Abstract
We have evaluated the use of an ultrasensitive reverse transcriptase (RT) activity assay to monitor plasma viremia in two human immunodeficiency virus type 1 (HIV-1) group O-infected patients treated with stavudine, lamivudine, and indinavir. After a initial decline in RT levels observed at 4 weeks of therapy, RT-based plasma viremia returned to baseline values at 28 or 44 weeks of treatment. The rebound in levels of RT activity was associated with the detection of phenotypic resistance to lamivudine and with the Met184Val mutation. Analysis of RT activity in plasma provides a sequence-independent means of monitoring virus loads in HIV-1 group O-infected patients.
Human immunodeficiency virus type 1 (HIV-1) strains categorized as group O (outlier) include highly divergent viruses that do not cluster with those belonging to the predominant group, M (major), or the recently described group N (9, 15). Group O infections are endemic in west central Africa but have also been identified in Europe and the United States (8). Like HIV-1 group M, group O strains cause AIDS, and therefore persons infected with group O viruses may benefit from antiretroviral therapy with reverse transcriptase (RT) and protease inhibitors (10).
Monitoring the responses of HIV-1 group O-infected persons to antiretroviral therapy is at present difficult because of the unavailability of group O-specific assays for quantitation of plasma viremia. Commercially available assays that measure HIV-1 RNA levels in plasma are based on subtype B sequences and are not adequate for quantitating divergent group O RNA genomes (6, 14). While several of these assays have now been modified to allow the detection of group O RNA sequences, their application to quantitation of levels of group O viruses in plasma has not been determined (13; M. P. De Baar, A. S. van der Schoot, F. Jacobs, K. H. M. van der Horn, M. Brok, P. Oudshoorn, S. Jurrians, and A. de Ronde, Prog. Abstr. 2nd Int. Workshop Drug Resist. Treatment Strategies, abstr. 66, p. 44, 1998).
In the absence of group O-specific assays, other, sequence-independent methods may be suitable to evaluate the responses of persons with group O infections to current antiretroviral drugs. RT is a virion-associated enzyme that can be generically detected and, therefore, is a suitable marker for HIV-1 group O in plasma. We have previously reported the development and use of an ultrasensitive PCR-based RT activity assay, named Amp-RT, to quantitate RT-based virus loads in the plasma of persons with HIV-1 subtype B infections (4, 7). We report here the application of this assay to the analysis of virus loads in HIV-1 group O-infected patients. The Amp-RT assay detects RT activity by using a known nonretroviral heteropolymeric RNA template derived from the encephalomyocarditis virus (EMCV) genome. The RT-generated EMCV cDNA is detected by PCR amplification and probing with an internal oligonucleotide (7). We have previously demonstrated that the dynamics of plasma viremia measured by RT activity were similar to those measured by levels of RNA during either primary infection or following antiretroviral therapy (4, 17). These findings support the use of Amp-RT for measuring changes in virus load in plasma of persons with group O infections.
Levels of RT activity in longitudinal plasma samples from two HIV-1 group O-infected patients treated with RT and protease inhibitors were measured by Amp-RT. Levels of RT activity were measured in virus pellets from 2 μl of plasma as previously described (5). Levels of RT activity were determined by enzyme-linked immunosorbent assay, using an EMCV-specific probe, and were quantitated by comparison to a standard curve generated with known RT activity units from a reference HIV-1 virus stock (4). The results are expressed as units of RT activity per milliliter and reflect the averages of values from duplicate Amp-RT reactions (4).
The first patient studied, ESP2, was a 35-year-old Spanish woman who was diagnosed with AIDS in March 1995. Serologic and molecular evidence of infection with HIV-1 group O in this woman has been previously reported (3). The patient started antiretroviral treatment in April 1996 after being diagnosed with Pneumocystis carinii pneumonia. The patient's history of antiretroviral therapy included treatment with the following combinations: (i) zidovudine (250 mg twice a day [b.i.d.]) and didanosine (200 mg b.i.d.); (ii) stavudine (40 mg b.i.d.) and didanosine (200 mg b.i.d.); (iii) stavudine (40 mg b.i.d.), lamivudine (150 mg b.i.d.), and indinavir (1,200 mg b.i.d.); and (iv) stavudine (40 mg b.i.d.), lamivudine (150 mg b.i.d.), ritonavir (400 mg b.i.d.), and saquinavir (400 mg b.i.d.).
