofsson A., Humbert M., Hansson L., Österborg A., Bergman P., et al. Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat. Med. 2022 doi: 10.1038/s41591-022-01700-x. Published online January 14, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.GeurtsvanKessel C.H., Geers D., Schmitz K.S., Mykytyn A.Z., Lamers M.M., Bogers S., Scherbeijn S., Gommers L., Sablerolles R.S.G., Nieuwkoop N.N., et al. Divergent SARS CoV-2 Omicron-reactive T- and B cell responses in COVID-19 vaccine recipients. Sci. Immunol. 2022 doi: 10.1126/sciimmunol.abo2202. Published online February 3, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keeton R., Tincho M.B., Ngomti A., Baguma R., Benede N., Suzuki A., Khan K., Cele S., Bernstein M., Karim F., et al. T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature. 2022 doi: 10.1038/s41586-022-04460-3. Published online January 23, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J., Chandrashekar A., Sellers D., Barrett J., Jacob-Dolan C., Lifton M., McMahan K., Sciacca M., VanWyk H., Wu C., et al. Vaccines Elicit Highly Conserved Cellular Immunity to SARS-CoV-2 Omicron. Nature. 2022 doi: 10.1038/s41586-022-04465-y. Published online January 31, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marco L.D., D’Orso S., Pirronello M., Verdiani A., Termine A., Fabrizio C., Capone A., Sabatini A., Guerrera G., Placido R., et al. Preserved T cell reactivity to the SARS-CoV-2 Omicron variant indicates continued protection in vaccinated individuals. bioRxiv. 2021 doi: 10.1101/2021.12.30.474453. Preprint at. [DOI] [Google Scholar]
- 46.Naranbhai V., Nathan A., Kaseke C., Berrios C., Khatri A., Choi S., Getz M.A., Tano-Menka R., Ofoman O., Gayton A., et al. T-cell reactivity to the SARS-CoV-2 Omicron variant is preserved in most but not all individuals. Cell. 2022;185 doi: 10.1016/j.cell.2022.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tarke A., Coelho C.H., Zhang Z., Dan J.M., Yu E.D., Methot N., Bloom N.I., Goodwin B., Phillips E., Mallal S., et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell. 2022;185 doi: 10.1016/j.cell.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lipsitch M., Krammer F., Regev-Yochay G., Lustig Y., Balicer R.D. SARS-CoV-2 breakthrough infections in vaccinated individuals: measurement, causes and impact. Nat. Rev. Immunol. 2022;22:57–65. doi: 10.1038/s41577-021-00662-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J., et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le Bert N., Clapham H.E., Tan A.T., Chia W.N., Tham C.Y.L., Lim J.M., Kunasegaran K., Tan L.W.L., Dutertre C.-A., Shankar N., et al. Highly functional virus-specific cellular immune response in asymptomatic SARS-CoV-2 infection. J. Exp. Med. 2021;218:e20202617. doi: 10.1084/jem.20202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szabo P.A., Dogra P., Gray J.I., Wells S.B., Connors T.J., Weisberg S.P., Krupska I., Matsumoto R., Poon M.M.L., Idzikowski E., et al. Longitudinal profiling of respiratory and systemic immune responses reveals myeloid cell-driven lung inflammation in severe COVID-19. Immunity. 2021;54:797–814.e6. doi: 10.1016/j.immuni.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., et al. Yale IMPACT Team Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meckiff B.J., Ramírez-Suástegui C., Fajardo V., Chee S.J., Kusnadi A., Simon H., Eschweiler S., Grifoni A., Pelosi E., Weiskopf D., et al. Imbalance of Regulatory and Cytotoxic SARS-CoV-2-Reactive CD4+ T Cells in COVID-19. Cell. 2020;183:1340–1353.e16. doi: 10.1016/j.cell.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maier H.E., Balmaseda A., Ojeda S., Cerpas C., Sanchez N., Plazaola M., van Bakel H., Kubale J., Lopez R., Saborio S., et al. An immune correlate of SARS-CoV-2 infection and severity of reinfections. medRxiv. 2021 doi: 10.1101/2021.11.23.21266767. Preprint at. [DOI] [Google Scholar]
- 55.Gilbert P.B., Montefiori D.C., McDermott A.B., Fong Y., Benkeser D., Deng W., Zhou H., Houchens C.R., Martins K., Jayashankar L., et al. Immune Assays Team§. Moderna, Inc. Team§. Coronavirus Vaccine Prevention Network (CoVPN)/Coronavirus Efficacy (COVE) Team§. United States Government (USG)/CoVPN Biostatistics Team§ Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2022;375:43–50. doi: 10.1126/science.abm3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feng S., Phillips D.J., White T., Sayal H., Aley P.K., Bibi S., Dold C., Fuskova M., Gilbert S.C., Hirsch I., et al. Oxford COVID Vaccine Trial Group Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:2032–2040. doi: 10.1038/s41591-021-01540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Danier J., Callegaro A., Soni J., Carmona A., Kosalaraska P., Rivera L., Friel D., Pu W., Vantomme V., Dbaibo G., et al. Association Between Hemagglutination Inhibition Antibody Titers and Protection Against Reverse-Transcription Polymerase Chain Reaction-Confirmed Influenza Illness in Children 6-35 Months of Age: Statistical Evaluation of a Correlate of Protection. Open Forum Infect. Dis. 2022;9:ofab477. doi: 10.1093/ofid/ofab477. [DOI] [PMC free article] [PubMed] [Google Scholar]