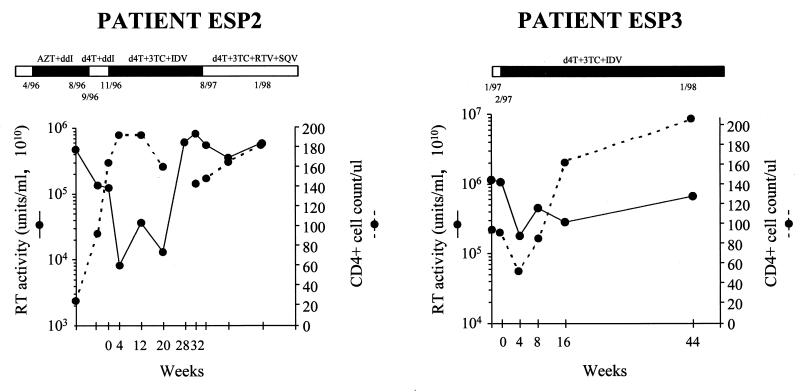
Figure 1 shows the virion-associated RT activity determined by Amp-RT and CD4+ cell counts before and during antiretroviral therapy. A moderate decrease in RT levels (∼0.6 log10 units of RT activity) was observed during treatment with stavudine and didanosine and was associated with an increase in CD4+ cell concentration from 24 to 164/μl. Didanosine administration was discontinued because of drug toxicity, and triple antiretroviral therapy with stavudine, lamivudine, and indinavir was initiated. A maximal decrease in plasma RT activity of 1.2 log10 units was seen at 4 weeks of therapy and was associated with an increase in CD4+ cell concentration to 192/μl. The RT activity observed at the nadir of viremia (8.2 × 10−7 U/ml) was equivalent to that expressed by ∼8,000 HIV-1 particles/ml based on previous estimates (4), suggesting partial suppression of virus replication. RT activity in plasma returned to baseline values at 28 weeks of therapy, while the CD4+ cell concentration decreased to 140/μl at 32 weeks of therapy. Because the patient developed several episodes of nephrolitiasis, two protease inhibitors (ritonavir and saquinavir) were used instead of indinavir. No significant decrease in plasma RT levels was observed (<0.5 log10), although the CD4+ cell concentration increased from 148 to 182/μl (Fig. 1). During the follow-up period, the patient reported good adherence to therapy except for a voluntary discontinuation for 2 weeks in December 1996. RNA was undetectable by RT-PCR (Amplicor Monitor Assay; Roche Molecular Systems), branched DNA assay (bDNA; Chiron Corp.), and nucleic acid sequence-based amplification (NASBA; Organon Technika) in all samples except the baseline sample, which had 12,000 RNA copies/ml by the bDNA assay. However, the RT-based virus load observed in this particular sample (1.31 × 10−5 U/ml) suggests the presence of 136,800 virions/ml, indicating that the virus load measured by the bDNA assay was an underestimation (4). These results confirm the inability of these group M-specific methods to reliably quantitate group O RNA levels because of sequence divergence (6, 14).
FIG. 1.
Histories of antiretroviral therapy, RT-based plasma viral loads, and CD4+ lymphocyte counts for two HIV-1 group O-infected patients (ESP2 and ESP3). AZT, zidovudine; ddI, didanosine; d4T, stavudine; 3TC, lamivudine; IDV, indinavir; RTV, ritonavir; SQV, saquinavir.
The second patient studied, ESP3, was a 46-year-old African man from Equatorial Guinea who was diagnosed with HIV-1 group O infection in 1997 (12). Antiretroviral treatment was initiated with triple combination therapy (stavudine at 40 mg b.i.d., lamivudine at 150 mg b.i.d., and indinavir at 1,200 mg b.i.d.). A rapid decrease in plasma RT activity of 0.8 log10 units was observed at 4 weeks of treatment. However, RT levels increased over time, returning to baseline values after 44 weeks of treatment (Fig. 1). Similarly to patient ESP2, the nadir in plasma RT activity did not result in undetectable RT levels; RT activity was equivalent to that expressed by ∼150,000 HIV-1 particles/ml (4). The CD4+ cell concentration increased from 96 to 209/μl during this period of time. The patient admitted missing 2 to 5 of the 14 weekly doses of indinavir during the first 6 months of treatment.
The results observed for both patients indicate treatment failures, as defined by the lack of suppression of virus replication to undetectable levels at 16 to 24 weeks of treatment and a rise in viral load of >0.6 log units above the patient's nadir values (1). However, despite the absence of sustained virus suppression, a significant clinical and immunologic benefit was evident for both patients. Patient ESP2 gained 13 kg of weight and was able to resume work. During follow-up, it was determined that she had not developed major or minor opportunistic infections, and suppressive prophylaxis for P. carinii pneumonia was discontinued in October 1997. Patient ESP3 remained asymptomatic during the follow-up period, did not develop major or minor opportunistic infections, and was able to resume work. Similar findings have been recently observed in subtype B-infected patients who have been monitored for up to 36 to 48 weeks of treatment (D. Kaufmann, G. Pantaleo, P. Sudre, and A. Telenti, Letter, Lancet 351:723–724). The duration of the clinical and immunologic improvements seen in both patients cannot be predicted, but transience is likely given the observed lack of virus suppression.
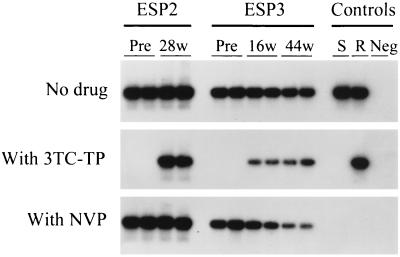
We also determined whether the virus rebound observed in both patients by Amp-RT was associated with the emergence of lamivudine-resistant HIV-1. We screened for phenotypic resistance to lamivudine by using a novel Amp-RT-based assay which measures the susceptibility of plasma RT activity to inhibition by lamivudine 5′-triphosphate (3TC-TP) (5). As illustrated in Fig. 2, the RT activities in pretherapy samples and wild-type (WT) controls were completely inhibited by 3TC-TP, demonstrating WT susceptibility to lamivudine. In contrast, resistance to lamivudine was observed for RT from plasma samples obtained from patient ESP2 at week 28 of therapy and from patient ESP3 at weeks 16 and 44 of therapy (Fig. 2). Quantitative analysis of the Amp-RT signal provides information on the level of resistance (5). This analysis indicated an increase in the proportion of lamivudine-resistant viruses in patient ESP3 over time (RT inhibition by 3TC-TP, 81% at week 16 of treatment versus 54% at 44 weeks). To confirm the specificity of the results obtained with 3TC-TP, we also analyzed the plasma RT from both patients for susceptibility to nevirapine, a drug to which HIV-1 group O is naturally resistant (2). As expected, all samples from the two patients exhibited nevirapine resistance, which was indicated by the lack of complete RT inhibition (17). In contrast, the two control WT subtype B viruses were completely inhibited by nevirapine (Fig. 2).
FIG. 2.
Determination of phenotypic resistance to 3TC of HIV-1 from plasma of two HIV-1 group O-infected patients (ESP2 and ESP3) by measuring levels of RT inhibition by 5 μM 3TC-TP (5). Results of duplicate tests of plasma samples obtained before (Pre) and during treatment with lamivudine, stavudine, and indinavir (week 28 [28w] in patient ESP2 and weeks 16 [16w] and 44 [44w] in patient ESP3) are shown. Samples were adjusted to similar levels of RT activity before testing. Signal from Amp-RT reactions containing 50 μM nevirapine (NVP) are also shown. S, lamivudine-sensitive HIV-1 reference virus (xxBRUpitt); R, lamivudine-resistant HIV-1 reference virus (M184Vpitt); Neg, uninfected control supernatant.
Genotypes of RT from both patients were also determined by sequence analysis of proviral DNA as described (11). In pretherapy samples from both patients, the RT gene encoded WT Met at codon 184. In contrast, the analysis of RT sequences after 28 (patient ESP2) and 16 (patient ESP3) weeks of treatment showed the presence of the Met184Val mutation and a mixture of WT and Met184Val, respectively. The presence of mixtures of WT and Met184Val in patient ESP3 correlated with partial RT inhibition by 3TC-TP (Fig. 2). Sequences of RT in samples obtained from patient ESP2 before therapy and after 28 weeks of treatment with stavudine, lamivudine, and indinavir have been previously reported (12).
The association between phenotypic resistance to lamivudine and the Met184Val mutation strongly suggests that this substitution confers resistance to that drug in HIV-1 group O, a finding consistent with that for subtype B infections. This finding is also similar to that previously seen in another group O-infected patient and confirms the role of this conserved amino acid in lamivudine resistance in this divergent virus (2). Taken together, our results indicate that the observed rebound in RT-based plasma viremia was associated with the emergence of lamivudine-resistant virus, thus indicating that selection of lamivudine-resistant virus was partially responsible for the observed virologic treatment failure.
The moderate decline in plasma viremia observed in both patients after treatment with stavudine, lamivudine, and indinavir may be related to a decreased susceptibility of group O viruses to the drugs and/or poor adherence to the treatment. Available sequence data for the pretreatment samples from both patients did not show any preexisting mutations (e.g., Q151M and T69SSA/S in the RT or L90M, M46I, and V82A/F/T in the protease) that are known to confer resistance to either stavudine or indinavir in subtype B infections (12, 16). While phenotypic analysis of baseline samples is necessary to confirm the absence of reduced susceptibility to these drugs, the observed treatment failures may simply be related to the poor compliance to indinavir reported by patient ESP3 and the discontinuation of treatment by patient ESP2 (R. Rodriguez-Rosado, I. Jiménez-Nacher, V. Soriano, P. Anton, and J. González-Lahoz, Letter, AIDS 12:1112–1113, 1998).
Our data highlight the usefulness of Amp-RT-based assays for monitoring virus loads in plasma and emergence of phenotypic resistance to lamivudine in HIV-1 group O-infected patients. These assays should facilitate future studies aimed at evaluating the responses of HIV-1 group O-infected patients to combination therapy with RT and protease inhibitors.
Nucleotide sequence accession numbers.
The nucleotide sequences of RT from patient ESP3 have been submitted to the GenBank database and assigned accession no. AF081821 (pretherapy) and AF081822 (at week 16 of treatment).
Acknowledgments
We thank Diane Havlir for critical reading of the manuscript and Raymond F. Schinazi for providing 3TC-TP.
REFERENCES
- 1.DeGruttola V, Hughes M, Gilbert P, Phillips A. Trial design in the era of highly effective antiviral drug combinations for HIV infection. AIDS. 1998;12(Suppl. A):S149–S156. [PubMed] [Google Scholar]
- 2.Descamps D, Collin G, Letourneur F, Apetrei C, Damond F, Loussert-Ajaka I, Simon F, Saragosti S, Brun-Vézinet F. Susceptibility of human immunodeficiency virus type 1 group O isolates to antiretroviral agents: in vitro phenotypic and genotypic analyses. J Virol. 1997;71:8893–8898. doi: 10.1128/jvi.71.11.8893-8898.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.García Lerma J G, Gutierrez M, Mas A, Bravo R, Aguilera O, Soriano V. Report of the first two cases of HIV-1 group O infection in Spain. Med Clin. 1996;107:418–421. [PubMed] [Google Scholar]
- 4.García Lerma J G, Yamamoto S, Gómez-Cano M, Soriano V, Green T A, Busch M P, Folks T M, Heneine W. Measurement of human immunodeficiency virus type 1 plasma virus load based on reverse transcriptase (RT) activity: evidence of variabilities in levels of virion-associated RT. J Infect Dis. 1998;177:1221–1229. doi: 10.1086/515272. [DOI] [PubMed] [Google Scholar]
- 5.García Lerma J G, Schinazi R F, Juodawlkis A S, Soriano V, Lin Y, Tatti K, Rimland D, Folks T M, Heneine W. A rapid non-culture-based assay for clinical monitoring of phenotypic resistance of human immunodeficiency virus type 1 to lamivudine (3TC) Antimicrob Agents Chemother. 1999;43:264–270. doi: 10.1128/aac.43.2.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gobbers E, Fransen K, Oosterlaken T, Janssens W, Heyndrickx L, Ivens T, Vereecken K, Schoones R, van de Wiel P, van der Groen G. Reactivity and amplification efficiency of the NASBA HIV-1 RNA amplification system with regard to different HIV-1 subtypes. J Virol Methods. 1997;66:293–301. doi: 10.1016/s0166-0934(97)00072-4. [DOI] [PubMed] [Google Scholar]
- 7.Heneine W, Yamamoto S, Switzer W M, Spira T J, Folks T M. Detection of reverse transcriptase by a highly sensitive assay in sera from persons infected with human immunodeficiency virus type 1. J Infect Dis. 1995;171:1210–1216. doi: 10.1093/infdis/171.5.1210. [DOI] [PubMed] [Google Scholar]
- 8.Jaffe H W, Schochetman G. Group O human immunodeficiency virus-1 infections. Infect Dis Clin N Am. 1998;12:39–46. doi: 10.1016/s0891-5520(05)70407-4. [DOI] [PubMed] [Google Scholar]
- 9.Leitner T. Genetic subtypes of HIV-1, sect. III. In: Myers G, Korber B, Foley B, et al., editors. Human retroviruses and AIDS 1996. Los Alamos, N. Mex: Los Alamos National Laboratory; 1996. p. 28. [Google Scholar]
- 10.Nkengasong J N, Fransen K, Willens B, Karita E, Vingerhoets J, Kestens L, Colebunders R, Piot P, van der Groen G. Virologic, immunologic, and clinical follow-up of a couple infected by the human immunodeficiency virus type one, group O. J Med Virol. 1997;51:202–209. [PubMed] [Google Scholar]
- 11.Quiñones-Mateu M E, Soriano V, Domingo E, Menéndez-Arias L. Characterization of the reverse transcriptase of a human immunodeficiency virus type 1 group O isolate. Virology. 1997;236:364–373. doi: 10.1006/viro.1997.8748. [DOI] [PubMed] [Google Scholar]
- 12.Quiñones-Mateu M E, Albright J L, Mas A, Soriano V, Arts E J. Analysis of pol gene heterogeneity, viral quasispecies, and drug resistance in individuals infected with group O strains of human immunodeficiency virus type 1. J Virol. 1998;72:9002–9015. doi: 10.1128/jvi.72.11.9002-9015.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Respess R A, Butcher A, Wang H, Chaowanachan T, Young N, Shaffer N, Mastro T D, Biryahwaho B, Downing R, Tanuri A, Schechter M, Pascu R, Zekeng L, Kaptué I, Gürtler L, Eberle J, Ellenberger D, Fridlund C, Rayfield M, Kwok S. Detection of genetically diverse human immunodeficiency virus type 1 group M and O isolates by PCR. J Clin Microbiol. 1997;35:1284–1286. doi: 10.1128/jcm.35.5.1284-1286.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segondy M, Ly T-D, Lapeyre M, Montes B. Evaluation of the Nuclisens HIV-1 QT assay for quantitation of human immunodeficiency virus type 1 RNA levels in plasma. J Clin Microbiol. 1998;36:3372–3374. doi: 10.1128/jcm.36.11.3372-3374.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simon F, Mauclère P, Rosques P, Loussert-Ajaka I, Müller-Trutwin M C, Saragosti S, Georges-Courbot M C, Barré-Sinoussi F, Brun-Vézinet F. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat Med. 1998;4:1032–1037. doi: 10.1038/2017. [DOI] [PubMed] [Google Scholar]
- 16.Vallejo A, Heredia A, Mas A, Lee S F, Epstein J S, Soriano V, Hewlett I K. Tropism, coreceptor use, and phylogenetic analysis of both the V3 loop and the protease gene of three novel HIV-1 group O isolates. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;18:417–425. doi: 10.1097/00042560-199808150-00002. [DOI] [PubMed] [Google Scholar]
- 17.Vázquez-Rosales J G, García Lerma J G, Yamamoto S, Switzer W M, Havlir D, Folks T M, Richman D D, Heneine W. Rapid screening of phenotypic resistance to nevirapine by direct analysis of HIV-1 reverse transcriptase activity in plasma. AIDS Res Hum Retrovir. 1999;15:1191–1200. doi: 10.1089/088922299310287. [DOI] [PubMed] [Google Scholar]